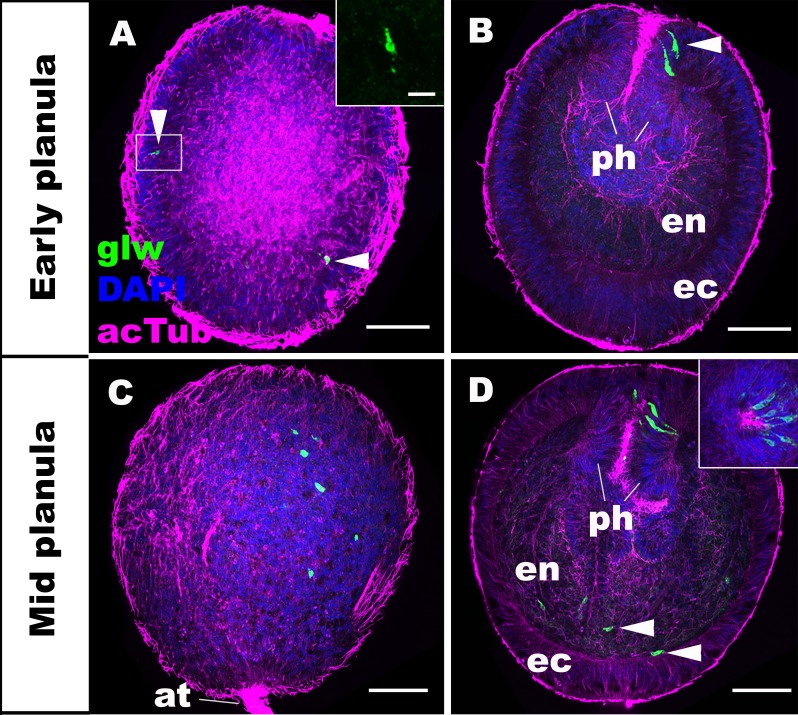
Figure 1. GLWamide precursor transcripts are expressed in ectodermal and endodermal epithelial cells of sea anemone planula larvae.
Z-projections of confocal sections of Nematostella vectensis at planula stages, labeled with an antisense riboprobe against GLWamide precursor transcript (‘glw’) and an antibody against acetylated ∂-tubulin (‘acTub’). Nuclei are labeled with DAPI. All panels show side views of animals with the blastopore/mouth facing up. The left columns (A and C) show superficial planes of section at the level of the ectoderm, and the right columns (B and D) show longitudinal sections through the center. (A, B) early planula. GLWamide transcripts are found in ectodermal sensory cells that are present in the outer epithelium and the pharynx (arrowheads in A and B). The inset in A show transcript-expressing epithelial cells in boxed regions with the apical side of the cell facing up. Note the spindle-shaped morphology characteristic of cnidarian sensory cells (Thomas and Edwards, 1991). (C, D) mid-planula. A subset of endodermal cells begin to express GLWamide transcript (arrowheads in D). The inset in D shows transverse sections of the pharynx, revealing the asymmetry in the spatial distribution of GLWamide-positive sensory cells. Abbreviations: ph pharynx; ec ectoderm; en endoderm; at apical tuft. Scale bar: 50 µm (A-D); 10 µm (inset in A).