ABSTRACT
The Arabidopsis GUT15 RNA belongs to a class of noncoding RNAs that are expressed from the intergenic regions of protein-coding genes. We show that the RNA polymerase II transcribed GUT15 transcript serves as a precursor for two stable RNA species, a tRNA-like molecule and GUT15-tRF-F5, which are both encoded by the final intron in the GUT15 gene. The GUT15-encoded tRNA-like molecule cannot be autonomously transcribed by RNA polymerase III. However, this molecule contains a CCA motif, suggesting that it may enter the tRNA maturation pathway. The GUT15-encoded tRNA-like sequence has an inhibiting effect on the splicing of its host intron. Moreover, we demonstrate that the canonical tRNA genes nested within introns do not affect the splicing patterns of their host protein-coding transcripts.
KEYWORDS: Arabidopsis, lncRNAs, GUT15, tRNA-like, splicing
Introduction
Recent advances in RNA sequencing technology and computational analyses have resulted in the identification of a notable number of long transcripts known as long noncoding RNAs (lncRNAs) that lack protein-coding capacity [1]. The heterogeneity in the biogenesis, expression levels, stability and evolution of this diverse class of molecules reflects the broad array of functions performed by lncRNAs in the cell. Based on animal studies, lncRNAs have emerged as novel factors regulating different aspects of gene expression, and these factors have been shown to participate in transcriptional silencing by recruiting chromatin remodeling complexes [2]. Several lines of evidence suggest that lncRNAs play a role in establishing and maintaining the architecture of nuclear compartments [2].
Numerous lncRNAs have been predicted and identified in several plant species [3]; however, our knowledge regarding their biogenesis and molecular functions remains limited to a few examples. Both AtCOOLAIR and AtCOLDAIR noncoding RNAs play roles in the epigenetic silencing of the FLC locus during vernalization [4,5]. The epigenetic landscape is also affected by the dual noncoding transcription of the APOLO locus (performed by both RNA polymerase II and V) that controls the auxin-driven chromatin loop dynamics to regulate the expression of the neighboring PID gene, which encodes an essential regulator of polar auxin transport [6]. The APOLO lncRNA-dependent oscillating chromatin topology further reflects an additional function ascribed to plant lncRNAs, i.e., the production of small interfering RNAs [7]. Generally, the biogenesis of 24 nt siRNAs requires noncoding transcription, which is driven by plant RNA polymerases IV and V, and leads to transcriptional gene silencing (TGS) by directing the DNA methylation of their locus of origin and adjacent genes [8]. In addition, plant lncRNAs have been associated with other biological processes. Phosphate homeostasis is regulated by the IPS1 lncRNA, which acts as an endogenous target mimic of miR399 [9]. Photomorphogenesis in Arabidopsis seedlings is regulated by the HID3 lncRNA, which is known to associate with PIF3 locus chromatin and repress its transcription [10]. In rice, the LDMAR noncoding transcript is required for normal male fertility under long-day conditions [11]. Plant lncRNAs are also involved in root nodule organogenesis; for example, the ENOD40 lncRNA plays a role in the cytoplasmic re-location of the nuclear protein RBP1 during nodulation in Medicago truncatula [12]. Bardou and coworkers [13] identified that the ASCO lncRNA is a novel player (competitor) in the regulation of alternative splicing. Recently, the ELENA1 noncoding transcript has been shown to be involved in the transcriptional regulation of plant innate immunity by interacting with Mediator and enhancing the expression of PR1 (PATHOGENESIS-RELATED 1) [14].
Long noncoding RNAs can be classified into the following five different classes according to their genomic location: natural antisense transcripts (NATs), intronic noncoding RNAs, enhancer RNAs, promoter-associated transcripts and large intergenic noncoding RNAs (lincRNAs) [1]. The lincRNA group is represented in A. thaliana by the GUT15 long noncoding transcript, which was first reported in tobacco as a short-lived RNA [15].
In this paper, we prove the existence of a tRNA-like structure nested within the gene encoding the Arabidopsis lncRNA GUT15. Interestingly, the biogenesis of this tRNA-like molecule relies on GUT15 transcript processing rather than on RNA polymerase III-driven transcription. Furthermore, the CCA trinucleotide, which is indicative of functional tRNAs, is added to the 3′-end of the tRNA-like molecule encoded by the final intron of the GUT15 primary transcript, and this molecule is most likely aminoacylated. Additionally, we show that the GUT15-encoded tRNA-like structure inhibits the splicing of its hosting intron.
Results
A tRNA-like molecule is encoded within the Arabidopsis lncRNA GUT15 locus
The re-annotation of the tRNA genes in the Arabidopsis genome performed in our laboratory revealed the presence of a tRNA-like sequence within the locus encoding the lncRNA GUT15 (Gene with unstable transcript 15; At2g18440) [16]. This tRNA-like sequence exhibits the conserved structural features present in canonical tRNAs. In particular, the length of the acceptor, the D- and T-stems, as well as the size of the D- and T-loops are consistent with the standard tRNA structure cloverleaf model (Fig. 1A, left panel) [17]. Interestingly, compared with bona fide tRNA sequences, the sequence of the T-loop (positions 3–9 of the B-box in the sequence logo) in the identified tRNA-like structure is very well conserved. This feature plays a role in the formation of the distinct L-shaped tertiary structure that is essential for tRNA function. In contrast, the A-box of the GUT15 tRNA-like structure differs from the consensus sequence due to two additional nucleotides (Fig. 1A, middle panel). Moreover, the region corresponding to the anticodon stem-loop in the tRNA-like structure cannot form the canonical 5-bp stem and 7-nucleotide loop. Another feature distinguishing the GUT15 tRNA-like structure from standard tRNA molecules is the presence of three rather than two unpaired bases at the junction between the acceptor and D-stem (Fig. 1A, left panel). The accumulation of the tRNA-like molecule encoded by GUT15 in Arabidopsis seedlings was validated by RNA sequencing (unpublished) and confirmed by Northern blotting and stem-loop RT-PCR (Fig. 1A, right panel). Importantly, the GUT15 tRNA-like molecule contains a CCA motif that is not encoded in the genome (Fig. 1A, left panel). Moreover, according to our tRNA-seq data, the tRNA-like RNA originating from GUT15 was detected in the pool of deacylated tRNA molecules, suggesting that this RNA can be aminoacylated.
Figure 1.
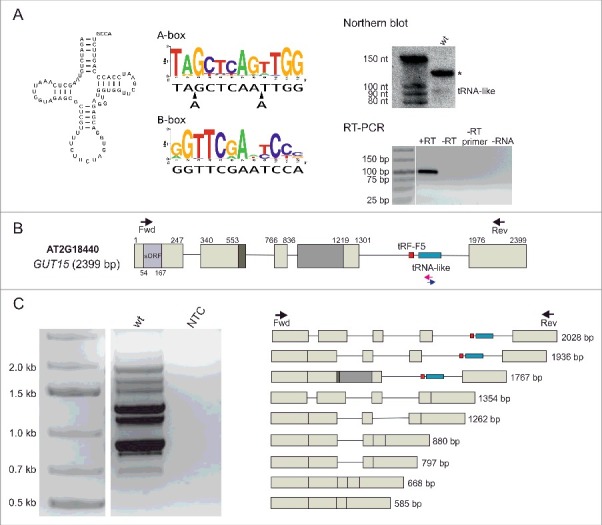
The Arabidopsis GUT15 lncRNA contains a tRNA-like sequence in its final intron. (A) The GUT15 tRNA-like secondary structure (left panel). Alignment of the GUT15-encoded tRNA-like molecule A-box and B-box to the consensus sequence profile of canonical tRNA promoter elements (middle panel). The tRNA-like sequence accumulation levels detected by Northern blotting and stem-loop RT-PCR in Arabidopsis seedlings (right panel). The asterisk corresponds to an unspecific band. +RT, the complete stem-loop RT-PCR reaction; -RT, no reverse transcriptase control; -RT primer, no stem-loop RT primer control; -RNA, no RNA control of the reverse transcription reaction. Marker in Northern, Decade™ Marker RNA (Thermo Fisher Scientific); marker in RT-PCR, Low Range DNA ladder (Thermo Fisher Scientific). (B) Structure of the GUT15 gene hosting the tRNA-like sequence within its final intron. Boxes represent exons, lines depict introns, and arrows represent the primers used in the RACE (blue and pink) and RT-PCR analyses (black). (C) RT-PCR analysis showing different GUT15 lncRNA splicing isoforms. Wt, cDNA template prepared from 10-day-old wild-type Arabidopsis seedlings; NTC, non-template control; marker, 1 kb plus (Thermo Fisher Scientific).
The GUT15 transcripts are capped, polyadenylated and undergo alternative splicing
To determine the structure of the transcripts originating from the Arabidopsis GUT15 locus, we performed 5′- and 3′-RACE experiments using primers that hybridize to the tRNA-like sequence, followed by RT-PCR amplification of the products obtained with primers designed to bind to the 5′- and 3′-ends of the longest RACE fragments. Using this approach, we identified polyadenylated GUT15 transcripts. Additionally, using the 5′-RLM-RACE procedure to selectively amplify only full-length capped RNAs, we showed that all analyzed GUT15 transcripts contained the cap structure, which, along with the poly(A) tail, constitutes the characteristics of transcription driven by RNA polymerase II (RNAPII). Based on the longest 3′- and 5′-RACE products, we calculated the length of the GUT15 lncRNA as 2399 bp (Fig. 1B). The presence of this full-length transcript was confirmed by RT-PCR. The experimentally established length of the GUT15 lncRNA from Arabidopsis seedlings was 100 bp longer than the GUT15 transcript described in the TAIR10 database [18]. However, this discrepancy may be due to differences in the regulation of GUT15 gene transcription in specific tissues or the different developmental stages during which the RNA samples were collected. The Arabidopsis GUT15 gene contains 4 introns. The discovered tRNA-like sequence is encoded by the final intron in the GUT15 transcripts (Fig. 1B). Interestingly, the RT-PCR amplification of the GUT15 cDNA revealed the presence of a series of splicing isoforms, and only three isoforms contained the tRNA-like-bearing intron (Fig. 1C).
The GUT15-encoded tRNA-like sequence affects the splicing of its host intron
The mapping of the small RNA-seq data to the GUT15 gene revealed a short sequence originating from the 5′-flanking region of the tRNA-like coding region in the final GUT15 intron (Fig. 1B). We called this new small RNA molecule GUT15-tRF-F5 according to our previously proposed nomenclature [19]. The abundance of this molecule in different small RNA libraries is summarized in Table 1.
Table 1.
sRNA-seq results (homemade libraries and public samples) for GUT15-tRF-F5 AAAAGAGAGTCAAAGAG.
| sample | count |
|---|---|
| cbc | 251 |
| cpl1-3 | 140 |
| serrate | 226 |
| hyl1-2 | 178 |
| drb2 | 83 |
| drb3 | 136 |
| drb4 | 112 |
| drb5 | 57 |
| dcl2-5_14 | 164 |
| dcl3-1_14 | 138 |
| dcl4-2_14 | 203 |
| rdr6-15 | 85 |
| sgs3-13 | 556 |
| xrn2-3 | 605 |
| xrn4-3 | 208 |
| rdr2-2 | 353 |
| rdr3b | 126 |
| rrp6l1-2 | 721 |
| rrp6l2-1 | 224 |
| rrp6l3-1 | 155 |
| rns2-2 | 12 |
| trz4 | 182 |
| wt31 | 83 |
| wt14 | 589 |
| heat_c1 | 1 |
| heat_c2 | 1 |
| heat_c3 | 1 |
| heat_30min_s1 | 1 |
| heat_30min_s2 | 1 |
| heat_6h_s3 | 1 |
| drought_30pct_c3 | 1 |
| cu_d_s1 | 1 |
| cu_d_s2 | 1 |
| cu_d_s3 | 1 |
| nacl_e_s1 | 5 |
| nacl_e_s3 | 1 |
| sulfur_d_s1 | 1 |
| cadmium_e_s1 | 1 |
| cadmium_e_s2 | 4 |
| SRR037653 | 20 |
| SRR037656 | 6 |
| SRR037657 | 17 |
| SRR037658 | 13 |
| SRR037659 | 6 |
| SRR037660 | 26 |
| SRR037663 | 18 |
| SRR037664 | 27 |
| SRR037665 | 20 |
| SRR037666 | 8 |
| SRR037667 | 24 |
| SRR037669 | 17 |
| SRR037670 | 4 |
| SRR037675 | 24 |
The close localization of the tRNA-like and GUT15-tRF-F5 sequences within the GUT15 lncRNA intron raised the following two possible scenarios regarding the biogenesis of the tRNA-like molecule and GUT15-tRF-F5: 1) GUT15-tRF-F5 and the tRNA-like molecule are generated from a tRNA-like precursor that is transcribed by RNA polymerase III (RNAPIII) or 2) alternatively, the GUT15-tRF-F5 and tRNA-like sequences are processed from GUT15 transcripts or already spliced introns carrying both the tRNA-like molecule and GUT15-tRF-F5. To examine the relationship between the proximity of the tRNA-like structure and biogenesis of GUT15-tRF-F5, we created genetic constructs encoding the following two versions of the GUT15 gene (both under the control of the CaMV 35S promoter): the full-length (35S-GUT15wt) version or a version lacking the tRNA-like sequence (35S-GUT15Δ); we introduced these constructs into Nicotiana benthamiana leaves using the agroinfiltration method (Fig. 2A). Using RT-PCR, we determined that the deletion of the tRNA-like sequence increases the splicing efficiency of its hosting intron (Fig. 2B). Moreover, the Northern blotting results showed an accumulation of processed GUT15-tRF-F5 from the mutated GUT15 gene variant lacking the tRNA-like sequence, suggesting that the proximity of the tRNA-like molecule negatively affects the expression of GUT15-tRF-F5 (Fig. 2C, middle panel). Importantly, this result excluded the possibility that GUT15-tRF-F5 is produced from the tRNA-like precursor because the deletion of the tRNA-like molecule resulted in GUT15-tRF-F5 overexpression. Overall, the intronic GUT15 tRNA-like sequence affects both the splicing of the final intron in the GUT15 lncRNA and the accumulation of GUT15-tRF-F5.
Figure 2.
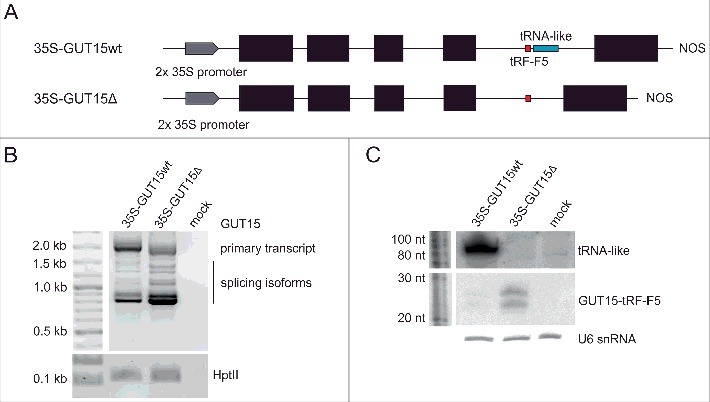
The tRNA-like sequence regulates the splicing of its host intron and the biogenesis of the GUT15-tRF-F5 small RNA. (A) Schematic representation of the GUT15 gene variants used. Black boxes represent exons, and lines depict introns. Positions of the tRNA-like and GUT15-tRF-F5 sequences are marked in blue and red, respectively. NOS, transcription terminator. (B) Primary transcripts and splicing isoforms of GUT15 detected by RT-PCR in N. benthamiana leaves infiltrated with the GUT15 gene variants shown in A. The expression of the hygromycin phosphotransferase gene (HptII) served as a positive control for the agroinfiltration experiment. Marker, 100 bp plus (Thermo Fisher Scientific). MOCK is a negative control for the agroinfiltration experiments (leaves infiltrated only with MES buffer). (C) The accumulation levels of the tRNA-like and GUT15-tRF-F5 sequences detected by Northern blotting in infiltrated leaves expressing the GUT15 gene variants shown in A. U6 snRNA serves as an RNA loading control. Marker, Decade™ Marker RNA (Thermo Fisher Scientific).
The GUT15-encoded tRNA-like molecule is generated from the GUT15 lncRNA
Thus far, we have shown that the tRNA-like sequence is transcribed as a part of the lncRNA GUT15, which is synthesized by RNAPII. However, this observation does not exclude that the tRNA-like locus can be independently transcribed also by RNAPIII. To address this issue, we transfected N. tabacum protoplasts with a GUT15 native construct lacking any upstream regulatory sequences (Δp-GUT15wt) to investigate the activity of the internal A-box and B-box tRNA promoter elements within the GUT15 tRNA-like sequence (Fig. 3A, upper panel, and Fig. 1A). The internal promoter elements within the tRNA-like coding sequence were not sufficient to drive its autonomous transcription by RNAPIII (Fig. 3B-C). In contrast, the GUT15 tRNA-like molecule was detected in tobacco protoplasts transfected with the UBQ promoter-driven GUT15 construct (Fig. 3B-C). Thus, the GUT15-encoded tRNA-like molecule can be processed from RNAPII-produced GUT15 transcripts independently of the promoter used, i.e., the native or UBQ promoter. Moreover, no differences were observed in the tRNA-like sequence expression levels when the 35S-GUT15Δ5′3′ss construct with mutated 5′- and 3′- splice sites in the tRNA-like hosting intron was transiently expressed in the tobacco leaves (Fig. 3A, lower panel, and 3D-E). Thus, the biogenesis of the tRNA-like molecule encoded within the GUT15 intron is reliant on RNAPII GUT15 transcription but independent of the GUT15 final intron splicing.
Figure 3.
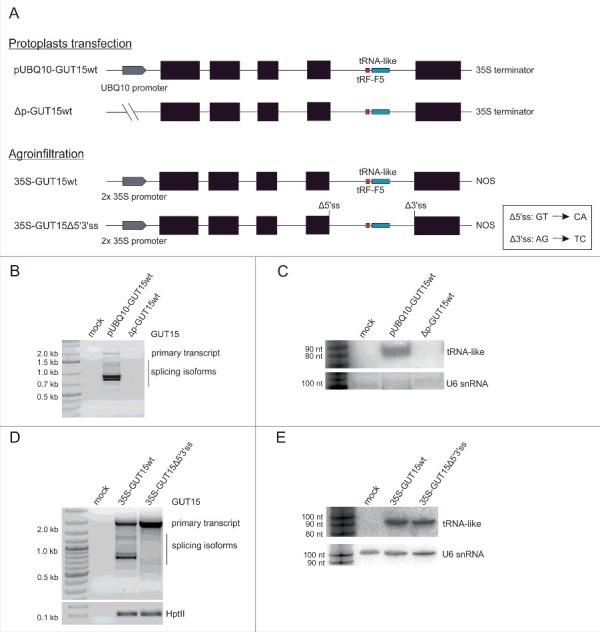
The tRNA-like molecule is co-expressed with only the GUT15 lncRNA. (A) Schematic representation of the GUT15 gene variants used, and the inactivating 5′ (Δ5′ss) and 3′ss (Δ3′ss) mutations are presented in the box. Black boxes represent exons, and lines depict introns. The splice sites and positions of the tRNA-like and GUT15-tRF-F5 sequences are marked in blue and red, respectively. NOS, transcription terminator; UBQ 10, ubiquitin promoter. (B) Primary transcripts and splicing isoforms of GUT15 recorded by RT-PCR in transfected tobacco protoplasts expressing the GUT15 gene variants shown in A (upper panel). MOCK is a negative control for the transfection. Marker, 1 kb plus (Thermo Fisher Scientific). (C) The levels of tRNA-like sequence detected by Northern blotting in tobacco protoplasts expressing the GUT15 variants shown in A (upper panel). U6 snRNA serves as an RNA loading control. Marker, Decade™ Marker RNA (Thermo Fisher Scientific). (D) Primary transcripts and splicing isoforms of GUT15 analyzed by RT-PCR in infiltrated N. benthamiana leaves expressing the GUT15 gene variants shown in A (bottom panel). The expression of the hygromycin phosphotransferase gene (HptII) served as a positive control for the agroinfiltration experiment. MOCK is a negative control for the agroinfiltration experiment (leaves infiltrated only with MES buffer). Marker, 100 bp plus (Thermo Fisher Scientific). (E) The levels of the tRNA-like molecule detected by Northern blotting in infiltrated N. benthamiana leaves expressing the GUT15 variants shown in A (bottom panel). U6 snRNA serves as an RNA loading control. Marker, Decade™ Marker RNA (Thermo Fisher Scientific).
Canonical tRNA genes nested within introns of protein-coding genes do not affect the splicing patterns of their host transcripts
Because the tRNA-like structure in the GUT15′s final intron affects its splicing, we globally examined the occurrence of tRNAs in genes across the A. thaliana genome. The computational analyses identified 19 protein-coding genes containing tRNA sequences in a sense context (in introns and untranslated regions (UTRs)) (Table 2). RT-PCR performed for 3 of these genes revealed that introns containing tRNAs are efficiently spliced in Arabidopsis seedlings (Fig. S3). Subsequently, we selected a gene encoding oligosaccaryltransferase (At3g12587) as an example to study the influence of tRNA sequences on the splicing of their host introns using a transient expression assay in N. benthamiana leaves (Fig. 4A-B). In contrast to the GUT15 transcript, no notable differences were detected in the intron splicing efficiency when the naturally occurring tRNA-ArgCCG was removed from its host intron (Fig. 4C). Thus, the standard tRNA molecules embedded in the introns of protein-coding genes do not have a negative effect on the splicing of their host introns. To further validate this observation, we prepared the following two variants of the original GUT15 intron 4 (pGUT15): in one variant, the tRNA-like sequence was removed (pGUT15Δ), and in the other variant, the GUT15 tRNA-like sequence was replaced with the tRNA-AlaAGC gene (At1g06610; pGUT15(Ala)) (Fig. 5A-B, left panel). These constructs were introduced into N. tabacum protoplasts, and their intron excision efficiency was tested using RT-qPCR. All analyzed versions of the GUT15 intron were efficiently spliced in the tobacco protoplast transient expression system. However, according to the investigation of the specific copy numbers of the isoforms using an qPCR-based absolute quantification method, the GUT15 intron lacking the tRNA-like sequence (pGUT15Δ construct) was excised more efficiently than the other two intron variants (pGUT15 and pGUT5(Ala)) (Fig. 5C, left panel). These observed changes are consistent with our previous tobacco agroinfiltration experimental results.
Table 2.
Protein-coding genes overlapping annotated tRNA genes in the sense orientation in Arabidopsis genome.
| Gene symbol / annotation | tRNA | Context |
|---|---|---|
| AT1G64130 – polyketide cyclase/dehydrase and lipid transport superfamily | LeuCCA | CDS/intron |
| AT1G68760 – nudix hydrolase 1 hydrolyzes 8-oxo-(d)GTP to 8-oxo-(d)GMP | ProAGG | 3′-UTR |
| AT1G71697 – choline kinase 1 increased in response to wounding | GlyGCC | 3′-UTR |
| AT2G07771 – cytochrome C assembly protein | IleAAU | CDS/3′-UTR |
| AT4G24030 – unknown protein | GlyCCC | CDS/3′-UTR |
| AT2G24390 – AIG2-like (avirulence induced gene) family protein | ArgACG | 5′-UTR (longest mRNA isoform) |
| AT4G03410 – peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein | GlnCUG | 5′-UTR |
| AT4G25640 – detoxifying efflux carrier 35, multidrug and toxin efflux family transporter | LeuCAA | CDS (mRNA isoform 2) |
| AT5G07630 – lipid transporter | LeuCAA | CDS/intron |
| AT5G45720 – AAA-type ATPase family protein DNA polymerase III complex | GluUUC | 3′-UTR |
| AT2G07681 – cytochrome C assembly protein | IleAAU | intron |
| AT2G07706 – unknown protein | MetCAU | intron |
| AT2G07815 – cytochrome C biogenesis | GlyGCC | intron |
| AT3G11402 – Cys/His-rich C1 domain family protein | ValCAC | intron |
| AT3G12587 – oligosaccaryltransferase | ArgCCG | intron |
| AT4G02060 – minichromosome maintenance (MCM2/3/5) family protein | HisGUG | intron (longest isoform, alternative transcription initiation) |
| AT2G36145 – unknown protein | AsnGUU | intron |
| AT5G39530 – unknown function (DUF1997) | ProAGG | intron |
| AT5G57880 – multipolar spindle 1 involved in meiotic spindle organization | MetCAU | intron |
potential pseudogenes
Figure 4.
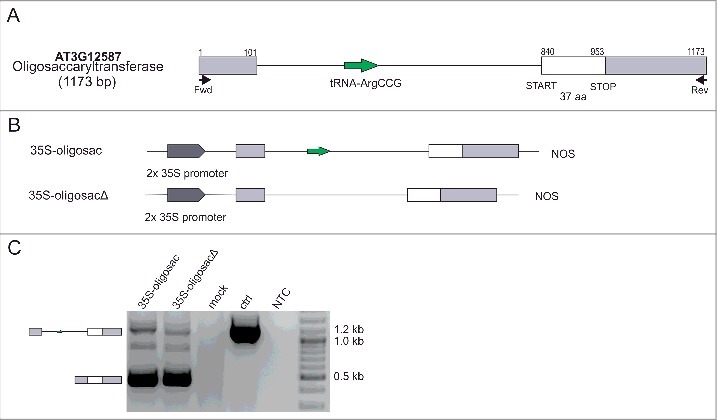
Standard tRNAs do not affect the splicing of their hosting genes. (A) Schematic structure of the At3g12587 hosting tRNA-ArgCCG within its intron. The white box represents an exon, the gray boxes represent UTRs, the lines depict introns, and the arrow corresponds to the tRNA (adapted from TAIR10). (B) Schematic representation of the oligosaccaryltransferase gene variants used. NOS, transcription terminator. (C) Splicing isoforms of the oligosaccaryltransferase transcript recorded by RT-PCR in infiltrated N. benthamiana leaves expressing the oligosaccaryltransferase gene variants shown in B. Schematic structures of the identified isoforms are presented on the left. MOCK is a negative control for the agroinfiltration experiment (leaves infiltrated only with MES buffer). ctrl, amplification of genomic DNA; NTC, non-template control; and marker, 100 bp plus (Thermo Fisher Scientific).
Figure 5.
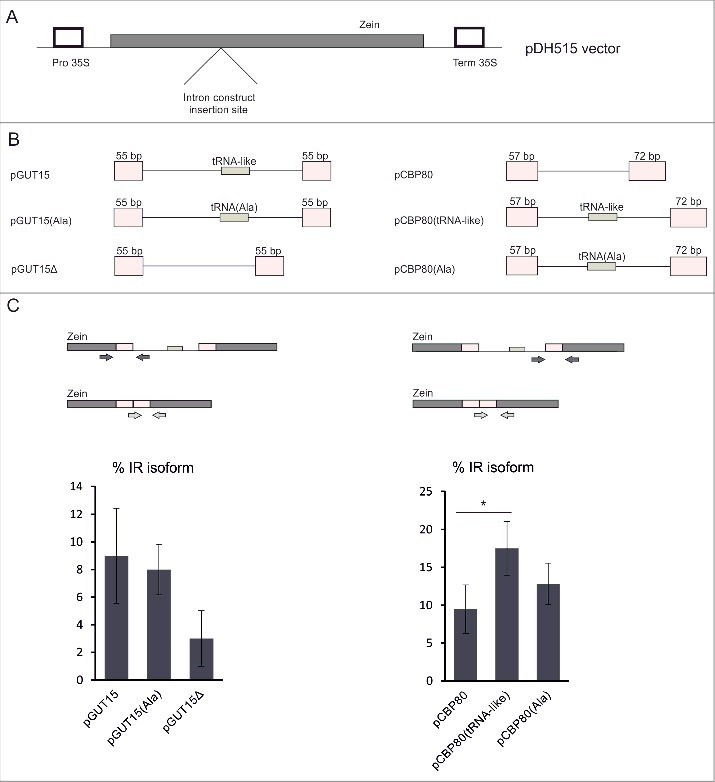
Schematic representation of the intron constructs used to examine the splicing of the tRNA/tRNA-like carrying introns. (A) Schematic structure of the pDH515 vector with marked positions of the inserted intron constructs. Closed box, the zein gene; open boxes, the CaMV 35S promoter and terminator regions. (B) Schematic diagrams of the intron constructs used. Boxes represent exons, and lines depict introns. The sizes of the original exon fragments used in the constructs are shown. (C) RT-qPCR analysis of the splicing efficiency of particular intron constructs. Arrows shown in the upper part of the panel with the intron mini-construct schemes depict the primers used. Intron retention isoform levels were calculated as a percentage of all splicing events (intron retained (IR) plus fully spliced (FS) transcripts, treated as 100%) identified within the analyzed intron. Error bars indicate SD (n = 3), and the asterisk indicates a significant difference in the splicing efficiency between the native construct and the mutated constructs (*p < 0.05).
To determine whether the tRNA-like molecule and tRNA-AlaAGC affect the splicing of intronic sequences, additional constructs were prepared and tested in transfected tobacco protoplasts. We selected CBP80 (CAP-BINDING PROTEIN 80; At2g13540) intron number 3 (similar in size to the GUT15 tRNA-like hosting intron) [20], which does not naturally possess any tRNA-resembling elements. The original intron construct was modified to contain either the GUT15 tRNA-like sequence or tRNA-AlaAGC (called pCBP80(tRNA-like) and pCBP80(Ala), respectively) (Fig. 5B, right panel). RT-qPCR analysis confirmed the inhibitory effect of the GUT15 tRNA-like molecule but not that of tRNA-AlaAGC on CBP80 intron 3 splicing (Fig. 5C, right panel). Therefore, the presence of tRNA loci within introns of RNAPII-dependent genes does not generally interfere with the splicing of their primary transcripts. However, the tRNA-like sequence from the GUT15 gene inhibits the splicing of the introns in which it is embedded.
Discussion
The GUT15 lncRNA was first identified in the tobacco BY-2 cell line as a transcript with a relatively short half-life [15]. In contrast, GUT15 RNA was found among highly stable transcripts in pollen from N. tabacum plants [21]. This differential stability suggests that the regulation of the GUT15 RNA life-time is tissue specific [21]. Interestingly, two Arabidopsis lncRNAs, the GUT15 and CR20-1, are hormonally regulated members of a family conserved among monocots and dicots [22,23]. Using a 3′-RACE approach, we confirmed that the AtGUT15 lncRNA is polyadenylated [23], which is similar to AtCR20-1 [22]. Furthermore, we demonstrated that AtGUT15 is capped and undergoes alternative splicing. The same mRNA-like features have also been observed in mammalian and other plant lncRNAs [4, 6, 24].
AtGUT15 and AtCR20-1 are known to share a highly similar segment that forms a stable secondary structure [25], and a part of this conserved region is present in another location of the Arabidopsis genome (chromosome 1), suggesting that this region could be potentially targeted by the sequence-specific activity of the GUT15/CR20 gene family [23]. Our computational analysis revealed the presence of a second conserved region within the Arabidopsis GUT15 locus. We identified a tRNA-resembling sequence in its final intron (intron 4). Notably, a highly similar sequence, i.e., lacking twelve nucleotides and differing by only a few nucleotides, is also located within the second exon of the Arabidopsis CR20-1 gene (Fig. S1A). Similar to the GUT15-encoded tRNA-like sequence, this sequence does not fold into a secondary structure with canonical anticodon stem-loop characteristics (Fig. S1B); however, this sequence was not identified among actively transcribed tRNAs or tRNA-resembling genes according to our tRNA-seq data. Importantly, the tRNA-like molecule from AtGUT15 is recognized by the tRNA maturation machinery, as evidenced by its 3′-end CCA motif that is not encoded in the genome. This posttranscriptional modification specific to mature tRNA molecules has recently been shown to be present (a complete or partial CCA-terminus) in certain non-tRNA substrates, such as the rps12, cox2 and atp9 mitochondrial mRNA transcripts in maize, human spliceosomal U2 snRNA and the 3′-end of the tobacco mosaic virus RNA [26]. The biological consequences of these unusual nucleotide incorporations remain unexplored and may reflect unspecific reactions of CCA-adding enzymes [26] because most of these transcripts contain a stem-loop structure at the 3′-end that resembles the tRNA minihelix, which is known to be an efficient substrate for CCA-addition in vitro [27,28].
In addition to having the CCA sequence, the GUT15-encoded tRNA-like molecule was detected only in the tRNA-seq library prepared from the deacylated tRNA fraction, suggesting that this molecule may be charged by an amino acid. The biological role of this newly identified tRNA-like sequence, notably its potential involvement in protein translation, remains to be elucidated. However, certain plant-specific RNA viral genomes contain tRNA-like structures that can be specifically aminoacylated by valine, histidine and tyrosine, and this tRNA mimicry is important for diverse aspects of viral infectivity [29]. In bacteria, it plays a role in tagging abnormal proteins for proteolysis [30].
Furthermore, the tRNA-like molecule from AtGUT15 is not expressed by RNAPIII and instead is processed from the GUT15 transcripts synthesized by RNAPII. A similar mechanism has been reported for two tRNA-like molecules generated from the mammalian lncRNAs MALAT1 and Menβ. In both cases, RNase P recognizes the tRNA-like structure in a nascent RNAPII transcript and cleaves it to simultaneously generate the 3′-end of the mature nuclear-retained lncRNA and the 5′-end of the tRNA-like small RNA [31, 32]. Recently, 132 genomic loci resembling the MALAT1 3′-end processing module have been identified among several vertebrate genomes [33]. Interestingly, both mascRNAs (MALAT1-associated small cytoplasmic RNAs) and the GUT15-tRNA-like molecule are marked with a CCA [31]. Furthermore, the RNAPII-mediated transcription of conventional tRNAs has been already reported for tRNASec in Trypanosoma brucei [34].
The AtGUT15 transcript serves as a precursor for another small noncoding RNA, the GUT15-tRF-F5. This small RNA is also generated from the final intron of the AtGUT15 lncRNA. Interestingly, the GUT15 tRNA-like sequence exerts an inhibitory effect on both the splicing of its host intron and GUT15-tRF-F5 biogenesis. Therefore, the GUT15-tRF-F5 fragment may be produced from the spliced intron. In contrast, the accumulation of GUT15 tRNA-like molecules is not influenced by GUT15 lncRNA splicing, suggesting that it is not produced from the intron excised from the AtGUT15 primary transcripts.
The GUT15 lncRNA has been previously characterized as a peptide-coding transcript, because both N. tabacum and A. thaliana genes contain putative short open reading frames (sORFs) of 78 and 75 amino acids, respectively [16]. In 2016, an sORF encoding a 37-aa peptide was identified in GUT15 in Arabidopsis roots using the Ribo-Seq method [35]. However, this peptide does not correspond to the 75-aa peptide predicted by van Hoof and coworkers [40]. This peptide has homologs in multiple species within the Brassicaceae family [35], but the function of this conserved short peptide is unknown. In the same study, additional 26 small ORFs were identified in annotated lncRNAs. These are not the only cases of transcripts believed to solely function as RNA molecules that do in fact code for small peptides [12,36]. In our studies we also detected the AtGUT15 transcripts to be attached to translating ribosomes. Interestingly, only the isoform lacking the tRNA-like sequence was associated with polyribosomal fractions (Fig. S2). Thus, we speculate that the spliced version of the GUT15 transcript can physically interact with ribosomes to produce a small peptide from its first exon, whereas primary GUT15 transcripts serve as a source of tRNA-like molecules. In contrast, Carlevaro-Fita et al. [37] have recently shown that ribosomes are the default destination of most cytoplasmic lncRNAs and may play a role in their degradation because blocking ribosomal elongation results in the stabilization of many associated lncRNAs, which may be also the case for the GUT15 lncRNA. In 1995, Taylor and coworkers found that the level of the shorter GUT15 transcript was significantly increased in an actinomycin D (inhibitor of RNAPII transcription)- and cycloheximide (translational inhibitor)-treated tobacco cell line [15]. Therefore, a complex regulatory mechanism underlies the biogenesis and functions of the noncoding RNAs and small peptides originating from the GUT15 lncRNA.
Our experiments shed light on the phenomena of tRNA and tRNA-like sequences embedded within introns of their host genes. By screening the TAIR10 database, we identified 19 host protein-coding genes containing annotated tRNAs in their introns and UTRs in a sense orientation. Additionally, our global approach revealed two Arabidopsis lncRNAs, GUT15 and CR20-1, that contain similar tRNA-like sequences in their intron and exon, respectively. Our transient expression experiments in tobacco protoplasts confirmed the negative effects of the GUT15 tRNA-like sequence on the splicing of both its original host and the CBP80 intron. Similarly, this influence was also observed when the tRNA-AlaAGC gene was inserted into the AtGUT15 intron, suggesting that this effect may be due to this specific intron. Moreover, the tRNA-ArgCCG embedded in the oligosaccaryltransferase gene does not affect the splicing pattern of its transcript. Accordingly, we conclude that the presence of a tRNA coding sequence in the host protein-coding genes does not interfere with gene splicing. Interestingly, Zhang and coworkers [38] revealed an additional role of tRNA sequences in systemic mRNA transport in plants. These authors found that mobile mRNAs in Arabidopsis are enriched in tRNA-like motifs or are transcribed from genes located in close proximity to annotated tRNA loci, which form di-cistronic mRNA-tRNA transcripts at a high frequency (according to paired-end RNA-seq data). However, the GUT15 lncRNA was not identified among the genes producing mobile transcripts [39], suggesting that the tRNA-related sequence derived from this lncRNA plays other roles.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana Columbia-0 wild-type plants were grown for 10 or 14 days as previously described [40].
N. benthamiana and N. tabacum var. Xanthi plants were grown for 4 and 8 weeks, respectively, at 22°C (16/8-hour light/dark cycle, 50% humidity, and 150–200 µmol m−2 s−1 photon flux density).
Construct preparation
For the agroinfiltration of the tobacco leaves, the genomic sequences of At2g18440 (GUT15) and At3g12587 (gene encoding oligosaccaryltransferase) were amplified (from A. thaliana genomic DNA isolated using the DNeasy Plant Mini Kit, Qiagen) and cloned into the pCR8 plasmid (Thermo Fisher Scientific) using the NotI and AscI restriction sites. The GUT15 and oligosaccaryltransferase constructs lacking the tRNA-like or tRNA sequences were created using a three-step PCR approach as previously described [40]. The mutagenesis of the GUT15 final intron splice sites was performed using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). To detect the expression in plants, the At2g18440 and At3g12587 gene versions were cloned into the pMDC32 Gateway binary vector [41] using the Gateway LR Clonase II Enzyme Mix (Thermo Fisher Scientific).
For transient expression in the tobacco protoplasts, Gateway LR Clonase II Enzyme Mix was used to generate expression clones carrying the genomic sequence of At2g18440 without or under the control of the UBQ10 promoter.
The intron mini-constructs were based on the following two Arabidopsis thaliana introns: the 4th intron from GUT15 (674 bp) and the 3rd intron from the CBP80 gene (485 bp). The intron sequences together with the fragments (>55 bp) from the original 5′- and 3′-exons were isolated from Arabidopsis genomic DNA by PCR and introduced to the plant expression vector pDH515 within an intronless zein gene using the unique BamHI restriction site [42,43]. For the mutant intron construct generation (with different tRNA types), a two-step PCR-based approach was used. First, the desired tRNA sequence was amplified from genomic DNA using primers containing intron-specific (CBP80 or GUT15) overhangs. Second, the PCR products were used as primers in the mutagenesis reaction using the appropriate original intron construct as a template and Pfu Ultra High Fidelity polymerase (Agilent Technologies). The sequences of all constructs were verified by Sanger sequencing. All oligonucleotide sequences are listed in Table 3.
Table 3.
Oligonucleotides used in this study.
| Primers used for constructs preparation | |||
| Name |
Sequence |
gene/cDNA fragment amplified using the primer pair |
Construct prepared using the amplified fragment |
| A01 A02 | ATTTGCGGCCGCATCACCGCCTCCATATTCTTTC TTTGGCGCGCCAATTAATCGTTCAACTATTATTCTATATTCTATATAACATG | GUT15 (At2g18440) | pUBQ10-GUT15wt, Δp-GUT15wt, 35S-GUT15wt, 35S-GUT15Δ, 35S-GUT15Δ5'3′ss |
| A03 | ATCACCGCCTCCATATTCTTTC | 1-1680 bp fragment of GUT15 | 35S-GUT15Δ |
| A04 | ATTTTAAAAATGACGAATAACATATCACGAATACCCTACCACTACG | ||
| A05 | TGGTAGGGTATTCGTGATATGTTATTCGTCATTTTTAAAATAAACTAAGTTATTC | 1762–2261 bp fragment of GUT15 | |
| A06 | AATTAATCGTTCAACTATTATTCTATATTCTATATAACATG | ||
| A07 | CCACAACAGATCTTTTGATCAAGATTGCTTTCAAGAACAAATCAACTTCAGA | mutagenesis of 5′ss in 35S-GUT15wt | 35S-GUT15Δ5'3′ss |
| A08 | TCTGAAGTTGATTTGTTCTTGAAAGCAATCTTGATCAAAAGATCTGTTGTGG | ||
| A09 | GAGTTCAGTGTGGGACAATCGAGTTGATCACAAAGAGCAATA | mutagenesis of 3′ss in 35S-GUT15wt | |
| A10 | TATTGCTCTTTGTGATCAACTCGATTGTCCCACACTGAACTC | ||
| A11 | ATTTGCGGCCGCCTGAGAGTGAGATCCAATACTTGTTCTGT | Oligosaccaryltransferase (At3g12587) | 35S-oligosac, 35S-oligosacΔ |
| A12 | TTTGGCGCGCCAAAGAAAAAAATTCAAAGTTTCATCAAACAT | ||
| A13 | ATTTGCGGCCGCCTGAGAGTGAGATCCAATACTTGTTCTGT | 1-369 bp fragment of Oligosaccaryltransferase | 35S-oligosacΔ |
| A14 | AAAGACCAAAAAAATGGGAACATTGGAGAACAAGTTGAGAATTTATG | ||
| A15 | TCAACTTGTTCTCCAATGTTCCCATTTTTTTGGTCTTTTTTTTTG | 443-1173 bp fragment of Oligosaccaryltransferase | |
| A16 | TTTGGCGCGCCAAAGAAAAAAATTCAAAGTTTCATCAAACAT | ||
| A17 | CGGGATCCTGAAGCTATGCAAAGCTGACGT | GUT15 intron with fragments of flanking exons | pGUT15, pGUT15(Ala), pGUT15Δ |
| A18 | CGGGATCCACACTAGTCACGTTAAGCAAATAGTACATC | ||
| A19 | AGTGGTAGGGTATTCGTGATATGGGGATGTAGCTCAGATGGTAG | At1g06610 (tRNA-AlaAGC) with GUT15 intron overhangs | pGUT15(Ala) |
| A20 | AGTTTATTTTAAAAATGACGAATAACTGGAGATGCGGGGTATCGAT | ||
| A21 | CGGGATCCTTTCTACTACAATGTGCTGAACAATTG | CBP80 (At2g13540) 3rd intron with flanking exons | pCBP80, pCBP80(tRNA-like), pCBP80(Ala) |
| A22 | CGGGATCCCTGGAAATTAGCGTGGACACTTTC | ||
| A23 | GTGTCTATTATATCATATATAACATGTGGGGATGTAGCTCAGATGGTAG | At1g06610 (tRNA-AlaAGC) with CBP80 intron overhangs | pCBP80(Ala) |
| A24 | AATACTATGAATAAATAGATTCCTACATGGAGATGCGGGGTATCGAT | ||
| A25 | GTGTCTATTATATCATATATAACATGTAGATCTGTAAGCTCAAATTGGTAGAGC | tRNA-like with CBP80 intron overhangs | pCBP80(tRNA-like) |
| A26 |
AATACTATGAATAAATAGATTCCTACACAGAACTGGGTGGATTCGAACCA |
|
|
| Primers used in RACE, RT-PCR and qPCR analyses | |||
| Name |
Sequence |
gene/cDNA fragment amplified using the primer pair |
Experiments in which the primer pair was used |
| B01 | TTCGAACCACCAACCTCTCGTCCAC | GUT15 (At2g18440) | 5' RLM-RACE |
| B02 | TCTAGTGGACGAGAGGTTGGTGGTTCG | GUT15 (At2g18440) | 3' RACE |
| B03 | ATCACCGCCTCCATATTCTTTC | GUT15 (At2g18440) | RT-PCR (RACE confirmation) |
| B04 | GGTTGAATGTATTATATATAGAAGTAACAGTACGAT | ||
| B05 | TTTGCTTCCTCCCTCTTTTTC | GUT15 (At2g18440) | RT-PCR (splicing isoforms analysis) |
| B06 | ACAATCCGCAATTCAAAAGC | ||
| B07 | CTGAGAGTGAGATCCAATACTTGTTCTGT | Oligosaccaryltransferase (At3g12587) | RT-PCR |
| B08 | AAAGAAAAAAATTCAAAGTTTCATCAAACAT | ||
| B09 | TCCACAACAGGCCACAATAA | GUT15 IR isoform amplification | qPCR |
| B10 | CCAATTGTTCAACCCTAC | ||
| B11 | TTTGTTCTTGAAAGGATTGTCC | GUT15 FS isoform amplification | qPCR |
| B12 | GGTAAGATGCCTGTTGCGATTGC | ||
| B13 | TCGAAGTTTGATGGATGTCTTTC | CBP80 IR isoform amplification | qPCR |
| B14 | GGTAAGATGCCTGTTGCGATTGC | ||
| B15 | TTTGTATGGGACTTTGATTGGTT | CBP80 FS isoform amplification | qPCR |
| B16 | GGTAAGATGCCTGTTGCGATTGC | ||
| B17 | ATTTCGGCTCCAACAATGTC | HptII | RT-PCR |
| B18 | GATGTTGGCGACCTCGTATT | ||
| B19 | GGTAACATTGTGCTCAGTGGTGG | Actin2 | RT-PCR |
| B20 | CTCGGCCTTGGAGATCCACATC | ||
| B21 | GGAAGAAATTGCTGGGGGTA | Zein | Reverse transcription (Zein_RT) |
| B22 | ACGCGACAGTAGGAAAATGG | At2g36145 | RT-PCR |
| B23 | CCTCATGTGACCCCAAACTT | ||
| B24 | TGTGACTATTTGTTGCCATGGG | At5g57880 | RT-PCR |
| B25 | CCAGTGACTCGGTCCATGTA | ||
| B26 | TCCAGATAAGCCCAGAAGTCA | At5g39530 | RT-PCR |
| B27 |
TCTAAAGGTACATCTGGCGGA |
|
|
| Oligonucteotides used as probes in Northern blot hybridization | |||
| Name |
Sequence |
Detection |
|
| C01 | GACTCTTTAACTCTATTTT | GUT15-tRF-F5 | |
| C02 | ACCAACCTCTCGTCCACTAGAAGA | GUT15-tRNA-like | |
| C03 |
TCATCCTTGCGCAGGGGCCA |
U6 snRNA |
|
| Oligunucletides used in stem-loop end-point RT-PCR analysis | |||
| Name |
Sequence |
Experiments in which the primers were used |
|
| D01 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGCAG | stem-loop reverse transcription primer | |
| D02 | TGGTAGAGCGCTCGTTTTCT | end-point RT-PCR of tRNA-like from GUT15 | |
| D03 | GTGCAGGGTCCGAGGT | ||
Transient expression in N. benthamiana leaves
The tobacco leaf agroinfiltration was performed as previously described [40,44].
Mesophyll protoplast transfection
The constructs were transfected into protoplasts of Nicotiana tabacum var Xanthi isolated from 8-week-old leaves using a previously described protocol for Arabidopsis protoplasts with the following modifications [40]: (1) 10 µg of the vector were used for the transfection, and (2) the transfected protoplasts were incubated overnight in the dark at 22°C before RNA isolation using a modified TRIzol method [45].
RNA isolation and cDNA preparation
For the Northern blot, RACE, qPCR and RT-PCR analyses, total RNA from 100 mg of tissue or protoplast suspensions was isolated using the TRIzol reagent (Thermo Fisher Scientific) or the Direct-zol™ RNA MiniPrep Kit (ZymoResearch). For the small RNA library construction and stem-loop reverse transcription, total RNA enriched for small RNAs was isolated from 100 mg of 14-day-old A. thaliana seedlings using a previously described protocol [46]. The integrity and quality of the NGS-dedicated RNA were verified using an Agilent RNA 6000 Nano Kit (Agilent Technologies).
For the cDNA preparation, total RNA was first treated with Turbo DNase I (Thermo Fisher Scientific); double treatment was required for RNA isolated from transfected protoplasts. The reverse transcription reactions were prepared from 0.5–3 μg of total DNase-treated RNA using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific) and either the oligo-dT(18) primer (Thermo Fisher Scientific) or the Zein-specific primer. The stem-loop pulsed reverse transcription was performed using 200 ng of total RNA enriched for small RNAs [47].
sRNA-seq, tRNA-seq and bioinformatics analyses
A. thaliana total RNA enriched for the small RNA species was used for the small RNA library generation (TruSeq Small RNA Library Prep Kit), and sequencing was performed using an Illumina HiSeq 2000 system. A bioinformatics analysis of the sRNA-seq data was performed as previously described [19].
For the tRNA sequencing, total RNA was isolated and deacylated as previously described [48]. After size fractionation in a PAA gel, the RNA molecules (65–100 nucleotides) were used for the library generation, followed by sequencing using an Illumina HiSeq Platform (Fasteris SA, Switzerland). The GUT15-encoded tRNA-like molecule was identified from the NGS data by mapping the obtained reads to the A. thaliana genome.
The sequence profiles of the A-box and B-box promoter elements were derived by analyzing all A. thaliana tRNA genes. The sequence logos in the profiles were produced using the WebLogo application using multiple sequence alignments of corresponding regions from tRNA genes [49].
Northern blot analyses
For the Northern blot analyses, 5–30 µg of total RNA were used to detect sRNA and tRNA-like sequences. The RNA electrophoresis, blot transfer and hybridization were performed as previously described [46,50]. All hybridizations were performed with three biological replicates.
RT-PCR analyses and quantitative real-time PCR (qPCR)
The RT-PCR amplifications were performed as previously described [51] using the gene-specific oligonucleotide primer pairs listed in Table 3. The amplification of the GUT15-encoded tRNA-like molecule was carried out as previously described [47]. The identity of the PCR product was verified by sequencing.
RT-qPCR was performed as previously described [40]. The amplification efficiency of each primer pair was calculated by making a 10-fold dilution series of the plasmid templates, calculating a linear regression based on the data points and estimating the efficiency from the line slope. The primer pairs with the highest almost equal amplification efficiency (max. difference of 10% was approved) and only one visible peak on the dissociation curve were used for the analysis (Table 3).
The expression levels of particular splicing isoforms were calculated by performing the absolute quantification method using standard curves obtained for a 10-fold dilution series of the plasmid bearing the appropriate isoform. To estimate the splicing efficiency, events identified for the analyzed intron (expressed in copy numbers) were summed and treated as 100%, and the contribution of the fully spliced and unspliced isoforms was then calculated.
All results were analyzed using the SDS 2.3 software (Thermo Fisher Scientific). Error bars were calculated using the SD Function in the Microsoft Excel software. The statistical significance of the presented results was estimated using Student′s t-test at a significance level of *p < 0.05.
3′-, 5′-RACE and 5′RLM-RACE experiments
The 5′- and 3′-RACE cDNA template synthesis and two-step RACE-PCR experiments were conducted using the SMARTer RACE cDNA Amplification Kit (Clontech) according to the manufacturer′s protocol. The cDNA template used in the 5′-RLM-RACE analysis was created using the GeneRacer™ Kit (Thermo Fisher Scientific) according to the manufacturer′s protocol. PCR reactions were performed using the Advantage 2 PCR Enzyme System (Clontech) on a Veriti thermal cycler (Applied Biosystems). PCR products were cloned into the pGEM T-Easy vector (Promega) and sequenced. The primer sequences are listed in Table 3.
Supplementary Material
Funding Statement
This work was supported by the National Science Center under grant numbers UMO-2013/11/N/NZ2/02511 (awarded to PP), UMO-2013/10/A/NZ1/00557 (awarded to AJ) and UMO-2011/03/B/NZ2/01416 (awarded to WMK); the Foundation for Polish Science under grants Mistrz 3.5/2014 (awarded to AJ) and START 32.2017 (awarded to KK); and the KNOW RNA Research Center in Poznan (01/KNOW2/2014).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Dr. Katarzyna D. Raczynska and Dr. Kamilla Bakowska-Zywicka for their help with the polysome profiling and Dr. Andrzej Pacak and Dr. Maria Barciszewska-Pacak for their help with the small RNA library construction.
References
- [1].Kung JTY, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193(3):651−669. doi: 10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17(12):756−770. doi: 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- [3].Liu X, Hao L, Li D, et al.. Long Non-coding RNAs and Their Biological Roles in Plants. Genomics Proteomics Bioinformatics. 2015;13(3):137−147. doi: 10.1016/j.gpb.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Swiezewski S, Liu F, Magusin A, et al.. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462(7274):799−802. doi: 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- [5].Heo JB, Sung S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science (80-). 2011;331(6013):76−79. doi: 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- [6].Ariel F, Jegu T, Latrasse D, et al.. Noncoding Transcription by Alternative RNA Polymerases Dynamically Regulates an Auxin-Driven Chromatin Loop. Mol Cell. 2014;55(3):383−396. doi: 10.1016/j.molcel.2014.06.011 [DOI] [PubMed] [Google Scholar]
- [7].Ben Amor B, Wirth S, Merchan F, et al.. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19(1):57−69. doi: 10.1101/gr.080275.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wierzbicki AT, Haag JR, Pikaard CS. Noncoding Transcription by RNA Polymerase Pol IVb/Pol V Mediates Transcriptional Silencing of Overlapping and Adjacent Genes. Cell. 2008;135(4):635−648. doi: 10.1016/j.cell.2008.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Franco-Zorrilla JM, Valli A, Todesco M, et al.. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39(8):1033−1037. doi: 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- [10].Wang Y, Fan X, Lin F, et al.. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci. 2014;111(28):10359−10364. doi: 10.1073/pnas.1409457111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ding J, Lu Q, Ouyang Y, et al.. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci U S A. 2012;109(7):2654−2659. doi: 10.1073/pnas.1121374109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Campalans A, Kondorosi A, Crespi M. Enod40, a Short Open Reading Frame-Containing mRNA, Induces Cytoplasmic Localization of a Nuclear RNA Binding Protein in Medicago truncatula. Plant Cell. 2004;16(4):1047−1059. doi: 10.1105/tpc.019406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bardou F, Ariel F, Simpson CG, et al.. Long Noncoding RNA Modulates Alternative Splicing Regulators in Arabidopsis. Dev Cell. 2014;30(2):166−176. doi: 10.1016/j.devcel.2014.06.017 [DOI] [PubMed] [Google Scholar]
- [14].Seo JS, Sun H-X, Park BS, et al.. ELF18-INDUCED LONG-NONCODING RNA Associates with Mediator to Enhance Expression of Innate Immune Response Genes in Arabidopsis. Plant Cell. 2017;29(5):1024−1038. doi: 10.1105/tpc.16.00886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Taylor CB, Green PJ. Identification and characterization of genes with unstable transcripts (GUTs) in tobacco. Plant Mol Biol. 1995;28(1):27−38.. [DOI] [PubMed] [Google Scholar]
- [16].Ambro van Hoof James P, Kastenmayer CBT and PJG . GUT15 cDNAs from Tobacco (Accession No. U84972) and Arabidopsis (Accession No. U84973) Correspond to Transcripts with Unusual Metabolism and a Short Conserved ORF. Plant Physiol. 1997;113:1004. [Google Scholar]
- [17].Söll D, RajBhandary U. TRNA: Structure, Biosynthesis, and Function. ASM Press; 1995. [Google Scholar]
- [18].Lamesch P, Berardini TZ, Li D, et al.. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40(Database issue):D1202−10. doi: 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Thompson A, Zielezinski A, Plewka P, et al.. tRex: A Web Portal for Exploration of tRNA-Derived Fragments in Arabidopsis thaliana. Plant Cell Physiol. 2018;59(1):e1−e1. doi: 10.1093/pcp/pcx173 [DOI] [PubMed] [Google Scholar]
- [20].Kmieciak M, Simpson CG, Lewandowska D, et al.. Cloning and characterization of two subunits of Arabidopsis thaliana nuclear cap-binding complex. Gene. 2002;283(1):171−183. doi: 10.1016/S0378-1119(01)00859-9 [DOI] [PubMed] [Google Scholar]
- [21].Ylstra B, McCormick S. Analysis of mRNA stabilities during pollen development and in BY2 cells. Plant J. 1999;20(1):101−108. doi: 10.1046/j.1365-313X.1999.00580.x [DOI] [PubMed] [Google Scholar]
- [22].Teramoto H, Toyama T, Takeba G, et al.. Noncoding RNA for CR20, a cytokinin-repressed gene of cucumber. Plant Mol Biol. 1996;32(5):797−808. doi:doi.org/ 10.1007/BF00020478 [DOI] [PubMed] [Google Scholar]
- [23].MacIntosh GC, Wilkerson C, Green PJ. Identification and Analysis of Arabidopsis Expressed Sequence Tags Characteristic of Non-Coding RNAs. Plant Physiol. 2001;127(November):765−776. doi: 10.1104/pp.010501.cient [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu J, Jung C, Xu J, et al.. Genome-Wide Analysis Uncovers Regulation of Long Intergenic Noncoding RNAs in Arabidopsis. Plant Cell. 2012;24(11):4333−4345. doi: 10.1105/tpc.112.102855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lu Cheng, Gustavo MacIntosh PG. Two Non-coding RNAs, AtGUT15 and AtCR20, modulate the ABA response at high temperature in Arabidopsis. 501707136 [Google Scholar]
- [26].Vörtler S, Mörl M. tRNA-nucleotidyltransferases: Highly unusual RNA polymerases with vital functions. FEBS Lett. 2010;584(2):297−302. doi: 10.1016/j.febslet.2009.10.078 [DOI] [PubMed] [Google Scholar]
- [27].Shi PY, Weiner AM, Maizels N. A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA. 1998;4(3):276−284.. [PMC free article] [PubMed] [Google Scholar]
- [28].Li Z, Sun Y, Thurlow DL. RNA minihelices as model substrates for ATP/CTP:tRNA nucleotidyltransferase. Biochem J. 1997;327(Pt 3):847−851.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dreher TW. Role of tRNA-like structures in controlling plant virus replication. Virus Res. 2009;139(2):217−229. doi: 10.1016/j.virusres.2008.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Keiler KC, Waller PRH, Sauer RT. Role of a Peptide Tagging System in Degradation of Proteins Synthesized from Damaged Messenger RNA. Science (80- ). 1996;271(5251):990−993. doi: 10.1126/science.271.5251.990 [DOI] [PubMed] [Google Scholar]
- [31].Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell . 2008;135(5):919−932. doi: 10.1016/j.cell.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sunwoo H, Dinger ME, Wilusz JE, et al.. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347−359. doi: 10.1101/gr.087775.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang B, Mao YS, Diermeier SD, et al.. Identification and Characterization of a Class of MALAT1 -like Genomic Loci. Cell Rep. 2017;19(8):1723−1738. doi: 10.1016/j.celrep.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aeby E, Ullu E, Yepiskoposyan H, et al.. tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans. Nucleic Acids Res. 2010;38(17):5833−5843. doi: 10.1093/nar/gkq345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hsu PY, Calviello L, Wu H-YL, et al.. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci. 2016;113(45):E7126−E7135. doi: 10.1073/pnas.1614788113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rohrig H, Schmidt J, Miklashevichs E, et al.. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc Natl Acad Sci U S A. 2002;99(4):1915−1920. doi: 10.1073/pnas.022664799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Carlevaro-Fita J, Rahim A, Guigó R, et al.. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA . 2016;22(6):867–882. doi: 10.1261/rna.053561.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang W, Thieme CJ, Kollwig G, et al.. tRNA-Related Sequences Trigger Systemic mRNA Transport in Plants. Plant Cell. 2016;28(6):1237−1249. doi: 10.1105/tpc.15.01056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Thieme CJ, Rojas-Triana M, Stecyk E, et al.. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants. 2015;1(4):15025. doi: 10.1038/nplants.2015.25 [DOI] [PubMed] [Google Scholar]
- [40].Knop K, Stepien A, Barciszewska-Pacak M, et al.. Active 5′ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res. 2016;323(5):gkw895. doi: 10.1093/nar/gkw895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462−469. doi: 10.1104/pp.103.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Simpson CG, Clark G, Davidson D, et al.. Mutation of putative branchpoint consensus sequences in plant introns reduces splicing efficiency. Plant J. 1996;9(3):369−380. doi: 10.1046/j.1365-313X.1996.09030369.x [DOI] [PubMed] [Google Scholar]
- [43].Lewandowska D, Simpson CG, Clark GP, et al.. Determinants of plant U12-dependent intron splicing efficiency. Plant Cell. 2004;16(5):1340−1352. doi: 10.1105/tpc.020743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bielewicz D, Kalak M, Kalyna M, et al.. Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 2013;14(7):622−628. doi: 10.1038/embor.2013.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat Protoc. 2006;1(2):581−585. doi: 10.1038/nprot.2006.83 [DOI] [PubMed] [Google Scholar]
- [46].Kruszka K, Pacak A, Swida-Barteczka A, et al.. Developmentally regulated expression and complex processing of barley pri-microRNAs. BMC Genomics. 2013;14(1):34. doi: 10.1186/1471-2164-14-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Varkonyi-Gasic E, Wu R, Wood M, et al.. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gaston KW, Rubio MAT, Alfonzo JD. OXOPAP assay: For selective amplification of aminoacylated tRNAs from total cellular fractions. Methods. 2008;44(2):170−175. doi: 10.1016/j.ymeth.2007.10.003 [DOI] [PubMed] [Google Scholar]
- [49].Crooks G, Hon G, Chandonia J, et al.. NCBI GenBank FTP Site\nWebLogo: a sequence logo generator. Genome Res. 2004;14:1188−1190. doi: 10.1101/gr.849004.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3(6):1077−1084. doi: 10.1038/nprot.2008.67 [DOI] [PubMed] [Google Scholar]
- [51].Raczynska KD, Ruepp M-D, Brzek A, et al.. FUS/TLS contributes to replication-dependent histone gene expression by interaction with U7 snRNPs and histone-specific transcription factors. Nucleic Acids Res. 2015;43(20):9711−9728. doi: 10.1093/nar/gkv794 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


