Abstract
Breast cancer (BC) is one of the most complex, diverse and leading cause of death in women worldwide. The present investigation aims to explore genes panel associated with BC in different African regions, and compare them to those studied worldwide.
We extracted relevant information from 43 studies performed in Africa using the following criteria: case-control study, association between genetic variations and BC risk. Data were provided on mutations and polymorphisms associated with BC without fixing a specific date. Case-only studies and clinical trials were excluded.
Our study revealed that the majority of African BC genetic studies remain restricted to the investigation of BRCA1 and BRCA2 genes and differences in their mutations spectrum. Therefore, it is necessary to encourage African researchers to characterize more genes involved in BC using methods generating global information such as next-generation sequencing in order to guide specific and more effective therapeutic strategies for the African community.
Key words: Africa, breast cancer, genetics, genetic variability
Introduction and background research
Breast cancer (BC) is one of the most complex and diverse diseases, it represents the leading cause of death in women worldwide among other cancers and infectious diseases [1]. Along with ovarian cancer, they constitute the most common forms of cancer in the developed and developing countries [2].
Since BC is considered as a public health problem in most countries as stated by the World Health Organization (WHO) [3]. Several studies have been conducted, either to establish the link between this malignant tumour and its host, also to understand its genesis and its response to treatments and drug effects [4]. According to the 2014 World Health Cancer report, they estimated 14 million new cases and 8.2 million deaths due to cancer worldwide [5]. Currently, 690 000 new cases are being diagnosed annually in the developed regions with around 92 000 new cases in Africa [6]. Even though the incidence for BC is high, the rate of mortality has been decreasing over the past 25 years [7]. The last update of the global burden of cancer – International Agency for Research on Cancer (GLOBOCAN-IARC) [8] also reported that the prevalence of BC in adult populations shows a high rate in Asia (36.7%), followed by Europe (29.1%), North America (17.2%), Africa (7.0%) and Latin America (8.8%). The incidence of mortality has been likely the same in Asia (39%), Europe (27.5%), North America (15.3%), Latin America (9.1%) and Africa (8%). By 2020, the forecast of BC will continue its increase slowly in the five continents from 1 651 872 in 2012 to 1 956 124 cases alongside with number of mortality going from 517 578 cases in 2012 to 617 479 [8]. The growing prevalence of BC, especially in Africa, incriminates many factors, including hormonal, toxic or genetic factors. In this context, many genes such as BRCA1/2,TP53, HER2 and CHEK2 have been studied to characterize mutations specific to each population.
The BC screening by mammography can reduce the cancer-specific mortality. The value of other screening methods such as the genetic detection of familial and non-familial mutations is relevant and explain a large part of BC heredity. Although, the lack of observations of such gene mutations, particularly in Africa, is one of the reasons for the growing disparity between the sensitivity of diagnosis and rate of variation. Besides genetic variation associated with inheritance of some genes with defined penetrance of BC [4], the determination of other risk factors leading to the genetic variability of the disease is an area that needs to be considered for accurate detection of BC in Africa.
Epidemiological studies have provided great support to understand the genetic variability and risk factors among populations around the world to facilitate the accuracy of diagnosis, medical support and drug response. Therefore, the aim of this work is to provide a panel of genes associated with BC in the African populations. This panel of genes will be extracted through a literature review. Our investigation could be useful both for future epidemiological studies and enable decisions on new research and management strategies for BC genetic studies.
Methods
We searched in the databases PubMed, Scopus and Web of Science for studies reporting genetic variations implicated in BC in Africa. The search was conducted based on specific Medical Subject Headings (MeSH) and keywords, which are BC, genetics, gene, mutation, association and Africa. All languages were searched initially, but only English language studies were selected.
We extracted relevant information from each study using a standardized form and we included studies in this review only if the following criteria were met: a case-control study on the association between a variation (SNP, InDel, CNV…) and BC risk, and data were provided on mutations and polymorphisms associated with BC. We excluded case-only studies and clinical trials. The study selection process and flow diagram for identifying studies is detailed in Fig. 1.
Fig. 1.
Flow diagram for identifying studies for assessment of breast cancer mutations in Africa.
In order to extract most of studies regarding BC and genetics, we identified published articles without fixing a specific date. The strategy adopted was applied for African populations to evaluate African BC specificities. We also searched the data concerning other continents, this served to compare and discuss African results.
Results
The African continent is commonly divided by the United Nations (UN) into five regions or sub-regions: Northern, Western, Central, Eastern and Southern Africa (http://www.un.org/esa/population/publications/worldageing19502050/pdf/96annexii.pdf). Therefore, we used this division to present the results retrieved from different databases.
Northern Africa region (NA)
For this region, we summarized studies on BC mutations in Algeria, Tunisia, Egypt and Morocco. Only Libya was excluded for lacking relevant genetic studies on BC. In total, 28 major studies have been conducted in NA region, including more than 3700 BC cases and showing that BRCA1/2 genes were the most investigated for their association with BC. Other genes such as HER2, P53, APOBEC3 and MTHFR were also studied.
In Algeria, three major studies have investigated the carried mutations on BRCA1 and BRCA2 genes in both sporadic and familial cases. Uhrhammer et al. [9], using complete gene sequencing found one BC founder mutation in BRCA1 gene (c.798_799delTT) in Algerian population with 9.8% familial cases and 36.4% sporadic cases. This mutation has been reported as the first non-Jewish founder mutation to be described in NA.
In another study performed by Cherbal et al. [10], over 101 individuals from 79 breast and ovarian cancer families were examined for unclassified variants (UVs) and polymorphisms in the BRCA1 and BRCA2 genes by Single-strand conformation polymorphism (SSCP) or High-Resolution Melting (HRM) curve analysis, followed by direct sequencing. The result revealed 168 UVs and polymorphisms in BRCA1/2 genes, 68 in BRCA1 and 100 in BRCA2. Cherbal and colleagues also performed BRCA1/2 mutation screening by HRM curve analysis and direct sequencing in 86 individuals from 70 Algerian BC families with history of BC, five mutations were detected in BRCA1 (c.83_84delTG, c.181T>G, c.798_799delTT, delEx2, delEx8) and 57 UVs/SNPs were revealed in both BRCA1 and BRCA2 [11]. In a recent study, Henouda et al. [12] found five mutations in 40 different Algerian families with early BC. Among them, five mutations were identified in BRCA1 (c.1817del, Del exon 2, c.4065_4068del, c.5332 + 1G>A, c.5117G>C) and nine in BRCA2 (c.7654dupA, c.1528G>T, Del exons 19-20, c.6450del, c.7462A>G, c.1504A>C, c.5117G>C, c.5939C>T, c.1627C>A).
In Tunisia, a BRCA1 study on nine Tunisian patients with hereditary BC was carried out in order to evaluate the implication of the BRCA1 and DNA mitochondrial mutations and revealed that the mitochondrial mutation 315.insC was strongly implicated in two unrelated patients [13]. Mahfoudh et al. [14] performed a screening for germline mutations of BRCA1 in 16 Tunisian high-risk BC families, where six families were found with BRCA1 mutations: three truncating mutations were described in BRCA1 (c.798_799delTT, c.3331_3334delCAAG, c.5266dupC), one specific to Tunisian population (c.212 + 2insG) and the c.798_799delTT was suggested to be a Tunisian founder mutation based on its frequency (18%). Mestiri et al. [15] screened 12 Tunisian women with familial or sporadic BC for BRCA1 gene mutations and the 1294del40 mutation of BRCA1 was found only in a patient with non-familial BC. In contrast, the 185delAG mutation was absent in all cases of BC. Troudi et al. [16] studied the BRCA1/2 genes in 36 Tunisian patients with breast and/or ovarian cancer. The results revealed 90% of cases with deleterious mutations. In BRCA1 four mutations (c.211dupA, c.4041delAG, c.2551delG and c.5266dupC) were detected in two unrelated patients, the c.5266dupC mutation was described for the first time in a non-Jewish Ashkenazi population. In addition, two frame-shift mutations (c.1309del4 and c.5682insA) were observed in BRCA2. Furthermore, Riahi et al. [17] performed a screening on BRCA1/BRCA2 genes in 48 patients by direct sequencing, the result revealed 12 mutations including three in BRCA1 (c.211dupA, c.5266dupC and c.1504_1508delTTAAA) and two novel mutations in BRCA2 (c.1313dupT and c.7654dupT). In a recent work, the same group proposed a cumulative mutation analysis with data from three Tunisian studies including 92 Tunisian women, the results revealed two recurrent mutations (c.211dupA and c.5266dupC) in 76% of BRCA1-related families and three recurrent mutations (c.1310_1313del, c.1542_1547delAAGA and c.7887_7888insA) in 90% of BRCA2-related families [18]. Hadiji-Abbes et al. [19] identified a novel in-frame deletion (5456del6 bp) in the BRCA2 gene in an early onset woman with BC without family history.
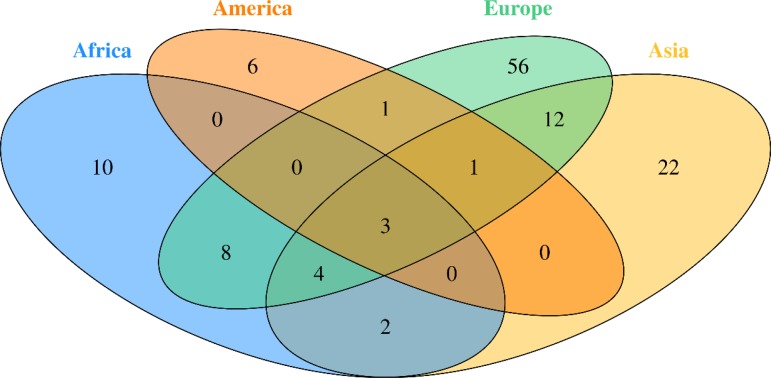
Fig. 2.
Venn diagram of studied Breast Cancer genes in different continent
HER2 gene has been reported as major factor in BC development and progression [20]. Therefore, Kallel et al. [20] focused on this gene by analysing three polymorphisms in 148 cases and 290 controls from the Tunisian population. The Ile(655)Val mutation was found to be significant in 90% of cases. The noncoding SNP (rs903506) and the H(AC)I4 mutation were also implicated. The association of Ile(655)Val with BC has been confirmed by a further meta-analysis from 27 case-control studies [21].
A study concerning the P53 genewas conducted in order to investigate the association of p53 Arg72Pro, Ins16 bp and G13964C polymorphisms and their haplotypes with BC risk in 159 Tunisian female patients. The results revealed that these mutations were associated with familial BC risk in this population [22].
The ESR1 and ESR2 genes were also studied by Kallel et al. [23] in 148 Tunisian BC patients and 303 controls using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. The ESR1 2014 G allele showed significant association with BC risk (p = 0.025).
In Egypt, Bensam et al. [24] screened BRCA1 and BRCA2 in 20 Egyptian patients with BC. Mutations were detected in 44% of the studied population, with 18% in the BRCA1 gene (185AGdel, 624C>T) and 26% attributed to BRCA2 (999TCAAAdel, 2256T>C, 8934G>A).
The P53 gene has also been studied in the Egyptian population by El-Ghannam et al. [25], their study focused on mutation detection in 30 patients with BC using flow-cytometry, PCR-SSCP and sequencing. P53 mutations including A218 T, R279G, S297X and Y159X were detected in 17% of patients.
In another study, Hussein et al. [26] examined the relation between the PON L55 M and Q192R polymorphisms with BC risk in Egyptian females and analyzed their relation to clinic-pathological parameters of BC. Both SNPs were shown to be significantly associated with an increased risk of BC. However, when they conducted a study in order to evaluate the association of two polymorphisms in XPD (Asp312Asn) and XRCC1 (A399G) on 100 BC Egyptian females, they did not detect an association between the XRCC1 gene mutation and BC [27].
In Morocco, three studies targeted BRCA1/2 mutations, the first one was conducted by Laarabi et al. [28], and was interested in analyzing BRCA1/2mutations in five healthy women belonging to three families with an elevated risk of BC, this investigation revealed that three asymptomatic women were found to be carriers of BRCA1/2 mutations. The second study was performed by Tazzite et al. [29] on a Moroccan cohort of 40 women, diagnosed with BC and a familial history of breast/ovarian cancer, or aged less than 40 years old showed that 25.64% of patients carried BRCA1/2 mutations. The third study, conducted by Laraqui et al. [30] on 121 Moroccan women diagnosed with BC, only BRCA1 status was investigated and revealed that mutations were found in 36.1% of familial cases and 1% (1/102) of early-onset sporadic. The result of these three studies showed 14 BRCA1/2 point mutations; nine in BRCA1 (c.68_69delAG, c.181T>G, c.798_799delTT, c.3279delC, c.2805delA, c.1016dupA, c.4942A>T, c.5062-5064delGTT, c.5095C>T) and five in BRCA2 (c.517-1G>A, c.3381delT, c.5073dupA, c.7110delA, c.7235insG).
Considering the potential function of driver and APOBEC3 genes in the process of tumorigenesis in BC, it is possible that germline variations and copy number variations (CNV) in those genes could influence the risk of BC. In this term, a case-control study was conducted in a group of Moroccan women targeting 36 SNPs in 13 genes (APOBEC3A, APOBEC3B, ARID1B, ATR, MAP3K1, MLL2, MLL3, NCOR1, RUNX1, SF3B1, SMAD4, TBX3, TTN). The analysis showed that 12 SNPs in eight driver genes, four SNPs in APOBEC3B and one SNP in APOBEC3A were associated with BC risk and/or clinical outcome at the significance level of 0.05. RUNX1_rs8130963 (p = 0.0005), TBX3_rs8853 (p = 0.0003), TBX3_rs1061651 (p = 0.0002), TTN_rs12465459 (p = 0.0009), were the most significantly associated SNPs with BC risk. A strong association with clinical outcome was detected for SMAD4_rs3819122 with tumour size (p = 0.009) [31].
Another study was conducted to investigate if MTHFR C677T polymorphism modulates the risk of developing BC in Moroccan women. Results showed a positive correlation between the MTHFR C677T polymorphism and progesterone receptor expression (p = 0.04). There was a significant association between C677T polymorphism and BC risk in both additive (p = 0.007) and dominant (p = 0.008) models [32].
Many studies were also conducted to investigate the implication of three genes described as a high penetrance BC susceptibility. The PIN3 Ins16 bp polymorphism of the TP53 gene [33], the ABCB1 C3435T polymorphism [32] and the +936C/T VEGF-A polymorphism [34]. However, there was no evidence of a significant association between these polymorphisms and risk of BC.
Southern Africa (SA) region
No founder genes have been described in BC patients of the four countries (Botswana, Lesotho, Namibia and Swaziland) included in SA's region. Whereas, the studies performed among South Africa's BC patients revealed variants in the two major genes BRCA1&BRCA2 and in five other genes deemed intermediate (CHEK2, PALB2) and minor (RAD50, MTHFR, hMLH).
Reeves et al. [35] were the first to report the role of BRCA1 in 90 SA BC families in 2004. Thereafter, Francies et al. [36] studied 108 sporadic patients with 78 women premenopausal and 30 triple negative breast cancer (TNBC) from the four South African ethnic groups, to determine the mutations frequency in BRCA1, BRCA2 and PALB2 and evaluate the presence of the CHEK2. This study used the next-generation sequencing (NGS) approach in combination with Multiplex Ligation-dependent Probe Amplification (MLPA) to detect large rearrangements in BRCA1 and BRCA2. The result revealed the BRCA2 (c.7934delG) Afrikaner founder mutation, the BRCA2 variant (c.9875C>T) and the CHEK2 mutation (c.1100delC). PALB2 variants (c.118A>G, c.2845T>C) were also described as probably damaging [36]. The two studies mentioned previously and carried out on the BRCA1 gene reported nine following mutations in a South African population: c.181T>G, c.212G>A, c.3593T>A, c.1155G>A, c.1953_1954insA, c.1843_1845delTCT, 1493delC, 185delAG, 4957insC, 5382insC, E881X and S451X [35, 36].
Sluiter et al. [37] also identified PALB2 as a BC susceptibility gene, and the related mutations double the BC risk with moderate to low penetrance. In a cohort of 48 young South African BC patients unselected for family history of BC, the authors determined the involvement of PALB2 mutations and they identified a novel truncating mutation, c.697delG (V233fs).
MTHFR, RAD50 and hMLH1 genes were also showed to be involved in the development of BC in this region of Africa [38, 39].
Western Africa region
In this region of Africa, few studies focused on the genetic risk factors for BC. These studies were performed only in Senegal and Nigeria, and were restricted on the description of the mutations in BRCA1, BRCA2 and LEPR genes.
In Nigeria, genetic studies revealed that BRCA1 and BRCA2 are the most prevalent cause of BC. The overall BRCA1/2 mutation rate is 11%, which is the highest reported rate for any BC cohort from a non-founder population unselected for family history, ethnicity or age of onset in Nigeria. Unexpectedly, only 7.1% of patients carried BRCA1 mutations and 3.9% individuals were BRCA2 mutation carriers. A total of 48 mutations were found in BRCA1/2 (31 in BRCA1 and 17 in BRCA2), including non-sense, missense, frame-shift and splice-site mutations. Deleterious mutations like Y101X, 1742insG, C64Y, 4241delTG and del Ex 21 were the most commonly carried BC-related mutations in exons 11, 12 and 21 of the BRCA1 gene. The BRCA2 gene in the Nigerian population carried the 1538delAAGA, 2630del11 and 9045delGAAA mutations in exons 10, 11 and 22, respectively [40].
The LEPR Gln223Arg polymorphism was also investigated in premenopausal Nigerian women carrying at least one LEPR 223Arg allele. This investigation revealed that the heterozygote Gln223Arg and mutant homozygote Arg223Arg have no association with postmenopausal BC risk (p = 0.68) [41]. Premenopausal Nigerian women carrying at least one LEPR 223Arg allele were at a modestly increased risk of BC (p = 0.07) [41].
In Senegal, BC studies revealed that the BRCA1 gene is the most common genetic risk factor for the disease development in Senegalese women. Indeed, a novel deleterious mutation (c.1949_1950delTA) has been described [42].
Eastern Africa region
In this region, our bibliographic research revealed that only Ethiopia and Sudan contributed to the determination of the genetic risk factors of BC.
In Sudan, two studies were performed, the first one reported 33 BRCA1 point mutations, found in 59 Central Sudanese premenopausal BC patients. [43]. The second study characterized germline BRCA1/2 mutations in patients (34 females, one male) selected by diagnosis within age 40 years or male gender. A total of 33/35 patient were found to carry 60 BRCA1/2 variants, among them, 17 were novel, 22 reported in populations from various geographic areas and 21 reported worldwide. The most frequent mutations found are in BRCA1 (c.3999delT, c.4065_4068delTCAA, c.557C>A, c.2458A>G, c.5090G>A) and in BRCA2 (c.3195_3198delTAAT, c.6406_6407delTT, c.8642_8643insTTTT, c.6101G>A, c.68-7delT) [44].
The most relevant studies in Ethiopia targeted the implication of the HER gene. This Proto-oncogene plays an important role in the carcinogenesis and the prognosis of BC [21]. Many studies have been conducted to explore the association between the HER2 Ile655Val polymorphism and BC risk, a significant association among Africans was found for Val/Val v. Ile/Ile genotypes: odds ratio = 78, 95% confidence interval = 1.94–39.72, p = 0.35 for heterogeneity; for the recessive model Val/Val v. Ile/Val + Ile/Ile: odds ratio = 8.60, 95% confidence interval = 1.92–38.48, p = 0.31 for heterogeneity [21].
Central Africa region
In this region, only Democratic Republic of Congo have studied the BRCA1/2 genes in a family with a severe history of BC at young ages. This genetic analysis revealed the presence of the c.2389_2390delGA mutation at the heterozygous state in all BC family members, the mutation leads to a frameshift at codon 797 of the BRCA1 gene (p.Glu797fs) [45].
A summary of the extracted data from our literature review, the panel of genes illustrated in Table 1 represents the genes associated with BC in the different African populations.
Table 1.
Panel of genes associated with BC in African populations
| Northern | Southern | Western | Eastern | Central | |||||
|---|---|---|---|---|---|---|---|---|---|
| Country | Algeria | Tunisia | Egypt | Morocco | South Africa | Nigeria | Senegal | Sudan | Democratic Republic of the Congo |
| Genes | BRCA1 BRCA2 |
BRCA1 BRCA2 HER2 ESR1 P53 |
BRCA1 BRCA2 P53 PON1 XPD |
BRCA1 BRCA2 APOBEC3A APOBEC3B ARID1B ATR MAP3K1 MLL2 MLL3 NCOR1 RUNX1 SF3B1 SMAD4 TBX3 TTN MTHFR HER2 |
BRCA1 BRCA2 CHEK2 PALB2 RAD50 MTHFR hMLH1 |
BRCA1 BRCA2 LEPR |
BRCA1 | BRCA1 BRCA2 |
BRCA1 BRCA2 |
Discussion
BC is the most commonly diagnosed cancer in African women and the main cause of death from cancer diseases. The evaluation of the BC genetic risk factor provided support to understand the relationship between genomics and cancer. Only nine of 54 countries in Africa have studied few genes involved in BC. In this review, data showed 27 distinct genes in Africa (Table 1). Mutations in BRCA1 and BRCA2 genes were identified in all five regions of the continent.
According to our literature review, the BRCA1 c.798_799delTT mutation was found recurrent in the three North African countries, Algeria [11] Tunisia [14] and Morocco [31] but absent in other African regions. Moreover, the mutation c.181T>G on BRCA1 was commonly reported only in the Algerian [11] and Moroccan [31] populations. Another BRCA1 mutation c.2612C>T was identified in Algeria [11] and Tunisia [17]. The BRCA2 mutations c.-26G>A, c.7242A>G and c.8503T>C were identified in two NA countries, Algeria [11] and Tunisia [14].
Furthermore, many common mutations in BRCA1/2 were found in other regions, especially in Western Africa, where three common mutations with BRCA1 (c.2082C>T, c.2311T>C, c.3548A>G) and two common BRCA2 mutations (c.3396A>G, c.3807T>C) were identified [40]. These inter-regions similarities were also observed between South Africa and two NA countries: the c.1504_1508del BRCA1 mutation is common between South Africa [46] and Tunisia [17], and the c.185delAG BRCA1 between South African [37] and Egyptian [24] populations. More details concerning mutations retrieved in African countries are listed in Table 2.
Table 2.
Mutation detection methods used in African Studies
| Northern | Southern | Western | Eastern | Central | |||||
|---|---|---|---|---|---|---|---|---|---|
| Country | Morocco | Algeria | Tunisia | Egypt | South Africa | Nigeria | Senegal | Sudan | Democratic Republic of the Congo |
| BRCA1 | c.68_69delAG c.181 T > G c.798_799delTT c.3279delC c.1016dupA c.4942A>T c.5062-5064delGTT c.5095C>T |
c.2082C>T c.2311 T > C c.2612C>T c.3418A>G c.3548A>G c.4308 T > C c.181 T > G c.798_799delTT |
c.798_799delTT c.1504_1508delTTAA c.211dupA c.2612C>T c.2311 T > C c.3022A>G c.4308 T > C |
c.185delAG c.624C>T |
c.185delAG c.448insA c.1127insA S451X c.4957insC c.1504_1508del |
IVS2 + 1G>A c.252delAA c.2311delAAGAA c.2082C>T c.2311 T > C c.3548A>G c.5123C>T |
c.1949_1950delTA | c.3999delT c.4065_4068delTCAA c.557C>A c.2458A>G c.1846_1848delTCT c.1088A>G |
c.2389_2390delGA |
| BRCA2 | c.517-1G>A c.3381delT c.5073dupA c.7110delA c.7235insG |
c.-26G>A c.865A>C c.3396A>G c.3807 T > C c.7242A>G c.7397C>T c.8503 T > C c.1310_1313delAAGA |
c.1313dupT c.7654dupT c.7242A>G c.-26G>A c.681 + 56C>T c.8503 T > C c.1310_1313del |
c.999delTCAAA c.2256 T > C c.8934G>A |
c.5771_5774del c.6447_6448dup c.7934delG c.582G > A |
c.1222delA c.1538delAAGA c.1590delA c.681 + 56C>T c.3396A>G c.3807 T > C c.8755–66 T > C |
c.296-7dupT c.5710C>G |
c.3195_3198delTAAT c.6406_6407delTT c.8642_8643insTTTT c.6101G>A c.68-7delT |
|
| Mutation detection methods | PCR, bi-directional sequencing, targeted direct sequencing. | PCR, PCR-SSCP, High-Resolution Melting (HRM) curve analysis, direct sequencing | PCR, direct sequencing. | PCR, single-strand conformational polymorphism (SSCP), Heteroduplex analysis (HDA), Cloning Vector, DNA Sequencing. | PTT and SSCP/HA analysis, PCR, Manual sequencing, Sanger sequencing, MLPA, Next-Generation Sequencing (NGS), Whole Exome Sequencing. | PCR, direct sequencing. | PCR, direct sequencing. | PCR, Protein truncation test (PTT), denaturing high performance liquid chromatography (DHPLC), direct sequencing. | |
| Reference | [29–31] | [11] | [14, 17, 18] | [24] | [37, 38, 46] | [40] | [42] | [43, 44] | [45] |
Our literature review revealed that the majority of genes were studied in the NA (18 genes) . Whereas, only six genes were studied in SA. We performed a functional analysis using the online tool ToppGene (https://toppgene.cchmc.org) to explore the biological function and pathways of the studied genes in both of NA (Table 3) and SA (Table 4). The functional analysis revealed a strong similarity between the candidate genes from NA and SA regions. Furthermore, the biological pathways ‘Role of BRCA1, BRCA2 and ATR in Cancer Susceptibility’ and ‘Breast Cancer’ were found in the top enriched terms for NA and SA regions with a p of 1.074×10−8 (2.09×10−10) and 2.804×10−5 (2.69×10−3), respectively.
Table 3.
Top enriched terms and biological pathways identified by functional analysis of breast cancer candidate genes studied in Northern Africa
| Enrichment term | ID | Source | p value |
|---|---|---|---|
| Enriched pathways | |||
| Role of BRCA1, BRCA2 and ATR in cancer susceptibility | M9703 | MSigDB C2 BIOCARTA (v6.0) | 1.074 × 10−8 |
| Cell Cycle: G2/M checkpoint | M8560 | MSigDB C2 BIOCARTA (v6.0) | 3.467 × 10−6 |
| Cell Cycle: G1/S check Point | M648 | MSigDB C2 BIOCARTA (v6.0) | 5.595 × 10−6 |
| Breast cancer | 1435207 | BioSystems: KEGG | 2.804 × 10−5 |
| Fanconi anemia pathway | 377262 | BioSystems: KEGG | 4.386 × 10−5 |
| Pancreatic cancer | 83108 | BioSystems: KEGG | 6.917 × 10−5 |
| Chronic myeloid leukemia | 83116 | BioSystems: KEGG | 9.437 × 10−5 |
| Enriched gene ontology term | |||
| Chromosome breakage | GO:0031052 | GO | 1.039 × 10−8 |
| Programmed DNA elimination | GO:0031049 | GO | 1.039 × 10−8 |
| Female sex differentiation | GO:0046660 | GO | 1.127 × 10−7 |
| Chromosome organization | GO:0051276 | GO | 1.3 × 10−7 |
| Positive regulation of chromosome organization | GO:2001252 | GO | 1.568 × 10−7 |
| Positive regulation of macromolecule biosynthetic process | GO:0010557 | GO | 2.852 × 10−7 |
| DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | GO:0006978 | GO | 2.894 × 10−7 |
Table 4.
Top enriched terms and biological pathways identified by functional analysis of breast cancer candidate genes studied in southern Africa
| Enrichment term | ID | Source | p value |
|---|---|---|---|
| Enriched pathways | |||
| Role of BRCA1, BRCA2 and ATR in Cancer Susceptibility | M9703 | MSigDB C2 BIOCARTA (v6.0) | 2.09 × 10−10 |
| ATM Signaling Pathway | M10628 | MSigDB C2 BIOCARTA (v6.0) | 1.24 × 10−7 |
| Homologous recombination | 83046 | BioSystems: KEGG | 1.15 × 10−6 |
| Fanconi anemia pathway | 377262 | BioSystems: KEGG | 2.82 × 10−6 |
| Cell Cycle: G2/M Checkpoint | M8560 | MSigDB C2 BIOCARTA (v6.0) | 7.44 × 10−5 |
| Platinum drug resistance | 1404797 | BioSystems: KEGG | 6.99 × 10−4 |
| Breast cancer | 1435207 | BioSystems: KEGG | 2.69 × 10−3 |
| Enriched gene ontology term | |||
| Double-strand break repair | GO:0006302 | GO | 2.53 × 10−9 |
| Intrinsic apoptotic signaling pathway in response to DNA damage | GO:0008630 | GO | 4.28 × 10−8 |
| Negative regulation of DNA metabolic process | GO:0051053 | GO | 4.76 × 10−8 |
| Regulation of DNA metabolic process | GO:0051052 | GO | 5.96 × 10−8 |
| Strand displacement | GO:0000732 | GO | 8.42 × 10−8 |
| Meiotic metaphase I plate congression | GO:0043060 | GO | 1.21 × 10−7 |
| DNA repair | GO:0006281 | GO | 3.21 × 10−7 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MSigDB, The Molecular Signatures Database.
In the remaining regions, a modest number of genes have been studied only in few countries (Nigeria, Senegal, Sudan, etc.), this may reflect the lack of clinical monitoring for BC patients and control of the disease.
The screening for different types of gene mutations to determine their origin, either somatic or germinal, or either founder or recurrent mutations helped to characterize the population specificities toward genes and mutations found in populations across the continent. Researchers in American continent have studied the genetic factors of BC through decades using different types of technologies, from RFLP-PCR to Whole Exome Sequencing, these studies have largely contributed to define major and minor genes and mutation incidence. Several mutations, mainly in BRCA1/2, have been described in both American and African populations, such 185delAG [46], 1742insG, 2630del11, 4241delTG, 9045delGAAA and delEx12 [47]. A wide spectrum of mutations in other genes associated with BC have been identified but have not been significantly reported in both continents [48, 49]. Many mutations described in the American studies have not been described or are rarely found in the African continent, such as VS511G.A, S955X, R1443X, E1644X, 943ins10, C64Y, M1775R, Q1090X and Y101X [49]. A common phylogenetic relation between American and African populations would explain the recurring mutations found among them, and the genetic history between women with BC found in North or South America. The diversity of ethnicity among the American populations has an important role in explaining these findings. Indeed, a significant number of women diagnosed with BC in North America have African or Latino-Hispanic origins and a relevant number of mutations found in these women have been described in African women with BC [49]. However, many detected mutations in the American women with African-related origins have not been described in African studies. Considering these undescribed mutations in African BC women, it would be relevant to complete the African genetic risk factor panel in forthcoming BC genetic studies.
Our literature search revealed that only nine African countries had studied the genes involved in BC. Unlike, in Europe, where the studies covered 15 countries, which gives a valuable information regarding BC genetics in Europe. Indeed, different studies have been carried out through the European continent to determine the role and the implication of BRCA1 and BRCA2 genes in BC, as well as detect novel mutations [50–72]. Other genes have been studied in both African and European continents and revealed to be associated with BC, such as CHEK2 [52, 65, 73–75], TP53 [73, 76, 77], MTHFR [78], PALB2 [79–81]. Whereas, other genes have been proved involved in BC development solely in Africa such as APOBEC3A, APOBEC3B, ARID1B, NCOR1, SMAD4, MAP3K1, HER2, HER and RAD50 or only in Europe such as RAD51 [82–84], COMT [85, 86], CYP17 [86], MRE11A [61, 87], CYP19 [87] and MDM2 [88].
The ethnic similarity between the different regions of Europe and Africa can guide African scientists to well investigate the genes involved in BC. For instance, research in North Africa should explore other genes, such as GSTM1, GSTT1 found in Portugal [89] and PALB2, ATM, TNFRSF11A found in Spain [80, 90, 91].
The investigation in Asia were more important. Indeed, many studies exploring the major and minor genes involved in BC were performed in different Asian countries. Several Meta-analyses and reviews were also conducted [21, 92, 93].
For BRCA1 and BRCA2 genes, several germline mutations were identified and listed in the National Comprehensive Cancer Network (NCCN) guidelines, however, no data were reported concerning African studies [94]. The genetic aetiology of hereditary BC has not yet been fully elucidated. Although germline mutations of high-penetrance genes such as BRCA1/2 were shown implicated in development of BC hereditary, at least half of all BC families are not linked to these genes [95]. For example, in China 42 deleterious germline mutations were identified in 21 genes, including 18.2% BRCA1 or BRCA2 mutations, 3% TP53 mutations, 5.1% DNA mismatch repair gene mutations, 1% CDH1 mutations, 6.1% Fanconi anemia pathway gene mutations, and 9.1% mutations in other genes [95].
CYP, CHEK2 and MTHFR were also studied and their involvement in BC showed that they may modify susceptibility to BC [70, 96, 97]. TNF-alpha, Enos, RAD50, CCND1, NBS1 and SULT1A1 genes were studied in Asia and the results exclude the possible role of RAD50 and NBS1 in familial BC [59]. SULT1A1 may be a low-penetrant risk factor for developing BC in the Asian population [66]. Although the clinical significance of newly identified low-penetration genetic mutations has not yet been fully appreciated in Asia, these new findings provide valuable epidemiological information for future studies of BC in the Asian population.
Unlike Africa, many studies in Asia have used recent techniques such as NGS and Whole Exome Sequencing that have contributed to the development of genetic screening panels [55, 98, 99]. For example, a test panel has been developed in accordance with NCCN, which has proposed guidelines for the genetic testing of the BRCA1 and BRCA2 genes, based on studies in western populations. However, like in Africa, there is still a gap in the availability of genetic counselling and genetic testing in Asian countries due to financial facilities, access and inaccurate reporting of a family history of cancer.
The variety of options now accessible for the patient and physician in making appropriate and timely decisions in hereditary breast and ovarian cancer has triggered a daily increase in the demand for mutation analysis of the BRCA1/2 genes on the other continents. Therefore, it is necessary for African scientists to implement these testing and analysis, to better characterize BC genetic risk factors.
The analysis of the African genetic studies reported in our literature search revealed that the screening of genes and mutations related to BC in African countries is less illustrative compared with other continents. Therefore, it is necessary to encourage researchers in Africa to characterize more genes involved in BC, to better target the diagnosis and guide specific and efficient therapeutic strategies for African community.
The insufficiency of reliable data on BC in Africa and the increase of mortality prevalence and incidence rates could be linked with precarious socioeconomic criteria. For instance, the weakness of the health system, the lack of health insurance and coverage, the limited access to medication, the scarcity of care facilities and counselling, the deficiency of genetic testing and low income. In addition, the lack of support for scientific research in several African countries also contributed to the spread of the disease.
Acknowledgements
We acknowledge Pr. Nicola Mulder's (University of Cape Town) support, advises and manuscript revision.
We acknowledge greatly H3Africa, H3ABioNet networks And the African Genomic Medicine Training (AGMT) committee
Declaration of interest
None.
References
- 1.Jemal A, et al. Global cancer statistics. CA: a Cancer Journal for Clinicians 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2.Abkevich V, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. British Journal of Cancer 2012; 107: 1776–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO). International Agency for Research on Cancer. World Cancer fact sheet. 2008.
- 4.Standish LJ, et al. Breast cancer and the immune system. Journal of the Society for Integrative Oncology 2008; 6: 158–168. [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization (WHO). International Agency for Research on Cancer. Report, W.C., World Cancer Report 2014.
- 6.World Health Organization (WHO). International Agency for Research on Cancer. Cancer incidence and mortality worldwide. 2008.
- 7.Frisby CM. Messages of hope: health communication strategies that address barriers preventing black women from screening for breast cancer. Journal of Black Studies 2002; 32: 489–505. [Google Scholar]
- 8.Ervik MFL,, et al. Cancer Today. Lyon, France: International Agency for Research on Cancer, 2016. [Google Scholar]
- 9.Uhrhammer N, et al. BRCA1 mutations in Algerian breast cancer patients: high frequency in young, sporadic cases. International Journal of Medical Sciences 2008; 5: 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherbal F, et al. BRCA1 and BRCA2 unclassified variants and missense polymorphisms in Algerian breast/ovarian cancer families. Disease Markers 2012; 32: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherbal F, et al. BRCA1 and BRCA2 germline mutations screening in Algerian breast/ovarian cancer families. Disease Markers 2010; 28: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henouda S, et al. Contribution of BRCA1 and BRCA2 germline mutations to early Algerian breast cancer. Disease Markers 2016; 2016: 7869095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Troudi W, et al. Penetrance of BRCA1 gene mutation and DNA mitochondrial in Tunisian breast cancer occurrence. La Tunisie Medicale 2009; 87: 494–498. [PubMed] [Google Scholar]
- 14.Mahfoudh W, et al. Hereditary breast cancer in middle eastern and north African (MENA) populations: identification of novel, recurrent and founder BRCA1 mutations in the Tunisian population. Molecular Biology Reports 2012; 39: 1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mestiri S, et al. Mutational analysis of breast/ovarian cancer hereditary predisposition gene BRCA1 in Tunisian women. Arch Inst Pasteur Tunis, 2000; 77: 11–15. [PubMed] [Google Scholar]
- 16.Troudi W, et al. Contribution of the BRCA1 and BRCA2 mutations to breast cancer in Tunisia. Journal of Human Genetics 2007; 52: 915–920. [DOI] [PubMed] [Google Scholar]
- 17.Riahi A, et al. Mutation spectrum and prevalence of BRCA1 and BRCA2 genes in patients with familial and early-onset breast/ovarian cancer from Tunisia. Clinical Genetics 2015; 87: 155–160. [DOI] [PubMed] [Google Scholar]
- 18.Riahi A, et al. Family history predictors of BRCA1/BRCA2 mutation status among Tunisian breast/ovarian cancer families. Breast Cancer 2017; 24: 238–244. [DOI] [PubMed] [Google Scholar]
- 19.Hadiji-Abbes N, et al. A novel BRCA2 in frame deletion in a Tunisian woman with early onset sporadic breast cancer. Pathol Biol (Paris) 2015; 63: 185–189. [DOI] [PubMed] [Google Scholar]
- 20.Kallel I, et al. HER2 polymorphisms and breast cancer in Tunisian women. Genetic Testing and Molecular Biomarkers 2010; 14: 29–35. [DOI] [PubMed] [Google Scholar]
- 21.Lu S, et al. HER2 ile655val polymorphism contributes to breast cancer risk: evidence from 27 case-control studies. Breast Cancer Research and Treatment 2010; 124: 771–778. [DOI] [PubMed] [Google Scholar]
- 22.Trifa F, et al. Haplotype analysis of p53 polymorphisms: Arg72Pro, Ins16bp and G13964C in Tunisian patients with familial or sporadic breast cancer. Cancer Epidemiology 2010; 34: 184–188. [DOI] [PubMed] [Google Scholar]
- 23.Kallel I, et al. Genetic polymorphisms in the EGFR (R521K) and estrogen receptor (T594T) genes, EGFR and ErbB-2 protein expression, and breast cancer risk in Tunisia. Journal of Biomedicine & Biotechnology 2009; 2009: 753683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bensam M, et al. Detection of new point mutations of BRCA1 and BRCA2 in breast cancer patients. Biochemical Genetics 2014; 52: 15–28. [DOI] [PubMed] [Google Scholar]
- 25.El-Ghannam DM, Arafa M, Badrawy T. Mutations of p53 gene in breast cancer in the Egyptian province of Dakahliya. Journal of Oncology Pharmacy Practice 2011; 17: 119–124. [DOI] [PubMed] [Google Scholar]
- 26.Hussein YM, et al. Association of L55M and Q192R polymorphisms in paraoxonase 1 (PON1) gene with breast cancer risk and their clinical significance. Molecular and Cellular Biochemistry 2011; 351: 117–123. [DOI] [PubMed] [Google Scholar]
- 27.Hussien YM, et al. Impact of DNA repair genes polymorphism (XPD and XRCC1) on the risk of breast cancer in Egyptian female patients. Molecular Biology Reports 2012; 39: 1895–1901. [DOI] [PubMed] [Google Scholar]
- 28.Laarabi FZ, et al. Genetic testing and first presymptomatic diagnosis in Moroccan families at high risk for breast/ovarian cancer. Oncology Letters 2011; 2: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tazzite A, et al. BRCA1 and BRCA2 germline mutations in Moroccan breast/ovarian cancer families: novel mutations and unclassified variants. Gynecologic Oncology 2012; 125: 687–692. [DOI] [PubMed] [Google Scholar]
- 30.Laraqui A, et al. Mutation screening of the BRCA1 gene in early onset and familial breast/ovarian cancer in Moroccan population. International Journal of Medical Sciences 2013; 10: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marouf C, et al. Analysis of functional germline variants in APOBEC3 and driver genes on breast cancer risk in Moroccan study population. BMC Cancer 2016; 16: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diakite B, et al. Methylenetetrahydrofolate reductase C677T polymorphism and breast cancer risk in Moroccan women. African Health Sciences 2012; 12: 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tazzite A, et al. Association between ABCB1 C3435T polymorphism and breast cancer risk: a Moroccan case-control study and meta-analysis. BMC Genetics 2016; 17: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahoui J, et al. The single nucleotide polymorphism+936 C/T VEGF is associated with human epidermal growth factor receptor 2 expression in Moroccan breast cancer women. Medical Oncology (Northwood, London, England) 2014; 31: 336. [DOI] [PubMed] [Google Scholar]
- 35.Reeves MD, et al. BRCA1 mutations in South African breast and/or ovarian cancer families: evidence of a novel founder mutation in Afrikaner families. International Journal of Cancer 2004; 110: 677–682. [DOI] [PubMed] [Google Scholar]
- 36.Francies FZ, et al. BRCA1, BRCA2 and PALB2 mutations and CHEK2 c.1100delC in different South African ethnic groups diagnosed with premenopausal and/or triple negative breast cancer. BMC Cancer 2015; 15: 912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sluiter M, Mew S, van Rensburg EJ. PALB2 sequence variants in young South African breast cancer patients. Familial Cancer 2009; 8: 347–353. [DOI] [PubMed] [Google Scholar]
- 38.Blokhuis MM, et al. The extracolonic cancer spectrum in females with the common ‘South African’ hMLH1 c.C1528T mutation. Familial Cancer 2008; 7: 191–198. [DOI] [PubMed] [Google Scholar]
- 39.van der Merwe N, et al. Exome sequencing in a family with luminal-type breast cancer underpinned by variation in the methylation Pathway22. International Journal of Molecular Sciences 2017; 18(2): 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fackenthal JD, et al. High prevalence of BRCA1 and BRCA2 mutations in unselected Nigerian breast cancer patients. International Journal of Cancer 2012; 131: 1114–1123. [DOI] [PubMed] [Google Scholar]
- 41.Okobia MN, et al. Leptin receptor Gln223Arg polymorphism and breast cancer risk in Nigerian women: a case control study. BMC Cancer 2008; 8: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diez O, et al. Novel BRCA1 deleterious mutation (c.1949_1950delTA) in a woman of Senegalese descent with triple-negative early-onset breast cancer. Oncology Letters 2011; 2: 1287–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biunno I, et al. BRCA1 point mutations in premenopausal breast cancer patients from central Sudan. Familial Cancer 2014; 13: 437–444. [DOI] [PubMed] [Google Scholar]
- 44.Awadelkarim KD, et al. BRCA1 and BRCA2 status in a central Sudanese series of breast cancer patients: interactions with genetic, ethnic and reproductive factors. Breast Cancer Research and Treatment 2007; 102: 189–199. [DOI] [PubMed] [Google Scholar]
- 45.Luyeye Mvila G, et al. From the set-up of a screening program of breast cancer patients to the identification of the first BRCA mutation in the DR Congo. BMC Public Health 2014; 14: 759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Leon Matsuda ML, et al. BRCA1 and BRCA2 mutations among breast cancer patients from the Philippines. International Journal of Cancer 2002; 98: 596–603. [DOI] [PubMed] [Google Scholar]
- 47.Villarreal-Garza C, et al. Significant clinical impact of recurrent BRCA1 and BRCA2 mutations in Mexico. Cancer 2015; 121: 372–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amemiya Y, et al. A comparative analysis of breast and ovarian cancer-related gene mutations in Canadian and Saudi Arabian patients with breast cancer. Anticancer Research 2015; 35: 2601–2610. [PubMed] [Google Scholar]
- 49.Ossa CA, Torres D. Founder and recurrent mutations in BRCA1 and BRCA2 genes in latin American countries: state of the Art and literature review. The Oncologist 2016; 21: 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adank MA, et al. CHEK2*1100delC homozygosity is associated with a high breast cancer risk in women. Journal of Medical Genetics 2011; 48: 860–863. [DOI] [PubMed] [Google Scholar]
- 51.Antoniou AC, et al. Common alleles at 6q25.1 and 1p11.2 are associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers. Human Molecular Genetics 2011; 20: 3304–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armaou S, et al. Contribution of BRCA1 germ-line mutations to breast cancer in Greece: a hospital-based study of 987 unselected breast cancer cases. British Journal of Cancer 2009; 101: 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balabas A, et al. Novel germline mutations in BRCA2 gene among breast and breast-ovarian cancer families from Poland. Familial Cancer 2010; 9: 267–274. [DOI] [PubMed] [Google Scholar]
- 54.Brozek I, et al. Prevalence of the most frequent BRCA1 mutations in polish population. Journal of Applied Genetics 2011; 52: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee HB, et al. The use of FNA samples for whole-exome sequencing and detection of somatic mutations in breast cancer surgical specimens. Cancer Cytopathology 2015; 123: 669–677. [DOI] [PubMed] [Google Scholar]
- 56.Gaudet MM, et al. Common genetic variants and modification of penetrance of BRCA2-associated breast cancer. PLoS Genetics 2010; 6: e1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Infante M, et al. Two founder BRCA2 mutations predispose to breast cancer in young women. Breast Cancer Research and Treatment 2010; 122: 567–571. [DOI] [PubMed] [Google Scholar]
- 58.Krupa R, et al. Association between polymorphisms of the BRCA2 gene and clinical parameters in breast cancer. Experimental Oncology 2009; 31: 250–251. [PubMed] [Google Scholar]
- 59.He M, et al. RAD50 and NBS1 are not likely to be susceptibility genes in Chinese non-BRCA1/2 hereditary breast cancer. Breast Cancer Research and Treatment 2012; 133: 111–116. [DOI] [PubMed] [Google Scholar]
- 60.Loizidou MA, et al. BRCA1 and BRCA2 mutation testing in Cyprus; a population based study. Clinical Genetics 2017; 91: 611–615. [DOI] [PubMed] [Google Scholar]
- 61.Loizidou MA, et al. DNA-repair genetic polymorphisms and risk of breast cancer in Cyprus. Breast Cancer Research and Treatment 2009; 115: 623–627. [DOI] [PubMed] [Google Scholar]
- 62.Minucci A, et al. High resolution melting analysis is very useful to identify breast cancer type 1 susceptibility protein (BRCA1) c.4964_4982del19 (rs80359876) founder calabrian pathogenic variant on peripheral blood and buccal swab DNA. Molecular Diagnosis & Therapy 2017; 21: 217–223. [DOI] [PubMed] [Google Scholar]
- 63.Mitrofanov DV, et al. Detection of 5382insC mutation in human BRCA1 gene using fluorescent labeled oligonucleotides. Molecular Biology (Mosk) 2009; 43: 999–1005. [PubMed] [Google Scholar]
- 64.Negura L, et al. Complete BRCA mutation screening in breast and ovarian cancer predisposition families from a north-eastern Romanian population. Familial Cancer 2010; 9: 519–523. [DOI] [PubMed] [Google Scholar]
- 65.Peixoto A, et al. International distribution and age estimation of the Portuguese BRCA2 c.156_157insAlu founder mutation. Breast Cancer Research and Treatment 2011; 127: 671–679. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z, et al. SULT1A1 r213h polymorphism and breast cancer risk: a meta-analysis based on 8,454 cases and 11,800 controls. Breast Cancer Research and Treatment 2010; 122: 193–198. [DOI] [PubMed] [Google Scholar]
- 67.Salgado J, et al. A novel BRCA2 mutation that segregates with breast and prostate cancer in a Spanish family. Breast Cancer Research and Treatment 2010; 121: 219–220. [DOI] [PubMed] [Google Scholar]
- 68.Schmidt MK, et al. Breast cancer survival of BRCA1/BRCA2 mutation carriers in a hospital-based cohort of young women. Journal of the National Cancer Institute 2017; 109. [DOI] [PubMed] [Google Scholar]
- 69.Serrano-Fernandez P, et al. Synergistic interaction of variants in CHEK2 and BRCA2 on breast cancer risk. Breast Cancer Research and Treatment 2009; 117: 161–165. [DOI] [PubMed] [Google Scholar]
- 70.Ma X, et al. Association between CYP19 polymorphisms and breast cancer risk: results from 10,592 cases and 11,720 controls. Breast Cancer Research and Treatment 2010; 122: 495–501. [DOI] [PubMed] [Google Scholar]
- 71.Uglanitsa N, et al. The contribution of founder mutations in BRCA1 to breast cancer in Belarus. Clinical Genetics 2010; 78: 377–380. [DOI] [PubMed] [Google Scholar]
- 72.Willems P, et al. Characterization of the c.190T>C missense mutation in BRCA1 codon 64 (Cys64Arg). International Journal of Oncology 2009; 34: 1005–1015. [DOI] [PubMed] [Google Scholar]
- 73.Bisof V, et al. The TP53 gene polymorphisms and survival of sporadic breast cancer patients. Medical Oncology (Northwood, London, England) 2012; 29: 472–478. [DOI] [PubMed] [Google Scholar]
- 74.Cybulski C, et al. Effect of CHEK2 missense variant I157T on the risk of breast cancer in carriers of other CHEK2 or BRCA1 mutations. Journal of Medical Genetics 2009; 46: 132–135. [DOI] [PubMed] [Google Scholar]
- 75.Cybulski C, et al. Estrogen receptor status in CHEK2-positive breast cancers: implications for chemoprevention. Clinical Genetics 2009; 75: 72–78. [DOI] [PubMed] [Google Scholar]
- 76.Bisof V, et al. TP53 gene polymorphisms and breast cancer in Croatian women: a pilot study. European Journal of Gynaecological Oncology 2010; 31: 539–544. [PubMed] [Google Scholar]
- 77.Zhang Z, et al. P53 codon 72 polymorphism contributes to breast cancer risk: a meta-analysis based on 39 case-control studies. Breast Cancer Research and Treatment 2010; 120: 509–517. [DOI] [PubMed] [Google Scholar]
- 78.Papandreou CN, et al. Evidence of association between methylenetetrahydrofolate reductase gene and susceptibility to breast cancer: a candidate-gene association study in a South-eastern European population. DNA and Cell Biology 2012; 31: 193–198. [DOI] [PubMed] [Google Scholar]
- 79.Dansonka-Mieszkowska A, et al. A novel germline PALB2 deletion in polish breast and ovarian cancer patients. BMC Medical Genomics 2010; 11: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garcia MJ, et al. Analysis of FANCB and FANCN/PALB2 fanconi anemia genes in BRCA1/2-negative spanish breast cancer families. Breast Cancer Research and Treatment 2009; 113: 545–551. [DOI] [PubMed] [Google Scholar]
- 81.Silvestri V, et al. Whole-exome sequencing and targeted gene sequencing provide insights into the role of PALB2 as a male breast cancer susceptibility gene. Cancer 2017; 123: 210–218. [DOI] [PubMed] [Google Scholar]
- 82.Gao LB, et al. RAD51 135g/C polymorphism and breast cancer risk: a meta-analysis from 21 studies. Breast Cancer Research and Treatment 2011; 125: 827–835. [DOI] [PubMed] [Google Scholar]
- 83.Romanowicz-Makowska H, et al. Single nucleotide polymorphisms in the homologous recombination repair genes and breast cancer risk in polish women. Tohoku Journal of Experimental Medicine 2011; 224: 201–208. [DOI] [PubMed] [Google Scholar]
- 84.Vuorela M, et al. Further evidence for the contribution of the RAD51C gene in hereditary breast and ovarian cancer susceptibility. Breast Cancer Research and Treatment 2011; 130: 1003–1010. [DOI] [PubMed] [Google Scholar]
- 85.Ding H, et al. COMT val158met polymorphism and breast cancer risk: evidence from 26 case-control studies. Breast Cancer Research and Treatment 2010; 123: 265–270. [DOI] [PubMed] [Google Scholar]
- 86.Ghisari M, et al. Polymorphism in xenobiotic and estrogen metabolizing genes, exposure to perfluorinated compounds and subsequent breast cancer risk: a nested case-control study in the Danish national birth cohort. Environmental Research 2017; 154: 325–333. [DOI] [PubMed] [Google Scholar]
- 87.Loizidou MA, et al. Genetic variation in genes interacting with BRCA1/2 and risk of breast cancer in the Cypriot population. Breast Cancer Research and Treatment 2010; 121: 147–156. [DOI] [PubMed] [Google Scholar]
- 88.Zhao E, et al. MDM2 SNP309 polymorphism and breast cancer risk: a meta-analysis. Molecular Biology Reports 2012; 39: 3471–3477. [DOI] [PubMed] [Google Scholar]
- 89.Ramalhinho AC, Fonseca-Moutinho JA, Breitenfeld L. Glutathione S-transferase M1, T1, and P1 genotypes and breast cancer risk: a study in a Portuguese population. Molecular and Cellular Biochemistry 2011; 355: 265–271. [DOI] [PubMed] [Google Scholar]
- 90.Grana B, et al. Germline ATM mutational analysis in BRCA1/BRCA2 negative hereditary breast cancer families by MALDI-TOF mass spectrometry. Breast Cancer Research and Treatment 2011; 128: 573–579. [DOI] [PubMed] [Google Scholar]
- 91.Bonifaci N, et al. Evidence for a link between TNFRSF11A and risk of breast cancer. Breast Cancer Research and Treatment 2011; 129: 947–954. [DOI] [PubMed] [Google Scholar]
- 92.Liede A, et al. Hereditary breast and ovarian cancer in Asia: genetic epidemiology of BRCA1 and BRCA2. Human Mutation 2002; 20: 413–424. [DOI] [PubMed] [Google Scholar]
- 93.Kim H, et al. Distribution of BRCA1 and BRCA2 mutations in Asian patients with breast cancer. Journal of Breast Cancer 2013; 16: 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.NCCN. National Comprehensive Cancer Network report, 2018.
- 95.Yang X, et al. Identification of a comprehensive spectrum of genetic factors for hereditary breast cancer in a Chinese population by next-generation sequencing. PLoS ONE 2015; 10: e0125571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baloch AH, et al. Novel nonsense variants c.58C>T (p.Q20X) and c.256G>T (p.E85X) in the CHEK2 gene identified in breast cancer patients from Balochistan. Asian Pacific Journal of Cancer Prevention 2016; 17: 3623–3626. [PubMed] [Google Scholar]
- 97.Qi X, et al. Methylenetetrahydrofolate reductase polymorphisms and breast cancer risk: a meta-analysis from 41 studies with 16,480 cases and 22,388 controls. Breast Cancer Research and Treatment 2010; 123: 499–506. [DOI] [PubMed] [Google Scholar]
- 98.Cai Q, et al. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nature genetics 2014; 46:886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guo X, et al. Use of deep whole genome sequencing data to identify structure risk variants in breast cancer susceptibility genes. Human Molecular Genetics 2018; Epub ahead of print 1; 27(5):853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]