ABSTRACT
Stromal/stem cell differentiation is controlled by a vast array of regulatory mechanisms. Included within these are methods of mRNA gene regulation that occur at the level of epigenetic, transcriptional, and/or posttranscriptional modifications. Current studies that evaluate the posttranscriptional regulation of mRNA demonstrate microRNAs (miRNAs) as key mediators of stem cell differentiation through the inhibition of mRNA translation. miRNA expression is enhanced during both adipogenic and osteogenic differentiation; however, the mechanism by which miRNA expression is altered during stem cell differentiation is less understood. Here we demonstrate for the first time that adipose-derived stromal/stem cells (ASCs) induced to an adipogenic or osteogenic lineage have differences in strand preference (-3p and -5p) for miRNAs originating from the same primary transcript. Furthermore, evaluation of miRNA expression in ASCs demonstrates alterations in both miRNA strand preference and 5'seed site heterogeneity. Additionally, we show that during stem cell differentiation there are alterations in expression of genes associated with the miRNA biogenesis pathway. Quantitative RT-PCR demonstrated changes in the Argonautes (AGO1-4), Drosha, and Dicer at intervals of ASC adipogenic and osteogenic differentiation compared to untreated ASCs. Specifically, we demonstrated altered expression of the AGOs occurring during both adipogenesis and osteogenesis, with osteogenesis increasing AGO1-4 expression and adipogenesis decreasing AGO1 gene and protein expression. These data demonstrate changes to components of the miRNA biogenesis pathway during stromal/stem cell differentiation. Identifying regulatory mechanisms for miRNA processing during ASC differentiation may lead to novel mechanisms for the manipulation of lineage differentiation of the ASC through the global regulation of miRNA as opposed to singular regulatory mechanisms.
KEYWORDS: miRNA, biogenesis, stem cell differentiation
Introduction
Adipogenesis and osteogenesis of adult stem cells occurs through the regulation and manipulation of stem cell proliferation and differentiation; this is achieved through transcriptional and post-transcriptional mechanisms [1,2]. Recent studies identify microRNAs (miRNAs) as a novel avenue for post-transcriptional regulation of stem cell differentiation [3,4]. Small changes in miRNA expression can have a large impact on a diverse array of cellular functions including metabolism, proliferation, apoptosis, and cell fate [5]. Having emerged as integral components of both adipogenesis and osteogenesis, miRNAs target and degrade key adipogenic and osteogenic genes (PPARG, CEBPα, DLK1, RUNX2, SMAD5) [6–10]. Additionally, miRNAs known to regulate pro-adipogenic genes are observed to be suppressed during adipogenesis (miR-27/PPARG, miR-31/CEBPα), while miRNAs that regulate anti-adipogenic genes demonstrate enhanced expression during adipogenesis (miR-15a/DLK1). Similarly, miRNAs that regulate pro-osteogenic genes are repressed during osteogenesis (miR-204/RUNX2, miR-135/SMAD5). Due to the integral role of miRNAs in the modulation of stem cell fate, miRNAs present a novel option for the induction of stem cells during tissue regeneration. Despite the potential therapeutic utility of miRNAs, barriers obstructing the deployment of miRNAs for stem cell induction include: 1) variations in miRNA expression profiles between different groups and 2) the incomplete understanding of miRNA expression during differentiation. Both of these issues can be addressed in part through greater understanding of miRNA processing as miRNA expression is regulated both transcriptionally and post-transcriptionally [11]. Post-transcriptional changes in miRNA expression will occur through alterations in the maturation of the miRNA from a primary (pri-) transcript to a mature functional miRNA [11]. Once transcribed, miRNAs are processed through either canonical or non-canonical mechanisms. Non-canonical methods for miRNA maturation involve the processing of a miRNA from the introns of a gene during gene splicing or through alternative cleavage events such as those that utilize lin28B, a suppressor of miRNA biogenesis [11]. During canonical processing a pri-miRNA transcript is cleaved in the nucleus by the double-stranded RNA-specific endoribonuclease (Drosha) in the microprocessor complex that contains Drosha and DiGeorge syndrome region 8 (DGCR8). Following the initial cleavage from pri- to precursor (pre-) miRNA, pre-miRNAs are exported from the nucleus to the cytoplasm by exportin5 (EXPO5) [11,12]. Once in the cytoplasm, pre-miRNAs are cleaved by Dicer to form a miRNA duplex that contains two functional miRNAs, designated as the -5p and -3p miRNA (Fig. 1A). Previous characterization of miRNAs encompassed within the same duplex defined one miRNA strand as the “mature” or “guide” strand as it was conventionally expressed in physiologically relevant levels while the other miRNA strand was designated as the “passenger” or “star (*)” strand as it was by convention believed to be degraded [12]. Recently instances have been reported by others and us where both miRNA strands of the miRNA duplex are expressed [13–15]. Additionally, the star (*) strand has been demonstrated in some cases to be favored with higher expression levels than the designated guide strand [16]. Identification of miRNA strand expression can be observed through compiled sequencing modalities such as miRBase [36–41], a small RNA sequencing database that identifies miRNA expression levels and variations in miRNA expression and sequence (Fig. 1B). While we have identified that miRNA duplexes have differential expression of guide and star miRNAs, we have not yet fully identified these expression differences in stem cells. Nor has a regulatory mechanism for miRNA strand preference been identified. Recently, knockout studies of Drosha and Dicer have shown that these proteins are required for adipogenesis and osteogenesis, demonstrating that canonical miRNA processing is integral to adult stem cell differentiation [17,18]. To date a role for proteins associated with the stability and biogenesis of miRNAs has yet to be determined during stem cell differentiation. In other biological processes such as cancer, the expression and activity of proteins that regulate miRNA stability have been identified [19–22]. These changes of miRNA processing and stability altered miRNA expression and in some instances altered strand preference [13]. This is achieved through 1) enhancing miRNA processing and increasing miRNA expression and mRNA targeting or 2) suppressing miRNA processing and decreasing miRNA expression and mRNA targeting [11,22]. Argonaute (AGO) proteins can preferentially change miRNA pools and expression levels [13,19,20]. In addition, AGO proteins are differentially regulated under different disease states. Here we aim to identify alterations in miRNA duplex strand preference through the evaluation of miRNA expression via qPCR and deep sequencing evaluation.
Figure 1.
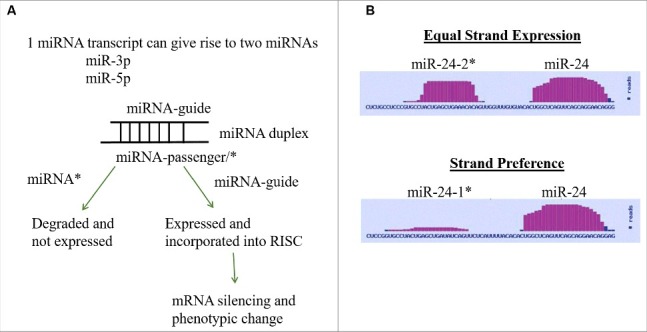
miRNA Strand Preference. (A) Depiction of conventional mechanism for miRNA strand expression where one miRNA strand (guide) is expressed and one miRNA strand (passenger/*) is repressed. (B) Depiction of differences in strand preference of miR-24 as viewed in the miRBase [37–41].
Results
Adult Adipose Derived Stem Cells (ASCs) demonstrate miRNA strand preference
Human ASCs obtained from three different donors were propagated individually in stromal media for 2 weeks and quantitative RT-PCR (qPCR) was performed for miRNAs known to be altered during stem cell differentiation. qPCR was performed for both miRNAs that encompass the miRNA duplex. Differences in the basal expression level were evaluated and compared between the two miRNAs in the duplex. For ease of convention miR-* is used to designate the less classically expressed miRNA in the duplex. Evaluation of miRNA expression patterns in stromal media demonstrated that miRNAs originating from the same miRNA duplex exhibited either strand preference (miR-335, miR-24, let-7b) or equal expression levels (miR-20a, miR-200c, miR-21) (Fig. 2A). Additionally, the miRNAs from the miR-335 duplex (miR-335 and miR-335*) demonstrated strand preference and the conventionally designated passenger/* strand at greater levels than the miR-335 guide strand (Fig. 2A). To determine the effects of stem cell differentiation on miRNA strand selection, ASCs were grown in either adipogenic or osteogenic differentiation media for two weeks. Adipogenic and osteogenic differentiation was confirmed through histochemical staining with Oil Red O or alizarin red, respectively (Fig. 2B). qPCR was performed on harvested cells and results demonstrated that ASC cultured in osteogenic media (Fig. 2B) maintained preference for expression of one miRNA strand over the other in a manner similar to ASC cultured in the presence of stromal media. In contrast, ASC induced with adipogenic medium displayed a loss of strand preference. Following adipogenic induction, miRNAs originating from the same miRNA duplex were expressed at equal levels (Fig. 2C). To gain a better understanding of miRNA strand preference and expression in ASCs at a global level, we used the University California Santa Cruz (UCSC) genome browser in silico to visualize and evaluate transcriptional specific expression levels of miRNAs that were altered during stem cell differentiation [23]. Small RNA sequencing data (GEO accession GSM977041) of human adipose stem cells was obtained and visualized through the genome browser [24–26]. Basal expression levels of miRNAs originating from the same miRNA duplex were evaluated for changes in convention strand preference, dual miRNA expression and alterations to the 5′ seed site. Of all miRNA, duplexes observed most demonstrated classical miRNA expression patterns where the traditional guide strand was solely expressed. However, some miRNAs demonstrated expression of either miRNAs (star and guide) or preference for the conventionally degraded star strand (Table 1 and Fig. 3). Among the miRNAs investigated, miR-17 and miR-17* (Fig. 3A, were expressed at equal levels, while miR-136*, favored the expression of the conventionally degraded miRNA over the conventional mature miRNA (Fig. 3B). Moreover, some miRNAs demonstrated alterations to the 5′ ends manifest as either addition or loss of nucleotides (Fig. 3C). Together, this data demonstrates that some miRNAs exist as isomiRs and conventional miRNA sequences and expression may be altered in ASCs.
Figure 2.
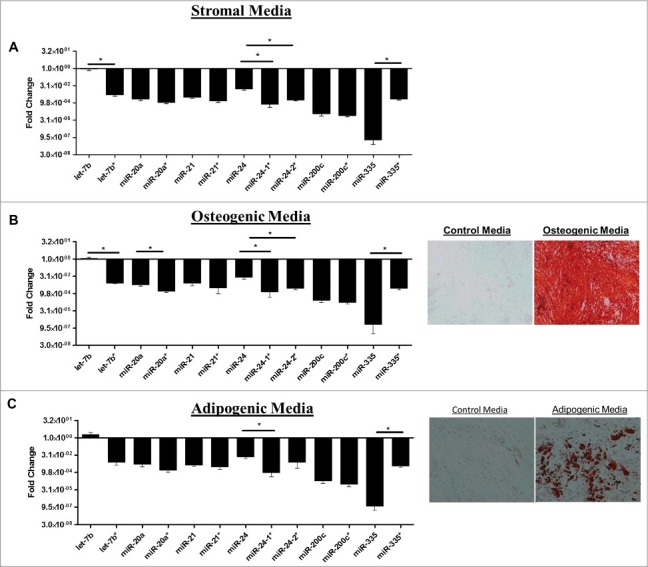
Adipose Derived Stem Cells Demonstrate differences in Strand Preferences during Adipogenic and Osteogenic Differentiation. (A-C) ASCs were grown in stromal, adipogenic, or osteogenic media for 2 weeks and then qPCR was performed for miRNA expression levels. Results represent fold change as relative gene expression (A) miRNA expression in stromal media. (B) miRNA expression and alizarin red stain following 2 weeks osteogenic differentiation. (C) miRNA expression and Oil Red O stain following 2 weeks adipogenic differentiation. Normalization was to u6 expression and error bars represent SEM. * significantly different p<0.05. Significance was evaluated for miRNA expression levels between miRNAs originating from the same miRNA duplex.
Table 1.
Identification of miRNA Strand Preference in Adipose Derived Stem Cells.
| miRNA | Guide | Star | Dominant Strand | Duplex Processing |
|---|---|---|---|---|
| let-7a | expressed | not expressed | guide | normal |
| let-7b | expressed | expressed | guide | normal |
| let-7c | expressed | not expressed | guide | normal |
| let-7d | expressed | not expressed | guide | normal |
| let-7e | expressed | not expressed | guide | normal |
| let-7f | expressed | not expressed | guide | normal |
| miR-100 | expressed | expressed | guide | normal |
| miR-103a | expressed | expressed | guide | normal |
| miR-106a | expressed | not expressed | guide | guide: loss of nucleotides 5' end |
| miR-107 | expressed | not expressed | guide | normal |
| miR-10a | expressed | expressed | guide | normal |
| miR-10b | expressed | expressed | guide | guide: normal, star: loss of nucleotides on 5′end |
| miR-125a | expressed | expressed | guide | unprocessed pre-miRNA |
| miR-125b (ch11) | expressed | expressed | guide | normal |
| miR-125b (ch21) | expressed | expressed | guide | star: loss of nucleotides at 5' end |
| miR-130a | expressed | not expressed | guide | normal |
| miR-130b | expressed | expressed | guide | normal |
| miR-132 | expressed | expressed | equal | normal |
| miR-136 | not expressed | expressed | star | star: loss of nucleotides at 5' end |
| miR-137 | expressed | not expressed | guide | normal |
| miR-138 | expressed | not expressed | guide | normal |
| miR-138 (chr3) | expressed | expressed | guide | star: loss of nucleotides 5' end |
| miR-139 | expressed | not expressed | guide | guide: loss of nucleotides at 5' end |
| miR-140 | expressed | expressed | guide | guide: loss of nucleotides at 5' end |
| miR-143 | expressed | not expressed | guide | normal |
| miR-148a | expressed | expressed | guide | guide: loss of nucleotides at 5' end |
| miR-152 | expressed | not expressed | guide | normal |
| miR-154 | expressed | expressed | guide | normal |
| miR-155 | expressed | not expressed | guide | normal |
| miR-17 | expressed | expressed | equal | normal |
| miR-181a | expressed | expressed | guide | normal |
| miR-181c | expressed | expressed | star | normal |
| miR-186 | expressed | not expressed | guide | normal |
| miR-191 | expressed | expressed | guide | guide: addition of nucleotides, star: loss of nucleotides 5' end |
| miR-193a | not expressed | expressed | star | normal |
| miR-193b | not expressed | expressed | star | normal |
| miR-199b | expressed | expressed | star | normal |
| miR-19b | expressed | not expressed | guide | normal |
| miR-20a | expressed | expressed | guide | normal |
| miR-21 | expressed | expressed | guide | normal |
| mir-210 | expressed | not expressed | guide | normal |
| miR-212 | expressed | not expressed | guide | normal |
| miR-218 (chr1) | expressed | expressed | guide | guide: normal, star: addition nucleotides 5' end |
| miR-218 (chr2) | expressed | not expressed | guide | normal |
| miR-22 | expressed | expressed | guide | normal |
| mir-222 | expressed | expressed | guide | star: addition nucleotides 5' end |
| miR-221 | expressed | expressed | star | normal |
| miR23a | expressed | no expression | guide | normal |
| miR-23b | expressed | no expression | guide | normal |
| miR-24 (chr9) | expressed | not expressed | guide | normal |
chr: chromosome, miRNA is designated as expressed if counts as observed in the UCSC Genome Browser are over 100. Star designation was defined by prior identifier through miRBase (http://www.mirbase.org).
Figure 3.
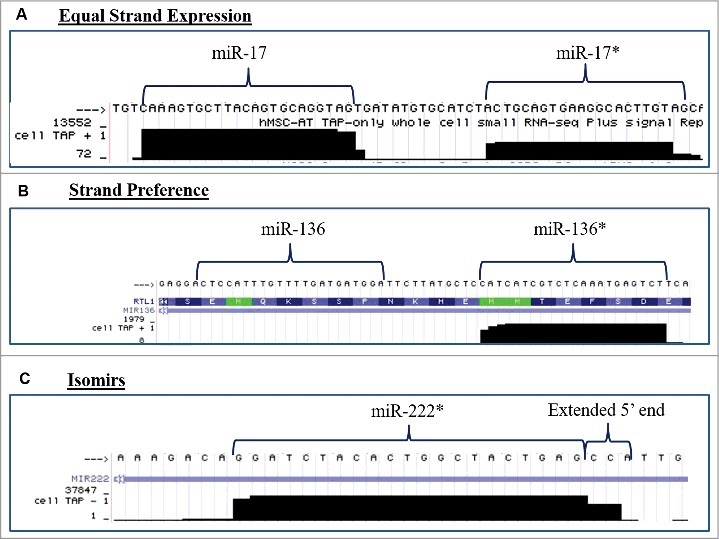
miRNA Duplexes Demonstrate Strand Preference. Evaluation of next generation sequencing of small RNA fraction of non-differentiated ASCs for alterations in strand preference and 5′heterogeneity and viewed in the genome browser. Representative pictures of miRNA demonstrating (A) equal miRNA expression levels of both miRNAs in a miRNA duplex, (B) miRNA strand preference for expression of the conventionally repressed miRNA (*), or (C) miRNA isoforms with changes to the 5'seed site of the miRNA. N = 2.
The miRNA biogenesis pathway is altered during adipose stem cell differentiation
Strand preference for one miRNA strand over the other miRNA in a duplex reflects either miRNA stability and/or biogenesis [13]. Based on this, our next studies evaluated genes associated with miRNA maturation. ASCs were induced with either adipogenic or osteogenic media for different intervals of time (4hr, 24hr, 48hr, 4 day, 1 week, and 2 weeks). Cells were then collected for RNA extraction and qPCR for genes associated with miRNA stability (AGO1, AGO2, AGO3, AGO4) and miRNA cleavage (Drosha, Dicer, TARBP). Results demonstrated that at 24hr post adipogenic induction, ASCs expressed enhanced Dicer gene expression and repressed AGO1 gene expression significantly relative to ASCs maintained in stromal culture media. At 4 days post adipogenic induction, ASCs expressed significantly enhanced AGO3 gene expression relative to ASC maintained in stromal medium. Similarly, at 1 week post adipogenic induction, ASCs expressed significantly enhanced Drosha and AGO2 gene expression relative to ASCs maintained in stromal medium (Fig. 4A and B). The ASCs maintained in osteogenic differentiation medium displayed enhanced gene expression levels for all four of the AGO genes (AGO1-4) throughout the time course relative to ASC maintained in stromal medium; however, osteogenic conditions did not modulate the levels of Drosha or Dicer gene expression (Fig. 4C and D).
Figure 4.
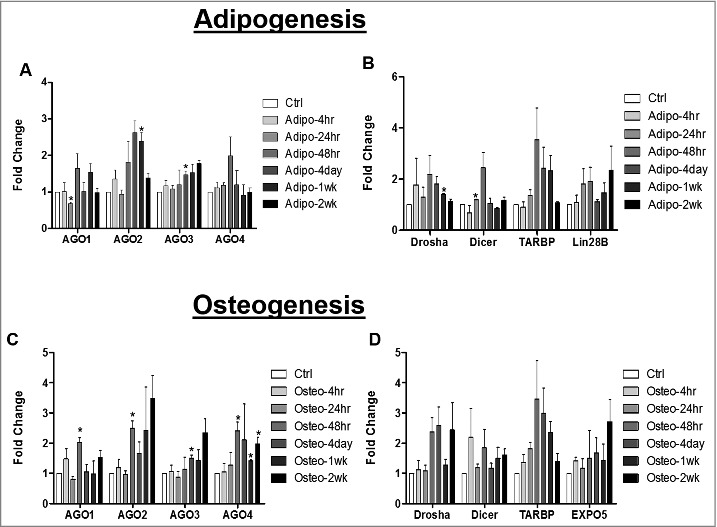
miRNA Biogenesis Pathway is Altered during Adipogenic and Osteogenic Differentiation of Adipose Derived Stem Cells. ASCs were grown in either stromal, osteogenic, or adipogenic media and were collected for RNA extraction at different intervals up to 2 weeks. qPCR was performed to evaluate expression of genes associated miRNA stability (A and C) or cleavage (B and D). Error bars represent SEM, n = 3, normalization was to stromal media, and Cycb gene expression. Fold change represents relative gene expression changes. * Significantly different p<0.05.
Since there were observed differences in changes in the AGO genes in both adipogenic and osteogenic differentiation, we next sought to determine if the AGO proteins were altered at 1 week and 2 weeks post adipogenic and osteogenic differentiation. At 1-week post adipogenic differentiation, ASC displayed significantly repressed protein levels for AGO1 and AGO3 relative to ASCs maintained in stromal media (Fig. 5A). At 2 weeks induction with adipogenic media, AGO1 protein levels were still significantly repressed while the other three AGOs (2-4) demonstrated no significant change in protein density relative to non-induced control (Fig. 5B). There was no significant change in AGO proteins during osteogenic differentiation throughout the time course (Fig. 5C and D).
Figure 5.
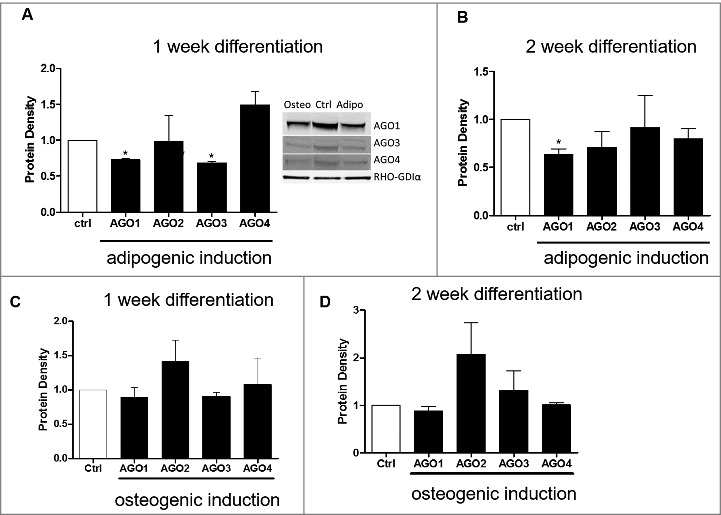
AGO1 Protein is repressed during Adipogenic and Osteogenic Differentiation of Adipose Derived Stem Cells. ASCs were grown in either stromal, osteogenic, or adipogenic media and collected for western blot for Ago1-4 protein expression levels at one and two weeks post differentiation. (A) 1 week and (B) 2 week adipogenic differentiation. (C) 1 week and (D) 2-week osteogenic differentiation. n = 3, error bars represent SEM. Normalization was to stromal media and GAPDH. * Significantly different p<0.05.
Discussion
This study demonstrates for the first time that lineage differentiation will alter both miRNA strand preference and miRNA biogenesis associated gene expression in adipose derived stromal/stem cells. miRNAs are well-documented mediators of differentiation, with some miRNAs being identified as crucial to regulating either adipogenesis or osteogenesis [31,32]. To date there is a large breadth of information on changes in the expression levels of specific miRNAs and their subsequent function in the promotion or inhibition of adipogenesis and osteogenesis [31]. Changes in miRNA expression are influenced by post-transcriptional alterations to the biogenesis of a pri-miRNA into a mature miRNA [11]. Previous knock out experiments of Dicer and Drosha have demonstrated that proper miRNA biogenesis is essential for adipogenic and osteogenic differentiation [18,33,34]. Loss of Dicer resulted in the inhibition of adipogenesis in murine pre-adipocytes and a similar effect was observed with Drosha knock out [33,34]. In accordance with this, our study demonstrated that both Dicer and Drosha gene expression were enhanced with adipogenic differentiation, suggesting that miRNAs under the control of Dicer and Drosha may be necessary for adipogenic differentiation. In contrast, there were no significant changes in Dicer or Drosha with osteogenic differentiation.
In addition to demonstrating changes in genes associated with miRNA cleavage, we also demonstrated that genes associated with the stability of miRNAs were altered. The AGO proteins act as the catalytic unit of mRNA silencing in the RISC complex [11]. These proteins facilitate the pairing of miRNAs and their complementary mRNAs to inhibit gene translation. In addition to regulating mRNA translation, the AGO proteins can stabilize miRNA expression. Previous studies demonstrated that enhanced AGO protein expression correlates with increased miRNA pools [35]. Furthermore, in humans, miRNAs may be stabilized by certain AGOs. This implies that enhanced expression of a particular AGO protein may facilitate the increased expression, stability, and mRNA targeting of specific miRNAs [36]. For example, in humans, it is demonstrated that AGO2 enhances the expression of miR-451 and AGO3 enhances let-7a-3p expression [28]. In the current study, we observed a repression in AGO1 at both the gene and protein levels. To date no prior studies had described such a preference for AGO1 and specific miRNAs. Indeed, the investigation of miRNA stability and AGO specific loading in humans is still in its infancy. Further insights into the alteration of the miRNA biogenesis pathway during stem cell differentiation will enhance our understanding and ability to manipulate adipogenesis and osteogenesis in human stem cells.
Previous convention has held that miRNAs deriving from the same miRNA duplex will select for one miRNA over the other, with one miRNA (guide strand) being incorporated into RISC to mediate mRNA expression while the other (passenger strand/*) will be degraded (Fig. 1). In the current study, ASCs grown in stromal medium expressed miRNA species from the same pre-miRNA duplex at either equal levels (miR-20a, miR-21, miR-200c) or selected for strand preference (miR-24-1, miR-24-2, miR-335, let-7b). Following adipogenic differentiation, miRNAs from the same duplex lost strand preference while osteogenic differentiation increased instances of strand preference (Fig. 2). Furthermore, we re-evaluated previously deposited small RNA sequencing of ASCs and demonstrated that baseline expression of some miRNAs resulted in dual expression of both miRNAs in a duplex, selection for the star strand, or loss of 5′ seed site (Fig. 3). These data are significant, as conventional identification of miRNAs altered in RNA sequencing does not normally distinguish miRNA strand selection in a duplex. This adds a new paradigm to miRNA function, demonstrating that for some miRNAs both miRNAs in the pre-miRNA duplex are expressed at functionally relevant levels. Consequently, this highlights the need to validate and evaluate both miRNAs in a pre-miRNA duplex, particularly in the context of stem cell differentiation. Importantly others have shown that miRNAs derived from the same pre-miRNA duplex may have compensatory effect, targeting different genes in the same pathway [14,15]. Conversely, some miRNAs such as miR-21-3p and miR-21-5p have opposing effects [28]. To date, such studies have been limited to pathological states such as cancer, but have yet to be examined under cell physiological conditions, such as stem cell differentiation [37]. The current study shows that some miRNA duplexes favor expression of the miRNA strand that is conventionally predicted to be degraded. Through our analysis of previously published small RNA sequencing of ASCs, we determined that miRNAs, such as miR-222*, were expressed as truncated isomiRs with alternative 5′ends. As the 5′ end of the miRNA dictates mRNA target selection, this may account for differences in miRNA/mRNA target selection; however, further studies are necessary [29,30].
Materials and methods
Cell culture and ASC donors
ASCs were derived from subcutaneous abdominal adipose tissue donated with written informed consent under a Western Institutional Review Board approved protocol by subjects undergoing elective liposuction as previously described [36]. All donors were male, lean, and had an age range of (18-35yrs). All experiments with ASCs were performed with pooled donor samples, n = 3 using the ASC at cell passages of 2 to 4. Osteogenic qPCR and drug (R&D Systems, Minneapolis, MN) stimulation experiments were done with individual donors, n = 3. ASCs were maintained in alpha-MEM supplemented with 10% FBS (Hyclone, Marlborough, MA), 1% pen/strep antibiotic, and 1% L-glutamine (CCM). For osteogenic differentiation alpha-MEM media was supplemented with 10% fetal bovine serum, 100 units/mL of penicillin and 100µg/mL of streptomycin (antibiotic), 10 nM dexamethasone, 10 mM b-glycerolphosphate and 50 μg/mL ascorbate-2-phosphate (Sigma-Aldrich, St. Louis, MO). Adipogenic induction media was supplemented with 10% fetal bovine serum, 100 units/mL of penicillin and 100µg/mL of streptomycin (antibiotic), 0.25 IBMX, 66uM biotin, 34uM D-pantothenate, 5uM rosiglitazone, 1uM dex and 200nM human insulin.
Adipogenic and osteogenic differentiation assays
Pooled ASCs were seeded 3.5 × 105 cells/well in a 6well plate in 10% FBS complete culture media (CCM). Upon confluence, media was changed to either adipogenic or osteogenic induction media. Wells were fed every 3 days with adipogenic media or CCM. At designated intervals, cells were washed with PBS and either formalin (10%) fixed or collected for RNA extraction and Western blot. For fixed cells, formalin was removed and washed with PBS and stained with Adipo Q (Sigma-Aldrich, St. Louis, MO). Adipo Q was removed, cells washed and images were taken at 10x objective. Wells were fed every 3 days with osteogenic media or CCM. Cells were then washed with water and formalin (10%) fixed. Alizarin red stain (Sigma-Aldrich, St. Louis, MO) was added for 1 hour and then wells were washed with water 3 times. Cells were imaged at 20x magnification.
Quantitative PCR analysis
Cells were plated as induced with adipogenic media as described above. At appropriate time points (4hr, 24hr, 48hr 4day, 1wk, 2wk) cells were harvested for total RNA extraction using mRneasy kit (Qiagen, Valencia, CA). cDNA synthesis was performed with 1ug of RNA with the iScript cDNA synthesis kit (BioRad, Hercules, CA). 150ng of cDNA was used for qPCR analysis with iQ5 SYBR green (BioRad, Hercules, CA). Primer sequences listed in supplemental table S1. Normalization was to CYCB and 3 biological repeats were performed. miRNA was obtained during the total RNA extraction and qRT-PCR was performed using Qiagen miScript II kit as per the manufacturer's protocol (Qiagen, Valencia, CA) normalization was to U6.
Western blot analysis
ASCs were plated at 1 million cells per 10cm [2] dish and grown to confluence in CCM. Upon confluence, cells were induced with adipogenic media for 1 week or 2 weeks. Cells were fed every three days at appropriate end points cells were collected and lysed with MPER lysis buffer (Thermo Scientific, Waltham, MA) supplemented with 1% Halt protease and phosphatase (cocktail I and II) inhibitors (Thermo Scientific, Waltham MA). For each blot, total cell lysates were loaded on a 4–12% gel and then transferred to nitrocellulose. Blots were blocked in 5% licor blocking buffer (LI-COR, Lincoln, NE) for 1 hour then primary antibodies were added (diluted 1:1,000) (Cell Signaling, Beverly, MA). After 1 hour, blots were washed 3Xs in PBS for ten minutes each and secondary antibody (LI-COR, Lincoln, NE) was added (diluted 1:10,000).Following one hour, blots were washed 3Xs for ten minutes each with PBS. Band density was quantified with a LI-COR Odyssey fluorescent imager (LI-COR, Lincoln, NE). Loading control was to GAPDH and control was cells grown in CCM designated as “1”.
Next generation sequencing analysis
miRNA expression and evaluation of strand preference was observed through re-evaluation of previously aligned and deposited data files obtained through the UCSC Genome browser [24]. Data set evaluated was Release 3 (July 2012) of CSHL Small RNA-seq from the Gingeras lab GEO accession (GSM977041) and UCSC Accession: wgEncodeEH002888 and Lab producing data: Gingeras – CSHL [26,27]. Cell line used was hMSC-AT small RNA seq.
Statistics
Statistical analysis was performed with Graphpad5.0. Significance was determined by student t-test where statistically significant values displayed a p value of <0.05. All experiments were repeated in biological triplicates.
Supplementary Material
Funding Statement
U.S. Department of Defense (DOD) (W81XWH-13-2-0097).
Disclosure of potential conflicts of interest
Dr. Gimble is the co-founder, co-owner, and Chief Scientific Officer of LaCell LLC, a for-profit biotechnology company focusing on research tools and clinical translation of stromal/stem cell products and therapeutics.
Acknowledgments
The views expressed in this presentation are those of the author and do not reflect the official policy of the Department of the Navy, the Department of Defense or the United States Government. Some of the authors are military service members and employees of the US Government. This work was prepared as part of our official duties. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.' Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the US Government as part of that person's official duties. The study protocol was approved by the Walter Reed National Military Medical Center Institutional Review Board in compliance with all applicable Federal regulations governing the protection of human subjects. I certify that the document represents valid work; that if I used information derived from another source, I obtained all necessary approvals to use it and made appropriate acknowledgements in the document; and I take public responsibility for it”
References
- [1].Cinti S. The adipose organ. Fantuzzi G, Mazzone T ( ed.), Adipose Tissue and Adipokines in Health and Disease. Humana Press, Nutrition and Health; 2007. p. 3–19. [Google Scholar]
- [2].Chen Q, Shou P, Zheng C, et al.. Fate decision of Mesenchymal stem cells: Adipocytes or Osteoblasts. Cell Death Differ. 2016;23(7):1128–1139. doi: 10.1038/cdd.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huang C, Geng J, Jiang S. MicroRNAs in regulation of osteogenic differentiation of Mesenchymal stem cells. Cell Tissue Res. 2016 [DOI] [PubMed] [Google Scholar]
- [4].Shi C, Huang F, Gu X, et al.. Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget. 2016;7(26):40830–40845. doi: 10.18632/oncotarget.8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Martin EC, Qureshi AT, Dasa V, et al.. MicroRNA regulation of stem cell differentiation and disease of the bone and adipose tissue: Perspectives on miRNA biogenesis and cellular transcriptome. Biochimie. 2016;124:98–111. doi: 10.1016/j.biochi.2015.02.012. [DOI] [PubMed] [Google Scholar]
- [6].Karbiener M, Fischer C, Nowitsch S, et al.. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun. 2009;390(2):247–51. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- [7].McGregor RA, Choi MS. microRNAs in regulation of adipogenesis and obesity. Curr Mol Med. 2011;11(4):304–316. doi: 10.2174/156652411795677990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Andersen DC, Jensen CH, Schneider M, et al.. MicroRNA-15a fine-tunes the levels of delta-like 1 Homolog (DLK1) in Proliferating 3T3-L1 Preadipovytes. Exp Cell Res. 2010;316(10):1681–91. doi: 10.1016/j.yexcr.2010.04.002. [DOI] [PubMed] [Google Scholar]
- [9].Huang J, Zhao L, Xing L, et al.. microRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28(2):357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xu S, Santini GC, Veirman KD, et al.. Upregulation of miR-135b is involved in the impaired osteogenic differentiation of mesenchymal stem cells derived from multiple myeloma patients. PLOS One. 2013;8(11):e79752. doi: 10.1371/journal.pone.0079752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Winter J, Jung S, Keller S, et al.. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- [12].MacFarlane LA, Murphy PR. MicroRNA: Biogenesis, function, and role in cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Winter J, Diederichs S. Argonaute-3 Activates the let-7a Passenger Strand microRNA. RNA Biol. 2013;10(10):1631–1643. doi: 10.4161/rna.26424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Martin EC, Cogner AK, Yan TJ, et al.. MicroRNA-335-5p and -3p synergize to inhibit estrogen receptor alpha expression and promote tamoxifen resistance. FEBS Letters. 2017;591(2):382–392. doi: 10.1002/1873-3468.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rhodes LV, Martin EC, Segar HC, et al.. Dual Regulation by microRNA-200b-3p and microRNA-200b-5p in the inhibition of epithelial-to-mesenchymal transition in -triple-negative breast cancer. Oncotarget. 2015;6(18):16638–52. doi: 10.18632/oncotarget.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Martin EC, Elliott S, Rhodes LV, et al.. Preferential star strand biogenesis of pre-mir-24-2 targets PKC-alpha and suppresses cell survival in MCF-7 breast cancer cells. Mol Carcinog. 2014;53(1):38–48. doi: 10.1002/mc.21946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang Q, Li YC, Wang J, et al.. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci USA. 2008;105(8):2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gaur T, Hussain R, Mudhasani I, et al.. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol. 2010;340:10–21. doi: 10.1016/j.ydbio.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cogner AK, Martin EC, Yan TJ, et al.. Argonaute 2 expression correlates with a Luminal B breast cancer subtype and induces estrogen receptor alpha isoform variation. Non-Coding RNA. 2016;2(3):8. doi: 10.3390/ncrna2030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150(1):14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Melo SA, Moutinho C, Ropero S, et al.. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18(4):303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- [22].Huang JT, Wang J, Srivastava V, et al.. MicroRNA machinery genes as novel biomarkers for cancer. Front Oncology. 2014;4:113. doi: 10.3389/fonc.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Martin EC, Qureshi A, Dasa V, et al.. MicroRNA regulation of stem cell differentiation and diseases of the bone and adipose tissue: Perspectives on miRNA biogenesis and cellular transcriptome. Biochime. 2015;124:98–111. doi: 10.1016/j.biochi.2015.02.012 [DOI] [PubMed] [Google Scholar]
- [24].Kent WJ, Sugnet CW, Furey TS, et al.. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Karolchik D, Barber GP, Casper J, et al.. The UCSC Genome Browser Database:2014 Update. Nucleic Acid Res. 2014;42:D764–770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fejes-Toth K, Sotirova V, Sachidanandam R, et al.. Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature. 2009;457(7232):1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Langmead B, Trapnell C, Pop M, et al.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pink RC, Samuel P, Massa D, et al.. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynocol Oncol. 2015;137(1):143–51. doi: 10.1016/j.ygyno.2014.12.042. [DOI] [PubMed] [Google Scholar]
- [29].Tan GC, Dibb N. IsomiRs have functional importance. Malays Pathol. 2015;37(2):73–81. [PubMed] [Google Scholar]
- [30].Frith JE, Porrello ER, Cooper-White JJ. Concise review: New frontiers in microRNA-Base tissue regeneration. Stem Cells Transl Med. 2014;3(8):969–976. doi: 10.5966/sctm.2014-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yao S. MicroRNA biogenesis and their functions in regulating stem cell potency and differentiation. Biological Procedures Online. 2016;18:8. doi: 10.1186/s12575-016-0037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].SChoolmeesters A, Eklund T, Leake D, et al.. Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One. 2009;4(5):e5605. doi: 10.1371/journal.pone.0005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mudhasani R, Imbalzano AN, Jones SN. An essential role for dicer in adipocyte differentiation. J Cell Biochem. 2010;110(4): 812–816. doi: 10.1002/jcb.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang Q, Li YC, Wang J, et al.. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad S I USA. 2008;105(8):2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8(6):1149–57. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- [36].Strega-Roslan J, Koscianska E, Kozlowski P, et al.. The Role of the Precursor Structure in the Biogenesis of microRNA. Cell Mol Life Sci. 2011;68:2859–2871. doi: 10.1007/s00018-011-0726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. NAR. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. NAR. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Griffiths-Jones S, Saini HK, van Dongen S, et al.. miRBase: tools for microRNA genomics. NAR. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Griffiths-Jones S, Grocock RJ, van Dongen S, et al.. miRBase: microRNA sequences, targets and gene nomenclature. NAR. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Griffiths-Jones S. The microRNA registry. NAR. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


