ABSTRACT
Riboswitches are RNA regulators that control gene expression by modulating their structure in response to metabolite binding. The study of mechanisms by which riboswitches modulate gene expression is crucial to understand how riboswitches are involved in maintaining cellular homeostasis. Previous reports indicate that riboswitches can control gene expression at the level of translation, transcription or mRNA decay. However, there are very few described examples where riboswitches regulate multiple steps in gene expression. Recent studies of a translation-regulating, TPP-dependent riboswitch have revealed that ligand binding is also involved in the control of mRNA levels. In this model, TPP binding to the riboswitch leads to the inhibition of translation, which in turn allows for Rho-dependent transcription termination. Thus, mRNA levels are indirectly controlled through ribosome occupancy. This is in contrast to other riboswitches that directly control mRNA levels by modulating the access of regulatory sequences involved in either Rho-dependent transcription termination or RNase E cleavage activity. Together, these findings indicate that riboswitches modulate both translation initiation and mRNA levels using multiple strategies that direct the outcome of gene expression.
KEYWORDS: Genetic regulation, Riboswitch, Rho transcription termination, translation
Bacteria make use of sophisticated regulatory elements to finely control gene expression in response to environmental signals [1]. Such regulatory elements include riboswitches, RNA modules located in 5′ untranslated regions (UTR) that control gene expression upon the sensing of a specific metabolite [2,3]. Riboswitches are composed of two modular domains: an aptamer that is involved in metabolite binding, and an expression platform that undergoes structural changes which lead to altered gene expression [2]. Initial studies of riboswitches focused on elucidating the process of ligand recognition [4,5]. Further studies suggested that riboswitches regulate either at the level of transcription, by modulating the formation of an RNA structure that promotes intrinsic transcription termination, or at the level of translation, by controlling access to the ribosome binding site (RBS) [6]. This view has recently been challenged by the discovery of riboswitches that modulate mRNA decay and/or Rho-dependent transcription termination [7,8], thus expanding the list of known riboswitch regulatory mechanisms. Interestingly, it was also found that some of these riboswitches exert a dual level of regulation by controlling both mRNA levels and translation initiation. This is exemplified by the Escherichia coli lysC riboswitch, which adopts a structure that promotes RNase E cleavage within the riboswitch domain and simultaneously precludes ribosome access to the RBS [7]. Similarly, it was recently shown that the E. coli ribB and thiC riboswitches control both translation initiation and Rho-dependent transcription termination, by structural reorganization of their expression platforms [8,9]. Here, we discuss recently obtained data from our laboratories suggesting that such dual regulatory mechanisms are common among translationally-acting riboswitches [10].
Lessons from the past: Intimate relationships between the efficiency of translation, transcription, and mRNA decay
The absence of a nucleus in prokaryotic cells allows for coupling of transcription and translation. Thus, these processes are intimately connected, as exemplified by the process of ribosome-mediated transcriptional attenuation [11]. One of the first characterized examples of attenuation was the trp operon in E. coli [12]. In this regulatory system, the availability of tryptophan-charged tRNATrp is sensed by the translation of the TrpL leader peptide, which is involved in the modulation of transcription of the downstream genes of the operon. Specifically, attenuation at trp relies on a transcriptionally-paused RNA polymerase (RNAP) within the trpL leader transcript that allows time for a ribosome to initiate TrpL translation. When tRNATrp levels are high, efficient translation of TrpL favors the folding of an intrinsic terminator within the leader RNA, thereby attenuating transcription of the full trp operon. By contrast, when tRNATrp levels are limiting, ribosome stalling on tryptophan codons prevents terminator folding, thereby allowing RNAP to transcribe the complete trp operon. In other attenuation mechanisms, such as in the case of the E. coli tnaA operon [13], ribosome stalling allows Rho to load onto RNA and terminate transcription [14].
Translation efficiency can also impact mRNA levels post-transcriptionally, by controlling the rate of RNA degradation. Ribosome occupancy on mRNAs alters the accessibility for RNases [15]. In most cases, untranslated RNAs are more accessible, and hence translation levels are typically inversely correlated with mRNA stability. Consequently, modulation of translation can indirectly alter mRNA levels by affecting RNA degradation, as exemplified by sRNAs and RNA-binding proteins that repress translation [15,16].
Translationally-acting riboswitches modulate mRNA levels
As described above, many previous studies have revealed connections between translational regulation and regulation of mRNA levels either by modulating transcription or RNA stability. The fact that many riboswitches control translation initiation raises the possibility that their RNA levels are also regulated. We chose to investigate the role of Rho-dependent transcription termination in riboswitch regulation, since Rho is an important regulator of transcription in bacteria, and Rho only terminates transcription of untranslated RNA [16–18]. We determined the impact of Rho on transcription of riboswitch-controlled mRNAs by using chromatin immunoprecipitation to monitor the association of RNAP with DNA in riboswitch 5′ UTR regions, and within the corresponding ORFs. We chose to focus on riboswitches in E. coli, which are expected to function at the level of translation. We grew cells in rich medium to ensure that riboswitches would be bound to their respective metabolite. For genes that are regulated by premature Rho-dependent transcription termination, it is expected that local RNAP occupancy will be higher in the 5′ UTR than in the coding region owing to RNAP being released from the DNA within the coding region through the action of Rho. Remarkably, our data showed higher RNAP occupancy in the 5′ UTR region than in the ORF for six of the seven E. coli riboswitches: those responding to flavin mononucleotide (FMN) (ribB), thiamin pyrophosphate (TPP) (tbpA, thiC and thiM), lysine (lysC), and magnesium (mgtA). However, in a Rho mutant strain defective in the termination of many RNAs [19], RNAP occupancy was significantly increased within ORFs of genes controlled by FMN, TPP and lysine riboswitches. These data strongly suggest that Rho-dependent transcription termination is used by these riboswitches to modulate mRNA levels. Recently obtained RNA-seq data indicate that Rho is active within thiM and lysC riboswitch regions [20], suggesting that riboswitches can simultaneously modulate transcription termination and translation initiation. Surprisingly, our results showed that Rho is not involved in the regulation of the mgtA riboswitch as was found previously for the Salmonella Typhimurium homologue [8], although RNAP levels in the mgtA ORF are regulated by riboswitch activity. Additionally, we did not observe Rho-dependent termination associated with the btuB riboswitch, which is involved in the control of the btuB cobalamin transporter [21]. However, recently obtained data suggest that this may be due to adenosylcobalamin (AdoCbl) levels being sub-saturating for cells grown in LB medium (Bastet et al., unpublished observations). Overall, our data strongly suggest that most E. coli riboswitches rely on a dual regulatory mechanism whereby both translation initiation and Rho-dependent transcription termination are used to control gene expression. Interestingly, a dual regulatory mechanism has also been identified for the FMN riboswitch of Corynebacterium glutamicum [22], indicating that such dual-control mechanisms are not only restricted to E. coli.
Indirect and direct regulation of mRNA levels by riboswitches
To investigate how riboswitches control Rho-dependent transcription termination, we characterized the regulatory mechanism employed by the TPP-responsive E. coli thiM riboswitch [10]. It was previously observed using thiM-lacZ transcriptional fusions that the thiM riboswitch strictly regulates translation initiation and not mRNA levels [7,23]. At first glance, these results seem contradictory with our ChIP-qPCR data and Northern Blot experiments in which we observed the complete disappearance of thiMD mRNA following the TPP addition in the growth media [10]. However, transcriptional thiM-lacZ fusions revealed that a TPP-dependent reduction in the activity of thiM-lacZ transcriptional fusions required inclusion of >20 thiM codons [10]. Consistent with these data, in vitro transcription assays showed that Rho terminates transcription early in the thiM ORF (Fig. 1) [10]. Furthermore, RNase H assays revealed that the thiM region encompassing residues 201 to 210 was protected in the presence of Rho (Fig. 1), consistent with the presence of a Rho utilization site (rut) in this region. Accordingly, bioinformatic analysis showed that the thiM sequence encompassing the first 34 codons exhibits high and low proportions of cytosine and guanine [10], respectively, expected features of a rut [18]. Together, these observations support a model in which inhibition of translation indirectly controls thiM mRNA levels by allowing Rho binding to nascent RNA, leading to transcription termination early within the thiM ORF. Because rut and transcription termination sites are not located within the riboswitch domain but rather in the thiM ORF, it suggests that the activity of Rho should not be dependent on the sequence and specific architecture of the riboswitch but solely on its ability to prevent translation. Indeed, we found that Rho-dependent transcription termination occurred even when using chimeric btuB-thiM-lacZ fusions in which translation inhibition is performed by the AdoCbl-sensing riboswitch [10]. Such an indirect control mechanism using mRNA elements located early within a regulated ORF could serve as an efficient strategy to adjust RNA synthesis in the first steps of translation elongation [24].
Figure 1.
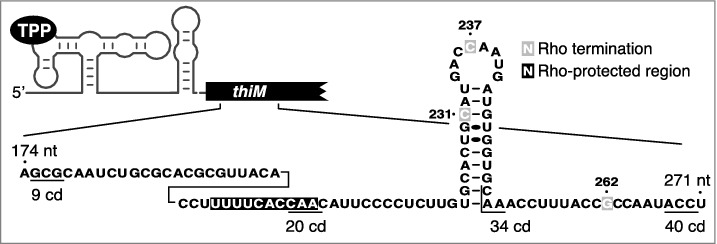
The expression of thiM is modulated through Rho-dependent transcription termination. RNase H cleavage assays show that a region (black) is protected in the presence of Rho, consistent with the presence of a rut. Rho-dependent transcription termination sites identified in vitro are shown in gray and codons (cd) 9, 20, 34 and 40 are underlined.
In contrast to the indirect mRNA control mechanism described above, the E. coli thiC, ribB and lysC riboswitches directly control RNA levels by modulating either Rho-dependent transcription termination or RNase accessibility [7–9]. In these cases, all necessary elements for mRNA regulation and translation inhibition are located within the riboswitch domain. Therefore, these riboswitches directly control mRNA levels by altering their structure upon metabolite binding, and not indirectly as a consequence of the inhibition of translation initiation (Fig. 2). For example, in addition to inhibiting translation initiation, ligand binding to the riboswitch domain was found to expose rut sequences (thiC and ribB) [8,9] or RNase E cleavage sites (lysC) [7]. In all cases, the regulation of mRNA levels is intimately linked to the identity of the riboswitch sequence, which is fundamentally different from the indirect control mechanism. Whether or not additional E. coli riboswitches [10] make use of direct or indirect mechanism to modulate Rho-dependent transcription termination will require further characterization.
Figure 2.
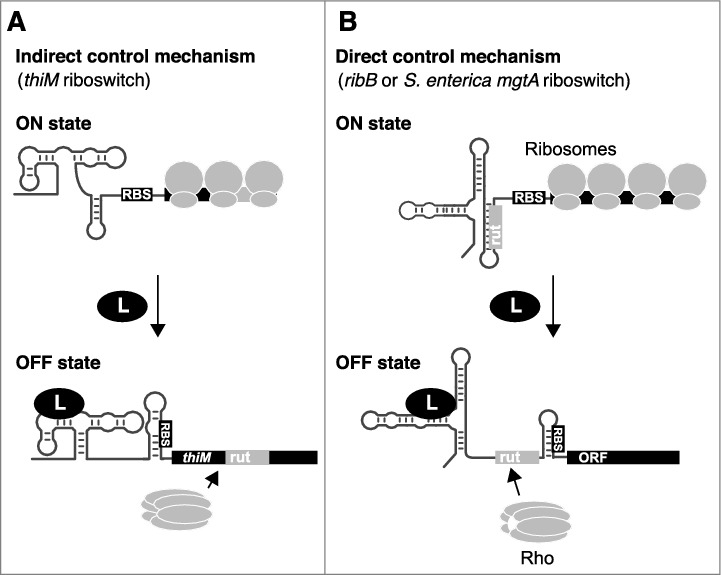
Indirect and direct control mechanisms used to regulate mRNA levels. A) The indirect control mechanism primarily relies on the modulation of translation initiation. Upon translation inhibition, a rut becomes accessible to Rho binding, leading to transcription termination. B) The direct control mechanism is performed by directly modulating the access of rut, which is dictated by ligand-dependent riboswitch conformational changes. Both indirect and direct control mechanisms rely on the inhibition of ribosome binding to the RBS in the ligand-bound state.
The level of regulatory control impacts expression kinetics
During the course of our study [10], we obtained evidence that the thiM riboswitch can perform TPP binding both co-transcriptionally and post-transcriptionally. However, Rho can only control expression if the ligand is bound co-transcriptionally, whereas inhibition of translation initiation can occur either co- or post-transcriptionally. Our finding suggests the possibility that the thiM riboswitch may also control RNA levels post-transcriptionally by promoting mRNA degradation as a result of keeping the mRNA ribosome-free in the TPP-bound state (Fig. 2A) [25]. Consistent with this possibility, our data indeed suggest that the thiM mRNA half-life is decreased in the presence of TPP [10]. Importantly, mRNA degradation is irreversible, whereas translational repression has the potential to be reversed if the ligand dissociates before the RNA is destroyed.
Concluding remarks
As described above, recent studies of riboswitch function have revealed that many riboswitches control gene expression at multiple levels. This is especially true for riboswitches that directly control translation initiation, since translation levels are often coupled to transcription termination and/or altered mRNA stability. Specifically, untranslated RNA is prone to Rho-dependent transcription termination and RNase-mediated degradation. Hence, we propose that Rho-dependent termination and RNA processing are likely to be secondary levels of regulation for most riboswitches that control translation. In some cases, riboswitches directly control Rho and/or RNase activity by modulating accessibility of rut and/or processing sites in the 5′ UTR. For these riboswitches, Rho-dependent transcription termination and RNase processing within the ORF are likely to be add-on regulatory mechanisms. By coupling translational repression to a reduction of mRNA levels, riboswitches ensure irreversible regulation that cannot be achieved through translational repression alone. Although it is now clear that riboswitches often regulate their target genes at multiple levels, there are still many factors that have not been well characterized in riboswitch regulation which will need to be addressed to obtain a complete picture regarding regulatory mechanisms. In particular, the recent characterization of orphan riboswitch candidates [26] suggests that new variations of genetic regulatory mechanisms are likely to be found.
Funding Statement
National Institutes of Health (1DP2OD007188). Canadian Institutes of Health Research (MOP828).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Lafontaine laboratory for discussion.
References
- [1].Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Breaker RR, McCown P, Corbino K, et al. Riboswitch diversity and distribution. RNA. 2017;23(7):995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jones CP, Ferré-D'Amaré AR. Long-range interactions in riboswitch control of gene expression. Annu Rev Biophys. 2017;46:455–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol. 2011;3:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–63. [DOI] [PubMed] [Google Scholar]
- [7].Caron MP, Bastet L, Lussier A, et al. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci U S A. 2012;109:E3444–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hollands K, Proshkin S, Sklyarova S, et al. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci U S A. 2012;109:5376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chauvier A, Picard-Jean F, Berger-Dancause J-C, et al. Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat Commun. 2017;8:13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bastet L, Chauvier A, Singh N, et al. Translational control and Rho-dependent transcription termination are intimately linked in riboswitch regulation. Nucleic Acids Res. 2017;45(12):7474–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–4. [DOI] [PubMed] [Google Scholar]
- [12].Yanofsky C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA. 2007;13:1141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Konan K V, Yanofsky C. Role of ribosome release in regulation of tna operon expression in Escherichia coli. J Bacteriol. 1999;181:1530–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stewart V, Landick R, Yanofsky C. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J Bacteriol. 1986;166:217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–33. [DOI] [PubMed] [Google Scholar]
- [16].Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: The RNA 3′-end chronicles. J Mol Biol. 2011;412:793–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kriner MA, Sevostyanova A, Groisman EA. Learning from the Leaders: Gene regulation by the transcription termination factor Rho. Trends Biochem Sci. 2016;41:690–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mitra P, Ghosh G, Hafeezunnisa M, et al. Rho protein: Roles and mechanisms. Annu Rev Microbiol. 2017;71:687–709. [DOI] [PubMed] [Google Scholar]
- [19].Martinez A, Burns CM, Richardson JP. Residues in the RNP1-like sequence motif of Rho protein are involved in RNA-binding affinity and discrimination. J Mol Biol. 1996;257:909–18. [DOI] [PubMed] [Google Scholar]
- [20].Sedlyarova N, Shamovsky I, Bharati BK, et al. sRNA-Mediated Control of Transcription Termination in E. coli. Cell. 2016;167:111–121.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci U S A. 2000;97:7190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Takemoto N, Tanaka Y, Inui M. Rho and RNase play a central role in FMN riboswitch regulation in Corynebacterium glutamicum. Nucleic Acids Res. 2015;43:520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ontiveros-Palacios N, Smith AM, Grundy FJ, et al. Molecular basis of gene regulation by the THI-box riboswitch. Mol Microbiol. 2008;67:793–803. [DOI] [PubMed] [Google Scholar]
- [24].Franklund C V, Kadner RJ. Multiple transcribed elements control expression of the Escherichia coli btuB gene. J Bacteriol. 1997;179:4039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dreyfus M. Killer and protective ribosomes. Prog Mol Biol Transl Sci. 2009;85:423–66. [DOI] [PubMed] [Google Scholar]
- [26].Weinberg Z, Lünse CE, Corbino KA, et al. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res. 2017;45:10811–23. [DOI] [PMC free article] [PubMed] [Google Scholar]


