Abstract
Primary cell models of human immunodeficiency virus (HIV) latency have become tools to both understand the mechanisms involved in establishment of latency and test preclinical strategies toward HIV-1 cure. These models rely on infection of CD4 T cells from healthy donors. As such, these models provide an opportunity to explore the role of biological sex, age, and HIV status on establishment and reactivation of latent HIV in vitro. We have used an established primary cell model of latency based on the generation of latently infected central memory CD4 T cells with the CXCR4 strain HIV-1NL4-3 to address whether these variables influence (i) HIV-1NL4-3 replication, (ii) establishment of latency, and (iii) latency reversal in CD4 T cells. Our results indicate that replication of HIV-1NL4-3, but not establishment of latency, is influenced by the age of female, but not male, donors. Moreover, the frequency of latently infected cells in this model is directly correlated with levels of productive infection in both male and female donors independent of age. We did not find differences in the ability of five different latency-reversing agents to reactivate latent HIV-1NL4-3. Finally, we have found that this model can be generated using cells from aviremic participants. In conclusion, we have further characterized the central memory T cell model of latency regarding biological sex and age and demonstrated that this model is suitable for use with cells isolated from aviremic participants, opening the opportunity to use this primary cell model to address cure approaches, including shock and kill, in HIV-infected individuals.
Keywords: : HIV, latency, reservoirs, primary cell models, biological sex, age
Introduction
Human immunodeficiency virus (HIV) has caused more than 35 million deaths worldwide. Management of the disease requires daily administration of a combination of antiretroviral therapy (ART) drugs for the life of the infected individual. This is due to the presence of an intact and inducible latent reservoir of HIV that rebounds after discontinuation of ART therapy.1–3 Elimination or reduction of this latent reservoir is crucial toward efforts to eradicate or control HIV. The frequency of the latent reservoir in vivo is limited (1–100 in a million resting CD4 T cells).4–6 The mechanisms by which HIV latency is established and maintained are not completely understood.
To understand the mechanisms that control HIV latency, researchers have developed several latency models using either tumoral cell lines or primary cells isolated from HIV-negative volunteers.7–22 These models have led to the discovery of several mechanisms governing HIV latency as well as development of therapeutic strategies currently under clinical evaluation (reviewed elsewhere23–25).
In this study, we used the cultured central memory T cell model (TCM model) of HIV latency19,20 to investigate whether the donor's biological sex, age, and HIV status influence establishment of latency or its reactivation with latency-reversing agents (LRAs) in vitro. We believe that these are important questions that have to be addressed as primary cell models of latency are becoming surrogates to both understand the mechanisms of persistence and test therapeutic strategies toward the latent reservoir.
First, it is important to address whether any of the proposed strategies will be affected by the biological sex and age of future participants in a clinical trial. Interestingly, our study has found that viral replication of HIV-1NL4-3, but not establishment of latency, is influenced by the age of donor cells only in female (but not male) donors. We further characterized the ability of five different LRAs to reactivate latent HIV-1NL4-3. Our analysis suggests that the activity of these LRAs is independent of biological sex. Furthermore, we demonstrate that this primary cell model can be performed in cells isolated from aviremic participants. This result suggests that the TCM model can be used to specifically evaluate LRAs in a patient-specific manner or to also evaluate kill strategies using syngeneic cytotoxic CD8 T cells or NK cells.26
Materials and Methods
Reagents
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID: Nelfinavir, Human recombinant IL-2 (rIL-2) from Dr. Maurice Gately, Hoffman-La Roche, Inc.,27 Raltegravir (Cat # 11680) from Merck and Company, Inc., and HIV-1NL4-3 from Dr. Malcolm Martin.28 Raltegravir was from Selleckchem (Houston, TX). rIL-2 was also obtained from the NCI Preclinical Repository. Pam3CKS4 was from InvivoGen (San Diego, CA), HODHBt was from AK Scientific, Inc. (Union City, CA), ingenol-3,20-dibenzoate and bryostatin-1 were from Enzo Life Sciences (Farmingdale, NY), and SAHA was from Cayman Chemical (Ann Arbor, MI).
Generation of latently infected TCM cells
Peripheral blood mononuclear cells (PBMCs) were isolated from blood of healthy donors. TCM cells and latently infected TCM cells were generated as previously described.19,20,29 Briefly, naïve CD4 T cells were isolated from healthy donors using the EasySep™ Human Naïve CD4+ T cell isolation kit (STEMCELL Technologies). After isolation, naïve cells were plated at a density of 0.5 × 106 cells/mL of RPMI (supplemented with 10% fetal bovine serum, l-glutamine, and penicillin/streptomycin) and activated with 12.5 μL of αCD3/αCD28-coated beads (human T-activator CD3/CD28 for T cell expansion and activation Dynabeads; Dynal/Invitrogen, Carlsbad, CA) in the presence of 10 ng/mL transforming growth factor-β1, 2 μg/mL anti-human IL-12, and 1 μg/mL anti-human IL-4 (all from PeproTech, Rocky Hill, NJ). Activation was performed in 96-well round plates with 100 μL/well to ensure homogeneous activation. After activation, cells were resuspended and Dynabeads were removed using a magnetic particle concentrator (Dynal MPC®-L; Invitrogen). Activated cells were kept at 1 × 106 cells/mL in complete medium with 30 IU/mL of IL-2. Media and IL-2 were replaced at days 4 and 5. To generate TCMs, media and IL-2 were replaced at days 7, 10, and 13. Cell density was maintained at 1 × 106. To generate latently infected TCMs, cells were infected at day 7 using NL4-3. One-fifth of the culture was left uninfected as a control. One-fifth of the culture was infected with a multiplicity of infection (MOI) of 0.3 by spinoculation at 2,900 rpm (1,741 × g) for 2 h at 37°C. After infection, cells were mixed with the other three-fifths of uninfected cells at 1 × 106 cells/mL in complete medium with 30 IU/mL of IL-2. At day 10, media and IL-2 were replaced and cells were cultured in 96-well round plates at 1 × 106 cells/mL with 100 μL/well to ensure cell-to-cell transmission. At day 13, cells were transferred to flasks, media and IL-2 were replaced, and 1 μM of raltegravir and 0.5 μM of nelfinavir were added to the cultures to stop viral replication. At day 17, CD4-positive cells were isolated using the Dynabeads® CD4 Positive Isolation Kit (Invitrogen 11331D), as indicated by the manufacturer. The amount of CD4 beads was increased threefold to allow efficient recovery.
Reactivation assays
A total of 1 × 105 cells were treated for each condition. As controls, cells were left unstimulated, treated with 30 IU/mL of IL-2, or reactivated with αCD3/αCD28-coated beads (one bead per cell). For LRA experiments, cells were incubated with 1 μM of Pam3CSK4, 335 nM SAHA, 100 nM ingenol-3,20-dibenzoate, 100 nM bryostatin-1, or 100 μM of HODHBt in the presence of IL-2. Viral reactivation was measured by assessing surface CD4 and intracellular p24Gag expression.
Flow cytometry analysis
For dual detection of CD4 and HIV-1 p24Gag, cells were first stained with the viability dye (Fixable Viability Dye eFluor 450), followed by staining with CD4 antibody (S3.5), APC conjugate (Molecular Probes™). After staining, cells were fixed, permeabilized, and stained for HIV-1 p24Gag, as previously described.20 In all experiments, CD4-positive HIV-1 p24Gag-negative staining regions were set with uninfected cells treated in parallel. Flow cytometry was performed with a BD FACSCanto II or BD LSRFortessa flow cytometer using FACSDiva acquisition software (Becton Dickinson, Mountain View, CA). Data were analyzed with FlowJo software (Tree Star, Inc., Ashland, OR).
Participant involvement
University of Utah—Donors 18 years and older served as volunteer blood donors. Written informed consent was obtained from all donors. These studies are covered under the institutional review board #67637 protocol approved by the University of Utah Institutional Review Board.
Gulf Coast Regional Blood Center—Volunteers 17 years and older served as blood donors. White blood cell concentrate (buffy coat) prepared from a single unit of whole blood by centrifugation was purchased.
Aviremic participants—Cells from aviremic participants were obtained through the Reservoir Characterization Section of the BELIEVE collaborative. Secondary use of samples was approved through George Washington University Institutional Review Boards. All subjects were adults and gave informed consent.
Statistics
For intrasex analysis, a two-tailed paired-samples nonparametric t-test analysis (Wilcoxon matched-pairs signed rank tests) was used to calculate p-values. For intersex analysis, unpaired two-tailed nonparametric t-test analysis (Mann–Whitney test) was used to calculate p-values. Pearson correlation coefficients and two-tailed p-values were calculated for correlations. Statistics were calculated using Prism 7 for Mac OS X software (GraphPad Software, Inc., La Jolla, CA).
Results
Biological sex and age as variables in HIV-1 replication
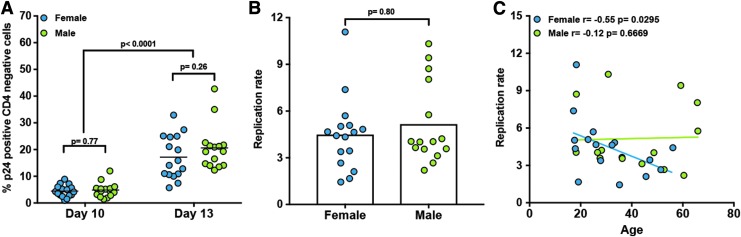
We first wanted to address whether biological sex and age were intrinsic variables impacting HIV-1NL4-3 replication in CD4 T cells. To address this question, we used the primary cell model of HIV infection and latency based on generation of TCMs.19,20 To that end, naïve CD4 T cells isolated from 16 females and 15 males ranging from ages 17.2 to 65.7 were activated under conditions that generate central memory CD4 T cells (TCM).19,30 After activation, cells were infected with the replication-competent HIV-1NL4-3 and subjected to the protocol previously described (Fig. 1A).20,31 First, we wanted to address whether biological sex and age influence viral replication in TCM cells in vitro. To that end, cells infected with replication-competent HIV-1NL4-3 were stained with fluorescently conjugated anti-CD4 antibodies, followed by an intracellular staining against p24Gag (Fig. 1B, day 10). At day 10, cells were crowded to allow cell-to-cell transmission for an additional 3 days and stained for CD4 and p24Gag (Fig. 1B, day 13). As shown in Figure 2A, the percentage of infected cells increased in all donors regardless of sex. To assess whether there was a difference in viral replication at the cellular level, we calculated the replication ratio as the percentage of p24Gag-positive CD4-negative cells at day 13 divided by those at day 10. This ratio allowed us to normalize viral replication to the initial spinoculation. There were no differences in viral replication in female versus male donors (Fig. 2B). Interestingly, there is a negative correlation between the ability of HIV to replicate in female donors and the age of the donor. This correlation was not observed in experiments using cells from male donors.
FIG. 1.
Outline of the TCM model of HIV latency. (A) Timeline of the assay. (B) Surface CD4 expression and intracellular p24Gag were measured at different time points after infection and after reactivation of latent HIV-1NL4-3. Analysis is representative of the experiments presented in this work.
FIG. 2.
Analysis of viral replication in the TCM model. (A) Percentage of infected cells measured as p24Gag positive CD4 negative at days 10 and 13 measured in CD4 T cells from 16 female and 15 male donors following the protocol in Figure 1. Replication ratio (B) was calculated from values in (A). (C) Correlation between the replication ratio and the age of donors stratified by biological sex. Two-tailed paired-samples nonparametric t-test analysis was used to calculate p-values between days. Unpaired two-tailed nonparametric t-test analysis (Mann–Whitney test) was used to calculate p-values between biological sexes. Correlations were determined using the Pearson correlation coefficient.
Biological sex and age as variables in establishment of HIV-1 latency
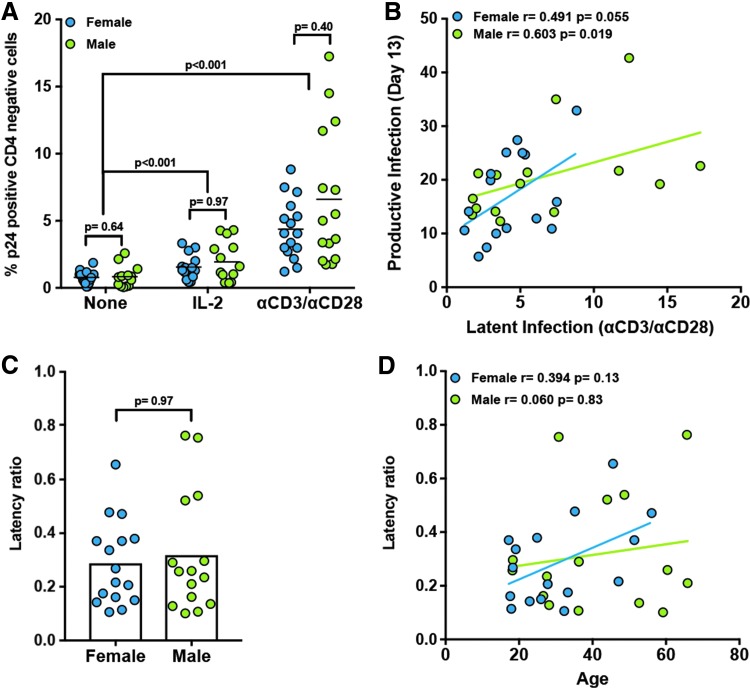
From day 13, a combination of 1 μM raltegravir and 500 nM nelfinavir was introduced in the cultures to block further viral replication (Fig. 1A, B). At day 17, cells that remained CD4 positive (containing both latently infected and uninfected cells) were sorted based on CD4 expression (Fig. 1B, day 17 postsort). After sorting, cells were left unstimulated for 48 h or stimulated with either IL-2 or αCD3/αCD28, and viral reactivation was measured by flow cytometry (Fig. 1B). IL-2 induced a small degree of viral reactivation relative to unstimulated controls in both male and female donors (Fig. 3A). Stimulation with αCD3/αCD28 reactivated latent HIV-1NL4-3 in all the donors tested (Fig. 3A). No differences were observed between female and male donors with any of these reactivation conditions (Fig. 3A).
FIG. 3.
Influence of biological sex and age in HIV-1NL4-3 latency and reactivation in TCMs. (A) Percentage of reactivated cells measured as p24Gag positive CD4 negative after treatment for 48 h with IL-2 or αCD3/αCD28 compared with unstimulated cells from latently infected cells generated in Figure 1. (B) Correlation between the percentage of p24Gag-positive CD4-negative cells during productive infection (day 13) and those during latent infection (αCD3/αCD28) stratified by biological sex. Latency ratio (C) was calculated from values in (B). (D) Correlation between the latency ratio and the age of donors stratified by biological sex. Two-tailed paired-samples nonparametric t-test analysis was used to calculate p-values between stimuli. Unpaired two-tailed nonparametric t-test analysis (Mann–Whitney test) was used to calculate p-values between sexes. Correlations were determined using the Pearson correlation coefficient.
In this model, there is a direct correlation between the levels of infection before ART at day 13, levels of integrated provirus in resting TCMs at day 17, and levels of cells that can reactivate latent HIV-1NL4-3 with αCD3/αCD28 at day 17.20,31 We thus compared the levels of viral infection at day 13 (productive infection) with levels of viral reactivation with αCD3/αCD28 (latent infection) in both female and male donors. There was a positive correlation between both productive and latent infections for both male and female donors (Fig. 3B). To compare whether biological sex had an influence on establishment of latency, we calculated the latency ratio as the percentage of latent infection divided by the percentage of productive infection. We did not observe a statistically significant difference between female and male donors (Fig. 3C). In contrast with the replication ratio, there was no correlation between the age of the donor and the latency ratio (Fig. 3D).
Biological sex as a variable in reactivation of HIV-1 latency
A panel of five LRAs with different mechanisms of action was tested for their ability to reactivate latent HIV-1NL4-3 in this model (Fig. 4). First, we have tested two protein kinase C (PKC) agonists, ingenol-3,20-dibenzoate and bryostatin-1 (Fig. 4A, B). The main mechanism of action of these two agonists is activation of NFκB in a PKC-dependent manner.32,33 In this primary cell model, ingenol-3,20-dibenzoate was able to reactivate latent HIV-1NL4-3 in both male and female donors at similar levels that were superior to αCD3/αCD28 (average of 140.2%) (Fig. 4A). This viral reactivation was independent of biological sex. As for ingenol-3,20-dibenzoate, bryostatin-1 was able to reactivate latent HIV in both male and female donors at similar levels (average of 79.8%) (Fig. 4B). We then tested HODHBt, a novel inhibitor of STAT5 SUMOylation that reactivates latent HIV by increasing STAT5 transcriptional activity within the HIV LTR.34 HODHBt also reactivated latent HIV in both male and female donors at similar levels (19.2% relative to αCD3/αCD28). Next, we tested the TLR-1/2 agonist Pam3CSK4, which reactivates latent HIV-1NL4-3 through activation of NFκB in an MyD88-mediated manner.35,36 This TLR-2 agonist reactivated an average of 7.3% relative to αCD3/αCD28 and no differences were observed between female and male donors (Fig. 4D). Finally, we tested the histone deacetylase inhibitor vorinostat (SAHA).37,38 Our data indicated that the ability of SAHA to reactivate latent HIV in this primary cell model is modest and an average of 4.2% relative to αCD3/αCD28, and no differences were observed between female and male donors (Fig. 4E). In conclusion, biological sex of donors did not influence the activity of the five LRAs tested in this model.
FIG. 4.
Influence of biological sex in latency reversal with five different latency-reversing agents. Latently infected cells were generated from healthy donors, and the ability of (A) 100 nM ingenol-3,20-dibenzoate; (B) 100 nM bryostatin-1; (C) 100 μM HODHBt; (D) 1 μM Pam3CSK4; and (E) 335 nM SAHA to reactivate latent HIV-1NL4-3 was assessed. Viral reactivation over IL-2 alone for each treatment was normalized to that of αCD3/αCD28. Unpaired two-tailed nonparametric t-test analysis (Mann–Whitney test) was used to calculate p-values between sexes.
Generating latently infected cells in cells isolated from aviremic participants
The TCM model is based on generation of latently infected cells in cells isolated from HIV-negative donors. We wanted to explore whether this model could be adapted to ex vivo infected cells from aviremic HIV participants. This will allow better evaluation of shock and kill strategies in cells isolated from aviremic HIV participants or to test the activity of LRAs before administration in clinical trials. To that end, naïve CD4 T cells from five male aviremic participants were isolated, activated, and infected ex vivo to generate latently infected cells, as previously described.19,20 We compared the ability of HIV-1NL4-3 to replicate and establish latency with that of male HIV-negative donors. As shown in Figure 5A, levels of infection were similar between participants and their male donor counterparts. No statistically significant differences were observed in the ability of HIVNL4-3 to replicate (Fig. 5B). Similar levels of latently infected cells were generated and able to reactivate in cells from aviremic participants and HIV-negative donors (Fig. 5C, D). Of interest, we were unable to detect any p24-positive cells in the ex vivo uninfected controls of cells isolated from aviremic participants (data not shown and Thomas et al.26). These results indicate that it is possible to generate latently infected TCMs from aviremic participants and open up the possibility to use the TCM model to evaluate different cure strategies toward the latent reservoir.
FIG. 5.
Generation of latently infected cells in cells isolated from aviremic participants. (A) Percentage of infected cells measured as p24Gag positive CD4 negative at days 10 and 13 in CD4 T cells from 5 male participants and 15 male donors following the protocol in Figure 1. Data from HIV-negative donors are the same as in Figure 1. Replication ratio (B) was calculated from values in (A). (C) Percentage of reactivated cells measured as p24Gag positive CD4 negative after treatment for 48 h with IL-2 or αCD3/αCD28 compared with unstimulated cells from latently infected cells generated in (A). Latency ratio (D) was calculated from values in (C). Unpaired two-tailed nonparametric t-test analysis (Mann–Whitney test) was used to calculate p-values between sexes.
Discussion
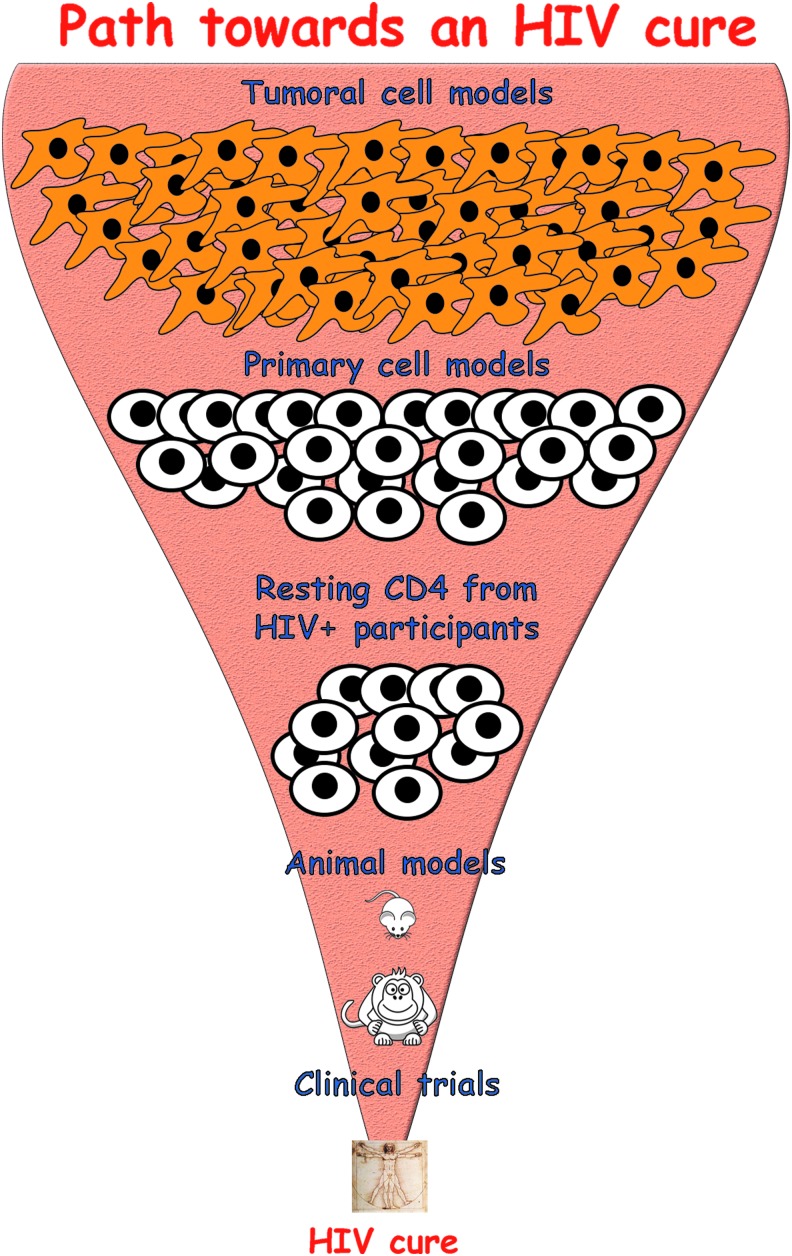
The search for an HIV cure is an uphill battle. Any strategy designed to target the latent reservoir must go through a series of validations in different models, from cell culture to animal models and, eventually, clinical trials. The more complex a model is, the fewer the strategies that can be tested before reaching clinical trials (Fig. 6). The path toward finding a cure is complex and primary cell models of latency can serve as reliable tools to help achieve this holistic goal.
FIG. 6.
Path toward an HIV cure. Therapeutic strategies toward an HIV cure go through a series of validations in different models before reaching clinical trials. These models include tumoral cells, primary cells, cells isolated from aviremic participants, and animal models. The increase in complexity of these models reduces the range of therapeutic strategies that can be tested in each one.
In this work, we have further characterized the TCM model of latency and answered two important questions relevant to the HIV cure field. First, we have characterized whether biological sex and age of the donor can influence establishment or reactivation of the latent reservoir using the CXCR4-tropic strain HIVNL4-3. We did not observe any significant difference between female and male donors in the ability of HIVNL4-3 to establish latency or to reactivate with 7 different stimuli: IL-2, αCD3/αCD28, ingenol-3,20-dibenzoate, bryostatin-1, HODHBt, Pam3CSK4, or SAHA. We did observe a negative correlation between replication of HIVNL4-3 and age, and this correlation was specific to female donors. It is worth noting that this work does not take into account any extrinsic factor that may affect viral replication and latency. For example, sex hormones have been shown to influence HIV infection and immune responses in general.39,40 Whether and how 17 β-estradiol (E2), progesterone (P4), or the androgens, dihydrotestosterone and testosterone, may affect our model of HIV infection are currently unknown. For example, androgens can repress the activity of NFκB in T cells.41 NFκB is one of the principal transcription factors involved in HIV transcription.42 Inhibiting NFκB by androgens may increase establishment of latency and/or block reactivation of latent HIV. In fact, 17β-estradiol has been shown to inhibit HIV replication through inhibition of HIV transcription in in vitro infected PBMCs.43 Further investigation is warranted to determine how sex hormones influence HIV infection in this primary cell model of latency. Second, we have shown that this model can be performed in cells isolated from aviremic participants. This has wide implications for cure research and opens the way to test novel cure strategies besides LRAs. It provides the opportunity to study relevant shock and kill strategies that use syngeneic CD8 or NK cells or evaluate LRAs, combinations, or chimeric antigen receptors in a patient-specific completely MHC-matched system. For example, we have recently shown that latently infected TCMs can be recognized by syngeneic Nef-specific CD8+ T cells.26
Primary cell models of HIV latency have made several important contributions to the field. In particular, the TCM model helps with identification of two novel pathways to target the latent reservoir. First, we found that TLR-2 agonists can directly reactivate latent HIV in resting CD4 T cells from both the TCM model and cells isolated from aviremic participants.35 This is in line with multiple studies showing that TLR agonists can reactivate the latent reservoir and have contributed to the designing of clinical trials to assess effects of these ligands on the latent reservoir.44–51 The second pathway identified involves targeting SUMOylation of STAT5.34 We found that HODHBt and derivatives reactivate latent HIV by inhibiting SUMOylation of STAT5. Interestingly, this LRA displays no activity in cell lines, therefore its activity had most likely been overlooked in previous efforts made to identify LRAs in tumoral cell models. Besides identifying novel LRAs, the TCM model has also been useful to characterize mechanisms involved in HIV persistence. We have previously shown that IL-7 can promote cell division in the absence of viral reactivation, involving homeostatic proliferation as a mechanism of persistence.52 This mechanism of persistence was first proposed by Chomont et al. and has been recently proposed to be one of the mechanisms involved in clonal expansion of latently infected cells.53–55 As clonal expansion of latently infected cells is becoming more apparent as a mechanism of persistence, it is important to understand the mechanisms that regulate this process.56,57 We have previously shown that the TCM model of latency supports generation of clonally expanded clones.58 Therefore, this model represents an ideal tool to also understand clonal expansion.
In spite of its advantages, the TCM model of latency has some caveats that will need further exploration. First, we are expanding the repertoire of molecular clones of HIV used beyond HIVNL4-3. We are in the process of optimizing this model to use founder viruses and viruses obtained from the latent reservoir to better represent in vivo infections. Second, this model only generates central memory CD4 T cells. As latent viruses are found in other subsets of memory CD4 T cells, it will be important to address whether the same mechanisms found in TCMs are found in other subsets. We had previously shown that latently infected cells can be generated when cells are polarized in TH1 and TH2.19 The laboratory of Jonathan Karn has used a similar model to understand latency in TH17 cells.17 Whether the mechanisms controlling latency and reactivation in these subsets are shared with TCMs warrants further investigation.
In summary, we have further characterized the TCM model of latency and address the influence of biological sex and age in this model. Additionally, we have demonstrated that this model can be generated using cells from aviremic participants. Our work expands on the multiple uses of this primary cell model of latency toward exploring the mechanisms involved in HIV persistence as well as to investigate therapeutic treatments to eradicate HIV.
Acknowledgments
The authors would like to specially thank Dr. Mathilde Krim and The Foundation for AIDS Research (amfAR) for their support to A.B. in 2009 with a Mathilde Krim Fellowship (Grant ID 107402) in Basic Biomedical Research to study HIV latency and further develop the primary cell model used in this work in V.P. laboratory. The authors would like to thank Thomas Zaikos for the insightful comments during preparation of the manuscript. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R21/R33 AI116212 and R01 AI124722 to A.B. and BELIEVE (NIH grant 1UM1AI26617) to D.F.N. M.A.S. was supported with a fellowship by the National Institutes of Health under Ruth L. Kirschtein National Research Service Award NIH 5T32 DK007115-40 from the National Institute of Diabetes and Digestive and Kidney Diseases. This research has been facilitated by services and resources provided by the District of Columbia Center for AIDS Research, an NIH-funded program (AI117970), which is supported by the following NIH Co-Funding and Participating Institutes and Centers: NIAID, NCI, NICHD, NHLBI, NIDA, NIMH, NIA, FIC, NIGMS, NIDDK, and OAR. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS: Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A 1998;95:8869–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finzi D, Hermankova M, Pierson T, et al. : Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 1997;278:1295–1300 [DOI] [PubMed] [Google Scholar]
- 3.Wong JK, Hezareh M, Gunthard HF, et al. : Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997;278:1291–1295 [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, Carruth L, Finzi D, et al. : Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997;387:183–188 [DOI] [PubMed] [Google Scholar]
- 5.Ho YC, Shan L, Hosmane NN, et al. : Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013;155:540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laird GM, Eisele EE, Rabi SA, et al. : Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog 2013;9:e1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS: Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 1987;238:800–802 [DOI] [PubMed] [Google Scholar]
- 8.Folks TM, Clouse KA, Justement J, et al. : Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci U S A 1989;86:2365–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoni BA, Rabson AB, Kinter A, Bodkin M, Poli G: NF-kappa B-dependent and -independent pathways of HIV activation in a chronically infected T cell line. Virology 1994;202:684–694 [DOI] [PubMed] [Google Scholar]
- 10.Jordan A, Bisgrove D, Verdin E: HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 2003;22:1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang HC, Xing S, Shan L, et al. : Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest 2009;119:3473–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marini A, Harper JM, Romerio F: An in vitro system to model the establishment and reactivation of HIV-1 latency. J Immunol 2008;181:7713–7720 [DOI] [PubMed] [Google Scholar]
- 13.Saleh S, Solomon A, Wightman F, Xhilaga M, Cameron PU, Lewin SR: CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: A novel model of HIV-1 latency. Blood 2007;110:4161–4164 [DOI] [PubMed] [Google Scholar]
- 14.Tyagi M, Pearson RJ, Karn J: Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol 2010;84:6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahu GK, Lee K, Ji J, Braciale V, Baron S, Cloyd MW: A novel in vitro system to generate and study latently HIV-infected long-lived normal CD4+ T-lymphocytes. Virology 2006;355:127–137 [DOI] [PubMed] [Google Scholar]
- 16.Lassen KG, Hebbeler AM, Bhattacharyya D, Lobritz MA, Greene WC: A flexible model of HIV-1 latency permitting evaluation of many primary CD4 T-cell reservoirs. PLoS One 2012;7:e30176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen K, Das B, Dobrowolski C, Karn J: Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. MBio 2017;8:e00133-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spina CA, Anderson J, Archin NM, et al. : An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog 2013;9:e1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosque A, Planelles V: Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 2009;113:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martins LJ, Bonczkowski P, Spivak AM, et al. : Modeling HIV-1 latency in primary T cells using a replication-competent virus. AIDS Res Hum Retroviruses 2016;32:187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNamara LA, Ganesh JA, Collins KL: Latent HIV-1 infection occurs in multiple subsets of hematopoietic progenitor cells and is reversed by NF-kappaB activation. J Virol 2012;86:9337–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaikos TD, Painter MM, Sebastian Kettinger NT, Terry VH, Collins KL: Class 1-selective histone deacetylase (HDAC) inhibitors enhance HIV latency reversal while preserving the activity of HDAC isoforms necessary for maximal HIV gene expression. J Virol 2018;92:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spivak AM, Planelles V: HIV-1 eradication: Early trials (and tribulations). Trends Mol Med 2016;22:10–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y, Anderson JL, Lewin SR: Getting the “Kill” into “Shock and Kill”: Strategies to eliminate latent HIV. Cell Host Microbe 2018;23:14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF: Latency reversal and viral clearance to cure HIV-1. Science 2016;353:aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas AS, Jones KL, Gandhi RT, et al. : T-cell responses targeting HIV Nef uniquely correlate with infected cell frequencies after long-term antiretroviral therapy. PLoS Pathog 2017;13:e1006629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahm HW, Stein S: Characterization of recombinant human interleukin-2 with micromethods. J Chromatogr 1985;326:357–361 [DOI] [PubMed] [Google Scholar]
- 28.Adachi A, Gendelman HE, Koenig S, et al. : Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 1986;59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosque A, Planelles V: Studies of HIV-1 latency in an ex vivo model that uses primary central memory T cells. Methods 2011;53:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messi M, Giacchetto I, Nagata K, Lanzavecchia A, Natoli G, Sallusto F: Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nat Immunol 2003;4:78–86 [DOI] [PubMed] [Google Scholar]
- 31.White CH, Moesker B, Beliakova-Bethell N, et al. : Transcriptomic analysis implicates the p53 signaling pathway in the establishment of HIV-1 latency in central memory CD4 T cells in an in vitro model. PLoS Pathog 2016;12:e1006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehla R, Bivalkar-Mehla S, Zhang R, et al. : Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor independent manner. PLoS One 2010;5:e11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang G, Mendes EA, Kaiser P, et al. : Reactivation of HIV latency by a newly modified ingenol derivative via protein kinase Cdelta-NF-kappaB signaling. AIDS 2014;28:1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosque A, Nilson KA, Macedo AB, et al. : Benzotriazoles reactivate latent HIV-1 through inactivation of STAT5 SUMOylation. Cell Rep 2017;18:1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novis CL, Archin NM, Buzon MJ, et al. : Reactivation of latent HIV-1 in central memory CD4(+) T cells through TLR-1/2 stimulation. Retrovirology 2013;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larson EC, Novis CL, Martins LJ, et al. : Mycobacterium tuberculosis reactivates latent HIV-1 in T cells in vitro. PLoS One 2017;12:e0185162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Contreras X, Schweneker M, Chen CS, et al. : Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem 2009;284:6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM: Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 2009;25:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Addo MM, Altfeld M: Sex-based differences in HIV type 1 pathogenesis. J Infect Dis 2014;209;Suppl 3:S86–S92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein SL, Flanagan KL: Sex differences in immune responses. Nat Rev Immunol 2016;16:626–638 [DOI] [PubMed] [Google Scholar]
- 41.Dunn SE, Ousman SS, Sobel RA, et al. : Peroxisome proliferator-activated receptor (PPAR)alpha expression in T cells mediates gender differences in development of T cell-mediated autoimmunity. J Exp Med 2007;204:321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nabel G, Baltimore D: An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 1987;326:711–713 [DOI] [PubMed] [Google Scholar]
- 43.Szotek EL, Narasipura SD, Al-Harthi L: 17beta-estradiol inhibits HIV-1 by inducing a complex formation between beta-catenin and estrogen receptor alpha on the HIV promoter to suppress HIV transcription. Virology 2013;443:375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bafica A, Scanga CA, Schito ML, Hieny S, Sher A: Cutting edge: In vivo induction of integrated HIV-1 expression by mycobacteria is critically dependent on Toll-like receptor 2. J Immunol 2003;171:1123–1127 [DOI] [PubMed] [Google Scholar]
- 45.Alvarez-Carbonell D, Garcia-Mesa Y, Milne S, et al. : Toll-like receptor 3 activation selectively reverses HIV latency in microglial cells. Retrovirology 2017;14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thibault S, Imbeault M, Tardif MR, Tremblay MJ: TLR5 stimulation is sufficient to trigger reactivation of latent HIV-1 provirus in T lymphoid cells and activate virus gene expression in central memory CD4+ T cells. Virology 2009;389:20–25 [DOI] [PubMed] [Google Scholar]
- 47.Tsai A, Irrinki A, Kaur J, et al. : Toll-like receptor 7 agonist GS-9620 induces HIV expression and HIV-specific immunity in cells from HIV-infected individuals on suppressive antiretroviral therapy. J Virol 2017;91:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlaepfer E, Speck RF: TLR8 activates HIV from latently infected cells of myeloid-monocytic origin directly via the MAPK pathway and from latently infected CD4+ T cells indirectly via TNF-alpha. J Immunol 2011;186:4314–4324 [DOI] [PubMed] [Google Scholar]
- 49.Scheller C, Ullrich A, McPherson K, et al. : CpG oligodeoxynucleotides activate HIV replication in latently infected human T cells. J Biol Chem 2004;279:21897–21902 [DOI] [PubMed] [Google Scholar]
- 50.Offersen R, Nissen SK, Rasmussen TA, et al. : A novel toll-like receptor 9 agonist, MGN1703, enhances HIV-1 transcription and NK Cell-mediated inhibition of HIV-1-infected autologous CD4+ T cells. J Virol 2016;90:4441–4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vibholm L, Schleimann MH, Hojen JF, et al. : Short-course toll-like receptor 9 agonist treatment impacts innate immunity and plasma viremia in individuals with human immunodeficiency virus infection. Clin Infect Dis 2017;64:1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V: Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog 2011;7:e1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chomont N, El-Far M, Ancuta P, et al. : HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009;15:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandergeeten C, Fromentin R, DaFonseca S, et al. : Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 2013;121:4321–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, Gurule EE, Brennan TP, et al. : Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc Natl Acad Sci U S A 2018;115:E2575–E2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maldarelli F, Wu X, Su L, et al. : HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014;345:179–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner TA, McLaughlin S, Garg K, et al. : HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014;345:570–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sunshine S, Kirchner R, Amr SS, et al. : HIV integration site analysis of cellular models of HIV latency with a probe-enriched next-generation sequencing assay. J Virol 2016;90:4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]