Abstract
Latent infection of CD4+ T cells is the main barrier to eradicating HIV-1 infection from infected patients. The cellular and molecular mechanisms involved in the establishment and maintenance of latent infection are directly linked to the transcriptional program of the different CD4+ T cell subsets targeted by the virus. In this review, we provide an overview of how T cell activation, T cell differentiation into functional subsets, and the mode of initial viral infection influence HIV proviral transcription and entry into latency.
Keywords: : HIV latency, HIV transcription, CD4 T cell differentiation
Introduction
HIV-1 is intricately linked to the biology of its preferred target host cell type, CD4+ T cells. This is particularly evident when considering the transcription of the HIV provirus and the combinatorial requirement for general cellular transcriptional machinery, chromatin regulators, and cell lineage-specific factors. The presence and absence of specific factors or repressive transcriptional mechanisms in different T cell subsets may promote the repression of proviral transcription thus establishing, maintaining, and biasing latent HIV-1 infection in different T cell subsets. HIV may encounter different cellular transcriptional conditions during initial entry and integration into a host cell or the transcriptional conditions can change after HIV has already integrated into the host DNA. Whether a host cell possesses a favorable or unfavorable transcriptional environment is directly dependent on T cell activation, cell cycle progression, maturation, differentiation status, and signals generated at the time of initial infection.
T cell maturation is driven in large part by the strength of signal through the T cell receptor (TCR)–major histocompatibility complex (MHC)–peptide interactions with antigen-presenting cells (APCs). However, it is also strongly influenced by other environmental cues in the tissue microenvironment, including cytokines, chemokines, and interactions with neighboring non-APCs. There have been several reviews that have focused on the general biochemical mechanism of transcriptional regulation, such as chromatin remodeling and RNAP II (RNA polymerase II complex) pausing that limit HIV transcription.1–3 In this review, we will focus on how intrinsic cell lineage-specific factors that are initiated by T cell maturation and the mode of infection may influence HIV replication and the establishment of latency.
Overview of T cell Activation and Maturation
Canonical antigen presentation to CD4+ T cells involves the direct cell–cell interaction between APCs and naive CD4+ T cells.4 APCs present MHC-II loaded with peptide to the TCR, whereas costimulatory molecules on naive T cells interact with their ligands on the surface of APCs. The macromolecular complex generated upon cell–cell interaction is known as the immunological synapse.5,6 Establishment of APC–T cell immunological synapses triggers cascades of positive and negative signaling that include nonreceptor tyrosine kinases, phosphorylation of downstream adaptor proteins, the assembly of multimolecular complexes that include lipid kinases, lipases, guanine nucleotide exchange factors, and small G-proteins, which culminate in increased intracellular calcium, activation of ERK/MAPK pathways, and induction of cellular transcription factors.7–9
T cell signals converge to remodel cortical actin and redistribute surface receptors and membrane domains of activated T cells.7–9 These signals control the immune response in part by influencing the generation and maintenance of T effector populations, memory cells, and tolerized T cells. They also polarize T cell responses, which in turn drive the activity and function of other immune cells, including but not limited to macrophages, dendritic cells (DCs), B cells, other CD4+ and CD8+ T cells.
The specific effector functions of stimulated T cells will largely depend on the strength of the signaling cascade. Binding avidity of the MHC-II/peptide complex to the TCR, the duration of this interaction, and engagement of costimulatory receptors and cytokines are the primary determinants of signaling strength.7 For example, differentiation of naive T cells to Th1 or Th2 effector functions can be modulated by stronger or weaker TCR signals, respectively.10–12 Strong signaling can also bias naive T cells toward effector subsets that have a relatively short half-life, whereas weaker signaling in response to self antigen can drive cells into anergy to establish immunological tolerance.10,13–16
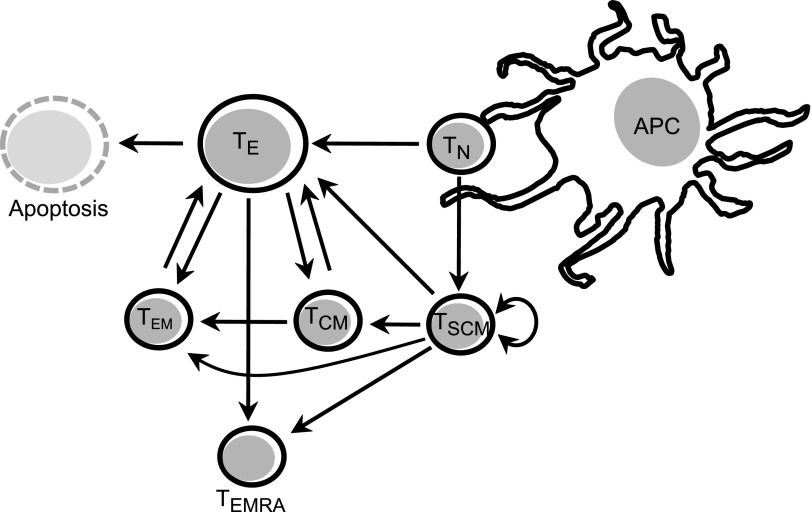
Upon resolution of the T cell response, most effector cells will turnover with relatively short half-lives, whereas a small subset will survive as memory cells (Fig. 1). Upon reencountering antigens, memory cells will respond more vigorously, thus forming the basis for immunological memory. Several subsets of memory cells have been recognized, including stem cell memory cells (TSCM), central memory cells (TCM), effector memory cells (TEM), and terminally differentiated memory cells (TEMRA or TTE).17 TSCM are a recently identified self-renewing subpopulation of memory cells that give rise to effector T cells and other memory cell populations.18–21 TCM have long half-lives and represent the primary cellular compartment responsible for long-lasting immunological memory.22,23 TEM cells home directly to inflamed tissues and are characterized by rapid response to antigen.22,23 TEMRA have low proliferative and functional capacity and express some markers of senescence.17 Homeostatic proliferation of these memory populations may require low-level signaling through the TCR and/or cytokines, including IL-7 and IL-15.24
FIG. 1.
Overview of T cell differentiation. Upon antigen presentation, TN are activated and differentiate in effector cells (TE). The specific effector functions that these cells will undertake largely depend on the stimulus and the cytokine milieu during antigen presentation. A fraction of TE cells will undergo apoptosis after resolution of the immune response while another fraction will return to a resting state to become memory cells (TEM, TCM, and TEMRA). TCM can differentiate into TEM or TE depending on the stimulus, whereas TEM can typically only differentiate into TE during subsequent stimulus. Some TN differentiate into TSCM after antigen presentation. These cells have self-renewal potential and can also differentiate into other memory subsets.
In the absence of professional APCs, T cells can be partially activated through alternative mechanisms. T cells have been shown to be activated by nonprofessional APCs, such as endothelial cells,25–27 stromal cells,28 and even activated CD4+ T cells.29–32 These atypical APCs express MHC-II but lack costimulatory molecules, such as CD28, which drives partial activation and biasing T cells toward an anergic or inactive state.33 T cell anergy is characterized by a significant reduction in cell proliferation and diminished release of cytokines upon subsequent antigen presentation. A similar inert T cell phenotype results from overstimulation or persistent signaling of T cells, as often occurs during chronic inflammatory diseases, including HIV infection and cancer. These exhausted T cells are defined by their loss of immune effector potential and proliferation.34 Both T cell anergy and T cell exhaustion have their own transcriptional profiles and are thought to be important mechanisms for controlling overactive immune responses and antigenic tolerance.15,35–37
In considering the generation of T cells into different functional subsets it is important to recognize that T cell maturation is a regulated process that involves differential T cell specific gene expression patterns.38,39 Effector and memory subsets as well as exhausted and anergic cells express distinct batteries of genes that maintain their phenotypes and function. Gene expression in these T cell subsets are actively maintained by combinatorial activities between cell-specific transcription factors, more general coactivator and corepressor complexes and chromatin remodeling factors. During HIV entry into different T subsets, the virus will encounter different transcriptional profiles and different intrinsic cellular factors, which will either support proviral transcription or drive proviral repression and latency.
T Cell Activation and Intrinsic Cellular Factors Regulate HIV Replication
HIV-1 can infect target cells through the dissemination of cell-free particles or through direct cell–cell contact.40,41 Similar to antigen presentation, efficient cell–cell contact-mediated transfer of HIV correlates with the redistribution of surface receptors, lipid rafts, reorganization of cortical actin, and the delivery of signals across a synaptic junction.42–46 Because of the physical and functional similarities to the immunological synapse, these structures have been termed infectious synapses (dendritic cell–T cell junctions) or virological synapses (T cell–T cell junctions).47–50 Whether the quality of signals emanating from this synapse support or alter HIV productive infection and the establishment of latency has not been fully investigated; although, HIV co-opts T cell activation to ensure efficient infection and replication.
For example, reports, including those from our laboratory,51–56 have demonstrated that tyrosine kinases, Lck,55,57,58 Fyn,59 ZAP-70,60,61 ITK,53,54,62 lipid kinases PI3K52,63–65 and PI4P5 kinase,66 and MAP kinase pathways67 regulate HIV entry, reverse transcription, proviral transcription, virus assembly, and release. We and others have also shown that signals emanating from CD28 positively and negatively regulate HIV transcription.51,52,65,68,69 Weaker signals, such as those that induce homeostatic proliferation of T cells, seem sufficient for cell division but not for stimulating HIV production. This is the basis for the expansion of latently infected cell clones in some HIV-infected individuals.70–76
In addition, the quality, duration, and magnitude of TCR and CD28-associated signals set a threshold for completing reverse transcription and productive HIV infection.65 For example, T cell activation has been shown to enhance the efficiency of reverse transcription since it is associated with low expression of the restriction factor SAMHD1 and increased availability of nucleotides.77–79 T cell activation will also influence integration site selection by reorganizing general chromatin organization and localization of transcriptionally active open chromatin to the nuclear periphery near nucleoporin structures.80–83 That antigen receptor-driven T cell activation influences HIV infection is supported by observations showing that superantigens increase susceptibility of CD4+ T cells to HIV infection84 and that in vitro and in vivo T cells specific for tetanus toxoid, Candida albicans,85 adenovirus,86 HSV-2,86 TB,87 and HIV88–90 are preferentially infected.
Once integrated, the provirus will be transcribed by the host transcriptional machinery. Efficient proviral transcription involves the binding of essential host transcription factors, such as NF-κB, AP-1, NFAT, and Sp1, and processive RNAP II.1–3 Repression of provirus transcription represents the primary mechanism of HIV latent infection. Insufficient signaling at the time of HIV infection may bias cells toward a latent infection. For example, efforts to establish primary models of latency suggest that minimal or partial activation either by polarizing cells toward a central memory phenotype,91 treating with chemokines,92, 93or infecting resting CD4+ T cells directly without additional stimulus94,95 biases in vitro infections toward latency.
Similarly, interactions with immature DCs96,97 or non-APCs, such as endothelial cells,98 and neighboring T cells may impact the establishment of latency. Ectopic cell–cell interactions or cytokine release alters the expression of intrinsic factors that control T cell maturation and potentially drive expression and repression of HIV transcription and latency in different T cell subsets through cell-specific transcriptional programs (Table 1). For example, effector memory T cells have increased expression of the transcription factors GATA-3 and c-Maf; these two factors are also essential for Th2 effector cell maturation.99–102 Both GATA-3 and c-Maf have been demonstrated to bind the LTR to co-operate with NF-κB and NFAT to facilitate transcription in Th2 and activated TEM.103–106
Table 1.
Cell Lineage Specific Intrinsic Factors and Their Effect on HIV Transcription and Latency
| Cell type | Repressive transcription factor | Activating transcription factor | References |
|---|---|---|---|
| Naïve Cells | |||
| TN | NDa | ||
| Effector Cells | |||
| Th1 | NDa | ||
| Th2 | GATA-3 | 1. Pereira LA, et al.103 | |
| cMaf | 2. Galio L, et al.104 | ||
| 3. Yang Z, et al.105 | |||
| 4. Zhang M, et al.106 | |||
| Th17 | RUNX1 | 1. Cleret-Buhot A, et al.153 | |
| PRC2 | 2. Klase Z, et al.154 | ||
| EHMT2 | 3. Nguyen K, et al.155 | ||
| Treg | FOXP3 | FOXP3 | 1. Grant C, et al.156 |
| 2. Selliah N, et al.157 | |||
| 3. Holmes D, et al.158 | |||
| 4. Holmes D, et al.159 | |||
| 5. Oswald-Richter K, et al.160 | |||
| TFH | Bcl-6 | Bcl-6 | 1. Baron BW, et al.161 |
| 2. Amet T, et al.112 | |||
| Memory Cells | |||
| TEM | Blimp-1, PRC2, EHMT2, G9a, SMYD2 | GATA-3 cMaf |
1. Kaczmarek Michaels K, et al.113 2. Nguyen K, et al.155 3. Boehm D, et al.162 |
| TCM | Blimp-1, PRC2, EHMT2, G9a, SMYD2 | ||
| TSCM | Blimp-1, PRC2, EHMT2, G9a, SMYD2 | ||
| TEMRA | Blimp-1, PRC2, EHMT2, G9a, SMYD2 |
ND = not determined.
An example of transcriptional repression by transcription factors in quiescent T cells is the Bcl6-Blimp-1 axis. These transcription factors are directly involved in the differentiation of effector T follicular helper cells (TFH) and T memory subsets.107–111 High Bcl6 expression and low Blimp-1 expression support TFH differentiation, whereas, elevated Blimp-1 expression is observed in quiescent memory T cell subsets. Both of these factors have been shown to directly and indirectly regulate HIV replication. Bcl6 was recently reported to inhibit several interferon-stimulated genes, thus likely contributing to the increased susceptibility of TFH cells to HIV replication.112 Blimp-1 on the other hand, directly interacts with interferon-stimulated response elements within the HIV proviral sequences and prevents transcription processivity, contributing to the establishment of latent infection in memory T cells.113 Blimp-1 is also upregulated in T cells that display exhausted phenotypes.36,114 Exhausted T cells have been shown to have provirus in HIV-infected individuals,115 but whether Blimp-1 is involved in the repression of HIV transcription in these cells has not been demonstrated.
Latent HIV infection appears to be preferentially contained within the central memory CD4+ T cell compartment.116–121 However, all T cell subsets that have been surveyed carry HIV provirus, including naive T cells.115–122 Latent infection of naive T cells has been largely ignored partly due to the very low frequency of these cells in HIV-infected individuals. It has been recently suggested that latent infection in naive CD4+ T cells is more difficult to reverse.123 This observation may suggest that intrinsic factors within naive cells may promote a “deep” latency that may be difficult to target by latency-reversing agents, thus having implications for strategies to reduce the size of the latent reservoir.
Cellular Mechanisms That Establish HIV Latent Populations
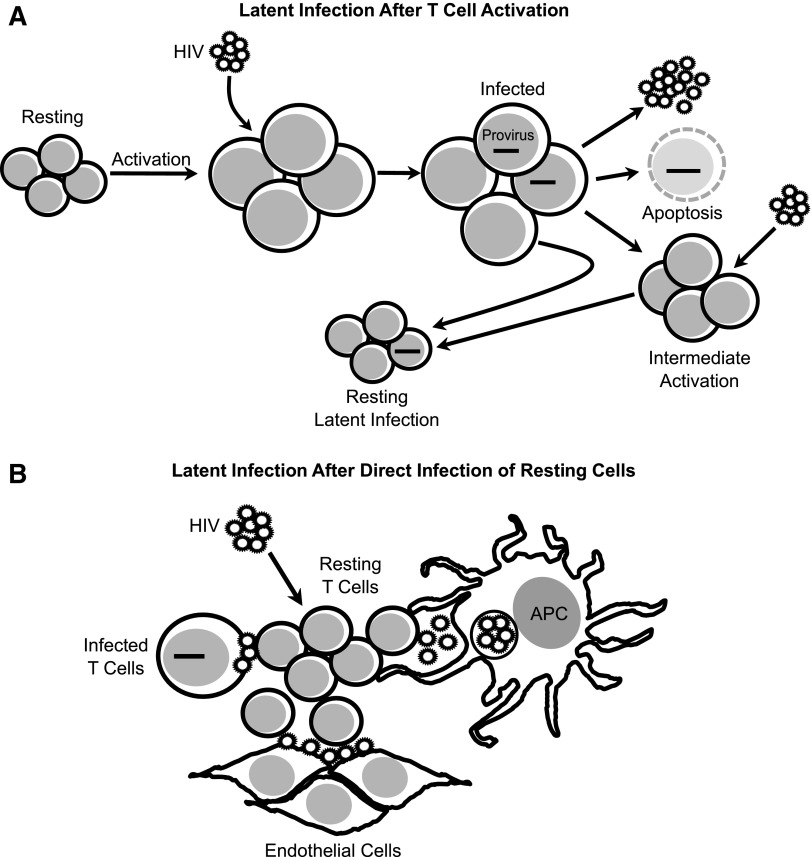
Latent infection has been suggested to be established by two mechanisms: (1) during the resolution of a T cell response when a subset of activated cells transition to a quiescent or resting state or (2) by direct infection of quiescent or resting T cells (Fig. 2). As mentioned in the previous section, most studies have found that the majority of latently infected T cells in treated individuals are memory T cells. This evidence suggests that the process of infected activated CD4+ T cells returning to a resting state is an important mechanism for the generation of latent infection in vivo.124
FIG. 2.
Cellular mechanisms for the generation of latent infection in CD4+ T cells. (A) Latent infection can be established during the transition of activated CD4+ T cells to a quiescent/resting state. When fully or intermediately activated, CD4+ T cells become infected with HIV, a proportion will undergo apoptosis due to the cytopathic effects of viral replication, and a proportion will survive infection and return to a resting state in which viral replication is suppressed.124,149 (B) Latent infection can also be established by direct infection of quiescent/resting CD4+ T cells with cell-free particles,94,136,137,150–152 or by cell-to-cell transmission from productively infected T cells (Agosto and Henderson, unpublished work), from APC,96,97,141 and from endothelial cells.98 APC, antigen-presenting cells.
Many in vitro models used for studying transcriptional regulation of latent infection are dependent on activated T cells returning to a resting state and extinguishing HIV proviral transcription.91,125–129 Such experimental models have shown that proviral transcription is limited in latently infected cells by three main biochemical mechanisms: (1) absence of positive transcription factors, such as NF-κB, to initiate proviral transcription, (2) epigenetic changes to chromatin and proviral DNA, and (3) repressive factors that prevent the processivity of the transcriptional machinery.130–133 However, it remains unclear if specific cellular factors are involved in the establishment and regulation of HIV transcription and latency when resting T cells are infected directly.134
Direct infection of quiescent or resting CD4+ T cells could explain the presence of proviral DNA in some quiescent T cell subsets such as naive cells and exhausted T cells.135–137 Interestingly, just as in activated CD4+ T cells, HIV integration in resting T cells is favored near transcriptionally active chromatin regions,138,139 yet infectious virus production is repressed. Despite the inability of infected resting CD4+ T cells to produce infectious particles, it has been observed that these cells do express some spliced HIV RNA.95 These RNAs in infected resting cells have been demonstrated to be translated into viral proteins, such as Gag and Nef, but little production of Tat, Rev, and Env is detected.95,140 Read-through of RNAP II from neighboring cellular genes or yet-to-be-identified cellular transcription factors may account for this pattern of HIV expression in latently infected resting cells.
Resting CD4+ T cells can also become infected through cell–cell contact. The best characterized mode of infection of resting cells by cell–cell contact is that mediated by dendritic cells.96,97,141 Dendritic cells capture particles through receptors such as Siglec-1/CD169 and DC-SIGN, which are then preserved in intracellular compartments.142–144 Upon interaction with T cells while probing for antigen-specific cells, particles are transferred and ultimately infect T cells. Dendritic cell maturation and functional subsets will directly influence whether this cell–cell interaction is productive or latent.96
Non-APCs have also been shown to transmit HIV directly to resting T cells, including endothelial cells98 and productively infected T cells. Although cell-to-cell transmission to resting CD4+ T cells has been suggested to be a highly inflammatory process resulting in the apoptosis of target and bystander resting T cells,145–147 recent work from our laboratory indicates that a proportion of target resting CD4+ T cells become latently infected through this process (Agosto and Henderson, unpublished observations). Interestingly, our work suggests that latent infection generated by cell-to-cell transmission between T cells is more difficult to reverse through TCR/CD28 signaling compared with latent infection generated by cell-free infection. This observation suggests that cell-to-cell transmission either modifies the transcriptional program in target T cells and a number of specific factors may be involved in tightly repressing HIV transcription or that this mode of viral transmission preferentially targets resting cells with a strongly repressive transcriptional program.
Conclusion
HIV transcription and the establishment of proviral latency are regulated by multiple biochemical and cellular mechanisms which will reflect how cells are activated, cell maturation, and differentiation. Two main strategies have been proposed for targeting the latent reservoir.148 The first strategy, known in the field as “shock and kill,” proposes to pharmacologically reactivate latent proviruses with the aim of inducing death of infected cells due to the cytopathic effects of viral replication or cytotoxic immune responses. The second strategy, known in the field as “block and lock,” proposes to suppress HIV transcription long term, thus eliminating the need for antiretroviral therapy. Regardless of which strategy will be used to target the latent reservoir, it will be critical that all populations that are harboring latent HIV are targeted to assure efficacy of a cure; thus, underscoring the importance of understanding the events of latency in multiple cell subsets in different tissue environments.
Acknowledgments
The authors would like to acknowledge the amfAR Mathilde Krim Fellowship in Basic Biomedical Research (award 109263-59-RKRL) and the amfAR Innovation Grant 109603-62-RGRL. The funding opportunity for postdoctoral research created to honor Dr. Mathilde Krim and her contributions to the advancement of HIV/AIDS research made an invaluable impact in our careers and research. The authors would also like to thank the NIH (awards AI097117 and AI118682) and the Providence–Boston CFAR (P30-AI042853) for additional funding.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Mbonye U, Karn J: The molecular basis for human immunodeficiency virus latency. Annu Rev Virol 2017;4:261–285 [DOI] [PubMed] [Google Scholar]
- 2.Agosto LM, Gagne M, Henderson AJ: Impact of chromatin on HIV replication. Genes 2015;6:957–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruelas DS, Greene WC: An integrated overview of HIV-1 latency. Cell 2013;155:519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith-Garvin JE, Koretzky GA, Jordan MS: T cell activation. Annu Rev Immunol 2009;27:591–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dustin ML, Chakraborty AK, Shaw AS: Understanding the structure and function of the immunological synapse. Cold Spring Harb Perspect Biol 2010;2:a002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumari S, Curado S, Mayya V, Dustin ML: T cell antigen receptor activation and actin cytoskeleton remodeling. Biochim Biophys Acta 2014;1838:546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Flies DB: Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 2013;13:227–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brownlie RJ, Zamoyska R: T cell receptor signalling networks: Branched, diversified and bounded. Nat Rev Immunol 2013;13:257–269 [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty AK, Weiss A: Insights into the initiation of TCR signaling. Nat Immunol 2014;15:798–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corse E, Gottschalk RA, Allison JP: Strength of TCR-peptide/MHC interactions and in vivo T cell responses. J Immunol 2011;186:5039–5045 [DOI] [PubMed] [Google Scholar]
- 11.Constant S, Pfeiffer C, Woodard A, Pasqualini T, Bottomly K: Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J Exp Med 1995;182:1591–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosken NA, Shibuya K, Heath AW, Murphy KM, O'Garra A: The effect of antigen dose on CD4+ T helper cell phenotype development in a T cell receptor-alpha beta-transgenic model. J Exp Med 1995;182:1579–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh NJ, Schwartz RH: The strength of persistent antigenic stimulation modulates adaptive tolerance in peripheral CD4+ T cells. J Exp Med 2003;198:1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards LJ, Evavold BD: T cell recognition of weak ligands: Roles of signaling, receptor number, and affinity. Immunol Res 2011;50:39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chappert P, Schwartz RH: Induction of T cell anergy: Integration of environmental cues and infectious tolerance. Curr Opin Immunol 2010;22:552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadegh-Nasseri S, Dalai SK, Korb Ferris LC, Mirshahidi S: Suboptimal engagement of the T-cell receptor by a variety of peptide-MHC ligands triggers T-cell anergy. Immunology 2010;129:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E: The who's who of T-cell differentiation: Human memory T-cell subsets. Eur J Immunol 2013;43:2797–2809 [DOI] [PubMed] [Google Scholar]
- 18.Gattinoni L, Speiser DE, Lichterfeld M, Bonini C: T memory stem cells in health and disease. Nat Med 2017;23:18–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Joe G, Hexner E, Zhu J, Emerson SG: Host-reactive CD8+ memory stem cells in graft-versus-host disease. Nat Med 2005;11:1299–1305 [DOI] [PubMed] [Google Scholar]
- 20.Gattinoni L, Lugli E, Ji Y, et al. : A human memory T cell subset with stem cell-like properties. Nat Med 2011;17:1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lugli E, Gattinoni L, Roberto A, et al. : Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat Protoc 2013;8:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A: Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999;401:708–712 [DOI] [PubMed] [Google Scholar]
- 23.Sallusto F, Geginat J, Lanzavecchia A: Central memory and effector memory T cell subsets: Function, generation, and maintenance. Annu Rev Immunol 2004;22:745–763 [DOI] [PubMed] [Google Scholar]
- 24.Surh CD, Sprent J: Homeostasis of naive and memory T cells. Immunity 2008;29:848–862 [DOI] [PubMed] [Google Scholar]
- 25.Hughes CC, Savage CO, Pober JS: Endothelial cells augment T cell interleukin 2 production by a contact-dependent mechanism involving CD2/LFA-3 interaction. J Exp Med 1990;171:1453–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiao SL, Kirkiles-Smith NC, Shepherd BR, McNiff JM, Carr EJ, Pober JS: Human effector memory CD4+ T cells directly recognize allogeneic endothelial cells in vitro and in vivo. J Immunol 2007;179:4397–4404 [DOI] [PubMed] [Google Scholar]
- 27.Shiao SL, McNiff JM, Pober JS: Memory T cells and their costimulators in human allograft injury. J Immunol 2005;175:4886–4896 [DOI] [PubMed] [Google Scholar]
- 28.Cohen JN, Guidi CJ, Tewalt EF, et al. : Lymph node-resident lymphatic endothelial cells mediate peripheral tolerance via Aire-independent direct antigen presentation. J Exp Med 2010;207:681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taams LS, van Eden W, Wauben MH: Antigen presentation by T cells versus professional antigen-presenting cells (APC): Differential consequences for T cell activation and subsequent T cell-APC interactions. Eur J Immunol 1999;29:1543–1550 [DOI] [PubMed] [Google Scholar]
- 30.Lamb JR, Skidmore BJ, Green N, Chiller JM, Feldmann M: Induction of tolerance in influenza virus-immune T lymphocyte clones with synthetic peptides of influenza hemagglutinin. J Exp Med 1983;157:1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidhu S, Deacock S, Bal V, Batchelor JR, Lombardi G, Lechler RI: Human T cells cannot act as autonomous antigen-presenting cells, but induce tolerance in antigen-specific and alloreactive responder cells. J Exp Med 1992;176:875–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kambayashi T, Laufer TM: Atypical MHC class II-expressing antigen-presenting cells: Can anything replace a dendritic cell? Nat Rev Immunol 2014;14:719–730 [DOI] [PubMed] [Google Scholar]
- 33.Schwartz RH: T cell anergy. Annu Rev Immunol 2003;21:305–334 [DOI] [PubMed] [Google Scholar]
- 34.Wherry EJ: T cell exhaustion. Nat Immunol 2011;12:492–499 [DOI] [PubMed] [Google Scholar]
- 35.Zheng Y, Zha Y, Gajewski TF: Molecular regulation of T-cell anergy. EMBO Rep 2008;9:50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crawford A, Angelosanto JM, Kao C, et al. : Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity 2014;40:289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crespo J, Sun H, Welling TH, Tian Z, Zou W: T cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr Opin Immunol 2013;25:214–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weng NP, Araki Y, Subedi K: The molecular basis of the memory T cell response: Differential gene expression and its epigenetic regulation. Nat Rev Immunol 2012;12:306–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang JT, Wherry EJ, Goldrath AW: Molecular regulation of effector and memory T cell differentiation. Nat Immunol 2014;15:1104–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong P, Agosto LM, Munro JB, Mothes W: Cell-to-cell transmission of viruses. Curr Opin Virol 2013;3:44–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sattentau Q: Avoiding the void: Cell-to-cell spread of human viruses. Nat Rev Microbiol 2008;6:815–826 [DOI] [PubMed] [Google Scholar]
- 42.Vasiliver-Shamis G, Cho MW, Hioe CE, Dustin ML: Human immunodeficiency virus type 1 envelope gp120-induced partial T-cell receptor signaling creates an F-actin-depleted zone in the virological synapse. J Virol 2009;83:11341–11355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasiliver-Shamis G, Tuen M, Wu TW, et al. : Human immunodeficiency virus type 1 envelope gp120 induces a stop signal and virological synapse formation in noninfected CD4+ T cells. J Virol 2008;82:9445–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agosto LM, Uchil PD, Mothes W: HIV cell-to-cell transmission: Effects on pathogenesis and antiretroviral therapy. Trends Microbiol 2015;23:289–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulpa DA, Brehm JH, Fromentin R, et al. : The immunological synapse: The gateway to the HIV reservoir. Immunol Rev 2013;254:305–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spear M, Guo J, Wu Y: The trinity of the cortical actin in the initiation of HIV-1 infection. Retrovirology 2012;9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDonald D, Wu L, Bohks SM, KewalRamani VN, Unutmaz D, Hope TJ: Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 2003;300:1295–1297 [DOI] [PubMed] [Google Scholar]
- 48.Jolly C, Kashefi K, Hollinshead M, Sattentau QJ: HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med 2004;199:283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed Z, Kawamura T, Shimada S, Piguet V: The role of human dendritic cells in HIV-1 infection. J Invest Dermatol 2015;135:1225–1233 [DOI] [PubMed] [Google Scholar]
- 50.Dale BM, Alvarez RA, Chen BK: Mechanisms of enhanced HIV spread through T-cell virological synapses. Immunol Rev 2013;251:113–124 [DOI] [PubMed] [Google Scholar]
- 51.Cook JA, Albacker L, August A, Henderson AJ: CD28-dependent HIV-1 transcription is associated with Vav, Rac, and NF-kappa B activation. J Biol Chem 2003;278:35812–35818 [DOI] [PubMed] [Google Scholar]
- 52.Cook JA, August A, Henderson AJ: Recruitment of phosphatidylinositol 3-kinase to CD28 inhibits HIV transcription by a Tat-dependent mechanism. J Immunol 2002;169:254–260 [DOI] [PubMed] [Google Scholar]
- 53.Readinger JA, Schiralli GM, Jiang JK, et al. : Selective targeting of ITK blocks multiple steps of HIV replication. Proc Natl Acad Sci U S A 2008;105:6684–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiralli Lester GM, Akiyama H, Evans E, Singh J, Gummuluru S, Henderson AJ: Interleukin 2-inducible T cell kinase (ITK) facilitates efficient egress of HIV-1 by coordinating Gag distribution and actin organization. Virology 2013;436:235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strasner AB, Natarajan M, Doman T, Key D, August A, Henderson AJ: The Src kinase Lck facilitates assembly of HIV-1 at the plasma membrane. J Immunol 2008;181:3706–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang P, Henderson AJ: Nef enhances c-Cbl phosphorylation in HIV-infected CD4+ T lymphocytes. Virology 2005;336:219–228 [DOI] [PubMed] [Google Scholar]
- 57.Pan X, Rudolph JM, Abraham L, et al. : HIV-1 Nef compensates for disorganization of the immunological synapse by inducing trans-Golgi network-associated Lck signaling. Blood 2012;119:786–797 [DOI] [PubMed] [Google Scholar]
- 58.van Wilgenburg B, Moore MD, James WS, Cowley SA: The productive entry pathway of HIV-1 in macrophages is dependent on endocytosis through lipid rafts containing CD4. PLoS One 2014;9:e86071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hohashi N, Hayashi T, Fusaki N, et al. : The protein tyrosine kinase Fyn activates transcription from the HIV promoter via activation of NF kappa B-like DNA-binding proteins. Int Immunol 1995;7:1851–1859 [DOI] [PubMed] [Google Scholar]
- 60.Hung CH, Thomas L, Ruby CE, et al. : HIV-1 Nef assembles a Src family kinase-ZAP-70/Syk-PI3K cascade to downregulate cell-surface MHC-I. Cell Host Microbe 2007;1:121–133 [DOI] [PubMed] [Google Scholar]
- 61.Sol-Foulon N, Sourisseau M, Porrot F, et al. : ZAP-70 kinase regulates HIV cell-to-cell spread and virological synapse formation. EMBO J 2007;26:516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarafdar S, Poe JA, Smithgall TE: The accessory factor Nef links HIV-1 to Tec/Btk kinases in an Src homology 3 domain-dependent manner. J Biol Chem 2014;289:15718–15728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Contreras X, Barboric M, Lenasi T, Peterlin BM: HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog 2007;3:1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doyon G, Sobolewski MD, Huber K, McMahon D, Mellors JW, Sluis-Cremer N: Discovery of a small molecule agonist of phosphatidylinositol 3-kinase p110alpha that reactivates latent HIV-1. PLoS One 2014;9:e84964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oswald-Richter K, Grill SM, Leelawong M, Unutmaz D: HIV infection of primary human T cells is determined by tunable thresholds of T cell activation. Eur J Immunol 2004;34:1705–1714 [DOI] [PubMed] [Google Scholar]
- 66.Barrero-Villar M, Barroso-Gonzalez J, Cabrero JR, et al. : PI4P5-kinase Ialpha is required for efficient HIV-1 entry and infection of T cells. J Immunol 2008;181:6882–6888 [DOI] [PubMed] [Google Scholar]
- 67.Furler RL, Uittenbogaart CH: Signaling through the P38 and ERK pathways: A common link between HIV replication and the immune response. Immunol Res 2010;48:99–109 [DOI] [PubMed] [Google Scholar]
- 68.Natarajan M, August A, Henderson AJ: Combinatorial signals from CD28 differentially regulate human immunodeficiency virus transcription in T cells. J Biol Chem 2010;285:17338–17347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Annibaldi A, Sajeva A, Muscolini M, et al. : CD28 ligation in the absence of TCR promotes RelA/NF-kappaB recruitment and trans-activation of the HIV-1 LTR. Eur J Immunol 2008;38:1446–1451 [DOI] [PubMed] [Google Scholar]
- 70.Hosmane NN, Kwon KJ, Bruner KM, et al. : Proliferation of latently infected CD4(+) T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J Exp Med 2017;214:959–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bui JK, Sobolewski MD, Keele BF, et al. : Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog 2017;13:e1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lorenzi JC, Cohen YZ, Cohn LB, et al. : Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc Natl Acad Sci U S A 2016:113:E7908–E7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simonetti FR, Sobolewski MD, Fyne E, et al. : Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 2016;113:1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vandergeeten C, Fromentin R, DaFonseca S, et al. : Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 2013;121:4321–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V: Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog 2011;7:e1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maldarelli F, Wu X, Su L, et al. : HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014;345:179–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plesa G, Dai J, Baytop C, Riley JL, June CH, O'Doherty U: Addition of deoxynucleosides enhances human immunodeficiency virus type 1 integration and 2LTR formation in resting CD4+ T cells. J Virol 2007;81:13938–13942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baldauf HM, Pan X, Erikson E, et al. : SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 2012;18:1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baldauf HM, Stegmann L, Schwarz SM, et al. : Vpx overcomes a SAMHD1-independent block to HIV reverse transcription that is specific to resting CD4 T cells. Proc Natl Acad Sci U S A 2017;114:2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marini B, Kertesz-Farkas A, Ali H, et al. : Nuclear architecture dictates HIV-1 integration site selection. Nature 2015;521:227–231 [DOI] [PubMed] [Google Scholar]
- 81.Lelek M, Casartelli N, Pellin D, et al. : Chromatin organization at the nuclear pore favours HIV replication. Nat Commun 2015;6:6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Di Primio C, Quercioli V, Allouch A, et al. : Single-cell imaging of HIV-1 provirus (SCIP). Proc Natl Acad Sci U S A 2013;110:5636–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Albanese A, Arosio D, Terreni M, Cereseto A: HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PLoS One 2008;3:e2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maier R, Bartolome-Rodriguez MM, Moulon C, Weltzien HU, Meyerhans A: Kinetics of CXCR4 and CCR5 up-regulation and human immunodeficiency virus expansion after antigenic stimulation of primary CD4(+) T lymphocytes. Blood 2000;96:1853–1856 [PubMed] [Google Scholar]
- 85.Hu H, Nau M, Ehrenberg P, et al. : Distinct gene-expression profiles associated with the susceptibility of pathogen-specific CD4 T cells to HIV-1 infection. Blood 2013;121:1136–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu H, Eller MA, Zafar S, et al. : Preferential infection of human Ad5-specific CD4 T cells by HIV in Ad5 naturally exposed and recombinant Ad5-HIV vaccinated individuals. Proc Natl Acad Sci U S A 2014;111:13439–13444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Geldmacher C, Ngwenyama N, Schuetz A, et al. : Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection. J Exp Med 2010;207:2869–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Demoustier A, Gubler B, Lambotte O, et al. : In patients on prolonged HAART, a significant pool of HIV infected CD4 T cells are HIV-specific. AIDS 2002;16:1749–1754 [DOI] [PubMed] [Google Scholar]
- 89.Douek DC, Brenchley JM, Betts MR, et al. : HIV preferentially infects HIV-specific CD4+ T cells. Nature 2002;417:95–98 [DOI] [PubMed] [Google Scholar]
- 90.Brenchley JM, Ruff LE, Casazza JP, Koup RA, Price DA, Douek DC: Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. J Virol 2006;80:6801–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bosque A, Planelles V: Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 2009;113:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saleh S, Wightman F, Ramanayake S, et al. : Expression and reactivation of HIV in a chemokine induced model of HIV latency in primary resting CD4+ T cells. Retrovirology 2011;8:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cameron PU, Saleh S, Sallmann G, et al. : Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc Natl Acad Sci U S A 2010;107:16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Swiggard WJ, Baytop C, Yu JJ, et al. : Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J Virol 2005;79:14179–14188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pace MJ, Graf EH, Agosto LM, et al. : Directly infected resting CD4+T cells can produce HIV Gag without spreading infection in a model of HIV latency. PLoS Pathog 2012;8:e1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kumar NA, Cheong K, Powell DR, et al. : The role of antigen presenting cells in the induction of HIV-1 latency in resting CD4(+) T-cells. Retrovirology 2015;12:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Evans VA, Kumar N, Filali A, et al. : Myeloid dendritic cells induce HIV-1 latency in non-proliferating CD4+ T cells. PLoS Pathog 2013;9:e1003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shen A, Baker JJ, Scott GL, Davis YP, Ho YY, Siliciano RF: Endothelial cell stimulation overcomes restriction and promotes productive and latent HIV-1 infection of resting CD4+ T cells. J Virol 2013;87:9768–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim JI, Ho IC, Grusby MJ, Glimcher LH: The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity 1999;10:745–751 [DOI] [PubMed] [Google Scholar]
- 100.Ho IC, Hodge MR, Rooney JW, Glimcher LH: The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell 1996;85:973–983 [DOI] [PubMed] [Google Scholar]
- 101.Zheng W, Flavell RA: The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 1997;89:587–596 [DOI] [PubMed] [Google Scholar]
- 102.O'Garra A, Gabrysova L: Transcription factors directing Th2 differentiation: Gata-3 plays a dominant role. J Immunol 2016;196:4423–4425 [DOI] [PubMed] [Google Scholar]
- 103.Pereira LA, Churchill MJ, Elefanty AG, et al. : Characterization of interactions between transcription factors and a regulatory region spanning nt -320 to -281 of the HIV-1 LTR in T-lymphoid and non-T-lymphoid cells. J Biomed Sci 2002;9:68–81 [DOI] [PubMed] [Google Scholar]
- 104.Galio L, Briquet S, Cot S, Guillet JG, Vaquero C: Analysis of interactions between huGATA-3 transcription factor and three GATA regulatory elements of HIV-1 long terminal repeat, by surface plasmon resonance. Anal Biochem 1997;253:70–77 [DOI] [PubMed] [Google Scholar]
- 105.Yang Z, Engel JD: Human T cell transcription factor GATA-3 stimulates HIV-1 expression. Nucleic Acids Res 1993;21:2831–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang M, Clausell A, Robinson T, et al. : Host factor transcriptional regulation contributes to preferential expression of HIV type 1 in IL-4-producing CD4 T cells. J Immunol 2012;189:2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Johnston RJ, Poholek AC, DiToro D, et al. : Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2009;325:1006–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oestreich KJ, Mohn SE, Weinmann AS: Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol 2012;13:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martins GA, Cimmino L, Shapiro-Shelef M, et al. : Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol 2006;7:457–465 [DOI] [PubMed] [Google Scholar]
- 110.Fu SH, Yeh LT, Chu CC, Yen BL, Sytwu HK: New insights into Blimp-1 in T lymphocytes: A divergent regulator of cell destiny and effector function. J Biomed Sci 2017;24:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kallies A, Hawkins ED, Belz GT, et al. : Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol 2006;7:466–474 [DOI] [PubMed] [Google Scholar]
- 112.Amet T, Son YM, Jiang L, et al. : BCL6 represses antiviral resistance in follicular T helper cells. J Leukoc Biol 2017;102:527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kaczmarek Michaels K, Natarajan M, Euler Z, Alter G, Viglianti G, Henderson AJ: Blimp-1, an intrinsic factor that represses HIV-1 proviral transcription in memory CD4+ T cells. J Immunol 2015;194:3267–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shin H, Blackburn SD, Intlekofer AM, et al. : A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity 2009;31:309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fromentin R, Bakeman W, Lawani MB, et al. : CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog 2016;12:e1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chomont N, El-Far M, Ancuta P, et al. : HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009;15:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hiener B, Horsburgh BA, Eden JS, et al. : Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep 2017;21:813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Soriano-Sarabia N, Bateson RE, Dahl NP, et al. : Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol 2014;88:14070–14077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ostrowski MA, Chun T-W, Justement SJ, et al. : Both memory and CD45RA(+)/CD62L(+) naive CD4(+) T cells are infected in human immunodeficiency type 1-infected individuals. J Virol 1999;73:6430–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pierson T, Hoffman TL, Blankson J, et al. : Characterization of chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type-1. J Virol 2000;74:7824–7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pinzone MR, O'Doherty U: Measuring integrated HIV DNA ex vivo and in vitro provides insights about how reservoirs are formed and maintained. Retrovirology 2018;15:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tran TA, de Goer de Herve MG, Hendel-Chavez H, et al. : Resting regulatory CD4 T cells: A site of HIV persistence in patients on long-term effective antiretroviral therapy. PLoS One 2008;3:e3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsunetsugu-Yokota Y, Kobayahi-Ishihara M, Wada Y, et al. : Homeostatically maintained resting naive CD4(+) T cells resist latent HIV reactivation. Front Microbiol 2016;7:1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shan L, Deng K, Gao H, et al. : Transcriptional reprogramming during effector-to-memory transition renders CD4(+) T cells permissive for latent HIV-1 infection. Immunity 2017;47:766–775.e763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Folks TM, Clouse KA, Justement J, et al. : Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci U S A 1989;86:2365–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Antoni BA, Rabson AB, Kinter A, Bodkin M, Poli G: NF-kappa B-dependent and -independent pathways of HIV activation in a chronically infected T cell line. Virology 1994;202:684–694 [DOI] [PubMed] [Google Scholar]
- 127.Yang HC, Xing S, Shan L, et al. : Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest 2009;119:3473–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tyagi M, Pearson RJ, Karn J: Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol 2010;84:6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sahu GK, Lee K, Ji J, Braciale V, Baron S, Cloyd MW: A novel in vitro system to generate and study latently HIV-infected long-lived normal CD4+ T-lymphocytes. Virology 2006;355:127–137 [DOI] [PubMed] [Google Scholar]
- 130.Natarajan M, Schiralli Lester GM, Lee C, et al. : Negative elongation factor (NELF) coordinates RNA polymerase II pausing, premature termination, and chromatin remodeling to regulate HIV transcription. J Biol Chem 2013;288:25995–26003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ping YH, Rana TM: DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J Biol Chem 2001;276:12951–12958 [DOI] [PubMed] [Google Scholar]
- 132.Zhang Z, Klatt A, Gilmour DS, Henderson AJ: Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J Biol Chem 2007;282:16981–16988 [DOI] [PubMed] [Google Scholar]
- 133.Jadlowsky JK, Wong JY, Graham AC, et al. : Negative elongation factor is required for the maintenance of proviral latency but does not induce promoter-proximal pausing of RNA polymerase II on the HIV long terminal repeat. Mol Cell Biol 2014;34:1911–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rezaei SD, Lu HK, Chang JJ, Rhodes A, Lewin SR, Cameron PU: The pathway to establishing HIV latency is critical to how latency is maintained and reversed. J Virol 2018;92:e02225–02217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dai J, Agosto LM, Baytop C, et al. : Human immunodeficiency virus integrates directly into naive resting CD4+ T cells but enters naive cells less efficiently than memory cells. J Virol 2009;83:4528–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tabler CO, Lucera MB, Haqqani AA, et al. : CD4+ memory stem cells are infected by HIV-1 in a manner regulated in part by SAMHD1 expression. J Virol 2014;88:4976–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zerbato JM, Serrao E, Lenzi G, et al. : Establishment and reversal of HIV-1 latency in naive and central memory CD4+ T cells in vitro. J Virol 2016;90:8059–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brady T, Agosto LM, Malani N, Berry CC, O'Doherty U, Bushman F: HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS 2009;23:1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sherrill-Mix S, Lewinski MK, Famiglietti M, et al. : HIV latency and integration site placement in five cell-based models. Retrovirology 2013;10:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.DeMaster LK, Liu X, VanBelzen DJ, et al. : A subset of CD4/CD8 double-negative T cells expresses HIV proteins in patients on antiretroviral therapy. J Virol 2015;90:2165–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pope M, Betjes MG, Romani N, et al. : Conjugates of dendritic cells and memory T lymphocytes from skin facilitate productive infection with HIV-1. Cell 1994;78:389–398 [DOI] [PubMed] [Google Scholar]
- 142.Izquierdo-Useros N, Lorizate M, Puertas MC, et al. : Siglec-1 is a novel dendritic cell receptor that mediates HIV-1 trans-infection through recognition of viral membrane gangliosides. PLoS Biol 2012;10:e1001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kwon DS, Gregorio G, Bitton N, Hendrickson WA, Littman DR: DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity 2002;16:135–144 [DOI] [PubMed] [Google Scholar]
- 144.Puryear WB, Akiyama H, Geer SD, et al. : Interferon-inducible mechanism of dendritic cell-mediated HIV-1 dissemination is dependent on Siglec-1/CD169. PLoS Pathog 2013;9:e1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Monroe KM, Yang Z, Johnson JR, et al. : IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 2014;343:428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Doitsh G, Galloway NL, Geng X, et al. : Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 2014;505:509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Doitsh G, Cavrois M, Lassen KG, et al. : Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 2010;143:789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Darcis G, Van Driessche B, Van Lint C: HIV latency: Should we shock or lock? Trends Immunol 2017;38:217–228 [DOI] [PubMed] [Google Scholar]
- 149.Siliciano RF, Greene WC: HIV latency. Cold Spring Harb Perspect Med 2011;1:a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vatakis DN, Bristol G, Wilkinson TA, Chow SA, Zack JA: Immediate activation fails to rescue efficient human immunodeficiency virus replication in quiescent CD4+ T cells. J Virol 2007;81:3574–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lassen KG, Hebbeler AM, Bhattacharyya D, Lobritz MA, Greene WC: A flexible model of HIV-1 latency permitting evaluation of many primary CD4 T-cell reservoirs. PLoS One 2012;7:e30176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Saleh S, Solomon A, Wightman F, Xhilaga M, Cameron PU, Lewin SR: CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: A novel model of HIV-1 latency. Blood 2007;110:4161–4164 [DOI] [PubMed] [Google Scholar]
- 153.Cleret-Buhot A, Zhang Y, Planas D, et al. : Identification of novel HIV-1 dependency factors in primary CCR4(+)CCR6(+)Th17 cells via a genome-wide transcriptional approach. Retrovirology 2015;12:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Klase Z, Yedavalli VS, Houzet L, et al. : Activation of HIV-1 from latent infection via synergy of RUNX1 inhibitor Ro5-3335 and SAHA. PLoS Pathog 2014;10:e1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nguyen K, Das B, Dobrowolski C, Karn J: Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. mBio 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Grant C, Oh U, Fugo K, et al. : Foxp3 represses retroviral transcription by targeting both NF-kappaB and CREB pathways. PLoS Pathog 2006;2:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Selliah N, Zhang M, White S, et al. : FOXP3 inhibits HIV-1 infection of CD4 T-cells via inhibition of LTR transcriptional activity. Virology 2008;381:161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Holmes D, Knudsen G, Mackey-Cushman S, Su L: FoxP3 enhances HIV-1 gene expression by modulating NFkappaB occupancy at the long terminal repeat in human T cells. J Biol Chem 2007;282:15973–15980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Holmes D, Gao J, Su L: Foxp3 inhibits HDAC1 activity to modulate gene expression in human T cells. Virology 2011;421:12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Oswald-Richter K, Grill SM, Shariat N, et al. : HIV infection of naturally occurring and genetically reprogrammed human regulatory T-cells. PLoS Biol 2004;2:E198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Baron BW, Desai M, Baber LJ, et al. : BCL6 can repress transcription from the human immunodeficiency virus type I promoter/enhancer region. Genes Chromosom Cancer 1997;19:14–21 [PubMed] [Google Scholar]
- 162.Boehm D, Jeng M, Camus G, et al. : SMYD2-mediated histone methylation contributes to HIV-1 latency. Cell Host Microbe 2017;21:569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]