Abstract
A series of indeno[1,2-c]quinoline derivatives were designed, synthesized and evaluated for their anti-tuberculosis (anti-TB) and anti-inflammatory activities. The minimum inhibitory concentration (MIC) of the newly synthesized compound was tested against Mycobacterium tuberculosis H37RV. Among the tested compounds, (E)-N′-[6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene]isonicotino-hydrazide (12), exhibited significant activities against the growth of M. tuberculosis (MIC values of 0.96 μg/mL) with a potency approximately equal to that of isoniazid (INH), an anti-TB drug. Important structure features were analyzed by quantitative structure–activity relationship (QSAR) analysis to give better insights into the structure determinants for predicting the anti-TB activity. The anti-inflammatory activity was induced by superoxide anion generation and neutrophil elastase (NE) release using the formyl-l-methionyl-l-leucyl-l-phenylalanine (fMLF)-activated human neutrophils method. Results indicated that compound 12 demonstrated a potent dual inhibitory effect on NE release and superoxide anion generation with IC50 values of 1.76 and 1.72 μM, respectively. Our results indicated that compound 12 is a potential lead compound for the discovery of dual anti-TB and anti-inflammatory drug candidates. In addition, 6-[3-(hydroxymethyl)piperidin-1-yl]-9-methoxy-11H-indeno[1,2-c]quinolin-11-one (4g) showed a potent dual inhibitory effect on NE release and superoxide anion generation with IC50 values of 0.46 and 0.68 μM, respectively, and is a potential lead compound for the discovery of anti-inflammatory drug candidates.
Keywords: indeno[1,2-c]quinoline; antimycobacterial activity; anti-inflammatory activity
1. Introduction
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis which primarily affects the lungs, causing an intense local inflammatory response that is critical to the pathogenesis of tuberculosis [1,2]. Therefore, TB is a chronic inflammatory condition in which regulatory and pro-inflammatory processes occur that contribute to severe lung pathology, leading to lung tissue necrosis and the promotion of mycobacterial dissemination and transmission [3]. Anti-inflammatory drugs, especially corticosteroids, are currently used for adjunctive therapy in most severe life-threatening forms of tuberculosis, such as meningitis and pericarditis [4]. The importance of developing new drugs with dual anti-inflammatory and antimycobacterial activities is amplified by the emergence of multi-drug resistant (MDR) strains and the global human immunodeficiency virus (HIV) pandemic [5,6,7]. The anti-leprosy drug clofazimine [8] exhibited dual anti-inflammatory and antimycobacterial activities. However, its potency is not sufficient, motivating the design and search for novel agents.
Quinoline-based anti-TB compound bedaquiline is a highly potent anti-TB agent, has a novel mode of action and was recently approved for the treatment of multidrug-resistant tuberculosis (MDR-TB) [9]. Currently there is considerable interest in the synthesis of indeno[2,1-c]quinoline derivatives, because of their antimycobacterial activities [10,11,12]. However, the isomeric indeno[1,2-c]quinoline derivatives have attracted only limited attention. Over the past few years, we have been particularly interested in the synthesis of fluoroquinolone and benzofuroquinoline derivatives for anti-TB evaluations [13,14,15,16,17]. We have also synthesized certain polycyclic heterocycles such as 9-anilinoacridine, 9-phenoxyacridine, 4-phenoxyfuro[2,3-b]quinoline, quinolin-2(1H)-one, furo[3,2:3,4]naphtha[1,2-d]imidazole and benzo[f]indole-4,9-dione derivatives for the evaluation of their anti-inflammatory activities [18,19,20,21,22,23,24,25,26,27,28]. In continuation of our efforts to identify potential anti-TB agents with novel types of structures which are distinct from those of existing drugs, we herein describe the synthesis of certain indeno[1,2-c]quinoline derivatives for anti-inflammatory and anti-TB evaluations. The aim of our current study is to identify novel skeletons that exhibit dual anti-inflammatory and antimycobacterial activities with low cytotoxicities.
2. Results and Discussion
2.1. Chemistry
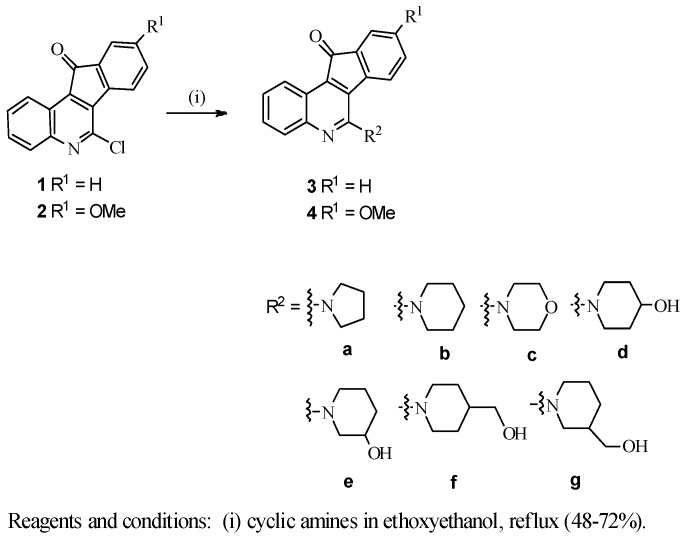
According to our previously reported procedures [29], 2-hydroxy-3-phenylquinoline-4-carboxylic acid was treated with POCl3 to afford 6-chloro-11H-indeno[1,2-c]quinolin-11-one (1), which was then reacted with cyclic amines to give 6-cycloamino-11H-indeno[1,2-c]quinolin-11-ones (3a–3g). Accordingly, compounds 4a–4g were prepared from the known 6-chloro-9-methoxy-11H-indeno [1,2-c]quinolin-11-one (2) [29] under similar reaction conditions as outlined in Scheme 1.
Scheme 1.
Synthesis of indeno[1,2-c]quinoline derivatives 3a–4g.
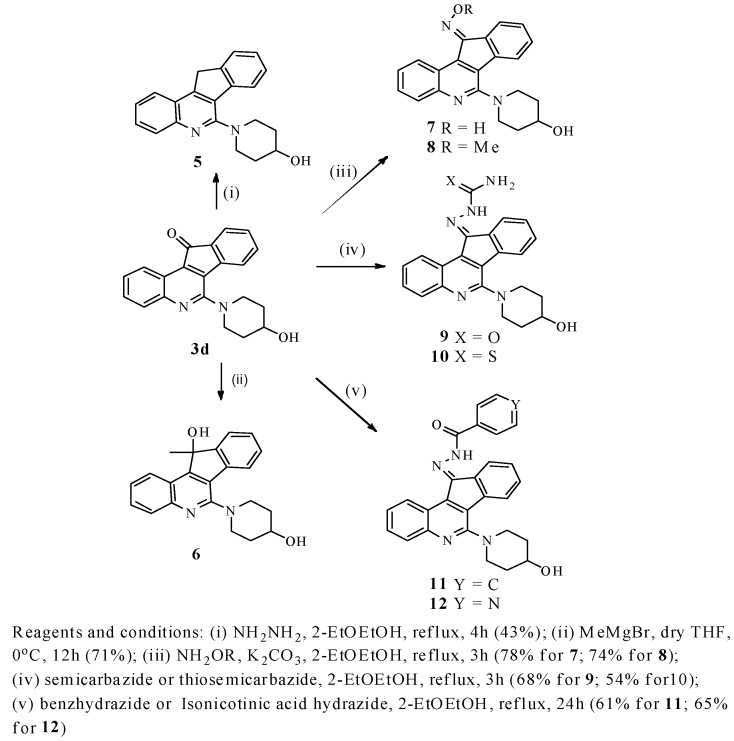
Among these compounds, 6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3d) exhibited the most potent activity against the growth of Mycobacterium tuberculosis (minimum inhibitory concentration (MIC) value of 2.09 μg/mL), and therefore, we decided to select 3d as a lead compound for structural optimization as described in Scheme 2. The Wolff–Kishner reduction of 3d with hydrazine afforded 1-(11H-indeno[1,2-c]quinolin-6-yl)piperidin-4-ol (5) in 43% yield. Treatment of 3d with methyl magnesium bromide (Grignard reagent) afforded 6-(4-hydroxypiperidin-1-yl)-11-methyl-11H-indeno[1,2-c]quinolin-11-ol (6) in 71% yield. (E)-6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one oxime (7) and (E)-6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one O-methyl oxime (8) were obtained from 3d by the treatment with NH2OH and NH2OMe, respectively. (E)-2-[6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene]hydrazinecarboxamide (9) and (E)-2-(6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene)hydrazinecarbothioamide (10) were obtained from 3d by the treatment with semicarbazide and thiosemicarbazide, respectively. (E)-N′-[6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene]benzohydrazide (11) and (E)-N′-(6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene)isonicotinohydrazide (12) were obtained from 3d by the treatment with benhydrazide and isonicotinic acid hydrazide, respectively.
Scheme 2.
Synthesis of indeno[1,2-c]quinoline derivatives 5–12.
2.2. Results and Discussion
The anti-TB activities of indeno[1,2-c]quinoline derivatives are summarized in Table 1. The 6-Cycloamino-11H-indeno[1,2-c]quinolin-11-ones (3a–3g) were found to be significantly inhibit the growth of M. tuberculosis, with MICs ranging from 2.09 to 4.97 μg/mL with >87% survival rate of Vero cells at a concentration of 20 μg/mL. Among them, 6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3d) was the most active, with MIC of 2.09 μg/mL. However, their respective 9-methoxy counterparts 4a–4g were inactive (MIC > 17 μg/mL) with an exception of 9-methoxy-6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one (4d) which exhibited the MIC of 7.74 μg/mL. Further structural derivatization of 3d resulted in two potential anti-TB agents, (E)-N′-[6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene]benzohydrazide (11) and (E)-N′-(6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene)isonicotinohydrazide (12) with MIC of 1.98 and 0.96 μg/mL. The anti-TB activity of 12 is equally active to the positive isoniazid (INH) with less cytotoxicity to the Vero cell. In addition, our results indicated that it is a feasible approach for the discovery of potential anti-TB agents through condensation of INH with an appropriate ketone.
Table 1.
Anti-Mycobacterium tuberculosis H37Rv activities of indeno[1,2-c]quinoline derivatives. Results expressed as minimum inhibitory concentration (MIC) compared to isoniazid (INH).
| Compounds | R1 | R2 | MIC (μg/ mL) | Survival Rate of Vero Cells @ 20 μg/mL (%) |
|---|---|---|---|---|
| 3a | H | 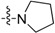 |
4.56 | 88.61 |
| 3b | H | 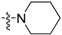 |
4.93 | 96.18 |
| 3c | H | 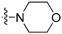 |
3.12 | 91.19 |
| 3d | H | 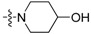 |
2.09 | 91.23 |
| 3e | H | 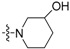 |
4.97 | 87.78 |
| 3f | H | 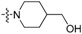 |
3.98 | 91.11 |
| 3g | H | 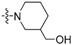 |
3.07 | 92.25 |
| 4a | OMe |  |
17 | 89.62 |
| 4b | OMe |  |
>20 | 99.55 |
| 4c | OMe |  |
>20 | 94.47 |
| 4d | OMe |  |
7.74 | 84.72 |
| 4e | OMe |  |
19.10 | 87.59 |
| 4f | OMe |  |
>20 | 90.26 |
| 4g | OMe |  |
>20 | 87.60 |
| 5 | 18.0 | 85.41 | ||
| 6 | >20 | 89.83 | ||
| 7 | >20 | 84.14 | ||
| 8 | >20 | 91.48 | ||
| 9 | >20 | 81.20 | ||
| 10 | >20 | 80.28 | ||
| 11 | 1.98 | 89.55 | ||
| 12 | 0.96 | 94.25 | ||
| INH | 0.8–1.2 | 84.80 | ||
These indeno[1,2-c]quinolin-11-one derivatives were also evaluated for their inhibitory effects on superoxide anion generation and neutrophil elastase (NE) release in formyl-l-methionyl-l-leucyl-l-phenylalanine (fMLF)-activated human neutrophils and results are shown in Table 2. Compounds 3a–3c were inactive while 3d–3g exhibited a potent dual inhibitory effect on neutrophil elastase (NE) release and superoxide anion generation with IC50 value in a range of 1.78 and 4.50 μM. These results indicated that the hydroxyl group substituted at the C6-piperidine ring is crucial for anti-inflammatory activities. Among them, 6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3d) was the most potent dual inhibitor on NE release and superoxide anion generation with IC50 values of 2.20 and 1.78 μM, respectively.
Table 2.
Anti-inflammatory activities of indeno[1,2-c]quinoline derivatives in formyl-Lmethionyl-l-leucyl-l-phenylalanine (fMLF)-activated human neutrophils (IC50 in μM) a.
| Compounds | Superoxide Anion | Elastase Release |
|---|---|---|
| 3a | >10 | >10 |
| 3b | >10 | >10 |
| 3c | >10 | >10 |
| 3d | 1.78 ± 0.44 | 2.20 ± 0.69 |
| 3e | 2.90 ± 0.11 | 2.80 ± 0.09 |
| 3f | 3.37 ± 0.17 | 4.50 ± 1.32 |
| 3g | 2.77 ± 0.47 | 2.12 ± 1.19 |
| 4a | >10 | >10 |
| 4b | >10 | >10 |
| 4c | >10 | >10 |
| 4d | 2.82 ± 0.30 | 2.05 ± 0.24 |
| 4e | 1.89 ± 0.65 | 2.93 ± 0.70 |
| 4f | 1.93 ± 0.92 | 0.88 ± 0.30 |
| 4g | 0.68 ± 0.14 | 0.46 ± 0.08 |
| 5 | 4.02 ± 1.08 | 5.29 ± 0.48 |
| 6 | >10 | >10 |
| 7 | 0.83 ± 0.47 | 1.39 ± 0.64 |
| 8 | 2.68 ± 0.95 | 2.13 ± 0.92 |
| 9 | 3.64 ± 1.43 | >10 |
| 10 | 2.41 ± 0.36 | >10 |
| 11 | 1.67 ± 0.87 | >10 |
| 12 | 1.72 ± 0.23 | 1.76 ± 0.49 |
| LY294002 b | 1.36 ± 0.33 | 2.21 ± 0.45 |
a Concentration necessary for 50% inhibition (IC50); Results are presented as mean ± SEM (n = 3). b LY294002 (a phosphatidylinositol-3-kinase inhibitor) was used as a positive control for superoxide anion generation and elastase release.
The same structure–activity relationships were observed for C9-methoxy derivatives in which compounds 4a–4c were inactive, while their respective C6-hydroxypiperidine derivative 4d–4g exhibited a potent dual inhibitory effect on NE release and superoxide anion generation. Among them, 9-methoxy-6-(3-piperidinemethano-1-yl)-11H-indeno[1,2-c]quinolin-11-one (4g) was the most potent dual inhibitor of NE release and superoxide anion generation, with IC50 values of 0.46 and 0.68 μM, respectively. Compound 5 was only weakly active, while compound 6 was inactive indicated the reduction of the keto group is unfavorable. The keto group of 3d can be converted into its oxime derivatives, compounds 7 and 8, and retain comparable anti-inflammatory activity. However, the semicarbazone derivatives, compounds 9 and 10, selectively inhibit superoxide anion generation with IC50 values of 3.64 and 2.41 μM, respectively. The benzohydrazide derivative 11 was capable of inhibiting superoxide anion generation with IC50 value of 1.67 μM but was inactive in the inhibition of NE release. (E)-N′-(6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene)isonicotino hydrazide (12) was found to be a potent dual inhibitor of NE release and superoxide anion generation, with IC50 values of 1.76 and 1.72 μM, respectively, approximately equal potent to the positive LY294002.
To provide a better understanding of the relationship between the structure descriptors and anti-TB activities, a quantitative structure–activity relationship (QSAR) model was constructed to analyze important features and evaluate prediction performance. First, a total of 13 chemicals with determined MIC values as shown in Table 1 were randomly divided into a training dataset for model construction and a test dataset for independent test of the constructed model. The training and test datasets consist of 9 (12, 11, 3g, 3c, 3e, 3a, 4d, 4a and 5) and 4 (3d, 3f, 3b and 4e) chemicals, respectively. Subsequently, each chemical was transformed into a vector of 1648 descriptors representing its structure features. The sequential feature selection algorithms developed by our group [30,31] were applied to simultaneously select a minimum number of relevant descriptors maximizing the R2 performance and develop multiple regression models for QSAR analysis. Please note that only the three most important features were considered due to the small number of chemicals in the dataset. In the QSAR analysis, pMIC values were utilized in that the MIC values in μg/mL were converted toμM/mL and then converted to −log MIC (pMIC). The model is expected to be useful for chemicals within the pMIC range from 1.24 to 2.67. The final QSAR model with high performance with R2 = 0.96, R2adj = 0.94, root-mean-squared error (RMSE) = 0.085, and Q2 = 0.86 could well explain the variation of pMIC values representing the anti-TB activity. The constructed QSAR model and three important descriptors are shown in Table 3. SCH-6 (simple chain, order 6) epresents the Chi chain descriptor of order 6 [32] that directly correlates with pMIC values with a high correlation coefficient of 0.83 for the 13 chemicals. CrippenLogP is the Crippen’s LogP descriptor [33]. The hmax (representing the maximum hydrogen E-State value in a molecule) belongs to the descriptors of atom type electrotopological state [34,35,36].
Table 3.
The quantitative structure–activity relationship (QSAR) analysis result for pMIC values of anti-tuberculosis (TB) activity. hmax.
| Coefficient | Estimate Std. Error | t-Value | Pr (>|t|) | ||
|---|---|---|---|---|---|
| (Intercept) | 0.5035 | 1.1585 | 0.435 | 0.681929 | |
| SCH-6 | 11.463 | 1.0533 | 10.883 | 0.000114 | *** |
| CrippenLogP | −0.5711 | 0.1296 | −4.407 | 0.006977 | ** |
| hmax | −3.8978 | 1.4043 | −2.776 | 0.039106 | * |
*** p < 0.001, ** p < 0.01, * p < 0.05
While CrippenLogP and hmax show only a mild negative correlation to pMIC values with correlation coefficient values of −0.13 and −0.01, respectively, for the 13 chemicals, the combination of all three descriptors largely enhances the prediction performance of the QSAR model with a 15% improvement on R2. The observed and predicted pMIC values on the training dataset are shown in Table 4. To further evaluate the prediction ability of the QSAR model, the model was applied to predict the 4 chemicals in the test dataset. As shown in Table 5, the predicted and observed values for chemicals with exact MIC values are well correlated with a high correlation coefficient of 0.98. Altogether, the good performance of the QSAR models shows the importance of the three descriptors for determining the anti-TB activity. The model could be further tested and improved when more data are available.
Table 4.
The comparison of observed and predicted values for the training dataset.
| Compound | Observed pMIC | Predicted pMIC | Error |
|---|---|---|---|
| 12 | 2.67 | 2.64 | 0.03 |
| 11 | 2.35 | 2.37 | −0.02 |
| 3g | 2.05 | 1.86 | 0.19 |
| 3c | 2.01 | 2.08 | −0.08 |
| 3e | 1.82 | 1.80 | 0.02 |
| 3a | 1.82 | 1.88 | −0.06 |
| 4d | 1.67 | 1.73 | −0.06 |
| 4a | 1.29 | 1.38 | −0.10 |
| 5 | 1.24 | 1.18 | 0.07 |
Table 5.
The comparison of observed and predicted values for the test dataset.
| Compound | Observed pMIC | Predicted pMIC | Error |
|---|---|---|---|
| 3d | 2.20 | 2.00 | 0.20 |
| 3f | 1.94 | 1.95 | −0.01 |
| 3b | 1.80 | 1.87 | −0.07 |
| 4e | 1.28 | 1.58 | −0.30 |
3. Experimental Section
3.1. General
Melting points were determined on a Electrothermal IA9100 melting point apparatus and are uncorrected. Nuclear magnetic resonance (1H and 13C) spectra were recorded on a Varian Gemini 200 spectrometer or Varian-Unity-400 spectrometer. Chemical shifts were expressed in parts per million (δ) with tetramethylsilane (TMS) as an internal standard and coupling constant (J) in hertz (Hz). Thin-layer chromatography was performed on silica gel 60 F-254 plates purchased from E. Merck and Co. IR spectra were measured using a FTIR Perkin-Elmer system-2000 spectrometer (Perkin-Elmer, Waltham, MA, USA). The elemental analyses were performed in the Instrument Center of National Science Council at National Cheng-Kung University using Heraeus CHN-O Rapid EA (Heraeus Co., Harau, Germany), and all values are within ± 0.4% of the theoretical compositions.
3.2. General Procedure for the Preparation of 6-Substituted-11H-indeno[1,2-c]quinolin-11-one Compounds 3a–g, 4a, 4d–g
A mixture of 6-chloro-11H-indeno[1,2-c]quinolin-11-one (1) or 6-chloro-9-methoxy-11H-indeno [1,2-c]quinolin-11-one (2), cyclic amine (3.0 mmol), and 2-ethoxyethanol (30 mL) was heated at reflux for 3–6 h (TLC monitoring). The solvent was removed in vacuo and the residue was suspended in H2O (20 mL). The resulting precipitate that separated was collected, washed with H2O, and dried to give a crude solid. The crude product was purified by crystallization from EtOH to afford compounds 3a–g, 4a, 4d–g. Preparation of compounds 4b and 4c had been described in our previous report [29].
3.2.1. 6-(Pyrrolidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3a)
Yield: 78%. m.p.: 163–164 °C. IR (KBr, cm−1): 2956, 1710. 1H-NMR (400 MHz, CDCl3) δ 1.98–2.01 (m, 4H, pyrrolidinyl-H), 3.58–3.62 (m, 4H, pyrrolidinyl-H), 7.23–7.27 (m, 1H, 9-H), 7.35–7.39 (m, 1H, 8-H), 7.44–7.50 (m, 2H, 3-, 10-H), 7.53–7.57 (m, 1H, 2-H), 7.64 (d, 1H, J = 7.6 Hz, 7-H), 7.78 (d, 1H, J = 8.8 Hz, 4-H), 8.71 (dd, 1H, J = 1.6, 8.8 Hz, 1-H). 13C-NMR (100 MHz, CDCl3) δ 24.93 (2C), 50.16 (2C), 120.10, 123.63, 123.97, 124.24, 125.66, 127.23, 128.29, 129.85, 130.60, 133.22, 134.52, 136.17, 143.95, 149.18, 155.44, 195.74. Anal. calcd. for C20H16N2O: C 79.98, H 5.37, N 9.33; found: C 79.93, H 5.39, N 9.28.
3.2.2. 6-(Piperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3b)
Yield: 75%. m.p.: 165–166 °C. IR (KBr, cm−1): 2934, 1715. 1H-NMR (400 MHz, CDCl3) δ 1.71 (br s, 2H, piperidinyl-H), 1.83–1.88 (m, 4H, piperidinyl-H), 3.34 (br s, 4H, piperidinyl-H), 7.26–7.30 (m, 1H, 9-H), 7.42–7.46 (m, 1H, 8-H), 7.48–7.52 (m, 1H, 3-H), 7.56–7.60 (m, 1H, 2-H), 7.64 (d, 1H, J = 7.2 Hz, 10-H), 7.72 (d, 1H, J = 7.2 Hz, 7-H), 7.84 (d, 1H, J = 8.4 Hz, 4-H), 8.74 (dd, 1H, J = 1.2, 8.0 Hz, 1-H). 13C-NMR (100 MHz, CDCl3) δ 24.23, 25.87 (2C), 51.10 (2C), 120.67, 123.22, 123.99, 124.19, 126.60, 127.84, 128.70, 129.77, 132.31, 133.07, 134.72, 136.16, 143.59, 149.27, 158.27, 195.64. Anal. calcd. for C21H18N2O∙0.1 H2O: C 79.77, H 5.80, N 8.86; found: C 79.77, H 5.74, N 8.75.
3.2.3. 6-(Morpholino-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3c)
Yield: 62%. m.p.: 171–173 °C. IR (KBr, cm−1): 2838, 1719. 1H-NMR (400 MHz, CDCl3) δ 3.41–3.44 (m, 4H, morpholinyl-H), 3.98–4.01 (m, 4H, morpholinyl-H), 7.28–7.32 (m, 1H, 9-H), 7.45–7.52 (m, 2H, 3-, 8-H), 7.58–7.63 (m, 1H, 2-H), 7.65–7.68 (m, 2H, 7-, 10- H), 7.86 (ddd, 1H, J = 0.8, 1.2, 8.4 Hz, 4-H), 8.75 (ddd, 1H, J = 0.8, 1.6, 8.4 Hz, 1-H). 13C-NMR (100 MHz, CDCl3) δ 50.24 (2C), 66.79 (2C), 120.89, 123.18, 124.03, 124.48, 127.04, 127.98, 128.92, 130.02, 131.67, 133.13, 134.80, 136.44, 143.06, 149.16, 157.04, 195.24. Anal. calcd. for C20H16N2O2: C 75.93, H 5.10, N, 8.86; found: C 76.33, H 5.15, N, 8.82.
3.2.4. 6-(4-Hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3d)
Yield: 68%. m.p.: 208-208 °C. IR (KBr, cm−1): 3542, 2919, 1710. 1H-NMR (400 MHz, CDCl3) δ 1.83–1.92 (m, 2H, piperidinyl-H), 2.13–2.17 (m, 2H, piperidinyl-H), 3.16–3.18 (m, 2H, piperidinyl-H), 3.70–3.75 (m, 2H, piperidinyl-H), 3.93–4.02 (m, 1H, piperidinyl-H), 7.27–7.31 (m, 1H, 9-H), 7.43–7.52 (m, 1H, 3-, 8-H), 7.57–7.61 (m, 1H, 2-H), 7.64–7.70 (m, 2H, 7-, 10- H), 7.86 (d, 1H, J = 8.4 Hz, 4-H), 8.73 (dd, 1H, J = 0.8, 8.4 Hz, 1-H). 13C-NMR (100 MHz, CDCl3) δ 34.37 (2C), 47.81 (2C), 67.90, 120.76, 123.06, 124.01, 124.36, 126.85, 127.76, 128.87, 129.96, 132.08, 133.02, 134.82 ,136.34, 143.32, 149.02, 157.35, 195.41. Anal. calcd. for C21H18N2O2: C 76.34, H 5.49, N 8.48; found: C 76.09, H 5.56, N 8.36.
3.2.5. 6-(3-Hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3e)
Yield: 62%. m.p.: 211–212 °C. IR (KBr, cm−1): 3383, 2932, 1717. 1H-NMR (400 MHz, DMSO-d6) δ 1.37–1.44 (m, 1H, piperidinyl-H), 1.71–1.80 (m, 1H, piperidinyl-H), 1.88–1.92 (m, 1H, piperidinyl-H), 1.99–2.01 (m, 1H, piperidinyl-H), 2.80 (br s, 1H, piperidinyl-H), 2.92 (br s, 1H, piperidinyl-H), 3.48 (d, 1H, J = 12.8 Hz, piperidinyl-H), 3.66 (d, 1H, J = 12.4 Hz, piperidinyl-H), 3.85 (br s, 1H, piperidinyl-H), 4.99 (br s, 1H, -OH), 7.37–7.41 (m, 1H, 9-H), 7.48–7.52 (m, 1H, 8-H), 7.61–7.70 (m, 4H, 2-, 3-, 7-, 10-H), 7.80 (d, 1H, J = 8.4 Hz, 4-H), 8.58 (dd, 1H, J = 0.8, 8.0 Hz, 1-H). 13C-NMR (100 MHz, DMSO-d6) δ 22.65, 32.81, 49.87, 56.95, 65.55, 119.89, 123.24, 123.49, 124.13, 126.77, 127.58, 129.22, 130.12, 131.57, 132.31, 135.47 ,135.54 , 142.55, 148.44, 157.10, 194.83. Anal. calcd. for C21H18N2O2∙1.2H2O: C 71.65, H 5.84, N 7.96; found: C 71.49, H 5.77, N 7.90.
3.2.6. 6-(4-Piperidinemethano-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3f)
Yield: 64%. m.p.: 223–224 °C. IR (KBr, cm−1): 3394, 2928, 1712. 1H-NMR(400 MHz, CDCl3) δ 1.58–1.62 (m, 4H, piperidinyl-H), 1.80 (br s, 1H, OH), 1.97 (d, 2H, J = 11.2 Hz, piperidinyl-H), 2.99 (m, 2H, piperidinyl-H), 3.66 (d, 2H, J = 6.4 Hz, 4′-CH2OH), 3.83 (d, 2H, J = 13.2Hz, piperidinyl-H), 7.27–7.31 (m, 1H, 9-H), 7.43–7.52 (m, 2H, 3-, 8-H), 7.56–7.61 (m, 1H, 2-H), 7.64–7.69 (m, 2H, 7-, 10-H), 7.85 (d, 1H, J = 8.4 Hz, 4-H), 8.74 (dd, 1H, J = 1.6, 8.0 Hz, 1-H). 13C-NMR (100 MHz, CDCl3) δ 28.72 (2C), 38.45, 50.09 (2C), 67.79, 120.72, 123.15, 124.00, 124.27, 126.71, 127.82, 128.78, 129.85, 132.20, 133.05, 134.76, 136.21, 143.46, 149.20, 157.10, 195.55. Anal. calcd. for C22H20N2O2∙0.2H2O: C 75.93, H 5.91, N 8.05; found: C 76.13, H 5.91, N 7.86.
3.2.7. 6-(3-Piperidinemethano-1-yl)-11H-indeno[1,2-c]quinolin-11-one (3g)
Yield: 54%. m.p.: 220–221 °C. IR (KBr, cm−1): 3355, 2936, 1715. 1H-NMR (400MHz, CDCl3) δ 1.31–1.40 (m, 1H, piperidinyl-H), 1.75–1.88 (m, 2H, piperidinyl-H), 1.92–1.99 (m, 1H, piperidinyl-H), 2.10–2.18 (m, 1H, piperidinyl-H), 2.52 (br s, 1H, 3′-CH2OH), 3.05–3.20 (m, 2H, piperidinyl-H), 3.59–3.66 (m, 3H, piperidinyl-H), 3.75–3.79 (m, 1H, piperidinyl-H), 7.25–7.29 (m, 1H, 9-H), 7.40–7.44 (m, 1H, 8-H), 7.46–7.51 (m, 1H, 3-H), 7.55–7.59 (m, 1H, 2-H), 7.62–7.65 (m, 2H, 7-, 10-H), 7.81 (d, 1H, J = 8.4 Hz, 4-H), 8.72 (dd, 1H, J = 1.2, 8.8 Hz, 1-H). 13C-NMR (100 MHz, CDCl3) δ 24.43, 26.73, 38.34, 51.58, 51.89, 65.23, 120.51, 123.18, 124.01, 124.28, 126.52, 127.38, 128.73, 129.97, 131.95, 132.97, 134.73, 136.46, 143.42, 149.03, 157.40, 195.46. Anal. calcd. for C22H20N2O2: C 76.72, H 5.85, N 8.13; found: C, 76.66, H 5.86, N 8.00.
3.2.8. 9-Methoxy-6-(pyrrolidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one (4a)
Yield: 79%. m.p.: 155–156 °C. IR (KBr, cm−1): 2934, 1715. 1H-NMR (400 MHz, CDCl3) δ 1.99 (m, 4H, pyrrolidinyl-H), 3.57 (m, 4H, pyrrolidinyl-H), 3.86 (s, 3H, 9-OMe), 6.92 (dd, 1H, J = 2.4, 8.4 Hz, 8-H), 7.20 (d, 1H, J = 2.4 Hz, 10-H), 7.34–7.39 (m, 2H, 2-, 7-H), 7.50–7.54 (m, 1H, 3-H), 7.76 (d, 1H, J = 8.4 Hz, 4-H), 8.66 (dd, 1H, J = 1.6, 8.4 Hz, 1-H). 13C-NMR (100 MHz, CDCl3) δ 24.84 (2C), 49.98 (2C), 55.72, 110.71, 118.59, 120.28, 123.67, 124.60, 125.67, 127.23, 129.33, 131.68, 135.19, 135.75, 135.91, 148.56, 155.25, 160.28, 195.55. Anal. calcd. for C21H18N2O2: C 76.34, H 5.49, N 8.48; found: C 76.04, H 5.56, N 8.38.
3.2.9. 6-(4-Hydroxypiperidin-1-yl)-9-methoxy-11H-indeno[1,2-c]quinolin-11-one (4d)
Yield: 72%. m.p.: 197–198 °C. IR (KBr, cm−1): 3295, 2939, 1713. 1H-NMR (400 MHz, CDCl3) δ 1.81–1.90 (m, 2H, piperidinyl-H), 2.12–2.16 (m, 2H, piperidinyl-H), 3.09–3.15 (m, 2H, piperidinyl-H), 3.67–3.72 (m, 2H, piperidinyl-H), 3.87 (s, 3H, 9-OMe), 3.95–3.99 (m, 1H, piperidinyl-H), 6.94 (dd, 1H, J = 2.4, 8.4 Hz, 8-H), 7.21 (d, 1H, J = 2.4 Hz, 10-H), 7.41–7.45 (m, 1H, 2-H), 7.53–7.57 (m, 2H, 3-, 7-H), 7.81 (d, 1H, J = 8.0 Hz, 4-H), 8.67–8.70 (m, 1H, 1-H). 13C-NMR (100 MHz, CDCl3) δ 34.41 (2C), 47.62 (2C), 55.77, 67.94, 110.78, 118.84, 120.93, 123.70, 124.08, 126.78, 127.86, 129.38, 132.99, 135.04, 135.33, 135.86, 148.59, 157.14, 160.68, 195.30. Anal. calcd. for C22H20N2O3: C 73.32, H 5.59, N 7.77; found: C 73.18, H, 5.80, N 7.52.
3.2.10. 6-(3-Hydroxypiperidin-1-yl)-9-methoxy-11H-indeno[1,2-c]quinolin-11-one (4e)
Yield: 66%. m.p.: 191–192 °C. IR (KBr, cm−1): 3408, 2937, 1714. 1H-NMR (400 MHz, CDCl3) δ 1.58 (br s, 1H, piperidinyl-H), 1.83–1.85 (m, 3H, piperidinyl-H), 3.36–3.52 (m, 3H, piperidinyl-H), 3.83 (s, 3H, 9-OMe), 3.94–3.97 (m, 1H, piperidinyl-H), 4.10 (s, 1H, piperidinyl-H), 6.87–6.92 (m, 1H, 10-H), 7.14–7.17 (m, 1H, 8-H), 7.37–7.44 (m, 2H, 2-, 3-H), 7.51–7.54 (m, 1H, 7-H), 7.74–7.77 (m, 1H, 4-H), 8.61–8.65 (m, 1H, 1-H). 13C-NMR (100 MHz, CDCl3) δ 20.26, 32.35, 51.75, 54.10, 55.70, 65.46, 110.69, 118.87, 120.85, 123.64, 124.29, 126.83, 127.93, 129.78, 133.39, 134.60, 134.80, 134.83, 147.44, 156.07, 160.67, 194.81. Anal. calcd. for C22H20N2O3∙1.5H2O: C 68.20, H 5.99, N 7.23; found: C 68.23, H 5.77, N 7.11.
3.2.11. 6-[4-(Hydroxymethyl)piperidin-1-yl]-9-methoxy-11H-indeno[1,2-c]quinolin-11-one (4f)
Yield: 68%. m.p.: 218–219 °C. IR (KBr, cm−1): 3352, 2928, 1711. 1H-NMR (400 MHz, DMSO-d6) δ 1.40–1.49 (m, 2H, piperidinyl-H), 1.62 (m, 1H, piperidinyl-H), 1.86 (d, 2H, J = 11.2 Hz, piperidinyl-H), 2.81–2.87 (m, 2H, piperidinyl-H), 3.39–3.44 (m, 2H, piperidinyl-H), 3.64 (d, 2H, J = 11.2 Hz, piperidinyl-H), 3.83 (s, 3H, 9-OMe), 4.59 (t, 1H, J = 5.2 Hz, -OH), 7.10–7.13 (m, 2H, 3-, 8-H), 7.44–7.49 (m, 2H, 2-, 7-H), 7.57–7.61 (m, 1H, 10-H), 7.74 (d, 1H, J = 8.0 Hz, 4-H), 8.50 (dd, 1H, J = 0.8, 8.4 Hz, 1-H). 13C-NMR (100 MHz, DMSO-d6) δ 28.49, 38.10, 49.68 (2C), 55.75 (2C), 65.84, 110.79, 118.93, 120.01, 122.92, 124.29, 126.70, 127.58, 129.50, 132.58, 134.31, 134.36, 134.91, 147.83, 157.18, 160.38, 194.52. Anal. calcd. for C23H22N2O3∙0.25H2O: C 72.89, H 6.00, N 7.39; found: C 72.54, H 5.96, N 7.33.
3.2.12. 6-[3-(Hydroxymethyl)piperidin-1-yl]-9-methoxy-11H-indeno[1,2-c]quinolin-11-one (4g)
Yield: 67%. m.p.: 243–244 °C. IR (KBr, cm−1): 3401, 2929, 1714. 1H-NMR (400 MHz, DMSO-d6) δ 1.11–1.17 (m, 1H, piperidinyl-H), 1.78–1.92 (m, 4H, piperidinyl-H), 2.58–2.63 (m, 1H, piperidinyl-H), 2.77–2.82 (m, 1H, piperidinyl-H), 3.28–3.32 (m, 2H, piperidinyl-H), 3.57 (d, 1H, J = 12.4 Hz, piperidinyl-H), 3.75 (d, 1H, J = 11.2 Hz, piperidinyl-H), 3.84 (s, 3H, 9-OMe), 4.58 (t, 1H, J = 4.8 Hz, -OH), 7.10–7.14 (m, 2H, 3-, 8-H), 7.45–7.53 (m, 2H, 2-, 7-H), 7.58–7.62 (m, 1H, 10-H), 7.75–7.77 (m, 1H, 4-H), 8.51–8.54 (m, 1H, 1-H). 13C-NMR (100 MHz, DMSO-d6) δ 24.44, 26.74, 50.61, 53.40, 55.78 (2C), 64.07, 110.79, 119.03, 120.03, 122.95, 124.54, 126.74, 127.60, 129.55, 132.67, 134.35 (2C), 134.97, 147.88, 157.34, 160.40, 194.59. Anal. calcd. for C23H22N2O3∙0.25H2O: C 72.89, H 6.00, N 7.39; found: C 72.94, H 5.96, N 7.40.
3.2.13. 1-(11H-Indeno[1,2-c]quinolin-6-yl)piperidin-4-ol (5)
A mixture of 3d (0.25 g, 1.0 mmol), and 80% hydrazine monohydrate (1 mL) in 2-ethoxyethanol (30 mL) was refluxed for 4 h (TLC monitoring). The solvent was removed in vacuo, and the residue was poured into H2O (100 mL). The resulting precipitate that separated was collected and crystallized from EtOH to give 5 (0.26 g, 82%). m.p.: 211–212 °C. IR (KBr, cm−1): 3384, 2926, 2928, 1573. 1H-NMR (400 MHz, CDCl3) δ 1.88–1.97 (m, 2H, piperidinyl-H), 2.14–2.19 (m, 2H, piperidinyl-H), 3.14 (br s, 2H, piperidinyl-H), 3.76–3.80 (m, 2H, piperidinyl-H), 3.97 (br s, 1H, piperidinyl-H), 4.18 (s, 2H, 11-CH2), 7.32–7.36 (m, 1H, 9-H), 7.40–7.47 (m, 2H, 2-, 8-H), 7.57–7.64 (m, 2H, 3-, 10-H), 7.89–8.00 (m, 3H, 1-, 4-, 7-H). 13C-NMR (100 MHz, CDCl3) δ 34.63 (2C), 35.96, 47.77 (2C), 68.41, 122.61, 123.58, 123.74, 124.51 (2C), 126.21, 126.96, 128.30, 128.39, 128.80, 140.65, 142.63, 146.07, 151.69, 157.95. Anal. calcd. for C21H20N2O∙0.1H2O : C 79.27, H 6.40, N 8.80; found: C 79.24, H 6.48, N 8.89.
3.2.14. 6-(4-Hydroxypiperidin-1-yl)-11-methyl-11H-indeno[1,2-c]quinolin-11-ol (6)
A mixture of 3d (0.25 g, 1.0 mmol), dry THF (20 mL) and 2 M methyl magnesium bromide (2 mL) was stirred at 0 °C for 12 h (TLC monitoring). The solvent was removed in vacuo, and the residue was poured into H2O (100 mL). The resulting precipitate that separated was collected and crystallized from EtOH to give 6 (0.25 g, 71%). m.p.: 146–147 °C. IR (KBr, cm−1): 3351, 2926, 1372. 1H-NMR (400 MHz, CDCl3) δ 1.76–1.99 (m, 2H, piperidinyl-H), 1.92 (s, 3H, 11-CH3), 2.10–2.19 (m, 2H, piperidinyl-H), 3.04–3.27 (m, 2H, piperidinyl-H), 3.75–3.78 (m, 2H, piperidinyl-H), 3.96 (m, 1H, piperidinyl-H), 7.36–7.48 (m, 3H, 2-, 8-, 9-H), 7.61–7.67 (m, 2H, 3-, 10-H), 7.87(d, 1H, J = 7.2 Hz, 7-H), 7.98 (br s, 1H, 4-H), 8.35–8.37 (d, 1H, J = 8.0 Hz, 1-H). 13C-NMR (100 MHz, CDCl3) δ 26.83, 34.51 (2C), 47.58 (2C), 67.98, 81.34, 118.87, 122.48 (2C), 122.85 (2C), 124.34 (2C), 124.67, 127.59, 128.74, 128.99 (2C), 129.26, 150.83. Anal. calcd. for C22H22N2O2∙0.7H2O: C 73.60, H 6.57, N 7.80; found: C 73.90, H 6.78, N 7.43.
3.2.15. (E)-6-(4-Hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one Oxime (7)
A mixture of 3d (0.27 g, 1.0 mmol), potassium carbonate (0.56 g,4.00 mmol) and NH2OH·HCl (0.14 g, 2.0 mmol) in 2-ethoxyethanol (30 mL) was refluxed for 3 h (TLC monitoring). The solvent was removed in vacuo, and the residue was poured into H2O (100 mL). The resulting precipitate that separated was collected and crystallized from EtOH to give 11 (0.27 g, 78%). m.p.: 155–156 °C. IR (KBr, cm−1): 3329, 2942. 1H-NMR (400 MHz, DMSO-d6) δ 1.68–1.76 (m, 2H, piperidinyl-H), 1.98–2.00 (m, 2H,piperidinyl-H), 2.99 (br s, 2H, piperidinyl-H), 3.55–3.58 (m, 2H,piperidinyl-H), 3.74 (br s, 1H, piperidinyl-H), 4.83 (br s, 1H, -OH), 7.40–7.49 (m, 2H, 8-, 9-H), 7.57–7.64 (m, 2H, 2-, 3-H), 7.80–7.90 (m, 2H, 7-, 10-H), 8.43 (d, 1H, J = 7.6 Hz, 4-H), 8.79 (dd, 1H, J = 1.2, 8.4 Hz, 1-H), 13.39 (s, 1H, NOH). 13C-NMR (100 MHz, DMSO-d6) δ 34.17 (2C), 48.04 (2C), 67.98, 120.79, 122.37, 125.30, 125.56, 126.19, 128.09, 128.25, 128.41, 128.87, 129.17, 130.87, 138.25, 139.59,146.94, 153.91, 157.40. Anal. calcd. for C21H19N3O2∙0.2H2O: C 72.27, H 5.60, N 12.04; found: C 72.67, H 5.72, N 11.63.
3.2.16. (E)-6-(4-Hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-one O-methyl Oxime (8)
Compound 8 was prepared from 3d and NH2OMe∙HCl by the same procedure as described for the preparation of 7. Yield: 74%. m.p.: 183–184 °C. IR (KBr, cm−1): 3350, 2938, 1025. 1H-NMR (400 MHz, DMSO-d6) δ 1.66–1.74 (m, 2H, piperidinyl-H), 1.96–1.99 (m, 2H, piperidinyl-H), 2.96 (br s, 2H, piperidinyl-H), 3.51–3.54 (m, 2H, piperidinyl-H), 3.72 (br s, 1H, piperidinyl-H), 4.32 (s, 3H, =NOCH3), 4.83 (br s, 1H, -OH), 7.35–7.48 (m, 2H, 8-, 9-H), 7.54–7.63 (m, 2H, 2-, 3-H), 7.77–7.81 (m, 2H, 7-, 10-H), 8.22 (d, 1H, J = 7.6Hz, 4-H), 8.72 (dd, 1H, J = 0.8, 8.4 Hz, 1-H). 13C-NMR (100 MHz, DMSO-d6) δ 34.19 (2C), 48.01 (2C), 64.39, 66.62, 120.60, 122.57, 125.21, 125.79, 126.62, 128.09, 128.34, 128.68, 129.33, 130.00, 131.54, 138.73,138.77, 147.05, 153.78, 157.28. Anal. calcd. for C22H21N3O2: C 73.52, H 5.89, N 11.69; found: C 73.24, H 5.93, N 11.49.
3.2.17. (E)-2-[6-(4-Hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene]hydrazine Carboxamide (9)
A mixture of 3d (0.27 g, 1.0 mmol) and semicarbazide (0.15 g, 2.0 mmol) in 2-ethoxyethanol (30 mL) was refluxed for 3 h (TLC monitoring). The solvent was removed in vacuo, and the residue was poured into H2O (100 mL). The resulting precipitate that separated was collected and crystallized from EtOH to give 9 (0.27 g, 68%). m.p.: 289–290 °C. IR (KBr, cm−1): 3468, 1725. 1H-NMR (400 MHz, DMSO-d6) δ 1.69–1.78 (m, 2H, piperidinyl-H), 1.99–2.02 (m, 2H, piperidinyl-H), 3.03 (br s, 2H, piperidinyl-H), 3.59–3.61 (m, 2H, piperidinyl-H), 3.76 (br s, 1H, OH), 7.00 (br s, 2H, NH2), 7.45–7.51 (m, 2H, 8-, 9-H), 7.58–7.65 (m, 2H, 2-, 3-H), 7.82 (d, 1H, J = 8.8 Hz, 10-H), 7.94 (d, 1H, J = 7.6 Hz, 7-H), 8.30 (d, 1H, J = 7.6 Hz, 4-H), 9.15 (d, 1H, J =8.4 Hz, 1-H), 10.45 (br s, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ 34.04 (2C), 47.79 (2C), 65.71, 120.65, 122.49, 124.98, 125.75, 125.87, 126.86, 127.80, 128.18, 129.44, 130.39, 138.37, 141.52 ,143.90, 145.40, 155.74, 156.57, 181.34. Anal. calcd. for C22H21N5O2∙0.5H2O: C 66.65, H 5.59, N 17.67; found: C 66.69, H, 5.70, N 17.31.
3.2.18. (E)-2-[6-(4-Hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene]hydrazine Carbothioamide (10)
Compound 10 was prepared from 3d and thiosemicarbazide by the same procedure as described for the preparation of 9. Yield: 54%. m.p.: 304–305 °C. IR (KBr, cm−1): 3408, 1601. 1H-NMR (400 MHz, DMSO-d6) δ 1.68–1.77 (m, 2H, piperidinyl-H), 1.99–2.01 (m, 2H, piperidinyl-H), 3.03 (br s, 2H, piperidinyl-H), 3.57–3.60 (m, 3H, OH, piperidinyl-H), 3.75 (br s, 1H, piperidinyl-H), 7.46–7.50 (m, 3H, 8-, 9-H), 7.59–7.65 (m, 2H, 2-, 3-H), 7.81 (d, 1H, J = 8.0 Hz, 10-H), 7.91 (d, 1H, J = 7.2 Hz, 7-H), 8.10 (d, 1H, J = 7.6 Hz, 4-H), 8.25 (br s, 1H, NH), 8.99 (br s, 1H, NH), 9.07 (d, 1H, J = 7.6 Hz, 1-H), 11.30 (br s, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ 34.45 (2C), 48.09 (2C), 65.56, 121.45, 121.91, 125.09, 125.50, 127.17, 127.64, 128.62, 130.25, 140.06, 156.12, 156.59, 157.61, 173.59. Anal. calcd. for C22H21N5OS∙0.6H2O: C 63.78, H 5.40, N 16.90; found: C 63.90, H 5.35, N 16.50.
3.2.19. (E)-N′-[6-(4-Hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene]benzohydrazide (11)
A mixture of 3d (0.27 g, 1.0 mmol) and benzohydrazide (0.27 g, 2.0 mmol) in 2-ethoxyethanol (30 mL) was refluxed for 24 h (TLC monitoring). The solvent was removed in vacuo, and the residue was poured into H2O (100 mL). The resulting precipitate that separated was collected and crystallized from EtOH to give 11 (0.28 g, 61%). m.p.: 258–259 °C. IR (KBr, cm−1): 3422, 2937, 1660. 1H-NMR (400 MHz, DMSO-d6) δ 1.70–1.78 (m, 2H, piperidinyl-H), 1.99–2.02 (m, 2H, piperidinyl-H), 3.03 (br s, 2H, piperidinyl-H), 3.58–3.61 (m, 2H, piperidinyl-H), 3.76 (br s, 1H, piperidinyl-H), 4.83 (br s, 1H, -OH), 7.45–7.52 (m, 2H, NH, 9-H), 7.61–7.72 (m, 5H, Ar-H), 7.81 (d, 1H, J = 8.4 Hz, 10-H), 7.93 (d, 1H, J = 7.6 Hz, 7-H), 8.07 (m, 2H, Ar-H), 8.26 (d, 1H, J = 8.0 Hz, 4-H), 9.17 (br s, 1H, 1-H), 12.09 (s, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ 34.20 (2C), 48.00 (2C), 65.71, 120.88, 122.53, 125.39, 125.76, 126.52, 126.94, 127.45, 127.98, 128.11, 128.27 (2C), 128.55, 128.69 (2C), 129.34, 131.48, 132.25, 133.05, 139.60, 140.64, 147.26, 157.17. Anal. calcd. for C28H24N4O2∙0.6H2O: C 73.22, H 5.53, N 12.20; found: C 73.09, H 5.43, N 12.21.
3.2.20. (E)-N′-[6-(4-Hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene]isonicotino Hydrazide (12)
Compound 12 was prepared from 3d and isonicotinic acid hydride by the same procedure as described for the preparation of 11. Yield: 65%. m.p.: 282–283 °C. IR (KBr, cm−1): 3365, 2948, 1649. 1H-NMR (400 MHz, DMSO-d6) δ 1.68–1.77 (m, 2H, piperidinyl-H), 1.99–2.01 (m, 2H, piperidinyl-H), 3.02 (br s, 2H, piperidinyl-H), 3.57–3.59 (m, 2H, piperidinyl-H), 3.75 (br s, 1H, piperidinyl-H), 4.84 (br s, 1H, -OH), 7.47–7.51 (m, 2H, 8-, 9-H), 7.61–7.65 (m, 2H, 2-, 3-H), 7.80 (d, 1H, J = 8.4 Hz, 10-H), 7.90–7.95 (m, 3H, Ar-H), 8.27 (d, 1H, J = 7.2 Hz, 4-H), 8.86 (d, 2H, J = 5.6 Hz, pyridinyl-H), 9.21 (br s, 1H, 1-H), 12.30 (s, 1H, NH). 13C-NMR (100 MHz, DMSO-d6) δ 34.17 (2C), 48.05 (2C), 65.54, 120.79, 122.00 (2C), 122.57, 125.18, 125.80, 126.75, 127.74, 128.00, 128.15, 128.41, 129.39, 131.77, 139.76, 140.47, 147.29, 150.39 (2C), 157.14. Anal. calcd. for C27H23N5O2∙1.8H2O: C 67.29, H 5.56, N 14.53; found: C 67.14, H 5.57, N 14.40.
3.3. Anti-Mycobacterium Activity
Primary screening is conducted against M. tuberculosis H37Rv (ATCC 27294) in BACTEC 12B medium (BD Microbiology Systems, Franklin Lakes, NJ, USA) using a broth microdilution assay to determine the actual minimum inhibitory concentration (MIC) using the Microplate Alamar Blue Assay (MABA) [37]. The MIC is defined as the lowest concentration effecting a reduction in fluorescence of 90% relative to controls. Vero cell (normal cell) viability is assessed on the basis of cellular conversion of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), into a formazan product using the Promega Cell Titer 96 Non-radioactive Cell Proliferation Assay (Promega, Madison, WI, USA).
3.4. Superoxide Generation and Elastase Release
Superoxide generation and elastase release were carried out according to the procedures described previously [38]. Neutrophils (6 × 105/mL) were equilibrated at 37 °C for 2 min and incubated with compounds for 5 min. Neutrophils were then activated by fMLF (100 nM) in the pretreatment of cytochalasin B (1 µg/mL for superoxide generation and 0.5 µg/mL for elastase release) for 10 min. Superoxide anion production was assayed by monitoring the superoxide dismutase-inhibitable reduction of ferricytochrome C. Elastase release was performed using MeO-Suc-Ala-Ala-Pro-Valp-nitroanilide as the elastase substrate.
3.5. Quantitative Structure–Activity Relationship (QSAR)
Structure features of 1648 1D and 2D descriptors and PubChem fingerprints for chemicals were firstly generated by using the PaDEL-Descriptor [39] software (http://padel.nus.edu.sg/software/padeldescriptor). The calculations are mainly based on the Chemistry Development Kit [40] with some additional descriptors and fingerprints including atom-type electrotopological state descriptors, McGowan volume, molecular linear free energy relation descriptors, ring counts, count of chemical substructures, binary fingerprints and count of chemical substructures. It has been extensively utilized in many QSAR studies such as non-genotoxic hepatotoxicity prediction [41] and inhibitor design [42]. Sequential feature selection algorithms developed by our group were subsequently applied to simultaneously identify important features and build multiple [30,31] regression models. For each run, the descriptor with highest R2 performance was iteratively appended into the multiple regression model. To avoid overfitting problems, only descriptors with a maximum correlation coefficient to selected descriptors ranging from −0.5 to 0.5 were considered. Due to the small number of chemicals, only the best three descriptors were considered. The descriptor set with the highest R2 performance is utilized to construct the final multiple regression model for QSAR analysis. The measurements of R2, Radj2, root-mean-squared error (RMSE), and leave-one-out cross-validated Q2 were applied to evaluate the performance of the QSAR model.
4. Conclusions
We have synthesized certain indeno[1,2-c]quinoline derivatives for anti-TB and anti-inflammatory evaluations. Among them, (E)-N′-[6-(4-hydroxypiperidin-1-yl)-11H-indeno[1,2-c]quinolin-11-ylidene]-isonicotinohydrazide (12), exhibited significant activities against the growth of M. tuberculosis (MIC values of 0.96 μg/mL) with a potency approximately equal to that of INH. Compound 12 has also demonstrated a potent dual inhibitory effect on NE release and superoxide anion generation with IC50 values of 1.76 and 1.72 μM, respectively. These results indicated that compound 12 is a potential lead compound for the discovery of dual anti-TB and anti-inflammatory drug candidates. In addition, 6-[3-(hydroxymethyl)piperidin-1-yl]-9-methoxy-11H-indeno[1,2-c]quinolin-11-one (4g) showed a potent dual inhibitory effect on NE release and superoxide anion generation with IC50 value of 0.46 and 0.68 μM respectively, and is a potential lead compound for the discovery of anti-inflammatory drug candidates. Further studies on the structural optimization and the molecular pharmacological mechanism of compounds 12 and 4g are ongoing.
Acknowledgments
Financial support of this work by the Minister of Science and Technology of the Republic of China (MOST 105-2320-B-037-007, MOST 105-2320-B-037-011, MOST 104-2320-B255-004-MY3, MOST 103-2320-B-037-011-MY3) and Kaohsiung Medical University (KMU-TP104E16, KMU-TP104E42, KMU-TP104H07, KMU-TP104H08, KMU-TP105E16, KMU-TP105E33, KMU-TP105H07, KMU-TP105H08, 105KMUOR02) are gratefully acknowledged. We also thank Center for Research Resources and Development at Kaohsiung Medical University for the instrumentation and equipment support.
Supplementary Materials
Author Contributions
Chih-Hua Tseng participated in synthesis, purification and characterization of the chemical compounds; Chun-Wei Tung participated in quantitative structure-activity relationship (QSAR) analysis; Chen-Hsin Wu and Cherng-Chyi Tzeng participated in synthesis, the interpretation of the results and in manuscript writing; Yen-Hsu Chen and Tsong-Long Hwang participated in the biological activity, the interpretation of the results and in manuscript writing; Yeh-Long Chen suggested the research idea, participated in the interpretation of the results and in manuscript writing.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds reported herein are available from the authors.
References
- 1.Scott H.M., Flynn J.L. Mycobacterium tuberculosis in chemokine receptor 2-deficient mice: Influence of dose on disease progression. Infect. Immun. 2002;70:5946–5954. doi: 10.1128/IAI.70.11.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters W., Scott H.M., Chambers H.F., Flynn J.L., Charo I.F., Ernst J.D. Chemokine receptor 2 serves an early and essential role in resistance to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2001;98:7958–7963. doi: 10.1073/pnas.131207398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Garra A., Redford P.S., McNab F.W., Bloom C.I., Wilkinson R.J., Berry M.P.R. The immune response in tuberculosis. Annu. Rev. Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 4.Dooley D.P., Carpenter J.L., Rademacher S. Adjunctive corticosteroid therapy for tuberculosis: A critical reappraisal of the literature. Clin. Infect. Dis. 1997;25:872–887. doi: 10.1086/515543. [DOI] [PubMed] [Google Scholar]
- 5.Worthington R.J., Melander C. Combination approaches to combat multi-drug-resistant bacteria. Trends Biotechnol. 2013;31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallis R.S., Maeurer M., Mwaba P., Chakaya J., Rustomjee R., Migliori G.B., Marais B., Schito M., Churchyard G., Swaminathan S., et al. Tuberculosis—Advances in development of new drugs, treatment regimens, host-directed therapies, and biomarkers. Lancet Infect. Dis. 2016;16:e34–e46. doi: 10.1016/S1473-3099(16)00070-0. [DOI] [PubMed] [Google Scholar]
- 7.Rayasam G.V., Balganesh T.S. Exploring the potential of adjunct therapy in tuberculosis. Trends Pharmacol. Sci. 2015;36:506–513. doi: 10.1016/j.tips.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Cholo M.C., Steel H.C., Fourie P.B., Germishuizen W.A., Anderson R. Clofazimine: Current status and future prospects. J. Antimicrob. Chemother. 2012;67:290–298. doi: 10.1093/jac/dkr444. [DOI] [PubMed] [Google Scholar]
- 9.Van Heeswijk R.P.G., Dannemann B., Hoetelmans R.M.W. Bedaquiline: A review of human pharmacokinetics and drug–drug interactions. J. Antimicrob. Chemother. 2014;69:2310–2318. doi: 10.1093/jac/dku171. [DOI] [PubMed] [Google Scholar]
- 10.Upadhayaya R.S., Shinde P.D., Sayyed A.Y., Kadam S.A., Bawane A.N., Poddar A., Plashkevych O., Földesi A., Chattopadhyaya J. Synthesis and structure of azole-fused indeno[2,1-c]quinolines and their anti-mycobacterial properties. Org. Biomol. Chem. 2010;8:5661–5673. doi: 10.1039/c0ob00445f. [DOI] [PubMed] [Google Scholar]
- 11.Upadhayaya R.S., Lahore S.V., Sayyed A.Y., Dixit S.S., Shinde P.D., Chattopadhyaya J. Conformationally-constrained indeno[2,1-c]quinolones—A new class of anti-mycobacterial agents. Org. Biomol. Chem. 2010;8:2180–2197. doi: 10.1039/b924102g. [DOI] [PubMed] [Google Scholar]
- 12.Upadhayaya R.S., Shinde P.D., Kadam S.A., Bawane A.N., Sayyed A.Y., Kardile R.A., Gitay P.N., Lahore S.V., Dixit S.S., Földesi A., et al. Synthesis and antimycobacterial activity of prodrugs of indeno[2,1-c]quinoline derivatives. Eur. J. Med. Chem. 2011;46:1306–1324. doi: 10.1016/j.ejmech.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 13.Sheu J.Y., Chen Y.L., Fang K.C., Wang T.C., Tzeng C.C., Peng C.F. Synthesis and antibacterial activity of 1-(substituted-benzyl)-6-fluoro-1,4-dihydro-4-oxoquinoline-3-carboxylic acids and their 6,8-difluoro analogs. J. Heterocycl. Chem. 1998;35:955–964. doi: 10.1002/jhet.5570350429. [DOI] [Google Scholar]
- 14.Sheu J.Y., Chen Y.L., Tzeng C.C., Hsu S.L., Fang K.C., Wang T.C. Synthesis, and antimycobacterial and cytotoxic evaluation of certain fluoroquinolone derivatives. Helv. Chim. Acta. 2003;86:2481–2489. doi: 10.1002/hlca.200390201. [DOI] [Google Scholar]
- 15.Zhao Y.L., Chen Y.L., Sheu J.Y., Chen I.L., Wang T.C., Tzeng C.C. Synthesis and antimycobacterial evaluation of certain fluoroquinolone derivatives. Bioorg. Med. Chem. 2005;13:3921–3926. doi: 10.1016/j.bmc.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y.L., Huang H.Y., Chen Y.W., Huang Z.Y., Tzeng C.C., Liu C.L., Yao C.W. Synthesis and antimycobacterial evaluation of metal-chelator bearing fluoroquinolones. Chin. Pharm. J. 2005;57:57–70. [Google Scholar]
- 17.Yang C.L., Tseng C.H., Chen Y.L., Lu C.M., Kao C.L., Tseng H.Y., Wu M.H., Tzeng C.C. Identification of benzofuro[2,3-b]quinoline derivatives as a new class of antituberculosis agents. Eur. J. Med. Chem. 2010;45:602–607. doi: 10.1016/j.ejmech.2009.10.050. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y.L., Lu C.M., Chen I.L., Tsao L.T., Wang J.P. Synthesis and antiinflammatory evaluation of 9-anilinoacridine and 9-phenoxyacridine derivatives. J. Med. Chem. 2002;45:4689–4694. doi: 10.1021/jm020102v. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y.L., Chen I.L., Lu C.M., Tzeng C.C., Tsao L.T., Wang J.P. Synthesis and anti-inflammatory evaluation of 9-phenoxyacridine and 4-phenoxyfuro[2,3-b]quinoline derivatives. Part 2. Bioorg. Med. Chem. 2003;11:3921–3927. doi: 10.1016/S0968-0896(03)00439-5. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y.L., Chen I.L., Lu C.M., Tzeng C.C., Tsao L.T., Wang J.P. Synthesis and anti-inflammatory evaluation of 4-anilinofuro[2,3-b]quinoline and 4-phenoxyfuro[2,3-b]quinoline derivatives. Part 3. Bioorg. Med. Chem. 2004;12:387–392. doi: 10.1016/j.bmc.2003.10.051. [DOI] [PubMed] [Google Scholar]
- 21.Kuan Y.H., Lin R.H., Chen Y.L., Tsao L.T., Tzeng C.C., Wang J.P. Effective attenuation of acute lung injury in vivo and the formyl peptide-induced neutrophil activation in vitro by CYL-26z through the phosphoinositide 3-kinase gamma pathway. Biochem. Pharmacol. 2006;72:749–760. doi: 10.1016/j.bcp.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y.L., Zhao Y.L., Lu C.M., Tzeng C.C., Wang J.P. Synthesis, cytotoxicity, and anti-inflammatory evaluation of 2-(furan-2-yl)-4-(phenoxy)quinoline derivatives, Part 4. Bioorg. Med. Chem. 2006;14:4373–4378. doi: 10.1016/j.bmc.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 23.Lin M.W., Tsao L.T., Chang L.C., Chen Y.L., Huang L.J., Kuo S.C., Tzeng C.C., Lee M.R., Wang J.P. Inhibition of lipopolysaccharide-stimulated NO production by a novel synthetic compound CYL-4d in RAW 264.7 macrophages involving the blockade of MEK4/JNK/AP-1 pathway. Biochem. Pharmacol. 2007;73:1796–1806. doi: 10.1016/j.bcp.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Tseng C.H., Lin C.S., Shih P.K., Tsao L.T., Wang J.P., Cheng C.M., Tzeng C.C., Chen Y.L. Furo[3, 2:3,4]naphtho[1,2-d]imidazole derivatives as potential inhibitors of inflammatory factors in sepsis. Bioorg. Med. Chem. 2009;17:6773–6779. doi: 10.1016/j.bmc.2009.07.054. [DOI] [PubMed] [Google Scholar]
- 25.Tseng C.H., Tzeng C.C., Shih P.K., Yang C.N., Chuang Y.C., Peng S.I., Lin C.S., Wang J.P., Cheng C.M., Chen Y.L. Identification of furo[3′,2′:3,4]naphtho[1,2-d]imidazole derivatives as orally active and selective inhibitors of microsomal prostaglandin E(2) synthase-1 (mPGES-1) Mol. Divers. 2012;16:215–229. doi: 10.1007/s11030-011-9347-9. [DOI] [PubMed] [Google Scholar]
- 26.Tsai Y.R., Huang L.J., Lee M.R., Chen Y.L., Kuo S.C., Tzeng C.C., Hsu M.F., Wang J.P. The signaling mechanisms mediating the inhibitory effect of TCH-1116 on formyl peptide-stimulated superoxide anion generation in neutrophils. Eur. J. Pharm. 2012;682:171–180. doi: 10.1016/j.ejphar.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Tseng C.H., Cheng C.M., Tzeng C.C., Peng S.I., Yang C.L., Chen Y.L. Synthesis and anti-inflammatory evaluations of â-lapachone derivatives. Bioorg. Med. Chem. 2013;21:523–531. doi: 10.1016/j.bmc.2012.10.047. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y.R., Tseng C.H., Chen Y.L., Hwang T.L., Tzeng C.C. Discovery of benzo[f]indole-4,9-dione derivatives as new type of anti-inflammatory agents. Int. J. Mol. Sci. 2015;16:6532–6544. doi: 10.3390/ijms16036532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng C.H., Chen Y.L., Lu P.J., Yang C.N., Tzeng C.C. Synthesis and antiproliferative evaluation of certain indeno[1,2-c]quinoline derivatives. Bioorg. Med. Chem. 2008;16:3153–3162. doi: 10.1016/j.bmc.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 30.Tung C.W. Prediction of pupylation sites using the composition of k-spaced amino acid pairs. J. Theor. Biol. 2013;336:11–17. doi: 10.1016/j.jtbi.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Tung C.W., Wu M.T., Chen Y.K., Wu C.C., Chen W.C., Li H.P., Chou S.H., Wu D.C., Wu I.C. Identification of biomarkers for esophageal squamous cell carcinoma using feature selection and decision tree methods. Sci. World J. 2013;2013:782031. doi: 10.1155/2013/782031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kier L.B., Hall L.H. Molecular Connectivity in Chemistry and Drug Research. Academic Press; New York, NY, USA: 1976. [Google Scholar]
- 33.Wildman S.A., Crippen G.M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 1999;39:868–873. doi: 10.1021/ci990307l. [DOI] [Google Scholar]
- 34.Gramatica P., Corradi M., Consonni V. Modelling and prediction of soil sorption coefficients of non-ionic organic pesticides by molecular descriptors. Chemosphere. 2000;41:763–777. doi: 10.1016/S0045-6535(99)00463-4. [DOI] [PubMed] [Google Scholar]
- 35.Hall L.H., Kier L.B. Electrotopological state indices for atom types: A novel combination of electronic, topological, and valence state information. J. Chem. Inf. Comput. Sci. 1995;35:1039–1045. doi: 10.1021/ci00028a014. [DOI] [Google Scholar]
- 36.Liu R., Sun H., So S.S. Development of quantitative structure—Property relationship models for early ADME evaluation in drug discovery. 2. Blood-brain barrier penetration. J. Chem. Inf. Comput. Sci. 2001;41:1623–1632. doi: 10.1021/ci010290i. [DOI] [PubMed] [Google Scholar]
- 37.Collins L., Franzblau S.G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang T.L., Li G.L., Lan Y.H., Chia Y.C., Hsieh P.W., Wu Y.H., Wu Y.C. Potent inhibition of superoxide anion production in activated human neutrophils by isopedicin, a bioactive component of the chinese medicinal herb Fissistigma oldhamii. Free Radic. Biol. Med. 2009;46:520–528. doi: 10.1016/j.freeradbiomed.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Yap C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011;32:1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 40.Steinbeck C., Han Y., Kuhn S., Horlacher O., Luttmann E., Willighage E. The Chemistry Development Kit (CDK): An open-source Java library for Chemo- and Bioinformatics. J. Chem. Inf. Comput. Sci. 2003;43:493–500. doi: 10.1021/ci025584y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tung C.W., Jheng J.L. Interpretable prediction of non-genotoxic hepatocarcinogenic chemicals. Neurocomputing. 2014;145:68–74. doi: 10.1016/j.neucom.2014.05.073. [DOI] [Google Scholar]
- 42.Chauhan J.S., Dhanda S.K., Singla D., The Open Source Drug Discovery. Agarwal S.M., Raghava G.P.S. QSAR-based models for designing quinazoline/imidazothiazoles/pyrazolopyrimidines based inhibitors against wild and mutant EGFR. PLoS ONE. 2014;9:e101079. doi: 10.1371/journal.pone.0101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.