OVERVIEW
The current standard of Response Evaluation Criteria in Solid Tumors (RECIST)–based tumor response evaluation is limited in its ability to accurately monitor treatment response. Radiomics, an approach involving computerized extraction of several quantitative imaging features, has shown promise in predicting as well as monitoring response to therapy. In this article, we provide a brief overview of radiomic approaches and the various analytical methods and techniques, specifically in the context of predicting and monitoring treatment response for non–small cell lung cancer (NSCLC). We briefly summarize some of the various types of radiomic features, including tumor shape and textural patterns, both within the tumor and within the adjacent tumor microenvironment. Additionally, we also discuss work in delta-radiomics or change in radiomic features (e.g., texture within the nodule) across longitudinally interspersed images in time for monitoring changes in therapy. We discuss the utility of these approaches for NSCLC, specifically the role of radiomics as a prognostic marker for treatment effectiveness and early therapy response, including chemoradiation, immunotherapy, and trimodality therapy.
The rapidly advancing therapeutic landscape in oncology is primarily reliant on the tremendous improvement in diagnostic tools and biomarkers for detecting cancer and specifically having the means to assess treatment response and detect adverse effects of treatment.1,2 The current gold standard in the case of solid tumors is the RECIST.3–5 RECIST was a guideline devised to standardize monitoring treatment based on imaging. The approach involves manually assessing differences in size of the target lesions on baseline and repeat CT scans (after therapy) to define various standardized categories3,6 of treatment response. However, even with modifications and refinements (Immune-Related Response Criteria [irRC]7 and modified RECIST [mRECIST]8), the current response evaluation criteria are limited, especially in the field of immunotherapy,9,10 because of the kinetics of tumor response to immuno-oncology treatment being unusual, with occasional radiographic progression as a result of inflammatory changes (pseudoprogression).11–14 Early response assessment would help oncologists modify or change treatments tailored individually to each patient under therapy.15
The aforementioned issues are especially critical and germane in the context of NSCLC, which makes up almost 85% of all lung cancers.16 NSCLC tumors have shown promising response to a multitude of treatment modalities, including a number of different experimental therapies. These include targeted therapies17 for common driver mutations, including EGFR,18 ALK,19 and KRAS.20,21 More recently, this has also come to include immune checkpoint inhibition therapy targeting the PD-1 receptor and its ligand, PD-L1.22
Unfortunately, for most of these therapies, there is a lack of accurate prognostic and predictive biomarkers to identify which patients will respond to a specific therapy. For instance, tissue-based markers of PD-L1 expression,23 currently the gold standard for selecting patients for immunotherapy, have not been particularly predictive.24
Consequently, there has been a push to develop and validate novel biomarkers and approaches for better character-ization and prediction of tumor response. Specifically, there has been interest in developing tools to provide better objective characterization of clinical benefit from among multiple therapeutic modalities, tools that provide potentially superior and complementary information to currently used measures, like RECIST.
RADIOMICS AS A NOVEL TOOL TO QUANTITATIVELY ANALYZE TUMOR IMAGING
Radiomics is the high-throughput extraction of quantitative imaging features from a radiographic image. Specifically, there has been a great deal of interest in relating radiomic measurements of regions of interest on radiographic images (e.g., MRI and CT scans) with presence and aggressiveness of disease.25,26
The field of radiomic analysis spans multiple approaches and modalities, including but not limited to computer-based measurements of the (1) shape of the nodule involved in capturing the surface irregularities, reflective of the differences in tumor growth; (2) intratumoral texture, which is used to characterize the intranodular heterogeneity patterns; (3) peritumoral texture that involves capturing heterogeneity patterns at the nodular interface; and (4) semantic features, which are visually discernible features described by radiologists, like presence of emphysema and nodule location. Delta-radiomic features or difference in radiomic features obtained from pre- and post-treatment scans are now being evaluated as a complement to RECIST for monitoring therapeutic response, with delta-radiomic features potentially being able to reveal subtle changes in the tumor that precede size or volumetric changes.27 Other features that have been recently shown to be promising include vessel tortuosity28 and three-dimensional volumetric features.29
Depending on the types of radiomic texture filters used (e.g., Gabor,30 Laws,31 Haralick,32 or Co-occurrence33,34), several hundreds of features might be extracted from a single scan. Typically, after feature extraction, the process of feature selection is used to filter out the number of features down to a smaller number. These features are then typically used to train a machine classifier to predict a specific clinical outcome (e.g., disease progression or recurrence).
Within the oncology space, radiomics has found applications as a means of diagnosis, a prognostic tool predicting response to therapy35 across organ systems, including but not limited to brain,36–39 head and neck,40–42 breast,43–50 lung,42,51–53 prostate,54–58 rectum,59–64 and liver.65–69 In the context of lung cancer, radiomics has successfully allowed detection of malignancies in screening CT scans,53 provided a means to differentiate between benign and malignant lesions,51 enabled the prediction of risk of recurrence post-therapy,52 and provided a means to noninvasively assess response to therapy70 as well as helped to identify patients who would most benefit from therapy.28
Radiomic Framework for Lung Cancer
In the context of lung cancer, radiomics represents a possible exciting complement to RECIST for monitoring therapeutic response on longitudinal serial imaging scans as well as helping to predicting response to treatment from baseline radiographic images. One of the most important advantages for radiomics is that it can be potentially integrated into the normal clinical workflow without being disruptive.
The underlying process of radiomic analysis can be broadly categorized into the following main categories: (1) image acquisition and preprocessing, (2) identification of the volume of interest, (3) feature extraction and selection of most discriminating features, and (4) use of the selected features to build a classifier to predict the outcome of interest. These modules are briefly described here below.
Image acquisition.
CT scans are the most commonly used diagnostic modality for identifying malignancy in a patient suspected to have lung malignancy.71 CT scans are also used in conjunction with the RECIST guidelines by radiologists and clinicians to monitor response to treatment as a function of changes in tumor volume. Current recommendations also suggest the use of low-dose CT (LDCT) as a means of screening in selected vulnerable populations. Radiomic analysis, meanwhile, is routinely done on noncontrast CT scans, because contrast can obscure the radiomic textural features.72 However, He et al73 found that the radiomic prediction accuracy in differentiating between benign and malignant nodules using intratumoral texture features only had a 5% difference in areas under the curve (AUCs; AUCnoncontrast was 0.75; AUCcontrast was 0.74).
One of the major limitations in quantitative image analysis and radiomics, however, is the wide variability and lack of uniformity across institutions, scanner platforms,74 slice thickness,75 reconstruction kernels,76 other acquisition parameters, and even nodule segmentations.9,77,78
He et al73 showed that the CT radiomic features to differentiate benign versus malignant nodules obtained on a thin slice (1.25 mm) performed better than those obtained on a thick slice (5 mm) with AUCthin of 0.75 versus AUCthick of 0.725 in an independent validation cohort (120 patients) of patients with solitary pulmonary nodules. On the same cohort, radiomics on a standard convolution kernel performed better (AUC of 0.725) compared with lung convolution kernel-based CT scan (AUC of 0.686). Mackin et al74 compared radiomic feature variability across four different CT scanner platiorms (Siemens, Philips, GE, and Toshiba) and found that the interscanner variability of features was large relative to the interpatient variability for a case of 20 patients with NSCLC.
Identification of the volume of interest and nodule segmentation.
The reconstructed images are ported into open source software that allows expert radiologists to place a boundary or a designated marker around the designated region of interest across the various slices where it appears. The identification of the volume of interest is first and the key step in additional radiomic analysis. Manual identification by a radiologist is the current gold standard in radiomic analysis. Additionally, quantitative techniques are being developed to identify regions with their own unique physiology on radio-graphic images by combining various imaging modalities.79
Meanwhile, nodule segmentation involves the process of separating the nodule from the entire CT scan and is often the most critical part of the radiomic feature analysis pipeline.35 This is primarily because a number of the radiomic parameters are extracted from the segmented volume itself. Manual annotations remain the gold standard in nodule segmentation, but semiautomated segmentation methods, often with a final check by a radiologist, have also been used as a means of establishing the “gold standard” for nodule boundary.80,81 With interobserver variability being reported to be present in manually segmented tumors,81,82 another (potentially more cumbersome) approach is to try to average or aggregate delineations from multiple individual readers.66
Feature extraction.
The different radiomic feature types are described below.
Semantic features.
These are characteristics of the tumor as described by the radiologist during the analysis of the image. These features tend to be more resilient to variations in scanner and acquisition parameters but suffer from subjective evaluation and interobserver variability.35,66 Some of these features include the location of the lung nodule, presence of emphysema, effusions, ground glass opacities, features in the lung parenchyma, nodule attenuation, and nodule marginal pafferns.72
First-order statistical features.
These features relate to statistical moments within the volume of interest and are calculated across the voxel intensities within the CT image. These statistics include energy, entropy, kurtosis, skewness, standard deviation, mean, median, range, and variance. For example, the standard deviation and variance reflect the degree to which the gray levels vary from the mean in the histogram. Kurtosis, meanwhile, measures the degree of histogram sharpness, whereas skewness reflects the asymmetry associated with the intensity distribution.83
Shape and volumetric features.
These features include margins, volume, minimum and maximum diameters, and surface area of the nodule; nodule volume was previously shown to be a predictor of therapy response.77,84,85 Shape features include width, height, depth, perimeter, area, eccentricity, compactness, radial distance, roughness, elongation equivalent diameter, and three-dimensional sphericity of the nodule.29,51,57 Other features include three-dimensional shape and margin sharpness.86,87
Texture features.
These are second-order and higher-order statistical measures to identify spatial and architectural relationships between the intensities of voxels in the area of interest. They are used to assess heterogeneous enhancement, a hallmark of malignant tumors. Below are some of the classes of texture features used for lung nodule characterization on CT scans.
Local binary patterns.
The local binary pattern feature vector involves dividing the area of interest into an array of pixels, each with a binary label. In each 3 × 3 matrix of pixels, the center pixel is compared with each of its neighboring eight pixels, and a binary value reflecting whether the center pixel is less or greater than the neighboring pixels is converted into a binary vector. Han et al88 showed the utility of local binary pattern radiomic features in distinguishing benign from malignant lung nodules on CT scans.
Gray-level co-occurrence matrices.
First described by Haralick and Shanmugam32 in conjunction with remote sensing image analysis in satellite data and eponymously called Haralick features, this class of features has been among the most popular set of radiomic measurements used in diagnosis, prognosis, and prediction of treatment response for NSCLC.52,88 Using higher-order statistics, these features provide information related to the spatial distribution and the relative position of various gray levels throughout the image.89
Laws’ texture energy measures.
Described by Laws,31 the texture energy approach measures the variations within a fixed window size. A set of nine 5 × 5 local texture masks is used to detect various texture types and then subsequently combined to calculate the texture energy. This is represented by a vector of nine numbers, each with a different texture attribute for the pixels analyzed. In a radiogenomic study,90 the Laws’ feature descriptor was also found to be the only radiomic feature descriptor to be significantly predictive for EGFR mutation (AUC of 0.67, p = .03).
Edge features.
These include Sobel91 and Canny92 filters, which determine the density and direction of the edges in the image as a means to characterize the texture pafterns. Edge detection methods have been used for lung nodule detection and segmentation from CT scans.93
Wavelet features.
These features provide a way to capture multiscale attributes across multiple different frequencies and wavelengths. These include the following attributes described below.
Fourier transforms.
This class of features involves calculating the contour of the tumor by evaluating the frequency and interval with which the tumor contour and margin change. This is accomplished by representing these changes as a wave with amplitude and frequency that can be modulated to reflect different degrees of feature granularity in the image.94
Gabor filter.
Named after Dennis Gabor and first used to describe the representation of an image in the visual cortex,95 the Gabor filters seek to capture gradients across different wavelengths and orientations.30,94,96 Gabor filters of various scales and rotations have been previously used to characterize textural pafterns within the intratumoral and peritumoral spaces.38,43
Co-occurrence of local anisotropic gradient orientations. The co-occurrence of local anisotropic gradient orientations radiomics descriptor (recently introduced in33) attempts to capture the entropy or disorderliness in pixel-level gradient orientations. Specifically, co-occurrence of local anisotropic gradient orientations has been shown to enable discrimination between treatment-related changes and disease recurrence on MRI scans and also distinguish between benign and malignant nodules on CT scans.37,38,43
Vessel torturosity features.
Alilou and colleagues28 first reported significant differences (AUC, 0.79, p < .01) in computer-extracted tortuosity of tumor-specific vasculature on non-contrast CT scans. Specifically, the study reported differences in vessel tortuosity between patients with NSCLC who did and did not respond to immunotherapy using nivolumab. Novel features extracted from the vessels supplying the tumor involved quantifying three key attributes: (1) torsion, (2) curvature, and (3) branching statistics and patierns. This feature seems to exploit the fact that increased angiogenesis tends to be observed in more aggressive tumors, leading to more tortuous vasculature.
Feature selection.
The massive number of computed radio-mic features results in the so-called “curse of dimensionality,” an issue where the number of features is much larger compared with the number of training instances. One of the ways to accomplish this is by capping the number of selected features to be approximately an order of magnitude smaller compared with the sample size.97 This is accomplished by a variety of feature selection algorithms, including univariate and multivariate models.98,99
Univariate algorithms.
These include chi square test,100 Fisher exact test,101 and Wilcoxon rank sum test,102 which involve comparing the association between the features and the chosen outcome variable to identify the most predictive features. For instance, to predict recurrence as an outcome, the chi square test would calculate the chi square coeticient between a feature variable and the recurrence outcome (recurrence labeled as one and no recurrence labeled as zero). Similarly, the Fisher exact test assigns high rank to features that have higher variance, whereas the Wilcoxon rank sum is a nonparametric univariate test to select features that are highly statistically significant in rejecting the null hypothesis (i.e., there were measurable differences between the cause and effect being studied; e.g., studying the relationship between treatment outcome and overall survival).
Multivariate algorithms.
Multivariate models solve the issues with univariate analysis by not only measuring feature associations with the chosen outcome but also, helping to address the interfeature associations. Multivariate models, which are used in radiomic analysis, include minimum redundancy maximum relevance,103 joint mutual information,104 and variable importance on projection measure for principal component analysis (PCA-VIP)105 to select the top features. For example, minimum redundancy maximum relevance helps to identify features that are not only discriminative but also, largely uncorrelated.
Other than discriminability, robustness and stability of the top-ranked features are important criteria in picking features. Various measures have been used to assess feature stability, including relative standard deviation,26 intraclass correlation coefficient,106 coefficient of variability,107 and latent and preparation-induced instability score.56 The availability of the National Cancer Institute–sponsored RIDER-CT data set, which comprises CT scans of patients with NSCLC taken 15 minutes apart, has also allowed for identification of those features that do not dramatically change between the test-retest scans.
Constructing a classifier.
There are primarily two different classification approaches, supervised and unsupervised, which in conjunction with the top identified features, can be used to predict the probability of an event or the outcome of interest.
Supervised classification models.
The supervised model is trained using a set of labels that represent the category of interest (e.g., differentiating between benign and malignant lung nodules or between responders and nonresponders). These approaches include support vector machines,108 random forest classifiers,109 linear discriminant analysis,110 quadratic discriminant analysis,111 and Adaboost.112
Unsupervised model.
In some instances, the outcome labels are not explicitly known. These scenarios are amenable to unsupervised clustering. This approach involves first clustering the given features into different categories without any predefined labels, with the goal being to discover the hidden categories. Unsupervised classification approaches have also been used recently in predicting treatment response in NSCLC.113–115 Clustering methods can include hierarchical-, Bayesian-, and partitioning-based approaches.116,117 In addition, unsupervised clustering might also be useful in evaluating the utility of the selected features, even with known outcome labels. If the most discriminating features fail to stratify the patients into the given outcomes of interest, it might call into question if the features are, in fact, suitable for predicting the chosen outcome of interest.
Analysis of the model performance.
Performance of radio-mic and delta-radiomic features in the context of supervised classification methods is typically done via a receiver operating characteristic curve obtained by ploting the true-positive rate against the false-positive rate while continuously varying the decision threshold. The results are then reported as the AUC, with a higher AUC reflecting higher classification performance. Other performance measures include accuracy, reliability, sensitivity, specificity, and true and false predictive rates. When using unsupervised approaches, like clustering, it becomes more complicated to evaluate performance without the presence of ground truth labels. External validity measures118,119 exist to evaluate clustering quality, evaluating how well the clustering matches the gold standard. These include purity, normalized mutual information, Rand index, and F measure. Internal quality criterion measures120 include atiaining a high intracluster similarity and a low intercluster similarity. Survival analysis is often a secondary measure reported to evaluate the performance of the prediction model using Kaplan–Meier and Cox proportional hazard analysis models121 to predict measures, like overall survival, disease-free survival, and progression-free survival.
APPLICATIONS OF RADIOMICS IN LUNG CANCER TREATMENT RESPONSE
Radiomics studies in the lung cancer space encompass both predicting response to therapy and monitoring response to a particular treatment. Below, we describe applications of radiomics in the context of radiation oncology and chemo-therapy as well as immune checkpoint inhibition therapy.
Radiation Oncology
In the radiation oncology space, radiomics is being shown to have substantial potential as a key noninvasive monitoring tool. Huynh et al122 showed that radiomic shape and tumor heterogeneity features from respiratory-gated CT scans enabled the prediction of treatment response to stereotactic body radiation therapy in patients with early-stage NSCLC. Also, Huynh et al123 used a principal component analysis feature selection algorithm to choose the most stable and discriminative features and built a multivariate model to predict response to stereotactic body radiation therapy in patients with NSCLC. Wavelet and textural features were found to be overexpressing in patients with distant metastasis (confidence interval of 0.67) that failed stereotactic body radiation therapy, whereas clinical and conventional parameters failed to be predictive in these patients. Meanwhile, Mationen et al124 showed the significance of radio-mic textural features, including gray-level co-occurrence matrices and gray-level features, which were intensified in patients who recurred even after radiation therapy in early-stage NSCLC. The radiomic features were predictive of recurrence (AUC of 0.85) after radiation. In contrast, six physician observers had a kappa value of 0.54 in predicting recurrence versus no recurrence in these patients. Coroller et al52 showed that radiomic wavelet features successfully predicted pathologic complete response in patients treated with chemoradiation alone in a cohort of 127 patients, whereas Jain et al125 found that radiomic textural features predicted pathologic complete response in 90 patients who underwent trimodality therapy using baseline CT scans. On an independent validation set of 45 patients, they obtained an AUC of 0.78 using a random forest–derived classifier comprising both intratumoral and peritumoral textural patterns. Fave et al27 showed that changes in radiomic intensity and texture features (delta-radiomics) from serial CT scans of patients with stage III NSCLC before, during, and after radiation therapy were strong indicators of tumor response to radiation therapy.
Chemotherapy
Rakshit et al126 showed that Haralick and Gabor textural features within the tumor were found to be overexpressed in patients who responded to pemetrexed chemotherapy in locally advanced NSCLC, and the radiomics predictor yielded an AUC of 81.33% on a blinded and independent validation set (22 patients). By including computer-extracted shape features, the AUC further improved to 83.44%. Interestingly, the most stable and discriminative features were obtained from the space immediately adjacent to the tumor (peritumoral space). Biologically, the peritumoral space is represented by the tumor microenvironment, which has been shown to be increasingly vital in characterizing tumor aggressiveness and the corresponding immune response.127 The increase in textural heterogeneity in the peritumoral region in chemotherapy responders can possibly be explained by subvisual microfibrosis as a result of chemotherapy response.
Immunotherapy
Low response rates (20%–40%) have been noted with immune checkpoint inhibitor drugs, even in patients having tissue-based high PD-L1 expression,22,128,129 the current gold standard in predicting response to immune therapy. Considering the gamut of adverse eftects130–132 that these drugs might potentially cause, radiomics from baseline CT scans is being explored as a biomarker for treatment response.
Radiomic features pertaining to vessel tortuosity extracted from baseline pretherapy CT scans have been recently identified as a potential independent predictor of response to Nivolumab in locally advanced NSCLC.28 Alilou and colleagues28 devised a novel radiomics descriptor to characterize the changes in the tumor microenvironment using vessel tortuosity metrics. In the training cohort (33 patients), the top three extracted vessel tortuosity features from pretreatment CT scans most predictive of outcome (responders vs. nonresponders determined clinically) were identified and then used for training a support vector machine classifier. The maximum curvature (f1), standard deviation of the torsion (f2), and mean curvature (f3) of the nodule vasculature were identified as the most discriminating features. Figures 1 and 2 illustrate the changes in vascular tortuosity for an IO responder and a nonresponder between pre- and post-treatment IO (2 weeks after the first cycle). The area (AUC) was 0.84 for the training and 0.73 for the test set (28 patients). Delta-radiomic features of changes in intraand peritumoral heterogeneity were leveraged by Xie et al70 using serial CT scans (pre- and post-treatment), which in turn, enabled early identification of early responders in 41 patients with NSCLC treated with Nivolumab. Tang et al133 presented an NSCLC radiomic signature using unsupervised classification approaches. Using tissue-based CD3 count and PD-L1 expression in an early-stage NSCLC cohort, they generated four distinct cohorts using a combination of CD3 and PD-L1 low/high expressions. Haralick texture features and intensity measures were used to correlate radiomics expression with these four clusters, and corresponding survival analysis was carried out. Multivariate survival models indicated the highest overall survival in the radiomic cluster that was strongly correlated with high CD3 and low PD-L1 expressions.
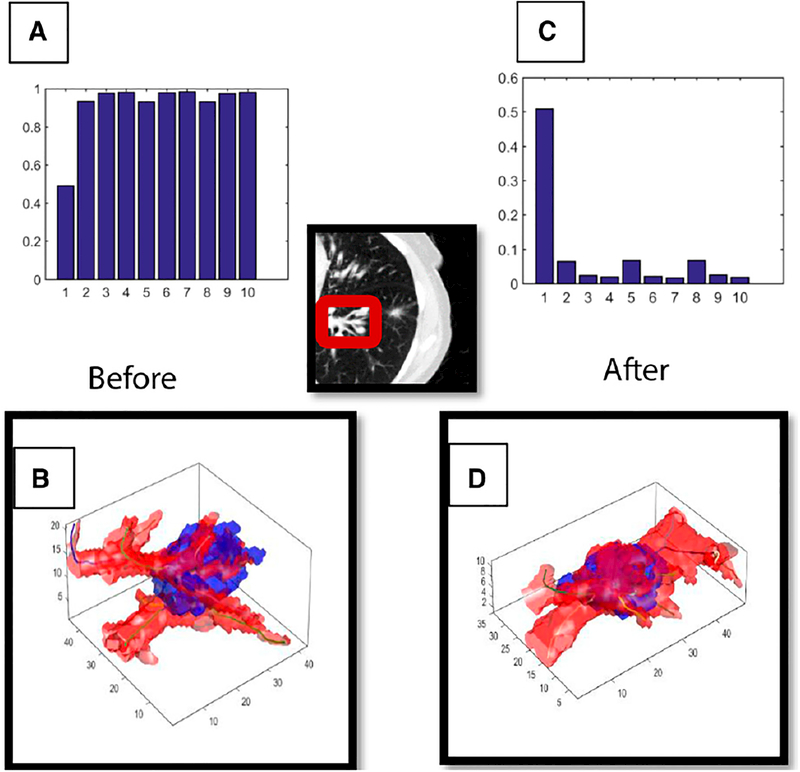
FIGURE 1. A Shows the Top 10 Tortuosity Radiomic Features in Responders Before Treatment, Whereas B Shows the Same 10 Features After Immunotherapy.
Meanwhile, visualization of vessel tortuosity surrounding the tumor is shown in B before treatment and in D after treatment. It is apparent from C and D that immunotherapy leads to both a visual and a radiomic decrease in tortuosity in patients who respond to immunotherapy after treatment.
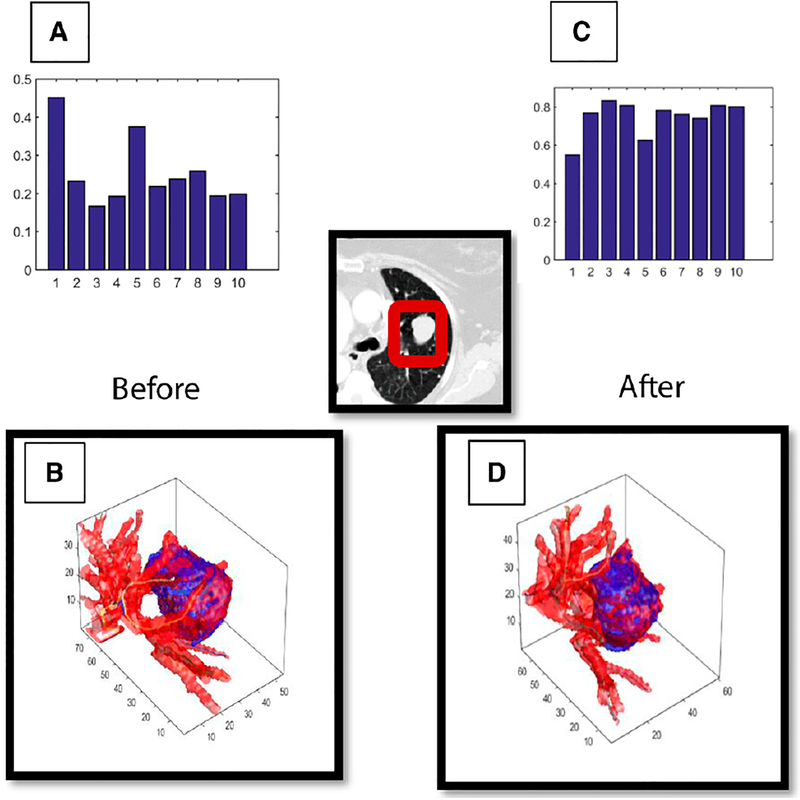
FIGURE 2. A Shows the Top 10 Tortuosity Radiomic Features in Nonresponders Before Treatment, Whereas B Shows the Same 10 Features After Immunotherapy.
Meanwhile, visualization of vessel tortuosity surrounding the tumor is shown in B before treatment and in D after treatment. It is apparent from C and D that patients who do not respond to immunotherapy have a visual and a radiomic increase (or remain stable) in tortuosity across all of the extracted vessel features.
CONCLUSION
In this article, we discussed the exciting and innovative space of radiomics and quantitative imaging techniques as a means to predict response to radiation therapy, different chemotherapeutic modalities, immune checkpoint inhibition treatment, and trimodality therapy as well as a prognostic indicator for early treatment response for these multiple regimens. These techniques and developments have shown tremendous promise in various domains, including recent explorations and forays in the expanding fields of immuno-therapy and targeted and molecular therapy in oncology. These novel imaging techniques have also shown better accuracy in monitoring treatment response compared with current RECIST-based guidelines.
We briefly overviewed the radiomic framework with a brief discussion about the various types of radiomics feature descriptors in use and selection of the most stable and discriminative features, and we constructed a predictive classifier and evaluated the performance of the developed model in predicting the outcome of interest as well as analyzing survival data. We also discussed how it could be seam-lessly integration into the clinical workflow with minimal disruption. We also spoke about the morphologic and histogenomic underpinning behind many of these computer-extracted features to provide a more intuitive reasoning behind its prediction capacity.
We also noted that the existence of a large number of sources of variance remains an impediment to accurate radiomic analysis. In conjunction with relatively small data sets and mostly retrospective analysis, these sources are some of the limitations that it needs to actively overcome. The development of new techniques to alleviate this variation needs to be validated in large-scale multisite and multi-ins tution prospective validation studies to confirm its usefulness. The groundwork, however, has been laid, and the time is opportune for radiomics to be tested as a biomarker in prospective cohort studies and randomized clinical trials. It is only after successful validation in large-scale trials that radiomics can be accepted as a part of routine clinical practice.
To be part of the routine clinical decision-making system, radiomics needs to be a joint collaboration of oncologists, radiologists, physicians, and computational scientists. We hope that this overview will help clinicians better appreciate the field of radiomics, especially its potential in playing an important part in predicting treatment response and monitoring ongoing therapy.
PRACTICAL APPLICATIONS.
Quantitative imaging biomarkers are urgently needed to move beyond tumor response evaluation by RECIST because of its widely acknowledged limitations.
Radiomics or computer-extracted novel imaging features from radiographic images have shown accuracy as a prognostic tool across the oncology space by identifying high-risk patients from low-risk ones.
Precision cancer therapy is another area where radiomics could enable personalization of therapy based on each tumor’s unique radiomic signature.
Radiomics on pretreatment images has shown promising results in also identifying patients that will respond to therapy and those that may not across various treatment modalities, including surgery, chemotherapy, immunotherapy, and molecular-targeted therapy.
Delta-radiomics on serial radiographic images could serve as an effective and noninvasive way to assess effectiveness of therapy as well as predict early treatment response.
ACKNOWLEDGMENT
Research reported in this publication was supported by National Cancer Institute of the National Institutes of Health Awards 1U24CA199374–01, R01 CA202752–01A1, R01 CA208236–01A1, R01 CA216579–01A1, R01 CA220581–01A1, and R21 CA195152–01; National Center for Research Resources Award 1 C06 RR12463–01; the Department of Defense (DOD) Prostate Cancer Idea Development Award; DOD Peer Reviewed Cancer Research Program W81XWH-16-1-0329; the Ohio Third Frontier Technology Validation Fund; and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering and the Clinical and Translational Science Award Program at Case Western Reserve University.
Footnotes
Disclosures of potential conflicts of interest provided by the authors are available with the online article at asco.org/edbook.
References
- 1.Nishino M, Jagannathan JP, Krajewski KM, et al. Personalized tumor response assessment in the era of molecular medicine: cancer-specific and therapy-specific response criteria to complement pitfalls of RECIST. AJR Am J Roentgenol. 2012;198:737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–247. [DOI] [PubMed] [Google Scholar]
- 4.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz LH, Litière S, de Vries E, et al. RECIST 1.1 update and clarification: from the RECIST committee. Eur J Cancer. 2016;62: 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishino M, Jagannathan JP, Ramaiya NH, et al. Revised RECIST guideline version 1.1: what oncologists want to know and what radiologists need to know. AJR Am J Roentgenol. 2010;195:281–289. [DOI] [PubMed] [Google Scholar]
- 7.Seymour L, Bogaerts J, Perrone A, et al. ; RECIST working group. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics.Lancet Oncol. 2017;18:e143–e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lencioni R, Llovet JM. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin Liver Dis. 2010;30:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erasmus JJ, Gladish GW, Broemeling L, et al. Interobserver and intraobserver variability in measurement of non-small-cell carcinoma lung lesions: implications for assessment of tumor response. J Clin Oncol. 2003;21:2574–2582. [DOI] [PubMed] [Google Scholar]
- 10.Nishino M, Hatabu H, Johnson BE, et al. State of the art: response assessment in lung cancer in the era of genomic medicine. Radiology. 2014;271:6–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiou VL, Burotto M. Pseudoprogression and immune-related response in solid tumors. J Clin Oncol. 2015;33:3541–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodi FS, Hwu W-J, Kefford R, et al. Evaluation of immune-related response criteria and RECIST v1.1 in patients with advanced melanoma treated with pembrolizumab. J Clin Oncol. 2016;34:1510–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solinas C, Porcu M, Hlavata Z, et al. Critical features and challenges associated with imaging in patients undergoing cancer immunotherapy. Crit Rev Oncol Hematol. 2017;120:13–21. [DOI] [PubMed] [Google Scholar]
- 14.Villaruz LC, Socinski MA. The clinical viewpoint: definitions, limitations of RECIST, practical considerations of measurement. Clin Cancer Res. 2013;19:2629–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tirkes T, Hollar MA, Tann M, et al. Response criteria in oncologic imaging: review of traditional and new criteria. Radiographics. 2013;33:1323–1341. [DOI] [PubMed] [Google Scholar]
- 16.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 17.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2015;4:36–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metro G, Finocchiaro G, Toschi L, et al. Epidermal growth factor receptor (EGFR) targeted therapies in non-small cell lung cancer (NSCLC). Rev Recent Clin Trials. 2006;1:1–13. [DOI] [PubMed] [Google Scholar]
- 19.Shaw AT, Solomon B. Targeting anaplastic lymphoma kinase in lung cancer. Clin Cancer Res. 2011;17:2081–2086. [DOI] [PubMed] [Google Scholar]
- 20.Matikas A, Mistriotis D, Georgoulias V, et al. Targeting KRAS mutated non-small cell lung cancer: a history of failures and a future of hope for a diverse entity. Crit Rev Oncol Hematol. 2017;110:1–12. [DOI] [PubMed] [Google Scholar]
- 21.Tomasini P, Walia P, Labbe C, et al. Targeting the KRAS pathway in non-small cell lung cancer. Oncologist. 2016;21:1450–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng X, Liu Y, Zhang J, et al. PD-1/PD-L1 checkpoint blockades in non-small cell lung cancer: new development and challenges. Cancer Lett. 2017;405:29–37. [DOI] [PubMed] [Google Scholar]
- 23.Kerr KM, Nicolson MC. Non–small cell lung cancer, PD-L1, and the pathologist. Arch Pathol Lab Med. 2016;140:249–254. [DOI] [PubMed] [Google Scholar]
- 24.Hersom M, Jørgensen JT. Companion and complementary diagnostics–focus on PD-L1 expression assays for PD-1/PD-L1 checkpoint inhibitors in NSCLC. Ther Drug Monit. 2018;40:9–16. [DOI] [PubMed] [Google Scholar]
- 25.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parmar C, Grossmann P, Bussink J, et al. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fave X, Zhang L, Yang J, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep. 2017;7:588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velcheti V, Alilou M, Khunger M. Changes in computer extracted features of vessel tortuosity on CT scans post-treatment in responders compared to non-responders for non-small cell lung cancer on immunotherapy. J Clin Oncol. 2017;35:11518–11518. [Google Scholar]
- 29.Ghose S, Shiradkar R, Rusu M, et al. Prostate shapes on pre-treatment MRI between prostate cancer patients who do and do not undergo biochemical recurrence are different: preliminary Findings. Sci Rep. 2017;7:15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones JP, Palmer LA. An evaluation of the two-dimensional Gabor filter model of simple receptive fields in cat striate cortex. J Neurophysiol. 1987;58:1233–1258. [DOI] [PubMed] [Google Scholar]
- 31.Laws KI. Rapid Texture Identification. Image Processing for Missile Guidance. 1980;0238: 10.1117/12.959169. [DOI] [Google Scholar]
- 32.Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybern B Cybern. 1973;SMC-3:610–621. [Google Scholar]
- 33.Prasanna P, Tiwari P, Madabhushi A. Co-occurrence of local anisotropic gradient orientations (CoLlAGe): a new radiomics descriptor. Sci Rep. 2016;6:37241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clausi DA. An analysis of co-occurrence texture statistics as a function of grey level quantization. Can J Remote Sens. 2002;28:45–62. [Google Scholar]
- 35.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotrotsou A, Zinn PO, Colen RR. Radiomics in brain tumors: an emerging technique for characterization of tumor environment. Magn Reson Imaging Clin N Am. 2016;24:719–729. [DOI] [PubMed] [Google Scholar]
- 37.Prasanna P, Patel J, Partovi S, et al. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi parametric MR imaging predict long versus short-term survival in glioblastoma multi forme: preliminary findings. Eur Radiol. 2017;27:4188–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiwari P, Prasanna P, Wolansky L, et al. Computer-extracted texture features to distinguish cerebral radionecrosis from recurrent brain tumors on multi parametric MRI: a feasibility study. AJNR Am J Neuroradiol. 2016;37:2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou M, Scott J, Chaudhury B, et al. Radiomics in brain tumor: image assessment, quantitative feature descriptors, and machine-learning approaches. AJNR Am J Neuroradiol. 2018;39:208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parmar C, Grossmann P, Rietveld D, et al. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front Oncol. 2015;5:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parmar C, Leijenaar RTH, Grossmann P, et al. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci Rep. 2015;5:11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gastounioti A, Conant EF, Kontos D. Beyond breast density: a review on the advancing role of parenchymal texture analysis in breast cancer risk assessment. Breast Cancer Res. 2016;18:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller BM, Chen J, Conant EF, et al. Breast Density and Parenchymal Texture Measures as Potential Risk Factors for Estrogen-Receptor Positive Breast Cancer. http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.2043710 doi:10.1117/12.2043710. Accessed February 11, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller BM, McCarthy AM, Chen J, et al. Associations between breast density and a panel of single nucleotide polymorphisms linked to breast cancer risk: a cohort study with digital mammography. BMC Cancer. 2015;15:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kontos D, Ikejimba LC, Bakic PR, et al. Analysis of parenchymal texture with digital breast tomosynthesis: comparison with digital mammography and implications for cancer risk assessment. Radiology. 2011;261:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giger ML, Karssemeijer N, Schnabel JA. Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer. Annu Rev Biomed Eng. 2013;15:327–357. [DOI] [PubMed] [Google Scholar]
- 49.Romo-Bucheli D, Janowczyk A, Gilmore H, et al. A deep learning based strategy for identifying and associating mitotic activity with gene expression derived risk categories in estrogen receptor positive breast cancers. Cytometry A. 2017;91:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romo-Bucheli D, Janowczyk A, Gilmore H, et al. Automated tubule nuclei quantification and correlation with oncotype DX risk categories in ER+ breast cancer whole slide images. Sci Rep. 2016;6:32706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alilou M, Beig N, Orooji M, et al. An integrated segmentation and shape-based classification scheme for distinguishing adenocarcinomas from granulomas on lung CT. Med Phys. 2017;44:3556–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119:480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu W, Parmar C, Grossmann P, et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol. 2016;6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghose S, Mitra J, Rivest-Hénault D, et al. MRI-alone radiation therapy planning for prostate cancer: automatic fiducial marker detection. Med Phys. 2016;43:2218–2228. [DOI] [PubMed] [Google Scholar]
- 55.Ginsburg SB, Algohary A, Pahwa S, et al. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: preliminary findings from a multi-institutional study. J Magn Reson Imaging. 2017;46:184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leo P, Lee G, Shih NNC, et al. Evaluating stability of histomorphometric features across scanner and staining variations: prostate cancer diagnosis from whole slide images. J Med Imaging (Bellingham). 2016;3:047502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rusu M, Purysko AS, Verma S, et al. Computational imaging reveals shape differences between normal and malignant prostates on MRI. Sci Rep. 2017;7:41261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shiradkar R, Podder TK, Algohary A, et al. Radiomics based targeted radiotherapy planning (Rad-TRaP): a computational framework for prostate cancer treatment planning with MRI. Radiat Oncol. 2016;11:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghose S, Denham JW, Ebert MA, et al. Multi-atlas and unsupervised learning approach to perirectal space segmentation in CT images. Australas Phys Eng Sci Med. 2016;39:933–941. [DOI] [PubMed] [Google Scholar]
- 60.Hu P, Wang J, Zhong H, et al. Reproducibility with repeat CT in radiomics study for rectal cancer. Oncotarget. 2016;7:71440–71446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Z, Zhang X-Y, Shi Y-J, et al. Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin Cancer Res. 2017;23:7253–7262. [DOI] [PubMed] [Google Scholar]
- 62.Lovinfosse P, Polus M, Van Daele D, et al. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur J Nucl Med Mol Imaging. 2018;45:365–375. [DOI] [PubMed] [Google Scholar]
- 63.Nie K, Shi L, Chen Q, et al. Rectal cancer: assessment of neoadjuvant chemoradiation outcome based on radiomics of multi parametric MRI. Clin Cancer Res. 2016;22:5256–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antunes J, Viswanath S, Brady JT, et al. Coregistration of preoperative MRI with ex vivo mesorectal pathology specimens to spatially map post-treatment changes in rectal cancer onto in vivo imaging: preliminary findings. Acad Radiol. Epub 2018 Jan 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cozzi L, Dinapoli N, Fogliata A, et al. Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric modulated arc therapy. BMC Cancer. 2017;17:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Echegaray S, Gevaert O, Shah R, et al. Core samples for radiomics features that are insensitive to tumor segmentation: method and pilot study using CT images of hepatocellular carcinoma. J Med Imaging (Bellingham). 2015;2:041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gewirtz AT. Deciphering the role of mesenteric fat in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2015;1:352–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hesketh RL, Zhu AX, Oklu R. Radiomics and circulating tumor cells: personalized care in hepatocellular carcinoma? Diagn Interv Radiol. 2015;21:78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Y, He L, Huang Y, et al. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepato-cellular carcinoma. Abdom Radiol (NY). 2017;42:1695–1704. [DOI] [PubMed] [Google Scholar]
- 70.Xie Y, Khunger M, Thawani R, et al. Evolution of radiomic features on serial ct scans as an imaging based biomarker for evaluating response in patients with non-small cell lung cancer treated with nivolumab. J Clin Oncol. 2017;35(suppl e14534):15. [Google Scholar]
- 71.Hochhegger B, Alves GRT, Irion KL, et al. PET/CT imaging in lung cancer: indications and findings. J Bras Pneumol. 2015;41:264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yip SSF, Liu Y, Parmar C, et al. Associations between radiologist-defined semantic and automatically computed radiomic features in non-small cell lung cancer. Sci Rep. 2017;7:3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He L, Huang Y, Ma Z, et al. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep. 2016;6:34921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mackin D, Fave X, Zhang L, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. 2015;50:757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu L, Ehmke RC, Schwartz LH, et al. Assessing agreement between radiomic features computed for multiple CT imaging settings. PLoS One. 2016;11:e0166550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim H, Park CM, Lee M, et al. Impact of reconstruction algorithms on CT radiomic features of pulmonary tumors: analysis of intra- and inter-reader variability and inter-reconstruction algorithm variability. PLoS One. 2016;11:e0164924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Labby ZE, Straus C, Caligiuri P, et al. Variability of tumor area measurements for response assessment in malignant pleural mesothelioma. Med Phys. 2013;40:081916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao B, Tan Y, Tsai W-Y, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6:23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parmar C, Rios Velazquez E, Leijenaar R, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One. 2014;9:e102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rios Velazquez E, Aerts HJ, Gu Y, et al. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists’ delineations and with the surgical specimen. Radiother Oncol. 2012; 105:167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Dam IE, van Sörnsen de Koste JR, Hanna GG, et al. Improving target delineation on 4-dimensional CT scans in stage I NSCLC using a deformable registration tool. Radiother Oncol. 2010;96:67–72. [DOI] [PubMed] [Google Scholar]
- 83.Kumar V, Gupta P. Importance of statistical measures in digital image processing. Int J Emerg Technol Adv Eng. 2012;2:56–62. [Google Scholar]
- 84.Labby ZE, Armato SG III, Dignam JJ, et al. Lung volume measurements as a surrogate marker for patient response in malignant pleural mesothelioma. J Thorac Oncol. 2013;8:478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee JH, Lee HY, Ahn M-J, et al. Volume-based growth tumor kinetics as a prognostic biomarker for patients with EGFR mutant lung adenocarcinoma undergoing EGFR tyrosine kinase inhibitor therapy: a case control study. Cancer Imaging. 2016;16:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guo W, Li H, Zhu Y, et al. ; TCGA Breast Phenotype Research Group. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging (Bellingham). 2015;2:041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ferreira JR Jr, Oliveira MC, de Azevedo-Marques PM. Integrating 3D image descriptors of margin sharpness and texture on a GPU-optimized similar pulmonary nodule retrieval engine. J Supercomput. 2017;73:3451–3467. [Google Scholar]
- 88.Han F, Wang H, Zhang G, et al. Texture feature analysis for computer-aided diagnosis on pulmonary nodules. J Digit Imaging. 2015;28:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Connor JPB, Rose CJ, Waterton JC, et al. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21:249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aerts HJWL, Grossmann P, Tan Y, et al. Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC. Sci Rep. 2016;6:33860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sobel I An isotropic 3 X 3 image gradient operator. Mach Vis Three-Demens Scenes. 1990;376–379. [Google Scholar]
- 92.Canny J A computational approach to edge detection. Read Comput Vis. 1987;184–203. [Google Scholar]
- 93.Campadelli P, Casiraghi E, Columbano S. Lung segmentation and nodule detection in postero anterior chest radiographs. http://homes.dsi.unimi.it/~campadel/Artcoli/GIRPR2004.pdf. Accessed April 18, 2018. [DOI] [PubMed]
- 94.Daugman JG. Complete discrete 2-D Gabor transforms by neural networks for image analysis and compression. IEEE Trans Acoust. 1988;36:1169–1179. [Google Scholar]
- 95.Marĉelja S Mathematical description of the responses of simple cortical cells. J Opt Soc Am. 1980;70:1297–1300. [DOI] [PubMed] [Google Scholar]
- 96.Mehrotra R, Namuduri KR, Ranganathan N. Gabor filter-based edge detection. Pattern Recognit. 1992;25:1479–1494. [Google Scholar]
- 97.Jain AK, Duin RPW, Mao J. Statistical pattern recognition: a review. IEEE Trans Pattern Anal Mach Intell. 2000;22:4–37. [Google Scholar]
- 98.Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–2517. [DOI] [PubMed] [Google Scholar]
- 99.Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. [Google Scholar]
- 100.Scheffe H The relation of control charts to analysis of variance and chi-square tests. J Am Stat Assoc. 1947;42:425–431. [PubMed] [Google Scholar]
- 101.Fisher RA. The Design of Experiments. Edinburgh, United Kingdom: Oliver and Boyd; 1937. [Google Scholar]
- 102.Wilcoxon F Individual comparisons by ranking methods. Biom Bull. 1945;1:80–83. [Google Scholar]
- 103.Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Patiern Anal Mach Intell. 2005;27:1226–1238. [DOI] [PubMed] [Google Scholar]
- 104.Yang H, Moody J. Proceedings of the International ICSC Symp Adv Intell Data Anal. New York, NY: Elsevier; 1999. [Google Scholar]
- 105.Ginsburg SB, Viswanath SE, Bloch BN, et al. Novel PCA-VIP scheme for ranking MRI protocols and identifying computer-extracted MRI measurements associated with central gland and peripheral zone prostate tumors. J Magn Reson Imaging. 2015;41: 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fleiss JL, Cohen J. The equivalence of weighted kappa and the intraclass correlation coefficient as measures of reliability. Educ Psychol Meas. 1973;33:613–619. [Google Scholar]
- 107.Leijenaar RT, Carvalho S, Velazquez ER, et al. Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013;52:1391–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hearst MA, Dumais ST, Osuna E, et al. Support vector machines. IEEE Intell Syst. 1998;13:18–28. [Google Scholar]
- 109.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 110.Lachenbruch PA, Sneeringer C, Revo LT. Robustness of the linear and quadratic discriminant function to certain types of non-normality. Commun Stat. 1973;1:39–56. [Google Scholar]
- 111.Mika S, Ratsch G, Weston J, et al. Neural Networks for Signal Processing IX, 1999. Proceedings of the 1999 IEEE Signal Processing Society Workshop. New York, NY: IEEE; 1999. [Google Scholar]
- 112.Rätsch G, Onoda T, Müller K-R. Soft margins for AdaBoost. Mach Learn. 2001;42:287–320. [Google Scholar]
- 113.Shen W, Zhou M, Yang F, et al. Multi-scale convolutional neural networks for lung nodule classification. Inf Process Med Imaging. 2015;24:588–599. [DOI] [PubMed] [Google Scholar]
- 114.Kuruvilla J, Gunavathi K. Lung cancer classification using neural networks for CT images. Comput Methods Programs Biomed. 2014;113:202–209. [DOI] [PubMed] [Google Scholar]
- 115.Jefferson MF, Pendleton N, Lucas SB, et al. Comparison of a genetic algorithm neural network with logistic regression for predicting outcome after surgery for patients with nonsmall cell lung carcinoma. Cancer. 1997;79:1338–1342. [DOI] [PubMed] [Google Scholar]
- 116.Au NHC, Cheang M, Huntsman DG, et al. Evaluation of immunohistochemical markers in non-small cell lung cancer by unsupervised hierarchical clustering analysis: a tissue microarray study of 284 cases and 18 markers. J Pathol. 2004;204:101–109. [DOI] [PubMed] [Google Scholar]
- 117.Taher F, Sammouda R. GCC Conference and Exhibition (GCC), 2011 IEEE. New York, New York: IEEE; 2011. Abstract 5752535. [Google Scholar]
- 118.Rand WM. Objective criteria for the evaluation of clustering methods. J Am Stat Assoc. 1971;66:846–850. [Google Scholar]
- 119.Halkidi M, Batistakis Y, Vazirgiannis M. Cluster validity methods: part I. SIGMOD Rec. 2002;31:40–45. [Google Scholar]
- 120.Rendón E, Abundez I, Arizmendi A, et al. Internal versus external cluster validation indexes. Int J Comput Commun. 2011;5:27–34. [Google Scholar]
- 121.Cox DR. Regression models and life-tables In Kotz S and Johnson NL (eds). Breakthroughs in Statistics. New York, NY: Springer; 1992; 527–541. [Google Scholar]
- 122.Huynh E, Coroller TP, Narayan V, et al. Associations of radiomic data extracted from static and respiratory-gated CT scans with disease recurrence in lung cancer patients treated with SBRT. PLoS ONE. 2017;12:e0169172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huynh E, Coroller TP, Narayan V, et al. CT-based radiomic analysis of stereotactic body radiation therapy patients with lung cancer. Radiother Oncol. 2016;120:258–266. [DOI] [PubMed] [Google Scholar]
- 124.Mattonen SA, Palma DA, Johnson C, et al. Detection of local cancer recurrence after stereotactic ablative radiation therapy for lung cancer: physician performance versus radiomic assessment. Int J Radiat Oncol Biol Phys. 2016;94:1121–1128. [DOI] [PubMed] [Google Scholar]
- 125.Jain P, Ahmad U, Murthy S, et al. MA 17.11 prediction of response to trimodality therapy using CT-derived radiomic features in stage III non-small cell lung cancer (NSCLC). J Thorac Oncol. 2017;12:S1875 (suppl 2). [Google Scholar]
- 126.Rakshit S, Orooji M, Beig N, et al. Evaluation of radiomic features on baseline CT scan to predict clinical benefit for pemetrexed based chemotherapy in metastatic lung adenocarcinoma. J Clin Oncol. 2016. (15_suppl; abstr 11582). [Google Scholar]
- 127.Hendry SA, Farnsworth RH, Solomon B, et al. The role of the tumor vasculature in the host immune response: implications for therapeutic strategies targeting the tumor microenvironment. Front Immunol. 2016;7:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Garon EB, Rizvi NA, Hui R, et al. ; KEYNOTE-001 Investigators. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. [DOI] [PubMed] [Google Scholar]
- 129.Sundar R, Cho B-C, Brahmer JR, et al. Nivolumab in NSCLC: latest evidence and clinical potential. Ther Adv Med Oncol. 2015;7:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tabchi S, Weng X, Blais N. Severe agranulocytosis in a patient with metastatic non-small-cell lung cancer treated with nivolumab. Lung Cancer. 2016;99:123–126. [DOI] [PubMed] [Google Scholar]
- 131.Spain L, Walls G, Julve M, et al. Neurotoxicity from immune-checkpoint inhibition in the treatment of melanoma: a single centre experience and review of the literature. Neurotoxicity from immune-checkpoint inhibition in the treatment of melanoma: a single centre experience and review of the literature. Ann Oncol. 2017;28:377–385. [DOI] [PubMed] [Google Scholar]
- 132.Linardou H, Gogas H. Toxicity management of immunotherapy for patients with metastatic melanoma. Ann Transl Med. 2016;4:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tang C, Hobbs B, Amer A, et al. Development of an immune-pathology informed radiomics model for non-small cell lung cancer. Sci Rep. 2018;8:1922. [DOI] [PMC free article] [PubMed] [Google Scholar]