ABSTRACT
The autophagy-lysosome pathway plays a fundamental role in the clearance of aggregated proteins and protection against cellular stress and neurodegenerative conditions. Alterations in autophagy processes, including macroautophagy and chaperone-mediated autophagy (CMA), have been described in Parkinson disease (PD). CMA is a selective autophagic process that depends on LAMP2A (lysosomal-associated membrane protein 2A), a mammal and bird-specific membrane glycoprotein that translocates cytosolic proteins containing a KFERQ-like peptide motif across the lysosomal membrane. Drosophila reportedly lack CMA and use endosomal microautophagy (eMI) as an alternative selective autophagic process. Here we report that neuronal expression of human LAMP2A protected Drosophila against starvation and oxidative stress, and delayed locomotor decline in aging flies without extending their lifespan. LAMP2A also prevented the progressive locomotor and oxidative defects induced by neuronal expression of PD-associated human SNCA (synuclein alpha) with alanine-to-proline mutation at position 30 (SNCAA30P). Using KFERQ-tagged fluorescent biosensors, we observed that LAMP2A expression stimulated selective autophagy in the adult brain and not in the larval fat body, but did not increase this process under starvation conditions. Noteworthy, we found that neurally expressed LAMP2A markedly upregulated levels of Drosophila Atg5, a key macroautophagy initiation protein, and that it increased the density of Atg8a/LC3-positive puncta, which reflects the formation of autophagosomes. Furthermore, LAMP2A efficiently prevented accumulation of the autophagy defect marker Ref(2)P/p62 in the adult brain under acute oxidative stress. These results indicate that LAMP2A can potentiate autophagic flux in the Drosophila brain, leading to enhanced stress resistance and neuroprotection.
Abbreviations: Act5C: actin 5C; a.E.: after eclosion; Atg5: autophagy-related 5; Atg8a/LC3: autophagy-related 8a; CMA: chaperone-mediated autophagy; DHE: dihydroethidium; elav: embryonic lethal abnormal vision; eMI: endosomal microautophagy; ESCRT: endosomal sorting complexes required for transport; GABARAP: GABA typeA receptor-associated protein; Hsc70-4: heat shock protein cognate 4; HSPA8/Hsc70: heat shock protein family A (Hsp70) member 8; LAMP2: lysosomal associated membrane protein 2; MDA: malondialdehyde; PA-mCherry: photoactivable mCherry; PBS: phosphate-buffered saline; PCR: polymerase chain reaction; PD: Parkinson disease; Ref(2)P/p62: refractory to sigma P; ROS: reactive oxygen species; RpL32/rp49: ribosomal protein L32; RT-PCR: reverse transcription polymerase chain reaction; SING: startle-induced negative geotaxis; SNCA/α-synuclein: synuclein alpha; SQSTM1/p62: sequestosome 1; TBS: Tris-buffered saline; UAS: upstream activating sequence.
KEYWORDS: Autophagy-lysosome pathway, Drosophila melanogaster, lysosomal-associated membrane protein 2A (LAMP2A), neuroprotection, Parkinson disease, synuclein alpha (SNCA)
Introduction
Autophagy is an essential cellular process conserved in all eukaryotic systems that degrades or recycles cellular components through delivery to lysosomes and plays essential function in development, cell physiology and protection against various stresses and diseases [1–4]. Three main autophagic pathways are described: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA). Macroautophagy involves the formation of a phagophore that sequesters parts of the cytoplasm, including protein aggregates or organelles such as damaged mitochondria (mitophagy); upon completion, the phagophore matures into a double-membrane autophagic vesicle (autophagosome). The autophagosomes then fuse with lysosomes leading to degradation of the vesicle content [4–6]. Microautophagy in contrast only implicates lysosomes or late endosomes that directly entrap cytoplasmic materials or proteins by membrane invagination [7–9].
Approximately 30% of cytosolic proteins contain a KFERQ-like pentapeptide motif [10] recognized by the chaperone HSPA8/Hsc70 (heat shock protein family A [Hsp70] member 8) [11,12]. These proteins are selectively degraded either by endosomal microautophagy (eMI), which involves engulfment by late endosomes [8,13,14], or, alternatively, by CMA, in which the substrate proteins are unfolded and translocated one by one through the lysosomal membrane [15–17]. CMA depends on the LAMP2A (lysosomal-associated membrane protein 2A) receptor, a membrane glycoprotein involved in protein translocation from the cytosol to the lysosomal lumen [18]. Structurally, LAMP2A consists of a heavily glycosylated large N-terminal lumenal domain, a single transmembrane spanning region and a short (12 amino acids) C-terminal tail exposed in the cytosol [19,20]. Alternative splicing of the human LAMP2 gene produces 3 protein isoforms (LAMP2A, LAMP2B and LAMP2C) that each carries out specific roles in autophagy [19,21], but only LAMP2A can function in CMA [22].
CMA alterations have been observed in several neuropathologies, including Parkinson disease (PD), Alzheimer disease and Huntington disease, in which neurodegenerative symptoms are associated with the accumulation of aberrant protein oligomers or aggregates [15,16,23–25]. Pathogenic mutations in 2 PD-associated genes, SNCA (synuclein alpha) and LRRK2 (leucine rich repeat kinase 2), have been reported to disrupt CMA-mediated cytosolic protein degradation [26,27]. Enhancing CMA through LAMP2A overexpression in the rat substantia nigra counteracts SNCA pathogenicity, leading to lower SNCA levels and decreased dopaminergic neuron degeneration [28]. This indicates that CMA is a potential target to prevent or treat PD and other neuropathologies.
Drosophila is a widely used model to study the mechanisms and regulations of autophagy in vivo [29–32]. In this organism, macroautophagy has prominent developmental functions [33] and plays a protective role against starvation and oxidative stress [34,35], whereas eMI is required for protein turnover at synapses [13] and is induced by prolonged starvation [14]. The LAMP2A protein contains in its C-terminal tail a specific peptide motif required for CMA that is known to be present only in mammals and birds. This suggests that CMA does not occur in Drosophila [13,14].
Here, in order to learn more about LAMP2A-mediated neuroprotective mechanisms, we studied whether this protein can act as a stress-protectant in the fly. We show that the pan-neuronal expression of human LAMP2A is sufficient to significantly enhance Drosophila resistance to various stresses, and to protect against mutant SNCA pathogenicity and prolong locomotor ability in aging flies. By using eMI fluorescent sensors and antibodies against autophagy-related proteins, we found that LAMP2A expression promoted both selective autophagy and autophagosome formation in the fly brain. LAMP2A also prevented Ref(2)P/SQSTM1/p62 (Refractory to sigma P) accumulation under oxidative stress, indicating that the neural expression of this lysosomal protein can stimulate autophagic flux in Drosophila.
Results
Neuronal LAMP2A expression protects flies from starvation and oxidative stress
To study the function of the human LAMP2A receptor in Drosophila, we generated UAS-LAMP2A strains. Pan-neuronal expression with elav-Gal4 (elav>LAMP2A) yielded viable fly progeny that developed normally to adults and did not have apparent behavioral defects. Reverse transcription polymerase chain reaction (RT-PCR) analysis performed on adult head RNA extracts from these flies, as well as whole-mount brain immunostaining with a specific antibody, confirmed the transgenic expression and stability of human LAMP2A in the fly (Figure S1a and b). We then examined the effects of the neural expression of this protein on Drosophila resistance to nutrient deprivation, a typical inducer of autophagy. A significant protection against prolonged (> 24 h) starvation was observed when LAMP2A was ectopically expressed in all Drosophila neurons (Figure 1a, see statistical data in Table S3). While the median survival was ~ 48 h for control starved flies, it rose to ~ 72 h for elav>LAMP2A flies in the same conditions. This higher resistance to starvation suggests that human LAMP2A expression may enhance autophagy in Drosophila.
Figure 1.
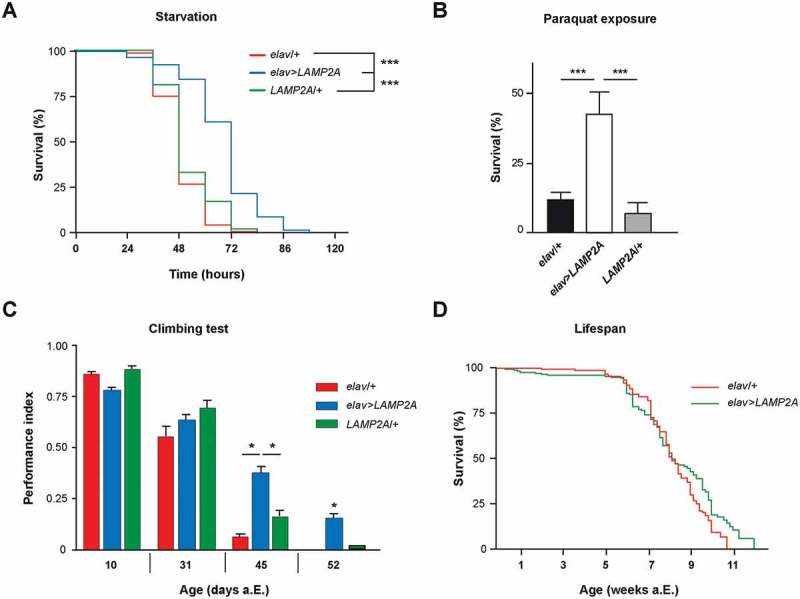
The LAMP2A receptor promotes stress resistance and neuroprotection in Drosophila. (a) Starvation resistance. Expression of the human LAMP2A protein in all neurons significantly extended Drosophila survival upon prolonged starvation (elav>LAMP2A flies) compared to elav-Gal4/+ (elav/+) driver and UAS-LAMP2A/+ (LAMP2A/+) effector controls. (b) Paraquat exposure. Survival of elav>LAMP2A flies fed with 20 mM paraquat for 72 h was markedly increased compared to the elav/+ and LAMP2A/+ controls. (c) Effect on age-related locomotor decline. Pan-neuronal LAMP2A expression (elav>LAMP2A flies) significantly delayed age-related decrements in climbing performance (SING assay) compared to driver and effector controls that behaved like the wild type. (d) Lifespan assay. elav/+ and elav>LAMP2A flies showed similar longevity curves (median lifespan 57 and 55 days, respectively) indicating that neuronal LAMP2A expression does not affect Drosophila lifespan.
Oxidative stress promotes protein damage and aggregation that can be mitigated by increased autophagy [15,35,36]. ROS produced mainly from mitochondrial source under starvation or other stressful conditions can directly induce autophagy by a mechanism that is not fully understood [37]. Increased LAMP2A transcription and CMA activation were previously demonstrated in rats under paraquat-mediated oxidative stress [38]. Here we find that flies with pan-neuronal LAMP2A expression showed much higher tolerance to paraquat compared to the driver and UAS controls: around 50% of LAMP2A flies were still alive after 72 h of paraquat exposure, at a time when all control flies were dead (Figure 1b). Therefore, human LAMP2A potently protects Drosophila neurons against acute oxidative stress.
LAMP2A delays locomotor senescence but does not prolong lifespan
Previous studies showed that restoring CMA by increasing LAMP2A levels could protect from age-related liver function decline in mice [39]. Here we monitored startle-induced negative geotaxis (SING), a frequently used behavioral paradigm to test for locomotor ability in Drosophila. SING progressively declines with age or in various mutant neuropathological conditions [40–45]. We observed that flies expressing human LAMP2A in neurons had a better preservation of climbing performance with age compared to controls, which was prominent after 6 weeks of adult life (Figure 1c). In contrast and interestingly, neuronal LAMP2A expression did not extend Drosophila longevity, indicating that it increased fly healthspan rather than lifespan (Figure 1d).
LAMP2A coexpression reduces neuronal SNCA accumulation and prevents SNCAA30P-induced locomotor defects
Accumulation of the chaperone protein SNCA plays a central role in PD pathogenesis [46]. This accumulation is normally mitigated by proteasomal degradation and lysosomal clearance, involving in particular CMA activation through LAMP2A and HSPA8 induction [47,48]. SNCA mutations promote protein aggregation and disrupt CMA [26], and this probably contributes to neuronal loss and subsequent motor symptoms in PD. Furthermore, regional variations in CMA activity and LAMP2A expression appear to correlate with differential vulnerability of brain regions to protein aggregation [49]. Both LAMP2A and HSPA8 mRNAs and proteins are reduced in the substantia nigra pars compacta of PD patients, leading to lower CMA activity and increased SNCA level [50,51].
In Drosophila, the expression of human wild-type or mutant SNCA in neurons causes progressive locomotor impairments in aging flies [40,42,52–54]. Remarkably, we observed that the SING defects induced by neuronal expression of PD-associated SNCA with alanine-to-proline at position 30 (SNCAA30P) were fully prevented when LAMP2A was coexpressed (Figure 2a). We also observed a reduced accumulation of SNCAA30P protein in 30-day-old flies when human LAMP2A was coexpressed with the pathogenic protein in all neurons (Figure 2b). Neither SNCAA30P nor LAMP2A mRNA levels were decreased when these transgenes were coexpressed compared to each protein alone (Figure S2a), indicating that the reduced accumulation of SNCAA30P protein was not related to the presence of 2 UAS transgenes in the genome. The lower SNCAA30P accumulation in elav>LAMP2A, SNCAA30P flies compared to elav>SNCAA30P is therefore likely to result from a faster degradation of the pathogenic mutant protein. This further indicates that human LAMP2A can activate autophagic clearance in flies. Taken together, these observations are reminiscent of the neuroprotective effect of LAMP2A overexpression in the rat substantia nigra, which prevents dopaminergic neurodegeneration induced by human SNCA and reduces SNCA levels in this PD model as well [28].
Figure 2.
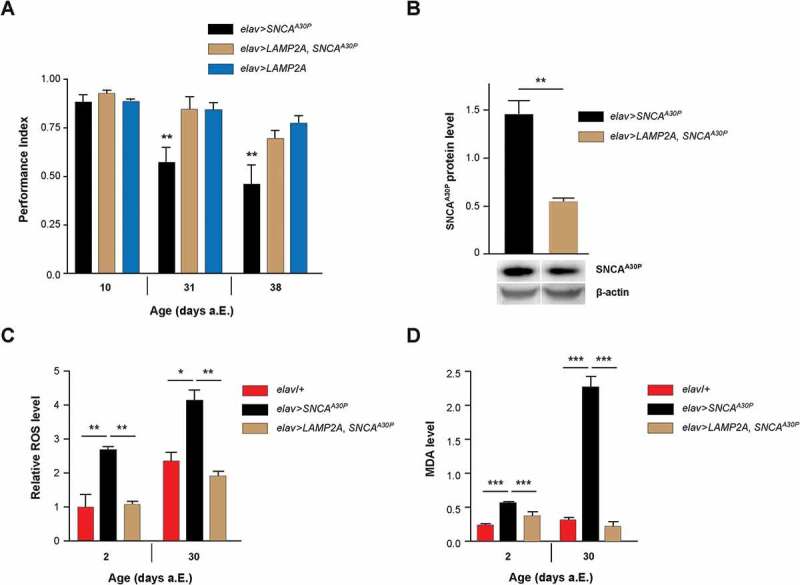
LAMP2A prevents SNCA-induced behavioral and oxidative defects in Drosophila. (a) LAMP2A coexpression fully prevented the progressive locomotor defects induced by pan-neuronal SNCAA30P. Climbing ability (SING assay) of elav>LAMP2A, SNCAA30P flies was compared to that of elav>SNCAA30P and elav>LAMP2A flies at 10, 31 and 38 days after a.E. (b) Human LAMP2A reduced neuronal SNCA accumulation. Western blots of head protein extracts from 30-day-old elav>SNCAA30P flies compared to elav>LAMP2A, SNCAA30P probed with anti-SNCA antibody. Act5C was used as a loading control. Quantification of SNCAA30P protein level from 3 independent experiments. Coexpression of LAMP2A reduced SNCAA30P accumulation without decreasing its mRNA level (see Fig. S2A). (c) ROS levels in the brain of elav>LAMP2A, SNCAA30P flies were lower than those of elav>SNCAA30Pand comparable to the elav/+ control at both 2 and 30 days a.E. Representative pictures of DHE-labeled brains are shown in Figure S2B. (d) MDA concentration assayed in the brain of 2- and 30-day-old adult Drosophila as an index of lipid peroxidation. Brain MDA level was markedly increased in elav>SNCAA30P flies but not in elav>LAMP2A, SNCAA30P flies that show similar levels as the elav/+ control.
LAMP2A prevents SNCA-induced ROS accumulation and oxidative defects
Previous reports in Drosophila and mouse suggest that the neurotoxicity of SNCA could be partly caused by an increase in oxidative stress associated with the abnormal accumulation of this protein [52,55–58]. This prompted us to analyze the levels of reactive oxygen species (ROS) in whole-mount Drosophila brains using dihydroethidium (DHE) dye fluorescence. Flies coexpressing SNCAA30P together with LAMP2A in all neurons showed significantly decreased ROS abundance compared to flies expressing SNCAA30P alone, both at 2 and 30 days after eclosion (a.E.) (Figure 2c and Figure S2b). We also determined the brain level of malondialdehyde (MDA), a compound used to assess the level of lipid peroxidation, as a test for oxidative stress-induced damage [59,60]. While the MDA amount in brain was found to be elevated in the presence of SNCAA30P, particularly in 30-day old flies, coexpression of LAMP2A in neurons fully restored brain MDA to normal levels (Figure 2d). This shows that LAMP2A can counteract the noxious pro-oxidant effect of mutant SNCAA30P in the Drosophila brain.
LAMP2A increases KFERQ motif-selective autophagy in the adult brain of fed flies but not in the larval fat body
In mammals, LAMP2A is a limiting step for CMA and changes in its levels modulate CMA activity [61]. All conditions known to activate CMA, including prominently starvation, up-regulate LAMP2A level at the lysosomal membrane [38,61,62]. Protection against starvation, oxidative stress and SNCAA30P conferred by LAMP2A expression in Drosophila suggests that this protein may be able to stimulate a KFERQ motif-selective autophagy process in this organism. A fluorescent biosensor containing a KFERQ peptide fused to photoactivable mCherry (KFERQ-PA-mCherry), has recently been shown to localize in the late endosome and lysosome compartment in larval fat body upon prolonged starvation, an effect dependent on Hsc70-4, the fly ortholog of HSPA8, and the ESCRT (endosomal sorting complexes required for transport) machinery [14]. This was interpreted as an evidence of eMI rather than CMA. We therefore used this biosensor to probe for the effect of LAMP2A on the accumulation of fluorescent puncta in late endosomes and lysosomes, either in fed or starvation conditions [14]. Although we observed an increase in biosensor puncta per cell in the larval fat body upon LAMP2A expression under fed conditions in some experiments, the effect was found not to be significant when all data were pooled (Fig. 3ai-ii’ and b). Starvation increased sensor puncta formation, as previously reported [14], but not further in LAMP2A-expressing larvae (Fig. 3aiii-iv’ and b). Similar results were obtained with another eMI biosensor, KFERQ-Split-Venus [13], in which the KFERQ recognition motif was fused to both the N– and C-terminal part of the yellow fluorescent protein variant Venus [63]. Split-Venus reconstitution occurs when both moieties concentrate upon endosomal targeting, leading to a fluorescent signal [13]. In the larval fat body, LAMP2A expression did not increase Split-Venus fluorescence nor puncta formation under fed or starvation conditions (Fig. S3a-c). Therefore, LAMP2A expression does not seem to induce KFERQ-dependent autophagy efficiently in the larval fat body.
To examine the effect of LAMP2A expression on selective autophagy in the brain, we expressed N- and C-KFERQ-Split-Venus together in all neurons, either with or without LAMP2A. Brains from fed or starved adult flies were dissected and mounted for confocal microscopy. Representative scans of whole brains are shown in Figure S4a. Fluorescent puncta were detected in the neuropil at high resolution, in particular in the calyx region of the mushroom bodies, where they could be most easily visualized and quantified (Figure 3c and Figure S4b). These puncta likely represent Split-Venus reconstitution events in the late endosome and lysosome compartments. A small but significant increase in overall fluorescence intensity in the brain neuropil of fed flies was observed at low resolution when LAMP2A was coexpressed (Figure 3d). We observed, remarkably, that the number of puncta in the calyx region was strongly increased (about twice as much) compared to controls without LAMP2A (Figure 3e). In contrast, we could not see significant puncta increments by LAMP2A expression under starvation condition (Figure 3d and e). This suggests that human LAMP2A can increase KFERQ motif-dependent selective autophagy in the fly nervous system. However, this effect does not occur in all tissues and has not been observed under starvation.
Figure 3.
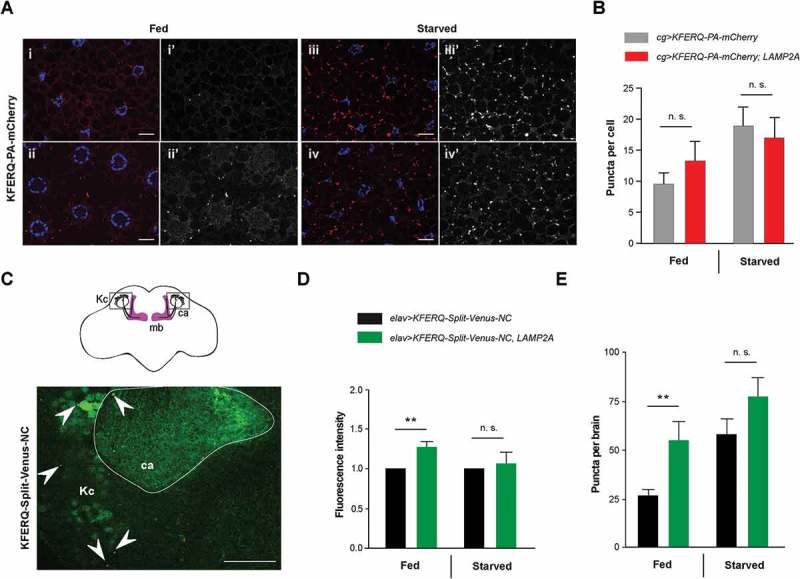
Effect of LAMP2A on selective autophagy in the larval fat body and adult brain. (a) In 3rd-instar larval fat body cells, LAMP2A expression (LAMP2A, bottom panels) did not increase KFERQ-PA-mCherry fluorescent sensor puncta formation 25 h after photoactivation, either under fed (i, ii) or starvation (iii, iv) conditions, compared to controls (top panels). In composite images, mCherry fluorescence is in red and DAPI-stained nuclei are in blue. i’-iv’ monochromatic images show the KFERQ-PA-mCherry single channel. Scale bars: 20 µm. (b) Quantification of puncta number per cell in larvae expressing photoactivated KFERQ-PA-mCherry in fat body with or without (control) LAMP2A under fed or starvation conditions. Starvation-induced sensor puncta formation was not further increased by LAMP2A expression. Similar results were obtained using a different eMI biosensor (KFERQ-Split-Venus) (shown in Figure S3). (c) Localization of reconstituted KFERQ-Split-Venus sensor fluorescent puncta (arrowheads) in a posterior region of the adult brain of elav>KFERQ-Split-Venus-NC flies expressing the eMI sensor in all neurons. The square in the scheme (top inset) shows localization of the magnified brain region that surrounds the calyx of the mushroom body, where fluorescence was prominent and in which puncta were scored. mb, mushroom body; Kc, cell bodies of the mushroom body Kenyon cells; ca, calyx. Representative scans of whole brain and calyx region for the different genotypes and feeding conditions are shown in Figure S4a and b, respectively. (d, e) Reconstitution of KFERQ-Split-Venus eMI sensor was increased in adult brain of fed, but not starved, flies expressing LAMP2A in all neurons (elav>KFERQ-Split-Venus-NC, LAMP2A) (LAMP2A, right panel), as indicated by higher overall fluorescence level (d) and increased density of eMI-positive puncta in the calyx region (e), compared to elav>KFERQ-Split-Venus-NC controls. Quantification from 3 independent experiments.
LAMP2A upregulates Atg5 expression and enhances macroautophagy in fly neurons
Suppression of macroautophagy in various models promotes neurodegeneration associated to the accumulation of polyubiquitinated protein aggregates [35,64–67]. Moreover, increasing basal macroautophagy is known to protect Drosophila against the deleterious consequences of oxidative stress [35,65,68]. We therefore considered macroautophagy as an alternative degradation process by which human LAMP2A could mediate neuroprotection. ATG5 is an E3-like ligase that together with ATG12 and ATG16L1 constitutes an elongation complex required for the formation of autophagosomes [69]. Here we found by western blot analysis that LAMP2A expression in neurons considerably increased levels of Drosophila Atg5 protein and of the Atg12-Atg5 complex (Figure 4a and b). Because it has recently been reported that Atg5 inactivation induces locomotor defects in flies [44], the upregulation of Atg5 expression could in part explain the improvement in locomotion ability we observed upon LAMP2A expression in PD-like conditions and in aged wild-type flies.
Figure 4.
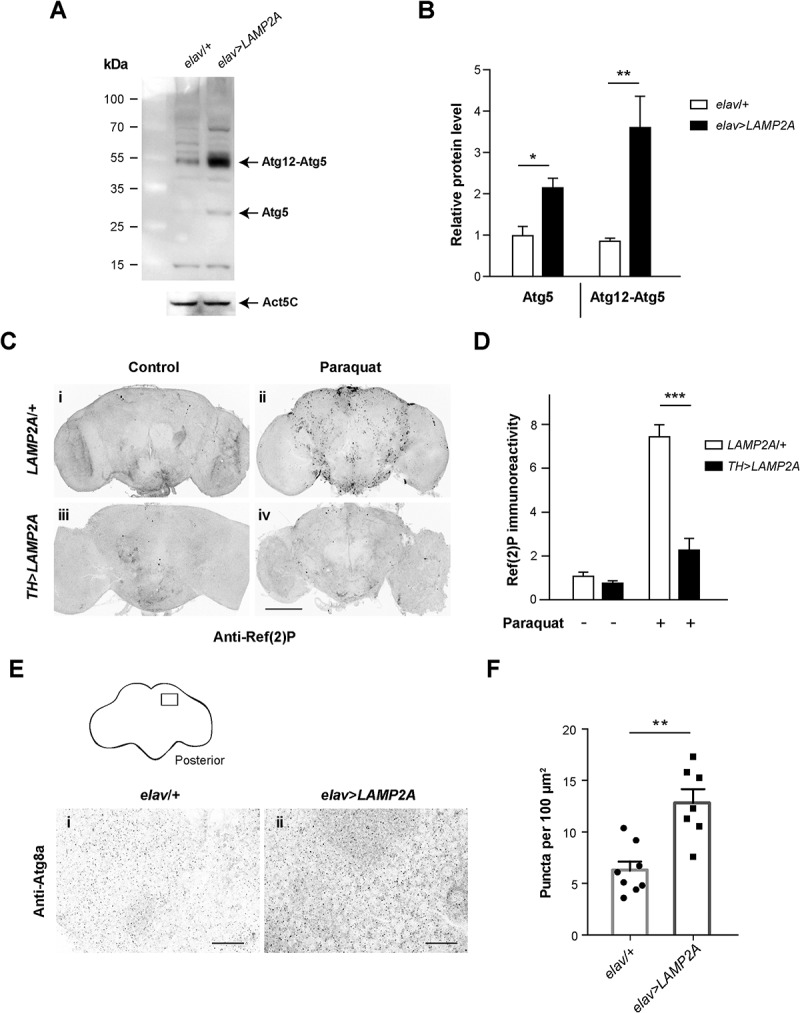
Human LAMP2A enhances macroautophagy in the Drosophila brain. (a, b) Effect of LAMP2A on Atg5 expression. (a) Western blot of head protein extracts from 10-day old control flies (elav/+) and flies expressing human LAMP2A in neurons (elav>LAMP2A) probed with anti-Atg5 antibody. LAMP2A expression markedly increased levels of Atg5 and of the Atg12-Atg5 complex that is required for autophagosome formation. Act5C served as a loading control. (b) Quantification of Atg5 protein and the Atg12-Atg5 complex from 3 independent western blot experiments. (c, d) Effect of LAMP2A on paraquat-induced Ref(2)P accumulation. (c) Anti-Ref(2)P immunostaining in whole-mount adult brains of LAMP2A/+ (panels i and ii) and TH>LAMP2A (panels iii and iv) flies exposed to paraquat (panels ii and iv) or not (panels i and iii). LAMP2A expression prevented paraquat-induced Ref(2)P accumulation (black puncta) suggesting that the human protein is able to maintain efficient autophagic flux under oxidative stress. Scale bar: 100 μm. (d) Quantification of Ref(2)P immunostaining in the central brain region normalized to LAMP2A/+ control not exposed to paraquat (n = 4 or 5 independent brains per condition). (e, f) Effect of LAMP2A on the number of Atg8a-positive puncta. (e) Anti-Atg8a immunostaining in whole-mount adult brains of elav/+ (panel i) and elav>LAMP2A (panel ii) flies. The inset scheme on top shows the posterior neuropil region that was magnified in panels i and ii and in which Atg8a puncta were counted. Scale bars: 10 µm. (f) Quantification of Atg8a-positive dots. Each black circle (elav/+) or square (elav>LAMP2A) represents the score for a different brain. The number of Atg8a puncta that reflect autophagosome formation was markedly increased in LAMP2A-expressing flies. Similar results were obtained in 3 independent experiments.
Ref(2)P, the Drosophila ortholog of human SQSTM1/p62 (sequestosome 1), is an autophagosome cargo protein contributing to the clearance of ubiquitinated proteins [66,67,70], which is commonly used as a marker of autophagic flux in fly tissues [71]. Dopaminergic neurons are the primary target of paraquat in Drosophila [72]. We therefore compared anti-Ref(2)P immunostaining in brains of oxidative stress-challenged flies expressing or not human LAMP2A in dopaminergic neurons. Whereas paraquat exposure increased Ref(2)P accumulation in neurons of control flies (Fig. 4ci-ii and d), this effect was strikingly reduced upon LAMP2A expression (Fig. 4ciii-iv and d). This indicates that human LAMP2A prevents paraquat-induced accumulation of Ref(2)P-containing aggregates in Drosophila neurons, most likely by enhancing clearance of these aggregates through Atg5-dependent macroautophagy.
Recruitment of Atg8a/LC3, the major Drosophila ortholog of human GABARAP (GABA type A receptor associated protein) and MAP1LC3/LC3 (microtubule associated protein 1 light chain 3) ortholog families of yeast Atg8, during autophagosome assembly is an essential step in the macroautophagy process [73,74]. To search for a direct demonstration that LAMP2A increases autophagosome formation in fly neurons, we analyzed the density of Atg8a-positive puncta by immunostaining. As shown in Figure 4e and f, we found that pan-neuronal expression of LAMP2A as a mean doubled the number of Atg8a-positive dots in the brain neuropil compared to control flies. This confirmed an expansion of the autophagosome compartment induced by LAMP2A expression in Drosophila neurons.
Discussion
Expression of the lysosomal receptor LAMP2A confers stress resistance and neuroprotection in Drosophila
The autophagy-lysosome pathway plays a fundamental role in cellular physiology. Impairment in this process, and particularly of macroautophagy and CMA, is often associated with the progression of neurodegenerative diseases, including PD [15,16,26,27,75–78]. These autophagic pathways are therefore major potential targets to improve the curative treatment of these neuropathologies. Selective protein degradation by CMA depends on the presence of a KFERQ peptide motif or related sequences in protein substrates [79]. These proteins individually interact with a complex containing the cytosolic chaperone HSPA8, the lysosomal membrane glycoprotein LAMP2A and other proteins, in which protein substrates are unfolded and translocated from the cytosol to the lysosomal lumen for degradation through LAMP2A [15,18].
Although CMA has been extensively characterized in mammals, it is currently considered that this process does not occur in invertebrates, including Drosophila. The only fly ortholog to human LAMP2A, named Lamp1, does not contain the amino-acid sequence present in the C-terminal tail of LAMP2A that is required for CMA, and it apparently does not form a complex with Drosophila Hsc70-4, the ortholog of human HSPA8 [13]. Another selective autophagy process, eMI, seems to be used instead in the fly [13,14]. eMI has been described in mammals as a process whereby endosomes engulf cytosolic material through the formation of multivesicular bodies that later fuse with lysosomes [8]. Cytosolic proteins bearing a KFERQ motif can be selectively targeted to this autophagy pathway by a LAMP2A-independent process that involves Hsc70-4 and the ESCRT machinery [8,13,14].
Here we investigated the potential activity of ectopically expressed human LAMP2A in Drosophila. We found that pan-neuronal expression of LAMP2A is harmless for behavior and survival, but rather is potently neuroprotective in flies, conferring increased resistance to various stresses: nutrient starvation, paraquat exposure, or mutant SNCA accumulation. A common feature shared by all these stressors is that they induce a significant rise in neuronal oxidative stress. LAMP2A appears therefore able to activate a powerful protective response preventing the deleterious consequences of ROS accumulation in the Drosophila nervous system, suggesting that this membrane protein can stimulate autophagic mechanisms in this organism.
LAMP2A expression in flies delays locomotor aging and SNCA-induced defects
Impairment of intracellular proteolysis could be largely responsible for the deficient removal of oxidized proteins in the aged organisms [80]. We observed that neuronal LAMP2A expression significantly improved the locomotor performance of aging flies without modifying Drosophila lifespan. This suggests that preventing the accumulation of oxidized or damaged proteins can prolong healthspan but is not sufficient to increase longevity in Drosophila. As mentioned above, these results agree with reports in other models suggesting that a lower cellular oxidative stress during aging cannot by itself promote prolonged lifespan [81,82]. It is suspected that other age-related genetic and physiological processes, which are still largely unresolved, play a more decisive role in lifespan regulation [83].
In humans, PD pathogenesis is associated with CMA impairments in the substantia nigra [50,51] and this may contribute to SNCA accumulation and neurotoxic effects in this disease. PD-associated SNCA and LRRK2 mutant proteins disrupt CMA-mediated cytosolic protein degradation [26,27], leading to the accumulation of SNCA oligomers and other damaged and potentially toxic proteins. Here we show in the Drosophila model that LAMP2A not only protected against SNCA-induced locomotor impairments, but also reduced ROS and MDA accumulation and so likely oxidative cellular damages, and at the same time decreased mutant SNCA protein level. Such a potent protection is quite comparable to the positive effects of virally-expressed LAMP2A against SNCA toxicity in the rat substantia nigra [28]. Our results indirectly suggest that LAMP2A selectively activates clearance of the pathogenic aggregated forms of SNCAA30P in Drosophila neurons, indicating again that an activation of autophagy is involved in its neuroprotective effects.
LAMP2A induces a selective autophagy process in the fly brain
We then assessed which type of autophagy can be activated by LAMP2A in Drosophila. It is known that starvation first activates macroautophagy within 1 h of nutrient deprivation [34,84]. Prolonged starvation then triggers LAMP2A-dependent CMA in mammalian cells [85,86] or ESCRT-dependent eMI in Drosophila [14]. Here we observed with 2 different eMI biosensors that LAMP2A expression did not alter either fluorescence intensity or puncta formation in the larval fat body under fed nor under starvation conditions. In contrast, when we expressed the KFERQ-Split-Venus eMI sensor in the adult brain, we observed that LAMP2A induced a significant increase in sensor fluorescence and puncta formation in fed but not in starved flies. This suggests that LAMP2A can induce a form of selective autophagy in the fly brain, but that this effect is unlikely to be involved in the increased starvation resistance induced by neuronal expression of this human protein. In mammals, CMA can be induced by overexpression of its receptor LAMP2A [28,39]. As mentioned above, CMA is not expected to occur in Drosophila owing to the lack of a LAMP2A counterpart in invertebrates [13,14]. However, other key CMA partners are expressed such as Hsc70-4 that shares 87% identity with human HSPA8 [13], and a KFERQ-like motif is present in about 43% of the fly proteins [14]. Further work is needed, therefore, to determine whether human LAMP2A can induce a CMA-like process in the Drosophila brain.
Human LAMP2A upregulates Atg5 and stimulates macroautophagy in Drosophila
Quite unexpectedly, we observed that human LAMP2A expression in fly neurons strongly increased levels of Atg5 and of the Atg12–Atg5 macroautophagy complex. This was apparently associated with an enhancement of autophagic flux because paraquat-induced Ref(2)P accumulation was prevented when LAMP2A was expressed in brain dopaminergic neurons. Finally, we observed that LAMP2A expression also markedly increased the density of Atg8a-positive puncta in the brain neuropil. Taken together, our results indicate that the ectopic expression of LAMP2A can increase autophagosome formation and enhance macroautophagy in the Drosophila nervous system. This is not contradictory to the effect we observed on LAMP2A-mediated clearance of mutant SNCAA30P, as it has been shown that SNCA can be degraded either by CMA or macroautophagy in mammalian neurons [47]. Accordingly, it was recently reported that paraquat-induced oxidative stress or MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) toxicity both impaired autophagic flux in human dopaminergic neuroblastoma cells and in zebrafish dopamine neurons, respectively, and that Atg5 appeared protective as well in these 2 PD models [68,87]. Our report also suggests a potential inductive effect of LAMP2A on the expression or activity of autophagy-related proteins. A recent article indicated that the lack of LAMP2 gene in mouse embryonic fibroblasts prevented STX17 (syntaxin 17) incorporation into autophagosomes, leading to a failure of their fusion with lysosomes [88], which is another indication that the LAMP2 proteins can contribute to autophagosome maturation.
In humans, LAMP2 gene deficiency leads to Danon disease, histologically characterized by an extensive accumulation of autophagic vacuoles in various tissues and defects in autophagosome-lysosome fusion [21,89,90]. A similar apparent block of autophagy could be reproduced in lamp2 knock-out mice, leading to the accumulation of autophagic vacuoles and of SQSTM1-positive aggregates in the brain [91–93], or by LAMP2 inactivation in mouse and human cultured cells [88,94–96]. The LAMP2 gene products appear to play a specific role in the fusion of autophagosomes with lysosomes [88,95,97]. A role for LAMP2 in macroautophagy in human cells was also previously indicated by functional mapping [98]. However, the matter is not simple as LAMP2 produces 3 protein isoforms with distinct roles in autophagy [19,21]. It would therefore be important to learn more about the respective functions of these LAMP2 isoforms in autophagy regulation, possibly by comparing their neuroprotective effects in Drosophila. Our present study suggests that LAMP2A can increase both KFERQ motif-dependent selective autophagy as well as macroautophagy in the fly brain. These effects could potentially reflect the diverse functions of this protein in the mammalian brain.
In future work, based on the highly positive effects of sustained LAMP2A expression reported here and the tractability of the Drosophila model, it should be possible to identify additional factors involved in LAMP2A-mediated neuroprotection. Such conserved factors or mechanisms could represent novel therapeutic targets potentially useful to improve human health during aging and the treatment of PD and other degenerative diseases through autophagy activation.
Materials and methods
Drosophila culture and strains
Flies were raised on a standard cornmeal-yeast-agar nutrient medium containing as an antifungal agent methyl 4-hydroxybenzoate (VWR International, 25,605.293), at 25°C and ~50% humidity with 12 h-12 h light-dark cycles. The following strains were used: w1118 as wild-type control, elav-Gal4 (elavC155) from the Bloomington Drosophila Stock Center, TH-Gal4 [99], cg-Gal4 [100], UAS-SNCAA30P [40] (kindly provided by Mel Feany, Harvard Medical School, Boston, MA), UAS-GFP::LAMP1; UAS-KFERQ-PA-mCherry [14], UAS-KFERQ-N-Venus, UAS-KFERQ-C-Venus [13] (kindly provided by Patrik Verstreken, KU Leuven, Center for Human Genetic, Leuven, The Netherlands) and UAS-LAMP2A (this report). A strain carrying both UAS-LAMP2A and UAS–SNCAA30P on the second chromosome was generated by meiotic recombination. For purpose of simplification, we used the driver>effector convention to indicate genotypes: elav-Gal4; UAS-LAMP2A flies, for example, were denoted as elav>LAMP2A.
DNA constructs
The human LAMP2A cDNA (clone H05D091G15) was obtained from Kabushiki Kaisha DNAFORM. The 1250 base pair LAMP2A insert was PCR amplified using primers with added restriction sites (Table S1). The sequence upstream of the ATG initiation codon in the forward primer was modified to match the Drosophila translation start consensus sequence. The PCR fragment was inserted into pUAST [101] and verified by sequencing (GATC Biotech). The UAS-LAMP2A plasmid was sent to BestGene Inc. for P-element transformation by random insertion. Non-recessive lethal second- and third-chromosome insertions that yielded strong expression were selected and used thereafter.
RNA detection
RT-PCR analysis was carried out and analyzed as previously described [72]. Total RNA was isolated by standard procedure generally from 20 heads of 10-day-old flies collected on ice and lysed in 600 µl QIAzol Reagent (Qiagen, 79,306). Total RNA (1 μg) was reverse transcribed using oligo(dT) primers with Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, K1671). Approximately 1 ng of the first strand cDNA was amplified in 25 μl of reaction mixture using PrimeStar Max DNA polymerase (Takara Bio, R045A). The program cycles included 10 s denaturation at 98°C, 10 s annealing at 55°C and 30 s elongation at 72°C, repeated 25 to 35 times. PCR product levels were estimated after electrophoresis by densitometry with the Fiji software [102]. Data were normalized to amplification level of the ribosomal RpL32/rp49 transcript. Sequences of the primers used are indicated in Table S2.
Adult brain immunostaining
Whole-mount brains from adult flies aged 8 days a.E. were dissected and processed for immunostaining as previously described [43,72]. The primary antibodies were: mouse monoclonal anti-human LAMP2 used at 1:200 dilutions (Developmental Studies Hybridoma Bank, H4B4), rabbit polyclonal antibody against Drosophila Ref(2)P [103] used at 1:100 (kindly provided by Sébastien Gaumer, Université Versailles-St-Quentin-en-Yvelines, France) and mouse monoclonal anti-human GABARAP (anti-Atg8) used at 1:500 (Santa Cruz Biotechnology, sc-377,300). Note that the amino acid sequences of human GABARAP and Drosophila Atg8a shares 92% identity and we also checked by western blot that this GABARAP antibody that cross-reacts with multiple species recognizes Drosophila Atg8a. Brains were mounted in Prolong Gold Antifade Mountant (Thermo Fisher Scientific, P36930) and images were acquired on a Nikon A1R confocal microscope (Nikon Instruments, Tokyo, Japan). Laser, filter and gain settings remained constant within each experiment. The number of Atg8a particles was scored after selecting the area of interest by using the particle analysis tool of the Fiji software.
Starvation and oxidative stress resistance
To monitor starvation resistance, 10-day-old female Drosophila were kept in vials containing Whatman blotting paper (Sigma-Aldrich, WHA3001917) soaked with water and no nutrient medium. Dead flies were scored every 12 h. At least 100 flies were tested per genotype. Oxidative stress resistance was assayed by exposure to paraquat (methyl viologen dichloride hydrate; Sigma-Aldrich, 856,177) using a previously described dietary ingestion procedure [72]. Non-virgin 8-day-old female Drosophila (~100 per genotype) were starved for 2 h in empty vials. They were then exposed to 20 mM paraquat diluted in 2% (wt:vol) sucrose (Euromedex, 200–301-B) (or sucrose only for controls) in 2-inch (5.2 cm) diameter Petri dishes (10 flies per dish) containing 2 layers of Whatman blotting paper soaked with the paraquat solution and incubated at 25°C in saturating humidity conditions. Fly survival was scored after 24, 48 and 72 h. In some experiments, brains from flies exposed to 20 mM paraquat for 24h were dissected and processed for immunostaining.
Locomotion assay
Locomotor decline during aging was monitored by a SING test as previously described [42,45]. For each genotype, 50 adult males divided into 5 groups of 10 flies were placed in a vertical column (25-cm long, 1.5-cm diameter) with a conic bottom end and left for approximately 25 min for habituation. They were tested individually by gently tapping them down (startle), and scoring the number of flies having reached the top of the column (above 22 cm) and remaining at the bottom end (below 4 cm) after 1 min. Each group was assayed 3 times at 15 min intervals. The performance index for each column was calculated as follows: ½[1 + (ntop-nbot)/ntot], where ntot is the total number of flies, and ntop and nbot the number of flies at the top and at the bottom, respectively. Results are the mean and SEM of the scores obtained with the 5 groups of flies per genotype.
Longevity studies
Lifespan analysis was performed as described [43]. Drosophila adult males (~50 animals/bottle in triplicate for each genotype) were collected within 24 h of emergence and incubated at 25°C. They were transferred into fresh bottles or vials every 2 or 3 days and the number of surviving flies was scored. Data are expressed as percentage of the initial fly number as a function of time.
Fluorescent biosensors
To monitor eMI activity in larvae, we first used a fluorescent sensor containing a KFERQ motif fused to photoactivable mCherry (KFERQ-PA-mCherry) as previously described [14]. Briefly, cg-Gal4, UAS-GFP::LAMP-1; UAS-KFERQ-PA-mCherry virgins were mated to w1118 or UAS-LAMP2A males. Early 3rd instar progeny larvae of these crosses were photoactivated and then incubated for 25h on 20% sucrose with heat-killed yeast paste for fed conditions, or 20% sucrose only for starvation. Following which, they were cut open, fixed in 4% paraformaldehyde (VWR International, 100,503–917), and processed for fat body tissue imaging on an ApoTome.2 system (Carl-Zeiss, Oberkochen, Germany). Around 150 cells were quantified in total per genotype in 4 independent experiments.
In other sets of experiments, we have used a Split-Venus eMI sensor [13], in which the recognition motif KFERQ is fused both to the N- or C-terminal portion of the yellow fluorescent protein variant Venus [63]. Split-Venus reconstitution occurs when both moieties concentrate upon eMI targeting, leading to a fluorescent signal [13]. UAS-KFERQ-N-Venus, UAS-KFERQ-C-Venus (here named UAS-KFERQ-Split-Venus-NC, control) or recombined UAS-KFERQ-Split-Venus-NC, UAS-LAMP2A flies were crossed to cg-Gal4 or elav-Gal4 for expression in fat body cells or in all neurons, respectively. 3rd instar larvae (3 to 5 per genotype in each experiment) were either fed on 20% sucrose plus yeast paste or starved for 4 h on 20% sucrose, before being processed for fat body tissue imaging. Whole-mount brains (3 to 7 per genotype and feeding condition in each experiment) were dissected from progeny adults aged 7 to 10 days a.E that were either normally fed or starved for 24 h. Brains were then fixed in 4% paraformaldehyde for 20 min and washed in 1X phosphate-buffered saline (PBS) pH 7.4 (Thermo Fisher Scientific, 70,011,044) for 15 min, twice. Split-Venus reconstitution in tissues was monitored by yellow fluorescence imaging (excitation at 515 nm, emission at 528 nm) on a Nikon A1R confocal microscope. Laser, filter and gain settings remained constant within one experiment. Overall fluorescence intensity was determined with the Fiji software by measuring relative mean density on Z projections of portions of fat body or on the whole brain, respectively, and the data were normalized to respective control values. The number of puncta was scored after selecting the area of interest by using the particle analysis tool of the Fiji software. Results are mean ± SEM of 3 independent experiments.
Western blot analyses
Fly heads (20) were homogenized in 100 μl RIPA buffer (Sigma-Aldrich, R0278) containing protease inhibitors (cOmplete mini Protease Inhibitor Cocktail tablets; Roche Diagnostics, 11,836,153,001) using a Minilys apparatus (Bertin Instruments, Montigny-le-Bretonneux, France). The lysates were incubated on ice for 30 min and centrifuged at 8,000 g for 10 min at 4°C. Protein samples were prepared by diluting the supernatant in 1 x final NuPAGE LDS Sample Buffer (Thermo Fisher Scientific, NP0001) and reducing agent (Thermo Fisher Scientific, NP0009), followed by 10-min denaturation at 70°C and 5 s centrifugation at 1,000 g. Proteins were separated in 4–12% NuPAGE Bis-Tris precast polyacrylamide gels (Thermo Fisher Scientific, NP0321BOX), using PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific, 26,619) as migration marker, and electrotransferred to Hybond ECL nitrocellulose (GE Healthcare Life Sciences, RPN3032D) or Amersham Hybond P 0.45 PVDF membranes (GE Healthcare Life Sciences, 10,600,023). Membranes were blocked after transfer for 2 h at room temperature in 1X Tris-buffered saline (TBS) pH 7.4 (Thermo Fisher Scientific, 10,648,973) containing 0.05% (v/v) Tween 20 (Sigma Aldrich, P1379) (TBS-Tween) supplemented with 5% skim milk. For immunodetection of human SNCA, transferred membranes were first fixed with 0.4% paraformaldehyde in PBS for 30 min to prevent the detachment of SNCA monomers as recently described [104]. The fixed membranes were then washed in TBS-Tween 3 times for 10 min and blocked for 2 h at room temperature in TBS-Tween + 5% skim milk. Membranes were then incubated overnight at 4°C in TBS-Tween containing 1% (w/v) bovine serum albumin (Euromedex, 04–100–812-C) with the following primary antibodies: rabbit polyclonal anti-SNCA used at 1:1,000 dilutions (Sigma-Aldrich, S3062), rabbit polyclonal anti-Atg5 at 1:500 (Novus Biologicals, NB110-53,818) and mouse monoclonal anti-ACTB (actin beta) at 1:5,000 (Abcam, ab20272) that cross-reacts with Drosophila Act5C (Actin 5C). Membranes were washed three times for 10 min in TBS-Tween and then incubated for 2 h at room temperature with horseradish peroxidase (HRP)-conjugated anti-rabbit (Abcam, ab7132) or anti-mouse (Jackson ImmunoResearch, 115–035–146) secondary antibodies at 1:5,000. Immunolabeled bands were revealed by using ECL RevelBlOt Intense (Ozyme, OZYB002-1000) as chemiluminescent HRP substrate and digitally acquired at different exposure times using ImageQuant TL software (GE Healthcare Life Science, Chicago, USA). Densitometry measures were made using Fiji software and normalized to Act5C measures as internal controls.
ROS detection and quantification
Reactive oxygen species (ROS) level was determined with the fluorescent dye dihydroethidium (DHE) (Thermo Fisher Scientific, D11347) following a published protocol [105] adapted to whole-mount Drosophila brains. DHE is preferentially oxidized by superoxide (O2.-) but it is also sensitive to other ROS, such as H2O2 and •OH, producing bright-red fluorescent products (ethidium, 2-hydroxyethidium, and dimers) measured at 555 nm as an index of intracellular oxidant formation [106]. 5 to 8 adult brains per genotype were dissected in Schneider insect medium (Sigma Aldrich, S0146) and incubated on an orbital shaker for 10 min at room temperature in 30 μM DHE diluted in the same medium. The brains were then washed 3 times in Schneider medium and fixed for 6 min in 7% formaldehyde (Thermo Fisher Scientific, 28,908) in PBS. After rinsing in PBS and mounting in Vectashield Antifade Mounting Medium (Vector Laboratories, H-1000), the brains were immediately scanned at constant gain setting on a Nikon A1R confocal microscope. Relative ROS levels were measured by quantification of whole brain average intensity level of the dye fluorescence using the Fiji software.
Estimation of lipid peroxidation
The quantification of MDA is an index of lipid peroxidation and ROS-induced damage [59]. MDA was determined in Drosophila brain extracts using a described procedure [107]. Ten brains were homogenized in 200 µl of ice-cold 20 mM Tris-HCl (Euromedex, EU0011C) (5 replicates of 10 brains per genotype) using a Minilys apparatus (Bertin Instruments, Montigny-le-Bretonneux, France) and the homogenate was microcentrifuged at 3,000 g for 20 min at 4°C. Then, 1 µl of the supernatant was added to 1.3 ml of acetonitrile-methanol 3:1 (vol:vol) solution containing 7.7 mM 1-methyl-2-phenylindole (Sigma Aldrich, 404,888) and 0.3 ml of 37% HCl was added. After vortexing, the tubes were incubated at 45°C for 40 min, cooled on ice and microcentrifuged at 1,500 g for 10 min at 4°C. Optical density of the supernatants was measured at 586 nm.
Statistical analysis
Data from locomotor assays and oxidative stress resistance have been analyzed by one-way ANOVA with the post-hoc Tukey-Kramer test. Survival curves for starvation resistance and longevity experiments were generated and compared using the log-rank test. Statistical analysis of DHE, MDA and eMI biosensor quantifications was performed by the Student t-test or one-way ANOVA with Tukey pairwise comparisons. All statistical analyses were performed with the Prism 6 software (GraphPad Software, San Diego, California). Significant values in all figures: *p < 0.05, **p < 0.01, ***p < 0.001.
Funding Statement
This work was supported by Fondation de France (to SB) [2012-00034435]; Labex MemoLife (to SB) [ANR-10-LABX-54 MEMOLIFE]; NIH (to AJ) [R01 GM119160]; Association France-Parkinson (to SB); PSL Research University (to SB).
Acknowledgments
We thank Drs. Mel Feany and Patrik Verstreken for Drosophila stocks, Dr. Sébastien Gaumer for antibody samples and Dr. Giorgio Matassi for helpful discussion about LAMP2A phylogenetics. This work was supported by funding from the Fondation de France, Association France-Parkinson, PSL Research University and Labex MemoLife (ANR-10-LABX-54 MEMOLIFE) to SB and NIH R01 GM119160 to AJ. ARI was recipient of a PhD fellowship from the Doctoral school 3C (ED3C) at University Pierre-et-Marie Curie, Paris, and then supported by a postdoctoral salary from the Labex MemoLife. JS, CP and AD are supported by PhD fellowships from the Chinese Scholarship Council, Association Lesch-Nyhan Action, and PSL Research University, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article shoud be accessed here.
References
- [1].Boya P, Reggiori F, Codogno P.. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15(7):713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20(3):460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16(6):345–357. [DOI] [PubMed] [Google Scholar]
- [4].Bento CF, Renna M, Ghislat G, et al. Mammalian autophagy: how does it work? Annu Rev Biochem. 2016;85:685–713. [DOI] [PubMed] [Google Scholar]
- [5].Feng Y, He D, Yao Z, et al. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ktistakis NT, Tooze SA. Digesting the expanding mechanisms of autophagy. Trends Cell Biol. 2016;26(8):624–635. [DOI] [PubMed] [Google Scholar]
- [7].Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7(7):673–682. [DOI] [PubMed] [Google Scholar]
- [8].Sahu R, Kaushik S, Cc C, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20(1):131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kawamura N, G-H S-W, Aoyama M, et al. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat Commun. 2012;3:1071. [DOI] [PubMed] [Google Scholar]
- [10].Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15(8):305–309. [DOI] [PubMed] [Google Scholar]
- [11].Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114(13):2491–2499. [DOI] [PubMed] [Google Scholar]
- [12].Horst M, Knecht EC, Schu PV. Import into and degradation of cytosolic proteins by isolated yeast vacuoles. Mol Biol Cell. 1999;10(9):2879–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Uytterhoeven V, Lauwers E, Maes I, et al. Hsc70-4 deforms membranes to promote synaptic protein turnover by endosomal microautophagy. Neuron. 2015;88(4):735–748. [DOI] [PubMed] [Google Scholar]
- [14].Mukherjee A, Patel B, Koga H, et al. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy. 2016;12(11):1984–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24(1):92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang G, Mao Z. Chaperone-mediated autophagy: roles in neurodegeneration. Transl Neurodegener. 2014;3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xilouri M, Stefanis L. Chaperone mediated autophagy in aging: starve to prosper. Ageing Res Rev. 2016;32:13–21. [DOI] [PubMed] [Google Scholar]
- [18].Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273(5274):501–503. [DOI] [PubMed] [Google Scholar]
- [19].Eskelinen E-L, Cuervo AM, Taylor MRG, et al. Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic. 2005;6(11):1058–1061. [DOI] [PubMed] [Google Scholar]
- [20].Wilke S, Krausze J, Büssow K. Crystal structure of the conserved domain of the DC lysosomal associated membrane protein: implications for the lysosomal glycocalyx. BMC Biol. 2012;10:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Endo Y, Furuta A, Nishino I. Danon disease: a phenotypic expression of LAMP-2 deficiency. Acta Neuropathol. 2015;129(3):391–398. [DOI] [PubMed] [Google Scholar]
- [22].Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113(24):4441–4450. [DOI] [PubMed] [Google Scholar]
- [23].Qi L, Zhang X-D. Role of chaperone-mediated autophagy in degrading Huntington’s disease-associated huntingtin protein. Acta Biochim Biophys Sin (Shanghai). 2014;46(2):83–91. [DOI] [PubMed] [Google Scholar]
- [24].Xilouri M, Stefanis L. Chaperone mediated autophagy to the rescue: A new-fangled target for the treatment of neurodegenerative diseases. Mol Cell Neurosci. 2015;66(A):29–36. [DOI] [PubMed] [Google Scholar]
- [25].Cai Z, Zeng W, Tao K, et al. Chaperone-mediated autophagy: roles in neuroprotection. Neurosci Bull. 2015;31(4):452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cuervo AM, Stefanis L, Fredenburg R, et al. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305(5688):1292–1295. [DOI] [PubMed] [Google Scholar]
- [27].Orenstein SJ, Kuo S-H, Tasset I, et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013;16(4):394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xilouri M, Brekk OR, Landeck N, et al. Boosting chaperone-mediated autophagy in vivo mitigates α-synuclein-induced neurodegeneration. Brain. 2013;136(7):2130–2146. [DOI] [PubMed] [Google Scholar]
- [29].Chang -Y-Y, Neufeld TP. Autophagy takes flight in Drosophila. FEBS Lett. 2010;584(7):1342–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mulakkal NC, Nagy P, Takats S, et al. Autophagy in Drosophila: from historical studies to current knowledge. Biomed Res Int. 2014;2014:273473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nagy P, Á V, Kovács AL, et al. How and why to study autophagy in Drosophila: it’s more than just a garbage chute. Methods. 2015;75:151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang H, Baehrecke EH. Eaten alive: novel insights into autophagy from multicellular model systems. Trends Cell Biol. 2015;25(7):376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tracy K, Baehrecke EH. The role of autophagy in Drosophila metamorphosis. Curr Top Dev Biol. 2013;103:101–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7(2):167–178. [DOI] [PubMed] [Google Scholar]
- [35].Simonsen A, Cumming RC, Brech A, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4(2):176–184. [DOI] [PubMed] [Google Scholar]
- [36].Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143(5):813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22(3):377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kiffin R, Christian C, Knecht E, et al. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15(11):4829–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14(9):959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404(6776):394–398. [DOI] [PubMed] [Google Scholar]
- [41].Jones MA, Grotewiel M. Drosophila as a model for age-related impairment in locomotor and other behaviors. Exp Gerontol. 2011;46(5):320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Riemensperger T, Issa A-R, Pech U, et al. A single dopamine pathway underlies progressive locomotor deficits in a Drosophila model of Parkinson disease. Cell Rep. 2013;5(4):952–960. [DOI] [PubMed] [Google Scholar]
- [43].Riemensperger T, Isabel G, Coulom H, et al. Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc Natl Acad Sci USA. 2011;108(2):834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kim M, Sandford E, Gatica D, et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife. 2016;5:e12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vaccaro A, Issa A-R, Seugnet L, et al. Drosophila clock is required in brain pacemaker neurons to prevent premature locomotor aging independently of its circadian function. PLoS Genet. 2017;13(1):e1006507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dehay B, Bourdenx M, Gorry P, et al. Targeting α-synuclein for treatment of Parkinson’s disease: mechanistic and therapeutic considerations. Lancet Neurol. 2015;14(8):855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vogiatzi T, Xilouri M, Vekrellis K, et al. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283(35):23542–23556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sk M, Al M, Ab M-B, et al. Lysosomal degradation of alpha-synuclein in vivo. J Biol Chem. 2010;285(18):13621–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Malkus KA, Ischiropoulos H. Regional deficiencies in chaperone-mediated autophagy underlie α-synuclein aggregation and neurodegeneration. Neurobiol Dis. 2012;46(3):732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Alvarez-Erviti L, Seow Y, Schapira AHV, et al. Influence of microRNA deregulation on chaperone-mediated autophagy and α-synuclein pathology in Parkinson’s disease. Cell Death Dis. 2013;4(3):e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, et al. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol. 2010;67(12):1464–1472. [DOI] [PubMed] [Google Scholar]
- [52].Barone MC, Sykiotis GP, Bohmann D. Genetic activation of Nrf2 signaling is sufficient to ameliorate neurodegenerative phenotypes in a Drosophila model of Parkinson’s disease. Dis Model Mech. 2011;4(5):701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Butler EK, Voigt A, Lutz AK, et al. The mitochondrial chaperone protein TRAP1 mitigates α-Synuclein toxicity. PLoS Genet. 2012;8(2):e1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Breda C, Nugent ML, Estranero JG, et al. Rab11 modulates α-synuclein-mediated defects in synaptic transmission and behaviour. Hum Mol Genet. 2015;24(4):1077–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Xu J, Kao S-Y, Lee FJS, et al. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med. 2002;8(6):600–606. [DOI] [PubMed] [Google Scholar]
- [56].Botella JA, Bayersdorfer F, Schneuwly S. Superoxide dismutase overexpression protects dopaminergic neurons in a Drosophila model of Parkinson’s disease. Neurobiol Dis. 2008;30(1):65–73. [DOI] [PubMed] [Google Scholar]
- [57].Alvarez-Fischer D, Fuchs J, Castagner F, et al. Engrailed protects mouse midbrain dopaminergic neurons against mitochondrial complex I insults. Nat Neurosci. 2011;14(10):1260–1266. [DOI] [PubMed] [Google Scholar]
- [58].Wang B, Liu Q, Shan H, et al. Nrf2 inducer and cncC overexpression attenuates neurodegeneration due to α-synuclein in Drosophila. Biochem Cell Biol. 2015;93(4):351–358. [DOI] [PubMed] [Google Scholar]
- [59].Del Rio D, Stewart AJ, Pellegrini N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr Metab Cardiovasc Dis. 2005;15(4):316–328. [DOI] [PubMed] [Google Scholar]
- [60].Hosamani R. Muralidhara. Acute Exposure Drosophila Melanogaster Paraquat Causes Oxidative Stress Mitochondrial Dysfunction Arch Insect Biochem Physiol. 2013;83(1):25–40. [DOI] [PubMed] [Google Scholar]
- [61].Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1(7):570–583. [DOI] [PubMed] [Google Scholar]
- [62].Dohi E, Tanaka S, Seki T, et al. Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem Int. 2012;60:431–442. [DOI] [PubMed] [Google Scholar]
- [63].Nagai T, Ibata K, Park ES, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20(1):87–90. [DOI] [PubMed] [Google Scholar]
- [64].Komatsu M, Waguri S, Chiba T. al. Loss Autophagy Central Nervous System Causes Neurodegeneration Mice Nature. 2006;441(7095):880–884. [DOI] [PubMed] [Google Scholar]
- [65].Juhász G, Erdi B, Sass M, et al. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21(23):3061–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bartlett BJ, Isakson P, Lewerenz J, et al. p62, ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy. 2011;7(6):572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gupta VK, Scheunemann L, Eisenberg T, et al. Restoring polyamines protects from age-induced memory impairment in an autophagy-dependent manner. Nat Neurosci. 2013;16(10):1453–1460. [DOI] [PubMed] [Google Scholar]
- [68].Garcia-Garcia A, Anandhan A, Burns M. Impairment of atg5-dependent autophagic flux promotes paraquat-and MPP+-induced apoptosis but not rotenone or 6-hydroxydopamine toxicity. Toxicol Sci. 2013;136(1):166–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kharaziha P, Panaretakis T. Dynamics of atg5-atg12-atg16l1 aggregation and deaggregation. Meth Enzymol. 2017;587:247–255. [DOI] [PubMed] [Google Scholar]
- [70].Komatsu M, Waguri S, Koike M. al. Homeostatic Levels P62 Control Cytoplasmic Inclusion Body Formation Autophagy-Deficient Mice Cell. 2007;131(6):1149–1163. [DOI] [PubMed] [Google Scholar]
- [71].DeVorkin L, Gorski SM. Monitoring autophagic flux using ref(2)P, the Drosophila p62 ortholog. Cold Spring Harb Protoc. 2014;2014(9):959–966. [DOI] [PubMed] [Google Scholar]
- [72].Cassar M, Issa A-R, Riemensperger T, et al. A dopamine receptor contributes to paraquat-induced neurotoxicity in Drosophila. Hum Mol Genet. 2015;24(1):197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Xu T, Kumar S, Denton D. Characterization of autophagic responses in Drosophila melanogaster. Meth Enzymol. 2017;588:445–465. [DOI] [PubMed] [Google Scholar]
- [74].Lőrincz P, Mauvezin C, Juhász G. Exploring autophagy in Drosophila. Cells. 2017;6(3):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xilouri M, Vogiatzi T, Vekrellis K, et al. Abberant alpha-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS ONE. 2009;4(5):e5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Liu X, Huang S, Wang X, et al. Chaperone-mediated autophagy and neurodegeneration: connections, mechanisms, and therapeutic implications. Neurosci Bull. 2015;31(4):407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Frake RA, Ricketts T, Menzies FM, et al. Autophagy and neurodegeneration. J Clin Invest. 2015;125(1):65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Martini-Stoica H, Xu Y, Ballabio A, et al. The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci. 2016;39(4):221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3(4):295–299. [DOI] [PubMed] [Google Scholar]
- [80].Vanhooren V, Navarrete Santos A, Voutetakis K, et al. Protein modification and maintenance systems as biomarkers of ageing. Mech Ageing Dev. 2015;151:71–84. [DOI] [PubMed] [Google Scholar]
- [81].Jang YC, Pérez VI, Song W, et al. Overexpression of Mn superoxide dismutase does not increase life span in mice. J Gerontol A Biol Sci Med Sci. 2009;64(11):1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in caenorhabditis elegans. PLoS Genet. 2009;5(2):e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gems D, Partridge L. Genetics of longevity in model organisms: debates and paradigm shifts. Annu Rev Physiol. 2013;75:621–644. [DOI] [PubMed] [Google Scholar]
- [84].Rusten TE, Lindmo K, Juhász G, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7(2):179–192. [DOI] [PubMed] [Google Scholar]
- [85].Cuervo AM, Knecht E, Terlecky SR, et al. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269(5–1):C1200–C1208. [DOI] [PubMed] [Google Scholar]
- [86].Massey AC, Kaushik S, Sovak G, et al. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci USA. 2006;103(15):5805–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hu Z-Y, Chen B, Zhang J-P, et al. Up-regulation of autophagy-related gene 5 (ATG5) protects dopaminergic neurons in a zebrafish model of Parkinson’s disease. J Biol Chem. 2017;292(44):18062–18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hubert V, Peschel A, Langer B, et al. LAMP-2 is required for incorporating syntaxin-17 into autophagosomes and for their fusion with lysosomes. Biol Open. 2016;5(10):1516–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Nishino I, Fu J, Tanji K, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature. 2000;406(6798):906–910. [DOI] [PubMed] [Google Scholar]
- [90].Rowland TJ, Sweet ME, Mestroni L, et al. Danon disease - dysregulation of autophagy in a multisystem disorder with cardiomyopathy. J Cell Sci. 2016;129(11):2135–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Tanaka Y, Guhde G, Suter A, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406(6798):902–906. [DOI] [PubMed] [Google Scholar]
- [92].Saftig P, Beertsen W, Eskelinen E-L LAMP-2. a control step for phagosome and autophagosome maturation. Autophagy. 2008;4(4):510–512. [DOI] [PubMed] [Google Scholar]
- [93].Rothaug M, Stroobants S, Schweizer M, et al. LAMP-2 deficiency leads to hippocampal dysfunction but normal clearance of neuronal substrates of chaperone-mediated autophagy in a mouse model for Danon disease. Acta Neuropathol Commun. 2015;3(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Eskelinen E-L, Illert AL, Tanaka Y, et al. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002;13:3355–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].R-A G-P, Boya P, Pauleau A-L, et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci. 2005;118:3091–3102. [DOI] [PubMed] [Google Scholar]
- [96].Morell C, Bort A, Vara-Ciruelos D, et al. Up-regulated expression of LAMP2 and autophagy activity during neuroendocrine differentiation of prostate cancer LNCaP cells. PLoS ONE. 2016;11:e0162977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Huynh KK, Eskelinen E-L, Scott CC, et al. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007;26:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Huttenhower C, Haley EM, Hibbs MA, et al. Exploring the human genome with functional maps. Genome Res. 2009;19:1093–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Friggi-Grelin F, Coulom H, Meller M, et al. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. [DOI] [PubMed] [Google Scholar]
- [100].Hennig KM, Colombani J, Neufeld TP. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J Cell Biol. 2006;173:963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. [DOI] [PubMed] [Google Scholar]
- [102].Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Nezis IP, Simonsen A, Sagona AP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Sasaki A, Arawaka S, Sato H, et al. Sensitive western blotting for detection of endogenous Ser129-phosphorylated α-synuclein in intracellular and extracellular spaces. Sci Rep. 2015;5:14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Owusu-Ansah E, Yavari A, Banerjee U. A protocol for in vivo detection of reactive oxygen species. Protocol Exchange. 2008. DOI: 10.1038/nprot.2008.23 [DOI] [Google Scholar]
- [106].Zielonka J, Kalyanaraman B. Hydroethidine-and mitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med. 2010;48:983–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Siddique YH, Mujtaba SF, Jyoti S, et al. GC-MS analysis of eucalyptus citriodora leaf extract and its role on the dietary supplementation in transgenic Drosophila model of Parkinson’s disease. Food Chem Toxicol. 2013;55:29–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


