Abstract
In the present work, 12 novel Schiff’s bases containing a thiadiazole scaffold and benzamide groups coupled through appropriate pharmacophore were synthesized. These moieties are associated with important biological properties. A facile, solvent-free synthesis of a series of novel 7(a–l) N-((5-(substituted methylene amino)-1,3,4-thiadiazol-2-yl)methyl) benzamide was carried out under microwave irradiation. Structures of the synthesized compounds were confirmed by IR, NMR, mass spectral study and elemental analysis. All the synthesized hybrids were evaluated for their in vitro anticancer activity against a panel of four human cancer cell lines, viz. SK-MEL-2 (melanoma), HL-60 (leukemia), HeLa (cervical cancer), MCF-7 (breast cancer) and normal breast epithelial cell (MCF-10A) using 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay method. Most of the synthesized compounds exhibited promising anticancer activity, showed comparable GI50 values comparable to that of the standard drug Adriamycin. The compounds 7k, 7l, 7b, and 7a were found to be the most promising anticancer agents in this study. A molecular docking study was performed to predict the probable mechanism of action and computational study of the synthesized compounds 7(a–l) was performed to predict absorption, distribution, metabolism, excretion and toxicity (ADMET) properties, by using QikProp v3.5 (Schrödinger LLC). The results showed the good oral drug-like behavior of the synthesized compounds 7(a–l).
Keywords: microwave-assisted synthesis, thiadiazoles, in vitro anticancer activity, molecular docking, ADMET
1. Introduction
Cancer is a disease in which cells grow and proliferate in an uncontrolled manner. Cancer evokes a high level of mortality regardless of the recent advances in the development of clinically authorized anticancer agents [1,2,3].
The development of resistance to chemotherapeutic agents and associated side effects are major obstacles to effectively treating cancer [3]. Thus, it is necessary to identify and develop new anticancer agents with improved efficacy and reduced side effects to complement the present chemotherapeutic strategies. Hence, further research towards the development of newer therapeutic drugs that more effectively combat cancer is needed.
One of the potential targets for new drugs is the matrix metalloproteinases (MMPs), a group of proteinases that have physiologic roles in degrading and remodeling the extracellular membrane. The MMPs are over-expressed in a variety of malignant tumor types, and their over-expression is associated with tumor aggressiveness and metastatic potential [4,5,6,7,8]. The specific alteration of the MMPs observed in malignant tissues and their participation in some of the major oncogenic mechanisms have fueled interest in the design and evaluation of MMP inhibitors as anticancer agents [9,10,11,12].
The class of organic compounds containing the azomethine (–HC=N–) group in their structure is called imine, or alternatively a Schiff base. Schiff bases, derived mostly from a variety of heterocyclic rings, were reported to possess a broad spectrum of pharmacological activities with a wide variety of biological properties, and development of new chemotherapeutic Schiff bases is now attracting the attention of medicinal chemists. They are known to exhibit a variety of potent biological activities.
Heterocyclic nucleus 1,3,4-thiadiazole constitutes an important class of compounds for new drug development because of its interesting biological properties, which include anticancer [13], antibacterial [14] and antifungal activity [15]. Thiadiazoles (e.g., 1,3,4-thiadiazoles) also exhibit a number of extremely interesting spectroscopic properties. One of the very interesting spectroscopic properties of thiadiazoles is the dual fluorescence effect [16,17,18,19]. In addition, 1,3,4-thiadiazole compounds are excellent ligands and a good skeleton for metallo-complexes [20]. Especially, the structure of ‘‘N–C–S’’ in 1,3,4-thiadiazole derivatives can work as the active center, chelate certain metal ions in vivo and show good tissue permeability. The synthesis of novel thiadiazole derivatives and investigation of their chemical and biological behavior have gained more importance in recent decades [21,22,23,24]. The 1,3,4-thiadozole compounds have been reported to inhibit MMPs [25]. Structure–activity relationship (SAR) studies have shown that thiadiazoles are selective for stromelysin-1(MMP-3) over gelatinase-A (MMP-2) with little or no affinity for collagenase (MMP-1) [25]. The dual roles that matrix metalloproteinases appear to play in cancer growth and metastasis [26,27] highlighted the need for studies that address a physical basis for selective inhibition of specific MMPs.
In the present work we planned to develop Schiff’s bases containing a thiadiazole ring and benzamide groups, because of the important biological properties associated with these groups. The coupling of these important moieties was achieved under microwave irradiation. Microwave-induced synthesis has various advantages over conventional synthesis, such as a highly accelerated reaction rate, reasonably good yields, a lesser solvent requirement, an eco-friendly method, a clean heating system and controlled reaction parameters [28].
All the synthesized hybrids were evaluated for their in vitro anticancer activity against a panel of four human cancer cell lines viz. against MCF-7 (human breast cancer cell line), HeLa (human cervical cancer cell line), SKMEL-2 (human melanoma cancer cell line) and HL-60 (human leukemia cancer cell line) by MTT assay using Adriamycin as the standard drug. A molecular docking study has also been performed to support the effective binding of the compound at the active site of the enzyme. Most of the synthesized compounds exhibited promising anticancer activity, with some compounds having GI50 values equivalent to that of the standard drug, Adriamycin. The compounds 7k, 7l, 7b and 7a were found to be the most promising anticancer agents in this study. All the synthesized compounds were screened for their in silico absorption, distribution, metabolism, excretion and toxicity (ADMET) study as well.
2. Result and Discussion
2.1. Chemistry
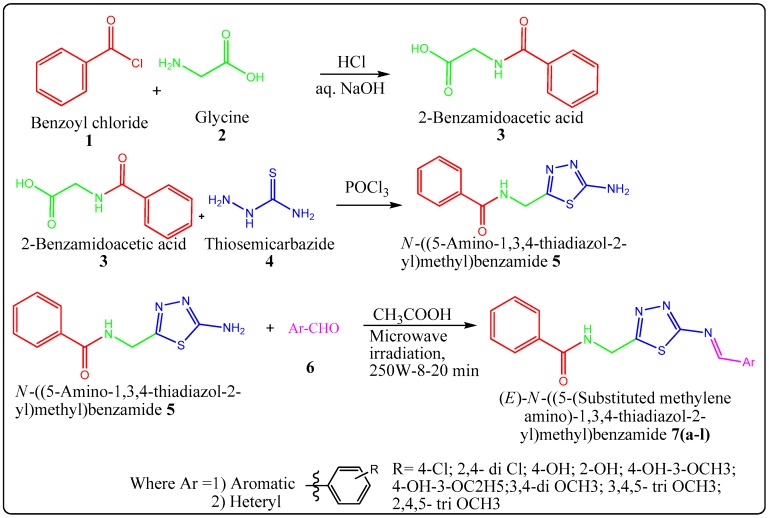
Herein, we report the facile synthesis of novel N-((5-(substituted methylene amino)-1,3,4-thiadiazol-2-yl)methyl) benzamide derivatives under microwave irradiation as shown in Scheme 1. The N-((5-(substituted methylene amino)-1,3,4-thiadiazol-2-yl)methyl) benzamide derivatives were synthesized from N-((5-amino-1,3,4-thiadiazol-2-yl)methyl) benzamide (0.01 mol) (5) and various suitable aldehydes (6) (0.01 mol) using conventional method and microwave method, as shown in Table 1. All the synthesized compounds were characterized by 1H-NMR, 13C-NMR, mass spectroscopy, IR and elemental analysis.
Scheme 1.
Synthesis of N-((5-(Substituted methylene amino)-1,3,4-thiadiazol-2-yl)methyl) benzamide derivatives 7(a–l).
Table 1.
Details of synthesis of N-((5-(Substituted methylene amino)-1,3,4-thiadiazol-2-yl)methyl) benzamide derivatives 7(a–l) using conventional method and microwave method.
| Entry | Conventional Method | Microwave Method | ||||
|---|---|---|---|---|---|---|
| Solvent | Temperature °C | Time (h) | Yield (%) | Time (min) | Yield (%) | |
| 7a | Ethanol | 80–90 | 3.30 | 78 | 8 | 95 |
| 7b | Ethanol | 80–90 | 5.10 | 77 | 12 | 92 |
| 7c | Ethanol | 80–90 | 5.00 | 78 | 12 | 94 |
| 7d | Ethanol | 80–90 | 7.20 | 66 | 15 | 94 |
| 7e | Ethanol | 80–90 | 7.25 | 52 | 15 | 92 |
| 7f | Ethanol | 80–90 | 8.00 | 48 | 18 | 92 |
| 7g | Ethanol | 80–90 | 5.25 | 76 | 12 | 95 |
| 7h | Ethanol | 80–90 | 7.30 | 66 | 15 | 88 |
| 7i | Ethanol | 80–90 | 8.00 | 68 | 20 | 86 |
| 7j | Ethanol | 80–90 | 8.00 | 48 | 20 | 88 |
| 7k | Ethanol | 80–90 | 4.15 | 44 | 10 | 85 |
| 7l | Ethanol | 80–90 | 4.25 | 56 | 10 | 84 |
2.2. In Vitro Anticancer Activity
The synthesized compounds 7(a–l) were evaluated for their in vitro anticancer activity against MCF-7 (human breast cancer cell line), HeLa (human cervical cancer cell line), SKMEL-2 (human melanoma cancer cell line) and HL-60 (human leukemia cancer cell line) cancer cell lines, as shown in Table 2. The most active compounds 7k, 7l, 7a and 7b were tested on non-tumorigenic breast epithelial cell lines (MCF-10A) to find out the selectivity towards the cancer cells. The GI50 values (the concentration required to achieve a growth inhibition of 50%) for the synthesized compounds were determined by MTT assay method. The in vitro anticancer evaluation results and GI50 values were listed in Table 2, and the well-known anticancer drug Adriamycin was used as positive control.
Table 2.
In vitro anticancer activity of the synthesized compounds 7(a–l).
| Entry | GI50 µM | ||||
|---|---|---|---|---|---|
| MCF-7 | HeLa | SKMEL-2 | HL-60 | MCF-10A | |
| 7a | 22.9 | 32.8 | 21.9 | 21.7 | >100 |
| 7b | 28.7 | 39.0 | 22.9 | 28.2 | 86.1 |
| 7c | 32.4 | 41.1 | 27.5 | 33.3 | ND |
| 7d | 36.7 | 52.4 | 34.0 | 40.2 | ND |
| 7e | 35.2 | 46.8 | 28.1 | 39.6 | ND |
| 7f | 38.4 | 49.2 | 30.0 | 37.5 | ND |
| 7g | 41.0 | 66.1 | 46.4 | 42.4 | ND |
| 7h | 46.2 | 71.7 | 49.1 | 48.2 | ND |
| 7i | 49.0 | 78.0 | 52.6 | 45.8 | ND |
| 7j | 51.4 | 78.8 | 55.7 | 49.9 | ND |
| 7k | 11.7 | 23.8 | 19.6 | 35.5 | > 100 |
| 7l | 19.0 | 28.8 | 22.0 | 29.9 | > 100 |
| ADR | < 10 | < 10 | < 10 | < 10 | ND |
ADR Adriamycin standard drug used.GI50 is the concentration exhibiting 50% inhibition of the growth, as compared to the growth of control.MCF-7 human breast cancer cell line, HeLa human cervical cancer cell line, SKMEL-2 human melanoma cancer cell line, HL-60 human leukemia cancer cell line, MCF-10A normal breast epithelial cell lines and ND = Not determined.
The results indicated that the compounds 7k, 7l, 7a and 7b exhibited significant cancer cell growth inhibition compared to reference standard Adriamycin against MCF-7, HeLa, SKMEL-2 and HL-60 cancer cell lines. From the in vitro anticancer activity results, it was observed that compound 7k, which has a furan ring, was found to have the GI50 values 11.7 µM, 23.8 µM, 19.6 µM and 35.5 µM for MCF-7, HeLa, SKMEL-2 and HL-60 cancer cell lines, respectively. Compound 7l, which has a thiophene ring, was found to have the GI50 values 19.0 µM, 28.8 µM, 22.0 µM and 29.9 µM for MCF-7, HeLa, SKMEL-2 and HL-60 cancer cell lines, respectively. Compound 7a containing 4-Chlorophenyl substituent (MCF-7 GI50 22.9 µM, HeLa GI50 32.8 µM, SKMEL-2 GI50 21.9 µM and HL-60 GI50 21.7 µM) and compound 7b 2,4 dichloro phenyl substituent (MCF-7 GI50 28.7 µM, HeLa GI50 39.0 µM, SKMEL-2 GI50 22.9 µM and HL-60 GI50 28.2 µM) were found to have good in vitro anticancer activity.
From the in vitro anticancer result data as shown in Table 2, the most active compounds 7k, 7l, 7a and 7b were tested on non-tumorigenic breast epithelial cell lines (MCF-10A) to find out the selectivity towards the cancer cells. Interestingly, compounds 7k, 7l and 7a were found to be selective towards cancer cells, since they did not exhibit cytotoxicity (GI50 >100 μM) on MCF-10A cells. However, compound 7b showed almost three times more selectivity for MCF-7 cancer cell lines in comparison to the MCF-10A normal breast epithelial cell lines.
Structure-activity relationship (SAR) studies for these compounds demonstrated that electron withdrawing groups such as chloro (7a, 7b) exhibited good in vitro anticancer activity compared to electron donating groups. Compound 7c with 4-hydroxyphenyl substituent (MCF-7 GI50 32.4 µM, HeLa GI50 41.1 µM, SKMEL-2 GI50 27.5 µM and HL-60 GI50 33.3 µM) was found to be more active than compound 7e with 4-hydroxy-3-methoxyphenyl substituent (MCF-7 GI50 35.2 µM, HeLa GI50 46.8 µM, SKMEL-2 GI50 28.1 µM and HL-60 GI50 39.6 µM) and compound 7f with 4-hydroxy-3-ethoxy phenyl substituent (MCF-7 GI50 38.4 µM, HeLa GI50 49.2 µM, SKMEL-2 GI50 30.0 µM and HL-60 GI50 37.5 µM). Replacement of the phenyl group in the parent compound by furan ring in 7k and thiophene ring in 7l showed a significant increase in the in vitro anticancer activity in comparison to the standard drug Adriamycin. From SAR, it can be considered that compounds containing electron-donating polar groups such as 7c, 7d, 7e, 7f, 7g, 7h, 7i and 7j are less active in comparison to the electron-withdrawing groups, such as 7a and 7b. It is also clear that the replacement of the phenyl ring with the furan ring (7k) and thiophene ring (7l) significantly increased the in vitro anticancer activity. Compounds having large, bulky di-substituted and tri-substituted groups on phenyl ring showed a decrease in in vitro anticancer activity (for example, 7e and 7f) than compounds having less bulky substituents on phenyl ring, such as 7a and 7b. Compounds 7k and 7l showed significantly good in vitro anticancer activity in comparison to the standard anticancer drug Adriamycin against MCF-7, HeLa, SKMEL-2 and HL-60 cancer cell lines and thus can be developed as anticancer agents in the future.
2.3. Molecular Docking
Unregulated or over-expressed matrix metalloproteinases (MMPs), including stromelysin, collagenase and gelatinase, were implicated in several pathological conditions, including arthritis and cancer. The 1,3,4-thiadozole compounds have been reported to inhibit selectively fibroblast stromelysin-1 [24,29]. In order to predict the binding affinity of our newly synthesized N-((5-(substituted methylene amino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7(a–l) derivatives into the binding site of fibroblast stromelysin-1, we decided to carry out a molecular docking study. The docking results indicated that compounds were held in the active pocket by a combination of various hydrogen bonds and hydrophobic interactions with fibroblast stromelysin-1. The docking results revealed that the compound 7k had the highest binding interaction with fibroblast stromelysin-1, with a docking score of −7.56, as shown in Table 3.
Table 3.
Docking score of all the synthesized derivatives 7(a–l).
| Compound | Docking Score | Compound | Docking Score |
|---|---|---|---|
| 7a | −6.61 | 7h | −6.33 |
| 7b | −6.56 | 7i | −6.61 |
| 7c | −5.93 | 7j | −5.10 |
| 7d | −5.82 | 7k | −7.56 |
| 7e | −5.84 | 7l | −7.32 |
| 7f | −5.66 | ADR | −4.33 |
| 7g | −5.26 |
On the basis of the docking results, it was found that compounds 7k, 7l, 7a and 7b had good potential to inhibit fibroblast stromelysin-1, which will be the probable mechanism of these derivatives for their anticancer activity. The docking score of all the synthesized derivatives is shown in Table 3.
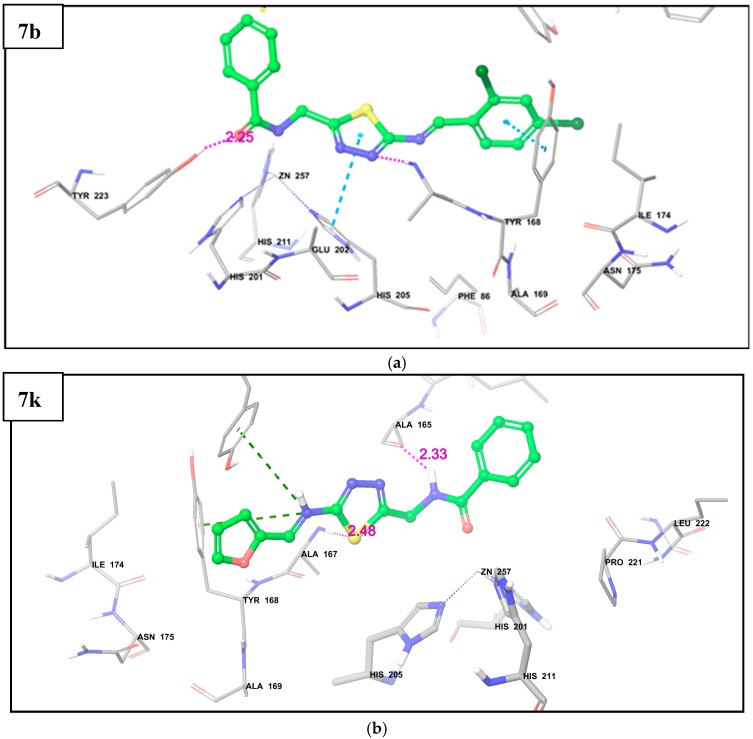
From the docking analysis, it was observed that compound 7b showed hydrogen bond interaction with amino acid residue Tyr223; while on the other hand, hydrogen bonding is seen with amino acid residues Ala165 and Ala167 in compound 7k (Figure 1). The hydrophobic interactions such as pi-pi stacking and pi-pi cation between compounds and surrounding hydrophobic amino acid residues of enzymes helped to stabilize the complex structure and thus enhanced the binding affinity of compounds towards fibroblast stromelysin-1 (Figure 1). It was also seen that the –CONH group of both compounds 7b and 7k makes a metal coordination with zinc ion present in the active site of the enzyme, which is a probable mechanism of inhibition of fibroblast stromelysin-1.
Figure 1.
Docking pose of: (a) Compound 7b; (b) Compound 7k in the active pocket of Fibroblast stromelysin-1. The pink-colored dotted line indicates H-bond, the cyan-colored dotted line indicates pi-pi stacking interaction, and the green-colored dotted line indicates pi-pi cation interaction between ligand and enzyme.
2.4. In Silico ADMET Prediction
The absorption, distribution, metabolism, excretion and toxicity (ADMET) play a critical role in helping to optimize the pharmacokinetic properties of new drugs, as it increases their success rate. Thus the prediction of the ADMET parameters prior to experimental studies is one of the most important aspects of both drug discovery and development of the drug molecule. The prediction of Lipinski’s rule-of-five was performed to indicate whether a chemical compound could be an orally active drug in humans. The prediction of ADMET properties was performed for all compounds and is shown in Table 4. It was observed that all the compounds exhibited a good predicted percentage of oral absorption (% ABS) ranging from 75–100%. The predicted aqueous solubility (Log S) (S in mol/L) and predicted partition coefficient (Log Po/w) value for all compounds was found within the range of −6.1 to −4.2 and 3.91–5.34 respectively, and thus is a probable reason for the predicted good percentage of oral absorption for all compounds. The efficiency and distribution of a drug may be affected by the degree to which it binds to the proteins present within the blood plasma. The log Khsa parameter is used for the prediction of binding to the human serum albumin. All compounds showed predicted log Khsa value ranges between −0.2 and 0.19, which indicated that a significant proportion of the compounds are likely to circulate freely in the blood stream and can reach the drug target sites.
Table 4.
In silico physicochemical pharmacokinetic parameters important for good oral bioavailability of the synthesized compounds 7(a–l).
| Entry | M.W a | LogP o/w b (−2.0–6.5) |
n-ON c (<10) | n-OHNH d (<5) | PSA e (7–200) | log Khsa f (−1.5–1.2) |
Log S g (−6–0.5) | % ABS h | #Meta i (1–8) | Log HERG j Below−5 |
Lipinski Rule of 5 (≤1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7a | 356.8 | 4.82 | 5.5 | 1 | 76.2 | 0.31 | −5.8 | 98 | 2 | −6.8 | 0 |
| 7b | 391.2 | 5.34 | 5.5 | 1 | 74.4 | 0.37 | −6.1 | 99 | 2 | −6.6 | 0 |
| 7c | 338.3 | 5.18 | 6.2 | 2 | 98.4 | 0.02 | −4.7 | 89 | 3 | −6.7 | 0 |
| 7d | 338.3 | 5.17 | 6 | 2 | 98.2 | 0.03 | −4.3 | 88 | 3 | −6.5 | 0 |
| 7e | 368.4 | 5.27 | 7.2 | 1 | 133.3 | −0.04 | −4.7 | 75 | 3 | −6.6 | 0 |
| 7f | 382.4 | 5.09 | 7 | 2 | 105.7 | 0.16 | −5.5 | 91 | 4 | −6.9 | 0 |
| 7g | 352.4 | 4.91 | 6.2 | 1 | 83.9 | 0.16 | −5.1 | 100 | 3 | −6.7 | 0 |
| 7h | 382.4 | 5.20 | 7 | 1 | 89.0 | 0.19 | −5.4 | 100 | 4 | −6.7 | 0 |
| 7i | 412.2 | 4.05 | 7.7 | 1 | 95.4 | 0.17 | −5.4 | 100 | 5 | −6.5 | 0 |
| 7j | 412.2 | 4.08 | 7.7 | 1 | 97.6 | 0.19 | −5.6 | 100 | 5 | −6.6 | 0 |
| 7k | 312.3 | 3.91 | 6 | 1 | 84.7 | −0.11 | −4.0 | 94 | 3 | −6.3 | 0 |
| 7l | 328.4 | 4.11 | 5 | 1 | 85.6 | −0.21 | −4.2 | 95 | 3 | −6.2 | 0 |
a Molecular weight of the molecule; b Predicted octanol-water partition coefficient (log Po/w) (−2.0 to 6.5); c n-ON number of hydrogen bond acceptors ≤10; d n-OHNH number of hydrogen bonds donors ≤5; e Polar surface area (PSA) (7.0–200.0); f Logarithm of predicted binding constant to human serum albumin (log Khsa) (−1.5 to 1.2); g Logarithm of aqueous solubility(Log S) (−6 to 0.5); h Percentage of human oral absorption (% ABS) (>80% is high, <25% is poor); #Metabolism I (1–8); j Predicted IC50 value for blockage of HERG K+ channels (log Khsa) (−1.5 to 1.2).
Human ether-a-go-go related gene (HERG) encodes a potassium ion (K+) channel that is implicated in the fatal arrhythmia known as torsade de pointes, or the long QT syndrome [30]. The HERG K+ channel, which is best known for its contribution to the electrical activity of the heart that coordinates the heart’s beating, appears to be the molecular target responsible for the cardiac toxicity of a wide range of therapeutic drugs [31]. HERG has also been associated with modulating the functions of some cells of the nervous system, and with establishing and maintaining cancer-like features in leukemic cells [32]. Thus, HERG K+ channel blockers are potentially toxic and the predicted IC50 values often provide reasonable predictions for the cardiac toxicity of drugs in the early stages of drug discovery [33]. Thus, based on predicted log HERG values, none of the synthesized compounds 7(a–l) were found to be toxic in nature. Similarly, it was also seen that none of the compounds have violated the Lipinski rule-of-five (M.W < 500, l Log Po/w<5, n-ON < 10, n-OHNH < 5). The overall ADMET prediction studies thus clearly suggested that all synthesized compounds have good pharmacokinetic properties and can be developed as good oral drugs in the future.
3. Materials and Methods
3.1. General Information
All the chemicals used for synthesis were of Merck, Sigma, Research lab, Qualigens make and Himedia. The reactions were carried out by conventional method and in synthetic microwave oven (Milestone micro synth). Melting points were determined in open capillaries using melting point apparatus and are uncorrected. All the reactions were performed in oven-dried glassware. Phosphorus oxychloride was used by distilling under reduced pressure. The synthetic protocol employed for the synthesis of N-((5-(substituted methylene amino)-1,3,4-thiadiazol-2-yl)methyl) benzamide derivatives 7(a–l) is presented in Scheme 1. The purity of the synthesized compounds was checked by TLC, and melting points were determined in open capillary tubes and are uncorrected. The NMR spectra of the final titled compounds N-((5-(substituted methylene amino)-1,3,4-thiadiazol-2-yl)methyl) benzamide were recorded on Brucker Advance II (400 MHz), and Infrared (IR) spectra were recorded for the compounds on JASCO FTIR (PS 4000, JASCO, Tokyo, Japan) using KBr pallet. The mass spectra were recorded on a Waters Micro Mass (ZQ 2000 spectrometer, UK). Elemental analyses were done with a FLASHEA 112 Shimadzu’ analyzer (Mumbai, Maharashtra, India) and all analyses were consistent (within 0.4%) with theoretical values.
3.1.1. Step I: General Process for Synthesis of 2-Benzamidoacetic Acid [34]
The first step involved dissolving 0.33 mol of 2-aminoacetic acid (2) in 10% NaOH solution contained in a conical flask. To this solution, 0.38 mol of benzoyl chloride (1) was added in five portions and shaken vigorously until the chloride reacted. The solution was transferred to a beaker containing crushed ice and dil. HCl was added until the solution was acidic to congo red paper. The resulting crystalline solid was collected and boiled with 10 mL of CCl4 for 10 min. The product was filtered and washed with CCl4. The solid product obtained was dried and recrystallized from ethanol. The melting point and yield were recorded.
3.1.2. Step II: General Procedure for Synthesis of N-((5-Amino-1,3,4-thiadiazol-2-yl)methyl) Benzamide [35]
Next, 2-benzamidoacetic acid (3) (0.05 mol) was refluxed with thiosemicarbazide (4) (0.05 mol) and phosphorus oxychloride (15 mL) for 1 h. The mixture was cooled and diluted with water (90 mL) and again refluxed for 4 h. Then the mixture was filtered, and the filtrate was basified with potassium hydroxide solution. The precipitate was filtered off and recrystallized from ethanol.
3.1.3. Step III: General procedure for synthesis of (E)-N-((5-(substituted methyleneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7(a–l)
A. Conventional method [36]
Equimolar quantities of N-((5-amino-1,3,4-thiadiazol-2-yl)methyl) benzamide (0.01 mol) (5) and various suitable aldehydes (6) (0.01 mol) were refluxed in the presence of glacial acetic acid (0.02 mol) in absolute ethanol (25 mL) for 3–8 h. The completion of the reaction was monitored by Thin Layer Chromatography (TLC). The reaction mixture was concentrated and cooled. The obtained solid was filtered and dried. The product was recrystallized from ethanol. The melting point and yield were recorded.
B. Microwave-assisted method [37]
Solvent-free synthesis of Schiff’s bases was achieved by cyclo addition of various suitable aldehydes (6) (0.01 mol) and N-((5-amino-1,3,4-thiadiazol-2-yl)methyl) benzamide (5) (0.01 mol) in the presence of a catalytic amount of glacial acetic acid under microwave irradiation at 250 W for 8–20 min, as shown in Scheme 1. The completion of the reaction was monitored by TLC. The synthesized products were recrystallized from ethanol. The same compounds were also synthesized using conventional approach. A comparative study in terms of yield and reaction period has been reported for both microwave-assisted and conventional methods. The conventional method required about 3–8 h, while the microwave-irradiation method required only 8–20 min, as shown in Table 1.
2-Benzamidoacetic acid 3. Yield: 95%; m.p.: 187–190 °C;IR (KBr νmax in cm−1): 3350.41 (NH), 3179.92 (OH), 2970.76 (C=H), 1810.26 (C=O of amide), 1725.01 (C=O of carboxylic acid); 1H-NMR (DMSO) δ ppm: 3.84 (s, 2H, CH2), 7.77–8.00 (m, 5H), 8.05 (s, 1H, NH), 11.35 (s, 1H, OH); 13C-NMR (DMSO) δ ppm: 41.34,127.31, 127.87, 128.00, 128.61, 132.89, 134.23, 167.87, 172.11; m/z: 179.06 (100.0%), 180.06 (10.3%), 181.06 (1.1%); Anal. Calcd. For C9H9NO3: C, 60.33; H, 5.06; N, 7.82; O, 26.79. Found: C, 60.35; H, 5.08; N, 7.80; O, 26.76.
N-((5-Amino-1,3,4-thiadiazol-2-yl)methyl) benzamide 5. Yield: 92%; m.p.: 218–220 °C; IR (KBr νmax in cm−1): 3360.45 (NH2), 3000 (C-H aromatic), 2970.76 (C=H), 1810.26 (C=O of amide), 1454.55 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.45 (s, 2H, CH2), 6.89 (s, 2H, NH2), 7.70–8.00 (m, 5H), 8.05 (s, 1H, NH); 13C-NMR (DMSO) δ ppm: 39.11,127.26, 127.69, 128.42, 128.76, 132.11, 134.21, 161.66, 167.89, 168.00; m/z: 234.06 (100.0%), 235.06 (11.8%), 236.05 (4.5%), 235.05 (1.5%); Anal. Calcd. For C10H10N4OS: C, 51.27; H, 4.30; N, 23.91; S, 13.69. Found: C, 51.29; H, 4.33; N, 23.89; S, 13.65.
(E)-N-((5-(4-Chlorobenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7a. Yield: 95%; m.p.: 126–128 °C; IR (KBr νmax in cm−1): 3355.41 (NH), 2970.76 (C=H), 1810.26 (C=O of amide), 1454.50 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.42 (s, 2H, CH2), 6.69–5.21 (m, 9H), 8.21 (s, 1H, NH), 10.33 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 39.11,127.36, 127.34, 127.51, 128.78, 128.98, 130.00, 130.63, 131.22, 133.23, 134.34, 134.51, 136.67, 160.00, 167.89, 168.00; m/z: 356.05 (100.0%), 358.05 (37.1%), 357.05 (20.7%), 359.05 (7.1%), 358.06 (1.6%), 360.04 (1.5%); Anal. Calcd. For C17H13ClN4OS: C, 57.22; H, 3.67; Cl, 9.94; N, 15.70; S, 8.99. Found: C, 57.24; H, 3.69; Cl, 9.97; N, 15.68; S, 8.98.
(E)-N-((5-(2,4-Dichlorobenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7b. Yield: 92%; m.p.: 112–114 °C; IR (KBr νmax in cm−1): 3352.41 (NH), 2975.76 (C=H), 1818.26 (C=O of amide), 1457.55 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.45 (s, 2H, CH2), 6.69–5.21 (m, 8H), 8.29 (s, 1H, NH), 10.33 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 40.11,127.36, 127.34, 127.51, 128.68, 128.98, 130.00, 130.53, 131.22, 133.13, 134.39, 134.41, 136.77, 161.00, 168.89, 169.00; m/z: 390.01 (100.0%), 392.01 (68.9%), 391.01 (20.7%), 393.01 (13.2%), 394.00 (13.1%), 395.01 (2.5%), 392.02 (1.8%), 394.01 (1.5%), 393.00 (1.0%); Anal. Calcd. For C17H12Cl2N4OS: C, 52.18; H, 3.09; Cl, 18.12; N, 14.32; S, 8.20. Found: C, 52.19; H, 3.11; Cl, 18.14; N, 14.30; S, 8.22.
(E)-N-((5-(4-Hydroxybenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7c. Yield: 94%; m.p.: 118–120 °C; IR (KBr νmax in cm−1): 3500.92 (OH), 3350.41 (NH), 2970.76 (C=H), 1810.26 (C=O of amide), 1458.50 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.44 (s, 2H, CH2), 5.43 (s,1H, OH), 6.69–5.20 (m, 9H), 8.21 (s, 1H, NH), 10.34 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 40.12,115.80, 116.19, 121.93, 125.20, 128.96, 129.41, 131.90, 134.60, 156.36, 163.36, 165.05, 168.54, 190.00; m/z: 338.08 (100.0%), 339.09 (18.6%), 340.08 (4.8%), 339.08 (2.3%), 340.09 (2.2%); Anal. Calcd. For C17H14N4O2S: C, 60.34; H, 4.17; N, 16.56; S 9.48. Found: C, 60.36; H, 4.19; N, 16.53; S, 9.45.
(E)-N-((5-(2-Hydroxybenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7d. Yield: 94%; m.p.: 106–108 °C; IR (KBr νmax in cm−1): 3500.90 (OH), 3350.58 (NH), 2970.66 (C=H), 1811.16 (C=O of amide), 1454.55 (CH of CH2); 1H-NMR (DMSO) δ ppm: 4.12 (s, 2H, CH2), 5.38 (s, 1H, OH), 7.02–7.71 (m, 4H), 7.81–8.10 (m, 5H), 8.16 (s, 1H, NH), 9.91 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 40.03,118.82, 120.52, 121.46, 126.89, 127.45, 127.91, 128.17, 129.80, 131.00, 132.41, 135.51, 160.55, 160.99, 169.09, 170.11; m/z: 338.08 (100.0%), 339.09 (18.6%), 340.08 (4.8%), 339.08 (2.3%), 340.09 (2.2%); Anal. Calcd. For C17H14N4O2S: C, 60.34; H, 4.17; N, 16.56; S, 9.48. Found: C, 60.36; H, 4.19; N, 16.53; S, 9.50.
(E)-N-((5-(4-Hydroxy-3-methoxybenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7e. Yield: 92%; m.p.: 124–126 °C; IR (KBr νmax in cm−1): 3500.90 (OH), 3350.31 (NH), 2971.76 (C=H), 1810.16 (C=O of amide), 1455.55 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.83 (s, 3H, OCH3), 4.10 (s, 2H, CH2), 5.35 (s, 1H, OH), 6.93 (d, J = 8.40 Hz,1H), 7.34 (d, J = 8.41 Hz,1H), 7.52 (s, 1H), 7.70–8.08 (m, 5H), 8.19 (s, 1H, NH), 10.00 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 39.87, 56.17, 113.17, 118.16, 122.15, 126.81, 127.01, 127.76, 128.28, 129.98, 130.16, 135.09, 149.71, 152.09, 159.99, 167.71, 169.18; m/z: 368.09 (100.0%), 369.10 (19.8%), 370.09 (4.8%), 370.10 (2.6%), 369.09 (2.3%); Anal. Calcd. For C18H16N4O3S: C, 58.68; H, 4.38;N, 15.21;S 8.70. Found: C, 58.70; H, 4.39; N, 15.19; S, 8.72.
(E)-N-((5-(3-Ethoxy-4-hydroxybenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7f. Yield: 92%; m.p.: 130–132 °C; IR (KBr νmax in cm−1): 3500.90 (OH), 3350.31 (NH), 2971.76 (C=H), 1810.16 (C=O of amide), 1457.55 (CH of CH2); 1H-NMR (DMSO) δ ppm: 1.32 (t, 3H, CH3), 4.09 (q, 2H, CH2), 4.46 (s, 2H, CH2), 5.35 (s, 1H, OH), 6.91–7.52 (m, 3H, CH), 7.63-8.09 (m, 5H, CH), 8.13 (s, 1H, NH), 10.00 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 14.87, 39.99, 64.57, 112.38, 116.59, 122.34, 127.00, 128.19, 128.88, 130.70, 132.55, 134.47, 148.83, 151.88, 161.66, 168.78; m/z: 382.11 (100.0%), 383.11 (22.9%), 384.11 (5.6%), 384.12 (2.1%); Anal. Calcd. For C19H18N4O3S: C, 59.67; H, 4.74; N, 14.65; S 8.38. Found: C, 59.69; H, 4.76; N, 14.61; S, 8.39.
(E)-N-((5-(4-Methoxybenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7g. Yield: 95%; m.p.: 116–118 °C; IR (KBr νmax in cm−1): 3352.18 (NH), 2971.66 (C=H), 1810.17 (C=O of amide), 1454.55 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.83 (s, 6H, OCH3), 4.19 (s, 2H, CH2), 7.06 (d, J = 8.40 Hz,1H), 7.16 (d, J = 8.41 Hz,1H), 7.60–8.06 (m, 7H), 8.17 (s, 1H, NH), 10.00 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 40.00, 55.89, 114.01, 114.45, 127.03, 127.52, 127.97, 128.01, 128.89, 130.71, 131.91, 132.96, 135.01, 161.00, 163.09, 169.99, 170.13; m/z: 352.10 (100.0%), 353.10 (21.8%), 354.10 (5.4%), 354.11 (1.8%); Anal. Calcd. For C18H16N4O2S: C, 61.35; H, 4.58; N, 15.90; S, 9.10. Found: C, 61.38; H, 4.59; N, 15.88; S, 9.13.
(E)-N-((5-(3,4-Dimethoxybenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7h. Yield: 88%; m.p.: 120–122 °C; IR (KBr νmax in cm−1): 3352.18 (NH), 2971.66 (C=H), 1810.17 (C=O of amide), 1456.75 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.83 (s, 6H, OCH3), 4.19 (s, 2H, CH2), 6.98–7.61 (m, 3H), 7.69–8.05 (m, 5H), 8.12 (s, 1H, NH), 9.98 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 39.96, 56.11, 108.92, 111.77, 124.69, 126.96, 127.58, 127.99, 128.81, 130.66, 132.10, 134.26, 149.92, 152.10, 159.91, 169.96, 170.09; m/z: 382.11 (100.0%), 383.11 (22.9%), 384.11 (5.6%), 384.12 (2.1%); Anal. Calcd. For C19H18N4O3S: C, 59.67; H, 4.74; N, 14.65; S 8.38. Found: C, 59.69; H, 4.76; N, 14.62; S, 8.36.
(E)-N-((5-(3,4,5-Trimethoxybenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7i. Yield: 86%; m.p.: 138–140 °C; IR (KBr νmax in cm−1): 3350.18 (NH), 2972.66 (C=H), 1810.17 (C=O of amide), 1456.87 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.85 (s, 9H, OCH3), 4.19 (s, 2H, CH2), 6.98–7.61 (m, 3H), 7.69–8.05 (m, 5H), 8.12 (s, 1H, NH), 9.98 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 39.96, 56.11, 108.92, 111.77, 124.69, 126.96, 127.58, 127.99, 128.81, 130.66, 132.10, 134.26, 149.92, 152.10, 159.91, 169.96, 170.09; m/z: 412.12 (100.0%), 413.12 (24.1%), 414.12 (5.9%), 414.13 (2.3%), 415.12 (1.1%); Anal. Calcd. For C20H20N4O4S: C, 58.24; H, 4.89; N, 13.58; S 7.77. Found: C, 58.27; H, 4.91; N, 13.54; S, 7.79.
(E)-N-((5-(2,4,5-Trimethoxybenzylideneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7j. Yield: 88%; m.p.: 140–142 °C; IR (KBr νmax in cm−1): 3350.18 (NH), 2972.66 (C=H), 1810.17 (C=O of amide), 1454.55 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.86 (s, 9H, OCH3), 4.20 (s, 2H, CH2), 6.99–7.65 (m, 3H), 7.67–8.07 (m, 5H), 8.17 (s, 1H, NH), 9.99 (s, 1H, N=CH); 13C-NMR (DMSO) δ ppm: 37.96, 55.35, 109.12, 110.97, 125.19, 126.96, 127.58, 128.99, 129.81, 131.66, 133.10, 135.26, 148.92, 153.10, 158.91, 168.96, 171.09; m/z: 412.12 (100.0%), 413.12 (24.1%), 414.12 (5.9%), 414.13 (2.3%), 415.12 (1.1%); Anal. Calcd. For C20H20N4O4S: C, 58.24; H, 4.89; N, 13.58; S 7.77. Found: C, 58.27; H, 4.92; N, 13.55; S, 7.78.
(E)-N-((5-(Furan-2-ylmethyleneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7k. Yield: 85%; m.p.: 124–126 °C; IR (KBr νmax in cm−1): 3350.41 (NH), 2970.76 (C=H), 1810.26 (C=O of amide), 1456.40 (CH of CH2); 1H-NMR (DMSO) δ ppm: 3.48 (s, 2H, CH2), 6.52 (t, 1H), 6.93 (d, J = 3.40 Hz,1H), 7.65 (d, J = 1.75 Hz, 1H), 7.80 (s, 1H, N=CH), 7.75–8.05 (m, 5H), 8.19 (s, 1H, NH); 13C-NMR (DMSO) δ ppm: 40.12, 112.26, 118.90, 126.99, 127.54, 128.04, 128.57, 131.11, 134.29, 144.48, 146.99, 150.44, 167.34, 168.09; m/z: 312.07 (100.0%), 313.07 (18.7%), 314.06 (4.5%), 314.07 (2.0%); Anal. Calcd. For C15H12N4O2S: C, 57.68; H, 3.87; N, 17.94; S, 10.27. Found: C, 57.69; H, 3.89; N, 17.92; S, 10.29.
(E)-N-((5-(Thiophen-2-ylmethyleneamino)-1,3,4-thiadiazol-2-yl)methyl) benzamide 7l. Yield: 84%; m.p.: 130–132 °C; IR (KBr νmax in cm−1): 3350.18 (NH), 2975.66 (C=H), 1815.17 (C=O of amide), 1455.67 (CH of CH2); 1H-NMR (DMSO) δ ppm: 4.46 (s, 2H, CH2), 7.17 (t, 1H, CH), 7.63–8.09 (m, 6H), 8.29 (s, 1H, NH); 13C-NMR (DMSO) δ ppm: 39.91, 127.00, 127.44, 127.99, 128.34, 128.89, 130.73, 132.77, 134.55, 142.98, 152.69, 167.98; m/z: 328.05 (100.0%), 329.05 (16.4%), 330.04 (9.1%), 329.04 (3.1%), 330.05 (2.0%), 331.04 (1.7%); Anal. Calcd. For C15H12N4O2S2: C, 54.86; H, 3.68; N, 17.06; S 19.53. Found: C, 54.88; H, 3.69; N, 17.03; S, 19.55.
3.2. In Vitro Anticancer Screening
The synthesized compounds 7(a–l) were evaluated for their in vitro anticancer activity against MCF-7 (human breast cancer cell line), HeLa (human cervical cancer cell line), SKMEL-2 (human melanoma cancer cell line) and HL-60 (human leukemia cancer cell line) cancer cell lines. The GI50 values (concentration required to Growth inhibition of 50%) for the synthesized compounds were determined using the MTT assays method.
Experimental procedure for MTT assay
The stock solutions of test compounds were prepared in DMSO. After 24 h incubation, different concentrations (2, 4, 6, 8 µM) of compounds, made by serial dilution in culture medium, were added in 48 h incubation. The final concentration of DMSO was 0.01% in each well. A separate well containing 0.01% DMSO only was run as DMSO control, which was found inactive under applied conditions. The cell growth was determined using MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (Sigma, St. Louis, MO, USA) reduction assay, which is based on the ability of viable cells to reduce a soluble yellow tetrazolium salt to blue formazan crystal [38,39]. Briefly, after 48 h of treatments, the 10 µl of MTT dye, prepared in phosphate buffered saline (PBS) was added to all wells. The plates were then incubated for 4 h at 37 °C. Supernatant from each well was carefully removed, formazon crystals were dissolved in 100 µL of DMSO and absorbance at 540 nm wavelength was recorded and each concentration was tested in threefold. The GI50 values were determined as concentration of compounds that inhibited cancer cell growth by 50%.
3.3. Computational Study
Molecular Docking and In Silico ADMET Prediction
Molecular docking studies were performed in Maestro 9.1 using Glide v6.8 (Schrodinger, LLC, New York, NY, USA). All the compounds were built using Maestro build panels and optimized to lower energy conformers using Ligprep v3.5.9 (Schrodinger, LLC). The X-ray crystal structure of the Fibroblast stromelysin-1 enzyme (PDB code 1USN) [21] was obtained from the protein data bank and prepared for docking using protein preparation wizards [40]. After preparation, the hydrogen bond network in the enzyme structure was optimized using OPLS_2005 force field. The minimization was terminated when the energy converged or the route mean square deviation (RMSD) reached a maximum cutoff of 0.30 Å. Grids were then defined around refined and minimized structures by centering on ligand using default box size. The final docking studies for all the compounds were performed using the extra-precision (XP) docking mode on generated grid of protein structure [41]. The evaluation of ligand-protein binding was done with Glide (docking) score.
The ADMET properties of lower-energy conformers of all compounds were predicted using QikProp v3.5 (Schrödinger LLC).
4. Conclusions
In summary, a new scaffold of 12 N-((5-(substituted methylene amino)-1,3,4-thiadiazol-2-yl)methyl) benzamide derivatives 7(a–l) has been synthesized in solvent free condition under microwave irradiation and evaluated for their potential in vitro anticancer activity on MCF-7, HeLa, SKMEL-2 and HL-60 human cancer cell lines by MTT assay using Adriamycin as a standard drug. The compounds 7k, 7l, 7b and 7a were found to be the most promising anticancer agents in this study. The compounds 7k, 7l and 7a were found to be selective towards cancer cells, since they did not exhibit cytotoxicity even at GI50 > 100 μM on MCF-10A normal breast epithelial cell lines. The compound 7b showed almost three times more selectivity for the MCF-7 cancer cell lines in comparison to the MCF-10A normal breast epithelial cell lines.
The good docking scores as well as anticancer activity of these compounds have been justified by investigating their interaction at the active site of Fibroblast stromelysin-1, as found in the docking studies. The ADMET studies predicted good oral drug-like behavior of the newly synthesized derivatives. The studies presented here provide innovative agents for the development of novel anticancer agents targeting MMPs with the goal of improving cancer care.
Acknowledgments
The authors are thankful to Fatima Rafiq Zakaria, Chairman, Maulana Azad Educational Trust and the Principal, Y.B. Chavan College of Pharmacy, Rafiq Zakaria Campus, Aurangabad 431001 (MS), India for providing the laboratory facility. The authors are grateful to Anti-Cancer Drug screening facility (ACDSF) at ACTREC, Tata Memorial Centre, Navi Mumbai for performing in vitro MTT assay for anti-cancer evaluation of our synthesized derivatives.
Author Contributions
A.P.G.N. research guide; S.V.T.performed the experiments and wrote the paper; S.S. performed the experiments; J.A.S. and M.P.V.-T. co-operated for spectral analysis; A.P.S. and D.K.L. performed molecular docking study.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Patrick G.L. An Introduction to Medicinal Chemistry. 4th ed. Oxford University Press Inc.; New York, NY, USA: 2009. p. 519. [Google Scholar]
- 2.Siegel R., Naishadham D., Jemal A. Cancer statistics. CA Cancer J. Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Hidalgo M., Gail Eckhardt S. Development of Matrix Metalloproteinase Inhibitors in Cancer Therapy. J. Natl. Cancer Inst. 2001;93:178–193. doi: 10.1093/jnci/93.3.178. [DOI] [PubMed] [Google Scholar]
- 5.Chambers A.F., Matrisian L. Changing views of the role of matrix metalloproteinases in metastasis. J. Natl. Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 6.Kahari V.M., Saarialho-Kere U. Matrix metalloproteinases and their inhibitors in tumour growth and invasion. Ann. Med. 1999;31:34–45. doi: 10.3109/07853899909019260. [DOI] [PubMed] [Google Scholar]
- 7.Ray J.M., Stetler-Stevenson W.G. Gelatinase A activity directly modulates melanoma cell adhesion and spreading. EMBO J. 1995;14:908–917. doi: 10.1002/j.1460-2075.1995.tb07072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleiner D.E., Stetler-Stevenson W.G. Matrix metalloproteinases and metastasis. Cancer Chemother. Pharmacol. 1999;43:S42–S51. doi: 10.1007/s002800051097. [DOI] [PubMed] [Google Scholar]
- 9.Denis L.J., Verweij J. Matrix metalloproteinase inhibitors: Present achievements and future prospects. Investig. New Drugs. 1997;15:175–185. doi: 10.1023/A:1005855905442. [DOI] [PubMed] [Google Scholar]
- 10.Wojtowicz-Praga S.M., Dickson R.B., Hawkins M.J. Matrix metalloproteinase inhibitors. Investig. New Drugs. 1997;15:61–75. doi: 10.1023/A:1005722729132. [DOI] [PubMed] [Google Scholar]
- 11.Brown P.D. Clinical studies with matrix metalloproteinase inhibitors. APMIS. 1999;107:174–180. doi: 10.1111/j.1699-0463.1999.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaplancikli Z.A., Altıntop M.D., Atli O., Sever B., Baysal M., Temel H.E., Demirci F., Ozdemir A. Synthesis and Evaluation of A New Series of Thiazole Derivatives as Potential Antitumor Agents and MMP Inhibitors. Anti-Cancer Agents Med. Chem. 2017;17:674–681. doi: 10.2174/1871520616666160802113620. [DOI] [PubMed] [Google Scholar]
- 13.Zou X.J., Zhang S.W., Liu Y., Liu Z.M., Gao J.W., Song Q.L., Pan Y., Zhang J.Z., Li X.J. Anti-tumor metastasis and Crystal Structure of N1-(1,3,4-thiadiazole-2-yl)-N3-m-chlorobenzoyl-urea. Chin. J. Struct. Chem. 2011;30:1001–1005. [Google Scholar]
- 14.Du H.T., Du H.J. Synthesis and Biological Activity of 6-(Substituted)-3-(3,4,5-trimethoxyphenyl)-1,2,4-triazolo[3,4-b][1,3,4]thiadiazole. Chin. J. Org. Chem. 2010;30:137–141. [Google Scholar]
- 15.Liu F., Luo X.Q., Song B.A., Bhadury P.S., Yang S., Jin L.H., Xue W., Hu D.Y. Synthesis and antifungal activity of novel sulfoxide derivatives containing trimethoxyphenyl substituted 1,3,4-thiadiazole and 1,3,4-oxadiazole moiety. Bioorg. Med. Chem. 2008;16:3632–3640. doi: 10.1016/j.bmc.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Matwijczuk A., Kluczyk D., Gorecki A., Niewiadomy A., Gagos M. Spectroscopic Studies of Fluorescence Effects in bioactive 4-(5-heptyl-1,3,4-thiadiazol-2-yl)benzene-1,3-diol and 4-(5-methyl-1,3,4-thiadiazol-2-yl)benzene-1,3-diol molecules Induced by pH Changes in Aqueous Solutions. J. Fluoresc. 2017:1–12. doi: 10.1007/s10895-017-2053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matwijczuk A., Kluczyk D., Górecki A., Niewiadomy A., Gagos M. Solvent Effects on Molecular Aggregation in 4-(5-Heptyl-1,3,4-thiadiazol-2-yl)benzene-1,3-diol and 4-(5-Methyl-1,3,4-thiadiazol-2-yl)benzene-1,3-diol. J. Phys. Chem. B. 2016;120:7958–7969. doi: 10.1021/acs.jpcb.6b06323. [DOI] [PubMed] [Google Scholar]
- 18.Kluczyk D., Matwijczuk A., Gorecki A., Karpinska M.M., Szymanek M., Niewiadomy A., Gagos M. Molecular Organization of Dipalmitoylphosphatidylcholine Bilayers Containing Bioactive Compounds 4-(5-Heptyl-1,3,4-thiadiazol-2-yl) Benzene-1,3-diol and 4-(5-Methyl-1,3,4-thiadiazol-2-yl) Benzene-1,3-diols. J. Phys. Chem. B. 2016;120:12047–12063. doi: 10.1021/acs.jpcb.6b09371. [DOI] [PubMed] [Google Scholar]
- 19.Matwijczuk A., Kamiński D., Górecki A., Ludwiczuk A., Niewiadomy A., Maćkowski S., Gagos M. Spectroscopic Studies of Dual Fluorescence in 2-((4-Fluorophenyl)amino)-5-(2,4-dihydroxybenzeno)-1,3,4-thiadiazole. J. Phys. Chem. A. 2015;119:10791–10805. doi: 10.1021/acs.jpca.5b06475. [DOI] [PubMed] [Google Scholar]
- 20.Karcz D., Matwijczuk A., Boroń B., Creaven B., Fiedor L., Niewiadomy A., Gagoś M. Isolation and spectroscopic characterization of Zn(II), Cu(II), and Pd(II) complexes of 1,3,4-thiadiazole-derived ligand. J. Mol. Struc. 2017;1128:44–50. [Google Scholar]
- 21.Shrivastava K., Purohit S., Singhal S. Studies of nitrogen and sulphur containing heterocyclic compound: 1,3,4-Thiadiazole. Asian J. Biomed. Pharm. Sci. 2013;3:6–23. [Google Scholar]
- 22.Siddiqui N., Ahujaa P., Ahsana W., Pandey S.N., Alama M.S. Thiadiazoles: Progress Report on Biological Activities. J. Chem. Pharm. Res. 2009;1:19–30. [Google Scholar]
- 23.Chaudhary D.K., Chaudhary R.P. Pharmacological Activities of 1,3,4 Thiadiazole Derivatives Review. Int. J. Pharm. Biol. Sci. Arch. 2013;4:256–264. [Google Scholar]
- 24.Chhajed M., Shrivastava A.K., Taile V. Synthesis of 5-arylidine amino-1,3,4-thiadiazol-2-[(N-substituted benzyol)]sulphonamides endowed with potent antioxidants and anticancer activity induces growth inhibition in HEK293, BT474 and NCI-H226 cells. Med. Chem. Res. 2014;23:3049–3064. doi: 10.1007/s00044-013-0890-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen E.J., Mitchell M.A., Hendges S.K., Belonga K.L., Skaletzky L.L., Stelzer L.S., Lindberg T.J., Fritzen E.L., Schostarez H.J., OSullivan T.J., et al. Synthesis of a series of stromelysin-selective thiadiazole urea matrix metalloproteinase inhibitors. J. Med. Chem. 1999;42:1525–1536. doi: 10.1021/jm9803222. [DOI] [PubMed] [Google Scholar]
- 26.Foda H.D., Zucker S. Matrix metalloproteinases in cancer invasion, metastasis and angiogenesis. Drug Discov. Today. 2001;6:478–482. doi: 10.1016/S1359-6446(01)01752-4. [DOI] [PubMed] [Google Scholar]
- 27.Coussens L.M., Fingleton B., Matrisian L.M. Matrix metalloproteinase inhibitors and cancer: Trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 28.Caddick S. Microwave Assisted Organic Reactions. Tetrahedron. 1995;5:10403–10432. doi: 10.1016/0040-4020(95)00662-R. [DOI] [Google Scholar]
- 29.Finzel B.C., Baldwin E.T., Bryant G.L., Hess G.F., Wilks J.W., Trepod C.M., Mott J.E., Marshall V.P., Petzold G.L., Poorman R.A., et al. Structural characterizations of nonpeptidic thiadiazole inhibitors of matrix metalloproteinases reveal the basis for stromelysin selectivity. Protein Sci. 1998;7:2118–2126. doi: 10.1002/pro.5560071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hedley P.L., Jorgensen P., Schlamowitz S., Wangari R., Moolman-Smook J., Brink P.A., Kanters J.K., Corfield V.A., Christiansen M. The genetic basis of long QT and short QT syndromes: A mutation update. Hum. Mutat. 2009;30:1486–1511. doi: 10.1002/humu.21106. [DOI] [PubMed] [Google Scholar]
- 31.Vandenberg J.I., Walker B.D., Campbell T.J. HERG K+ channels: Friend and foe. Trends Pharmacol. Sci. 2001;22:240–246. doi: 10.1016/S0165-6147(00)01662-X. [DOI] [PubMed] [Google Scholar]
- 32.Chiesa N., Rosati B., Arcangeli A., Olivotto M., Wanke E. A Novel Role for HERG K+Channels: Spike-Frequency Adaptation. J. Physiol. 1997;501:313–318. doi: 10.1111/j.1469-7793.1997.313bn.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aronov A.M. Predictive in silico modeling for HERG channel blockers. Drug Discov. Today. 2005;10:149–155. doi: 10.1016/S1359-6446(04)03278-7. [DOI] [PubMed] [Google Scholar]
- 34.Furniss B.S., Hannaford A.J., Smith P.W., Vogel A.I. Vogel’s textbook of practical organic chemistry. Longman Sci. Tech. 1989;5:1056. [Google Scholar]
- 35.Tomi I.H.R., Al-Daraji A.H.R., Al-Qaysi R.R.T., Hasson M.M., Al-Dulaimy K.H.D. Synthesis, characterization and biological activities of some azo derivatives of aminothiadiazole derived from nicotinic and isonicotinic acids. Arab. J. Chem. 2010;3:687–694. doi: 10.1016/j.arabjc.2010.12.003. [DOI] [Google Scholar]
- 36.Narayanan Moorthy N.S.H., Vittal U.B., Karthikeyan C., Thangapandian V., Venkadachallam A.P., Trivedi P. Synthesis, antifungal evaluation and in silico study of novel Schiff bases derived from 4-amino-5(3,5-dimethoxy-phenyl)-4H-1,2,4-triazol-3-thiol. Arab. J. Chem. 2014:244–252. doi: 10.1016/j.arabjc.2013.12.021. [DOI] [Google Scholar]
- 37.Miglani S., Mishra M., Chawla P. The rapid synthesis of schiff-bases without solvent under microwave irradiation and their antimicrobial activity. Der Pharm. Chem. 2012;4:2265–2269. [Google Scholar]
- 38.Mosman T.J. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 39.Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R. Feasibility of Drug Screening with Panels of Human Tumor Cell Lines Using a Microculture Tetrazolium Assay. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
- 40.Ertl P., Rohde B., Selzer P. Fast Calculation of Molecular Polar Surface Area as a Sum of Fragment-Based Contributions and Its Application to the Prediction of Drug Transport Properties. J. Med. Chem. 2000;43:3714–3717. doi: 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- 41.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]