Abstract
The evolutionary events that cause colorectal adenomas (benign) to progress to carcinomas (malignant) remain largely undetermined. Using multi-region genome/exome sequencing of 24 benign and malignant colorectal tumours, we probe the evolutionary fitness landscape occupied by these neoplasms. Unlike carcinomas, advanced adenomas frequently harbour sub-clonal driver mutations, which are considered to be functionally important in the carcinogenic process, that have not swept to fixation, and have relatively high genetic heterogeneity. Carcinomas are distinguished from adenomas by widespread aneusomies that are usually clonal and often accrue in a “punctuated” fashion. We conclude that adenomas evolve across an undulating fitness landscape, whereas carcinomas occupy a sharper fitness peak, probably owing to stabilising selection.
Introduction
The classical adenoma-carcinoma sequence of colorectal tumorigenesis1 postulates that a conventional colorectal adenoma (CRA) is initiated by “two hits” at APC2,3, and typically progresses to colorectal cancer (CRC) through a stepwise accumulation of driver mutations such as KRAS and TP53 and deletion of chromosome 18q4. The evolutionary dynamics presumed to underlie this process comprise a series of selective sweeps to (near) fixation, each triggered by an elevation in sub-clone fitness through the occurrence of a new, positively-selected driver mutation5. In this model, progression to an invasive lesion (carcinoma) is postulated to be prompted by the acquisition of a critical driver mutation burden, implying that adenomas and carcinomas should be distinguishable by specific driver mutations. CRCs can, however, develop without the full complement of driver mutations6,7, and some studies have suggested that sub-clonal evolution within established tumours is ‘effectively neutral’8,9, questioning whether selective sweeps occur at all, especially in established CRCs.
As part of a comprehensive assessment of colorectal tumour evolution, here we have attempted to re-assess the classical model and outline the evolutionary ‘fitness landscape’ of CRAs and CRCs. The fitness landscape, a concept, first introduced by Sewall Wright in 193210, is an abstraction to help visualise the relationship between genotypes and reproductive success (sub-clone fitness in this context). The X- and Y-axes can be thought of as the genotype ‘space’ (simplified to 2 dimensions) that can be occupied by adenomas and carcinomas. The Z-axis or height is proportional to genotype fitness: peaks represent particularly fit genotypes, valleys less fit genotypes, and ridges/plateaux equally fit genotypes. Individuals sampled from a population are likely to occupy (local) fitness peaks, because less fit individuals have been removed by negative (purifying or stabilising) selection. Herein we search for the genotypes associated with the fitness peaks occupied by CRAs and CRCs and probe peak shapes by quantifying intra-tumour heterogeneity (ITH). Transitions around the landscape are measured using phylogenetic and molecular clock analyses. These data provide a comprehensive understanding of the evolutionary trajectories underpinning the development of CRAs and CRCs.
Results
To map the evolutionary landscape of CRAs and CRCs, we performed multi-region whole-genome sequencing (WGS) or whole-exome sequencing (WES) on 2-16 regions (total 118) from 9 CRAs and 15 CRCs, each with constitutional DNA (Table S1 for sample details and S2 for sequencing statistics). Five CRCs, including four from Lynch syndrome patients, had microsatellite instability (MSI) owing to defective DNA mismatch repair, and these tumours were analysed as a distinct group unless otherwise stated. The remaining ten CRCs were microsatellite-stable (MSS) and of these, two were synchronous lesions from a single patient. Mutations in a subset of genes were validated using targeted molecular inversion probe sequencing (Online Methods).
Somatic single nucleotide alterations do not define CRC fitness peaks
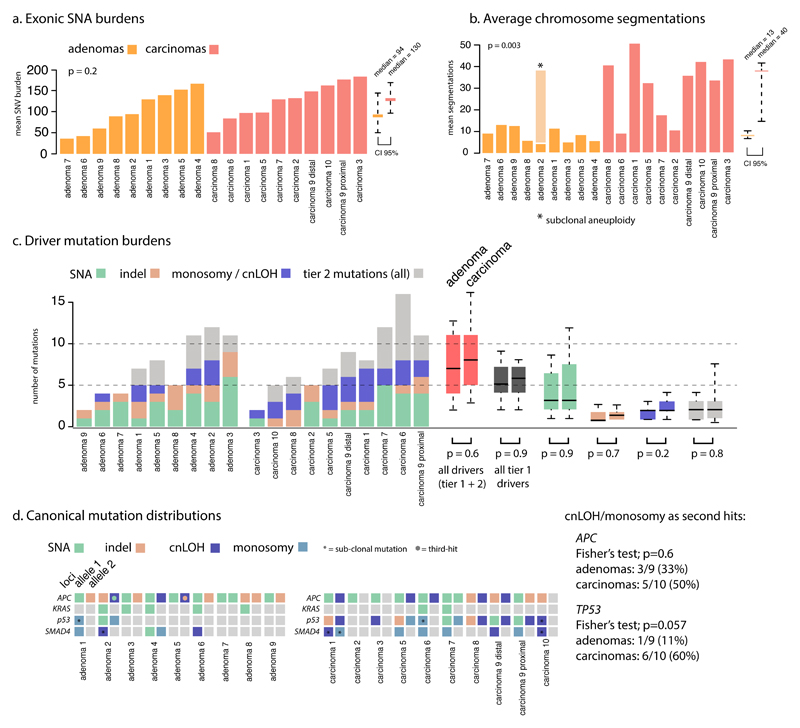
We first assessed how somatic single nucleotide alterations (SNAs) defined the co-ordinates of CRAs and CRCs in the fitness landscape. CRAs tended to have only slightly fewer SNAs than MSS CRCs CRAs: median exonic burden=94, 95% range [51-146]; MSS CRCs: median=130, 95% range [98-171]; p=0.29 Wilcoxon test; Figure 1A, Table S2). After sequencing coverage normalisation, the mutational frequency in CRAs remained very similar to that of MSS CRCs (CRA; 4.1/Mb [3.3-4.9], MSS CRC; 4.2/Mb [2.9-6.4], p=0.9).
Figure 1. Mutation burdens in CRAs and CRCs.
a. CRAs tended to have slightly fewer exonic SNAs than CRCs but the difference was not significant. The average burden and 95% range across these different tumours is shown by the rightmost bars. b. The number of individual CNAs (as measured by the number of segmentations) is significantly greater in CRCs than CRAs (p=0.003, 95% range shown by bars). c. SNA driver mutation burdens and allelic loss of 5q, 17p and 18q, are shown for each tumour. A comparison of all events is show by the red bars, while tier 1 driver changes exclusively are shown in dark grey, with tier 2 in light grey. d. Distribution of canonical driver mutations across tumours. APC is the only ubiquitous driver event. There is no significant enrichment of cnLOH mutations as second hits to APC or TP53 mutations in adenomas compared to carcinomas (though TP53 is borderline).
Next we compared the burden of driver mutations across CRAs and CRCs, and included SNAs and indels, and also cnLOH and monosomy (chromosome loss) events that are known to act as ‘second hits’ to the tumour suppressor genes APC and TP53, and also 18q allelic loss/imbalance (Figure 1C-D and Table S3). The burden of tier 1 mutations, which we defined as likely pathogenic changes in known CRC driver genes (see Online Methods, Table S4), was not significantly different in our cohort (CRAs: median=5 [2-9]; MSS CRCs: median=6 [2-8], p=0.9). We noted that the difference remained non-significant when comparing drivers across individual biopsies (p=0.19; Wilcoxon test). Individual tier 1 driver mutations were detected at similar frequencies across CRAs and CRCs, with the exception of TP53, which was more commonly mutated (possessing least one SNA, indel or copy change) in CRCs (Fisher test; p=0.005, see Figure 1D and Table S5). The frequency of tier 2 driver mutations (uncertain pathogenicity changes in CRC or pan-cancer driver genes) was also not discernably different in CRAs and MSS CRCs (CRAs: median=3 [2-4]; MSS CRCs: median=3 [1-7]; p=0.8). Several tier 2 driver mutations were specific to CRAs or CRCs, but most occurred infrequently; only KMT2C was notable, being mutated in 4 CRAs and no CRCs. The total driver mutation burdens (tier 1 and 2 combined; medians CRAs=7 [2-12]; CRCs=8 [3-15]; p=0.6; Figure 1C, Tables S3, S4) were also similar. Furthermore, when using an alternative definition of driver genes (the top 15 genes mutated in MSS CRCs [excluding TTN], according to the TCGA publication7, Table S4b), the burdens remained not statistically different in CRAs and CRCs (CRAs: median=5 [2-7]; MSS CRCs: median=5.5 [2-9], p=0.7).
The power to detect differences in mutation burden between CRAs and CRCs was limited by sample size, such that differences in exonic mutation burden smaller than approximately 30 mutations could not be detected with high power (see supplementary methods for post hoc power calculation). With this constraint in mind, SNA mutation burden (including tier 1 driver mutations) did not distinguish the relative co-ordinates of CRAs and CRCs in the evolutionary landscape.
Intra-tumour heterogeneity and phylogenetic analyses suggest that CRCs occupy sharper fitness peaks than CRAs
To broadly assess the shape of the fitness peaks occupied by CRAs and MSS CRCs, we measured the degree of ITH in each tumour. Excluding tumours with only two regions sampled (see Online Methods), a median 56% [53-70%] of all CRA SNAs were “sub-clonal” (variant not detected in all sampled regions). MSS CRCs had a significantly lower proportion of sub-clonal SNAs (45% [23-77%]; p=0.04; Figure 2 inset) than CRAs. The average pairwise genetic divergence between the regions of each tumour was then assessed following normalisation of sequencing coverage (Online Methods). CRAs showed significantly more divergence between biopsies than CRCs (CRA mean=2.0 versus CRC=1.7, divergent SNAs/Mb; p<2x10-16; Figure S1A), despite having the same average mutation burden. The measured values of ITH were unaffected by the number of biopsies available from each neoplasm (Figure S1B-C).
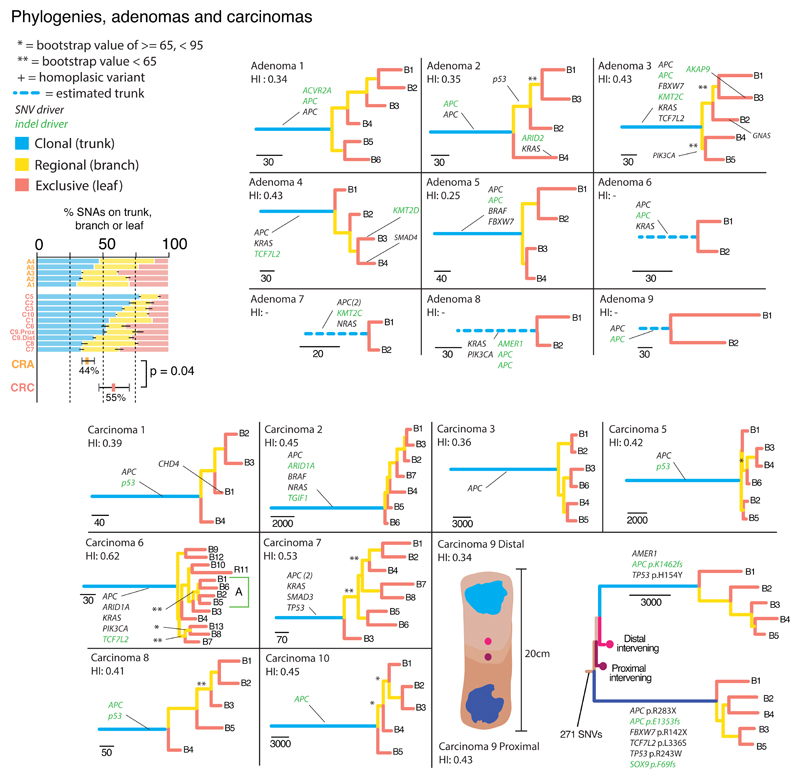
Figure 2. Phylogenetic analysis of CRAs and MSS CRCs.
Maximum parsimony construction of evolutionary trees. For tumours with only two regional biopsies, truncal mutations were simply those shared between the regions. Tier 1 driver mutations (Table S3) are shown, illustrating their enrichment on the trunks, especially in CRCs, indicating they are acquired early in evolutionary time. Phylogenetic trees showed were produced using all available SNAs. Tree shape robustness (branch support) was confirmed by bootstrapping. Branches had greater than 95% support unless otherwise stated (44/55 (80%) of branches had >95% support). The most parsimonious trees are shown except in carcinoma 6, where one clade could not be resolved (A: green box). Left Bar chart: Ubiquitous SNAs (found in all regional biopsies and on the trunk of the phylogenetic tree) are compared with sub-clonal SNAs on the phylogenetic tree branches (non-ubiquitous, but present in >1 region) and leaf (present in only one region). CRAs have a smaller proportion of ubiquitous variants than CRCs.
To further quantify ITH, we used SNAs to construct maximum parsimony phylogenetic trees (Figure 2). CRC topologies were often characterised by long trunks (variants ubiquitous across biopsies) with comparatively short branches and leaves (relatively few sub-clonal variants), thus appearing ‘palm tree-shaped’11. CRAs had proportionally shorter trunks, and thus longer branches/leaves than CRCs, albeit at borderline significance (average branch and leaf length as a proportion of the trunk: CRAs 82% versus MSS CRCs 50%; p=0.06; Figure 2, Table S6). The difference remained when the MSI+ CRCs were included in the analysis (CRAs 82% versus all CRCs 45%; p=0.05). CRAs are thus more genetically diverse than CRCs.
To investigate whether individual CRAs and CRCs occupied single or multiple fitness peaks, we compared the lengths of the phylogenetic tree branches/leaves. Large variations in branch length indicate that mutations accrue faster in some tumour regions than others, which can potentially be caused by selection on a new fitness peak. Average intra-tumour variation in relative branch/leaf length was generally low and similar across CRAs and CRCs (mean standard deviation: CRA 0.14 [0.06-0.24] versus CRC 0.2 [0.06-0.47]; p=0.68; Figure 3, Table S6). Formal assessment of unbalanced tree topologies could only be performed on one tumour (carcinoma 6) as high numbers of samples are needed for sufficient power12. Unbalanced trees occur when some ancestor clones produce more surviving lineages than another, another potential indicator of sub-clonal selection. We did not find any significant asymmetry in this single tumour analysis (Colless’ test, Yule model, p=0.3). Thus the available data were consistent with the idea that tumours occupied a single, potentially broad, fitness peak.
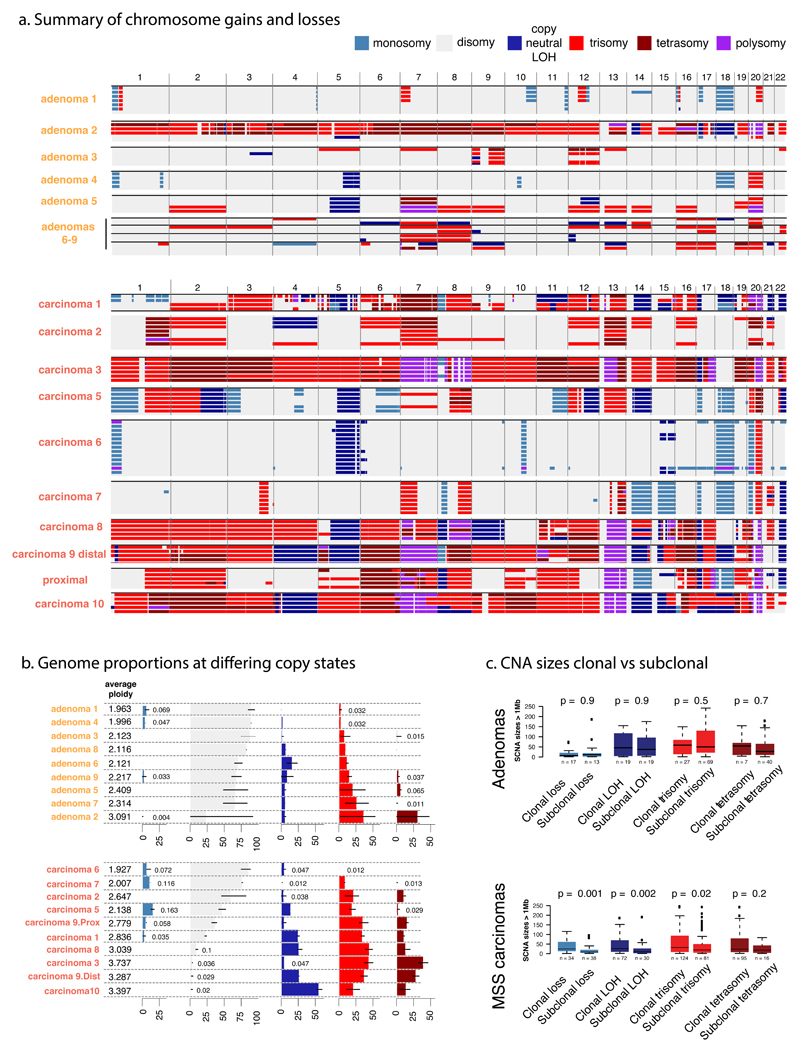
Figure 3. Copy number alterations in CRAs and MSS CRCs.
a. A genome-wide view of CNAs is shown for each region of CRAs (top) and CRCs (bottom). Cancers show a greater CNA burden than adenomas, and most CNAs are clonal in cancers, whereas CRAs show more frequent sub-clonal CNAs. Copy number ≥5 is shown as “polysomy”. b. The figure shows estimated ploidy and summarises the proportion of each tumour at different copy-states. Black bars show the range of biopsy copy-numbers. c. Size distributions of ubiquitous and sub-clonal (branch and leaf) CNAs demonstrate the preference of CRCs to have larger events. Boxplots show the median and inter quantile range (IQR), upper whisker is 3rd quantile + 1.5*IQR and lower whisker is 1st quantile - 1.5*IQR. The colour-coding of copy number states (top right) applies to all panels.
The SNA-based ITH and phylogenetic analyses suggested that CRAs were more heterogeneous than CRCs, consistent with the former occupying a broader fitness peak, under which several distinct genotype-phenotype combinations could co-exist. The lower ITH in CRCs could also, however, reflect a more recent selective sweep with a genetic bottleneck during the transition from an adenoma, and/or that CRCs were more spatially mixed than CRAs, causing variants at sub-clonal frequency in multiple samples to appear truncal. We therefore directly sought evidence of stronger selection in CRCs by examining the ratio of non-synonymous to synonymous mutations on tumour trunks and branches/leaves. This showed a reduction in non-synonymous mutations on the branches/leaves of CRCs relative to their trunks (Wilcoxon sign rank test, p=0.01; Figure S2), but no such reduction for CRAs (p=0.9), possibly representing on-going positive subclonal selection in CRAs. On the reasonable assumption that positive selection acted on the phylogenetic trunk - the location of almost all tier 1 driver mutations – together these results indicate that subclonal selection is absent (neutral dynamics) or weak within the established carcinoma, with possible negative (stabilising) selection also at play.
Mutational processes are not detectably associated with fitness advantages
Mutation signatures were identified de novo using the EMu program13. We recovered ageing, MSI-associated and molecular clock signatures14 (our Signatures A, B and C respectively), as expected (Figure S3A). Our Signature D, which resembles COSMIC Signature 17 (unknown aetiology, high CTT>CGT frequency15) was present at appreciable levels within carcinomas 2, 7, 9P and 10, with its activity often differing between the trunks and branches/leaves of the same lesion (Figure S3B,C). We explored whether signature D had any effect on sub-clonal evolution in CRCs with WGS. It appeared to increase the mutation burden in two CRCs 2 and 9P, but had no discernible effect on their evolution (details in Figure S3D). Carcinoma 9D, the synchronous partner of carcinoma 9P, showed low signature 17 activity, despite being located only 10cm apart in the bowel. These cancers also had different driver mutations, confirming that they essentially behaved as independent neoplasms, with no detectable effect of any shared microenvironment on mutagenic processes (Figure S4).
Major driver mutations can be sub-clonal in CRAs, but are very rarely so in MSS CRCs
Tier 1 driver mutations (defined above) were typically, but not always, clonal in CRAs, whereas in MSS CRCs drivers were more commonly clonal. However, these distributions were not significantly different between tumour types (CRAs=39/49, 80% versus CRCs=49/55, 89%; p=0.3). The clonal distributions of tier 2 clonal driver mutations were however, different; CRAs had significantly less clonal drivers than MSS CRCs (CRAs=7/15, 47% versus CRCs=21/26, 80%; Fisher’s exact test, p=0.03, Table S3). We noted that the clonality of tier 1 driver mutations was the same when using the second definition of driver mutations based on the TCGA publication7 (37/43, 86% versus 45/53, 85%; Fisher’s exact test, p>0.9, see Online Methods). The findings are consistent with a scarcity of sub-clonal expansions after the most recent common ancestor (MRCA) in CRCs. This trend seems to be similar in CRAs, though CRAs do show some evidence of sub-clonal driver mutations.
We additionally noted, however, that the most frequently mutated CRC driver genes, apart from the probable tumour-initiating mutations in APC, were sub-clonal in at least one CRA. Notably in adenoma 2, KRAS Q61H and an ARID2 frameshift mutation were present in one region, which was separate from the three regions of this tumour that contained a TP53 E219X mutation. Adenoma 3 had a PIK3CA E545K mutation in two tumour regions, GNAS R201H in another, and an AKAP9 frameshift in another. SMAD4 R496H in adenoma 4 was also present in a single region. There was no evidence from the phylogenetic analysis that these proven driver mutations were associated with differential sub-clonal expansion, suggesting that their selective benefits were relatively modest (Figure 2). NRAS G60V and PIK3CA H1047R were present in both regions of adenomas 7 and 8 respectively, but were putatively sub-clonal since their corrected allele frequencies were significantly lower (p<0.05) than those of other driver mutations, suggesting that biopsies crossed sub-clonal boundaries. By contrast, only one sub-clonal mutation with high-confidence pathogenicity (CHD1 R619X in carcinoma 1) was found in the MSS carcinomas. There was no evidence for parallel evolution of sub-clones based on recurrent known or novel drivers (details not shown).
We next we wished to relate the heterogeneity of mutational burdens to fundamental molecular processes. Immunohistochemistry for Ki-67 (proliferation) and β-catenin (activated Wnt-signalling; Figure S5; Online Methods) showed positive cell fractions of 53% [2-80%] and 82% [3-97%] respectively, with considerable variability between and within CRCs (Table S7). Neither Ki67 nor β-catenin expression was associated with regional SNA burden or ploidy (SNA burden, R2=0.2, p=0.2, ploidy, R2=0.9, p=0.08; Figure S5).
Genetic and spatial relationships between CRC sub-clones
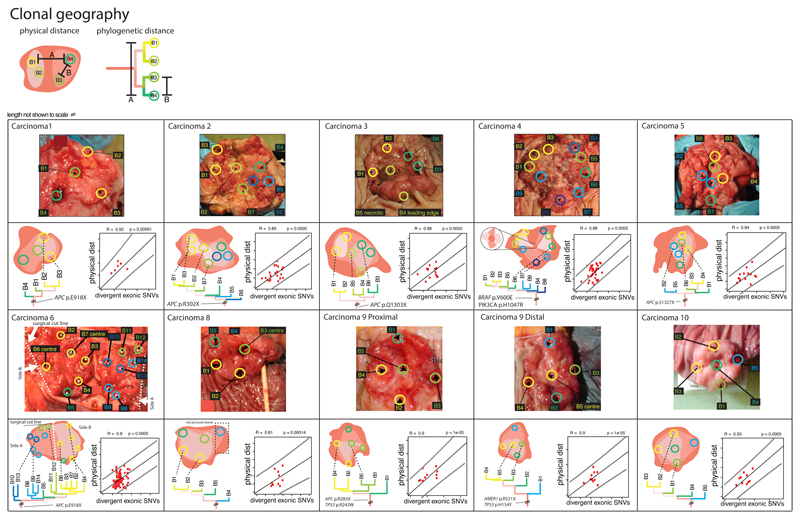
In all CRCs, physical and phylogenetic distances between biopsies were strongly correlated (R2=0.81-0.93, p<10-4 for all carcinomas measured; Figure 4). The invasive edge of CRCs and central regions had similar mutational burdens (exonic SNAs, edge versus central; p=0.76). We looked further for sub-clonal mixing within the sampled regions of the MSS CRCs with WGS data by clustering of SNA cancer cell fractions across related samples, using a Dirichlet process-based model (Figure S6). Only 10% of biopsy samples showed evidence of ≥1 sub-clonal population. Whilst we do not exclude a degree of sub-clonal intermingling, these results suggest that, given the depth of our sequencing data, sub-clonal expansions broadly occurred in a spatially contiguous, uniform and discrete fashion.
Figure 4. Geography of CRCs.
Photographs of the tumour specimens from histopathology departments are shown, with biopsy locations marked. The sporadic MSI+ cancer 4 is included here. The corresponding phylogenetic relationship between tumour regions is shown below the photograph of each tumour. The regression plots show pairwise physical and genetic separation for each biopsy from that cancer. There was a significant positive correlation between the phylogenetic (mutational) distance and physical distance in every case.
Copy number changes differ between CRAs and CRCs
We next assessed whether somatic copy number alterations (CNAs) might define the fitness peaks occupied by CRAs and MSS CRCs. Every region of every tumour carried at least one CNA, including cnLOH (see Online Methods). In a combined analysis of all regions from each tumour, as expected, adenomas had fewer CNAs (number of discrete CNA segments >1Mb) than carcinomas16,17 (CRAs median=13 [7-11] versus CRCs median=40 [15-42], p=0.003; Figure 1B). Correspondingly, the overall average proportion of the genome disrupted by CNAs (copy number ≠ 2, allelic ratio ≠ 1) was higher in CRCs (CRCs, 72% versus CRAs, 40%; p=0.05; Figure 3). These data show that despite carrying similar SNA burdens, CRCs display higher CNA levels than CRAs.
Driver CNAs are currently hard to identify with certainty in cancer18. In colorectal tumours, losses (deletions or cnLOH) on chromosomes 5q, 17p and 18q are often thought to be second hits involving tumour suppressors APC, TP53 and SMAD4 respectively (although 18q loss is more common than SMAD4 mutation). The status of other recurrent changes – such as 1q gain, 7 gain, 8p deletion, 13q gain and 20 gain – as drivers or passengers is less clear. Many recurrent, and hence potential driver7, CNAs were present at significantly higher frequencies in CRCs compared to CRAs (Figure 3). Notably, 17p loss occurred in 9/10 MSS CRCs, but only 2/9 CRAs (Fisher’s exact test, p=0.005), paralleling the TP53 SNA data. By comparison, loss at the APC locus (8/10 CRCs versus 5/9 CRAs; p=0.35) and the SMAD4 locus (7/10 CRCs versus 4/9 CRAs; p=0.37; Figure 2) occurred at similar frequencies in both lesion types.
Every tumour had at least 2 clearly sub-clonal CNAs (non-ubiquitous, present versus absent changes; Figure 3A) and no chromosome aberration was exclusively ubiquitous or sub-clonal across the tumours. Overall, 75% and 48% of gains were sub-clonal in CRAs and CRCs respectively (p=0.002), compared with 57% and 27% of losses/cnLOH (p=0.007; Figure S7). Thus, a greater proportion of CNAs were sub-clonal in CRAs than in CRCs.
We compared the size distribution of large (>1Mb) CNAs in early (truncal) versus late (sub-clonal) tumour evolution. In CRCs, sub-clonal CNAs were smaller than ubiquitous CNAs (p<0.001 ; Figure S3C), but this difference was not present in CRAs (p=0.45). The lower frequency of large CNAs later in evolutionary time in CRCs suggests that the cancers have obtained a near-optimal level of aneuploidy, with further large-scale CNAs subjected to negative/stabilising selection. In adenomas, since the overall CNA burden is lower, new large CNAs may still be tolerated.
MSS CRC evolution can involve either “punctuated” or more gradual CNA acquisition
Since CNAs were the principal genetic feature distinguishing CRAs and CRCs, we investigated their role in the transition between the benign and malignant fitness peaks. Utilising a similar strategy to Durink19 and Newman20 (details in Online Methods), we used the SNAs within informative chromosomal segments (copy number gains and cnLOH) as a molecular clock to time the occurrence of that CNA. SNAs present on a chromosome prior to gain, cnLOH or amplification increase in frequency (VAF) following the copy number change, whereas SNAs that accrue after the gain remain at their original, lower VAF. The ratio of higher to lower VAF SNAs therefore estimates the time of CNA occurrence.
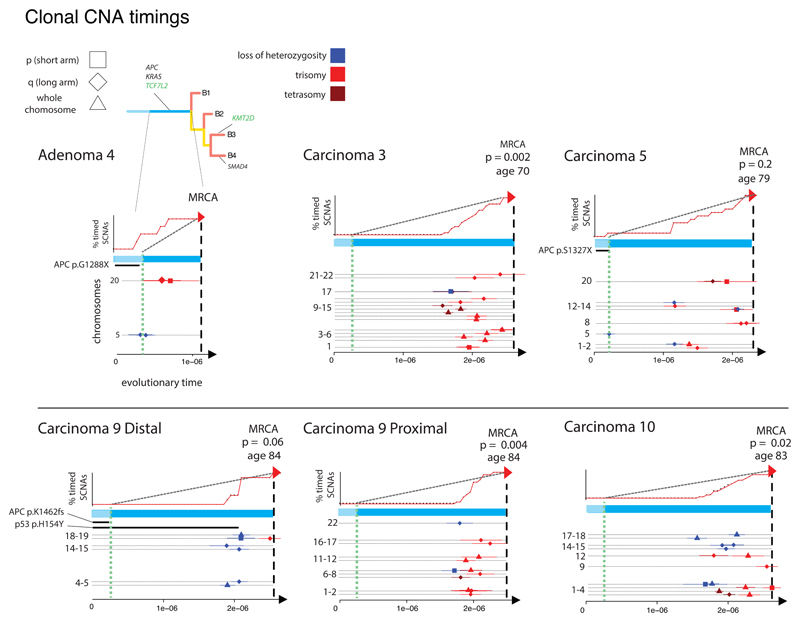
Sufficient SNAs for molecular clock analysis were only present in WGS data. Of the five MSS CRCs analysed by WGS, carcinomas 3, 9P and 10 showed a clustering of CNA timings shortly before the MRCA (Kolmogorov-Smirnov test against a uniform distribution of CNA timings, p<0.02 for all, Figure 5). A similar, borderline significant CNA cluster occurred in carcinoma 9D. Carcinoma 5 showed a more gradual accumulation of CNAs.
Figure 5. CNA timing.
The plots show the CNA timing results for the six neoplasms with WGS data. For each tumour, the X-axis represents inferred evolutionary time to the MRCA, since tumour initiation (unit of measurement is SNAs accrued per unit time). Green dashed line is inferred from the “second hit” at APC (and thus likely represents the time of initiation of the adenoma). The upper panels show the accumulation of CNAs (red, arrowed line) relative to a steady accumulation (black, dashed line); p-values are derived from Kolmogorov-Smirnov tests of inferred CNA time versus a uniform accumulation. The lower panel shows the estimated times of driver mutations, where these could be derived, for individual CNAs by chromosome arm and type of change. Bars indicate 95% confidence intervals for CNA timing estimates.
Since the timing method demonstrated a form of “punctuated” CNA evolution (rejection of null hypothesis of uniform accumulation), but did not distinguish between multiple gains of individual chromosomes and genome doubling followed by chromosomal gain or loss, we searched heuristically for evidence of genome doubling using a score based on the number of chromosome centromeres present at copy number 4 or above, with extra weight for allelic balance (Figure S8). Based on this measure, all of the CRCs with significantly or borderline significantly clustered CNA timings (n=4) were genome-doubled on this measure, as was the untimed carcinoma 8. The CNAs in these tumours are typically trisomies, judged to have arisen by chromosome (arm) loss subsequent to allele-balanced genome doubling. The other tumours (including carcinomas 1, 2, 4, 5, 6, 7) were scored as non-genome-doubled. Sub-clonal genome doubling was present in one CRA (adenoma 2). This tumour carried a TP53 mutation in its genome-doubled regions, and overall TP53 mutations (SNAs and/or CNAs) were associated with genome-doubling in MSS CRCs (Fisher’s Exact Test p=0.018). In addition, genome-doubled cancer regions had higher Ki67 expression (see above; p=0.04; Figure S5), hinting at the existence of a selective benefit of doubling.
The evolutionary landscape of microsatellite-unstable CRCs
The overall SNA burden of the 5 MSI+ CRCs was, as expected, far higher than in MSS CRCs (Figure S9A). More pointedly, the number of tier 1 CRC driver mutations was also higher (median=12 [4-14]) than in MSS CRCs, median=3, p=0.042, Figure S9B), whilst CNA burden was lower (Figure S9C). Of note, in MSI+ CRCs, the great majority of driver SNAs were truncal, the number of sub-clonal tier 1 drivers was only a little greater (median=1 [0-7]) than in MSS CRCs, and the proportion of all sub-clonal SNAs was not significantly increased (median MSI+ CRCs 34% versus MSS CRCs 42%; p=0.13; Figure S9D). In phylogenetic analysis, neither the average branch/leaf length as a proportion of the trunk nor its variability differed significantly between MSI+ and MSS CRCs (Figure S9E). Our signature B (COSMIC signature 6) predominated in MSI+ CRCs, especially on the branches/leaves, but the other COSMIC MSI-associated signatures14 were not detected. Overall, the data suggest that MSI+ CRCs evolve in a similar way to MSS CRCs, albeit with some limited evidence of sub-clonal selection.
Discussion
Here we have contrasted the patterns of evolution in colorectal carcinomas and their classical adenomatous precursor lesion, and our data begin to reveal the shape of the fitness landscape over which CRCs grow. CRAs tend to evolve through acquisition of major driver mutations in genes such as APC, KRAS, TP53 and via 18q loss as per the Kinzler and Vogelstein model4. More recently discovered cancer driver mutations are also present in many adenomas (Table S3 & S4). In fact, CRAs can harbour mutations in any of the major CRC driver genes, but those mutations do not necessarily occur in a stereotypic order. Driver mutation acquisition also does not necessarily cause selective sweeps (leading to ‘stepwise’ evolution of the tumour cell population), since sub-clones with additional major driver mutations may not displace sub-clones lacking those mutations, but instead may co-exist in spatially discrete areas. It follows that many driver mutations probably confer a relatively small selective advantage. This is reflected in several observations in CRAs, including a relatively high level of genetic diversity (both SNAs and CNAs), variation in the major driver mutation complement in different regions of individual tumours, and phylogenetic trees with relatively long branches/leaves. It is even possible that SNA accumulation is not an essential feature of tumorigenesis prior to malignancy, and we speculate that carcinomas need not arise from the sub-clone with the greatest number of driver mutations, thus explaining why some CRCs have a very small driver mutation complement7.
MSS CRCs have longer phylogenetic tree trunks than branches/leaves as compared to CRAs. These findings may reflect the influence of several factors, including not only selective constraints, but also time from the MRCA after an additional selective sweep, ploidy, sample purity and genomic instability. Overall, the lack of sub-clonal driver SNAs and reduction in non-synonymous SNAs on the branches and leaves of CRCs suggest that there is not strong positive sub-clonal selection for SNAs after the MRCA. CRAs on the other hand do show subclonal drivers and relatively high ITH together providing evidence of (perhaps relatively weak) subclonal selection.
Although present in CRAs, large CNAs and genome doubling are much more common in CRCs. CNAs on CRC tree branches/leaves are smaller than those on trunks. Whilst negative or stabilising selection remains difficult to measure, this is consistent with the relatively low genetic diversity in CRCs, based on SNAs and large CNAs. For most MSS CRCs, a near-triploid karyotype seems optimal, either through genome doubling followed by loss of some chromosomes, or through a gain of chromosomes that mostly occurs within a putatively short time window between malignant progression and the MRCA. In each case, one or more selective sweeps seem to occur, rendering the driver SNAs and most CNAs clonal. We do not exclude additional positive selection for specific sub-clonal CNAs in CRCs, but this remains unproven and indeed our data showed no evidence of sub-clonal selection. Although every CRC had at least one sub-clonal CNA, we found no evidence of parallel CNA evolution.
In all our MSI+ cases, defective MMR and most major driver mutations arose on the phylogenetic trunk, and the relative branch/leaf length was similar to that of MSS CRCs. Although the sporadic MSI+ cancer had a low driver mutation burden, as expected if driven in part by a methylator phenotype21, its evolution was otherwise similar to the Lynch syndrome CRCs. We speculate that MSI+ CRCs experience either multiple selective sweeps driven by individual SNAs, or, more intriguingly, by co-occurring or epistatically acting non-canonical driver SNAs (such as CTNNB1, SOX9, NF1 and CASP8).
A small number of ITH studies have been undertaken previously in CRC. Kim et al22 and Uchi et al23 performed multi-region WES of 5 primary and metastatic CRCs and 9 CRCs respectively, and Suzuki et al24 performed deep targeted sequencing of 799 genes in four CRCs. Similar to our study, these studies reported that major driver mutations, affecting APC, KRAS and TP53, were truncal, with the exception of PIK3CA. Uchi et al23 also reported that large copy gains were common on the trunk of the evolutionary tree, with focal deletions on branches.
Fewer studies of CRA evolution exist. Kim et al25 used WES to compare malignant and benign regions of 4 mixed cancer-in-adenoma polyps. They reported similar SNA burdens in cancer and adenoma regions, and thus suggested that the regions evolved in parallel, rather than the carcinoma progressing from a late adenoma. We note, however, that it is extremely hard to distinguish benign and malignant components of these lesions, since malignancy is defined not by cytology, but by invasion and hence the location of tumour cells. The different neoplastic components of such polyps may therefore, in reality, both be ‘cancerous’. For these reasons, in this study, we based our comparison between advanced CRAs and CRCs that were distinct lesions.
Previous work from our group10 examined single glands from 11 CRCs and 4 CRAs for CNAs, and for Ampliseq panels of SNAs that had been derived from bulk tumour WES. Although that manuscript and our present study had very different focuses, the findings are consistent. For example, one feature of the “Big Bang” model of sub-clonal intermixing expounded in the previous study is that after the MRCA, CRC sub-clones radiate outwards without notable differential sub-clonal expansion or selection of further advantageous variants; our present study is broadly consistent with those data. There are also, however, some differences between the studies that allow refinement of the “Big Bang” model. For example, our current study, which benefits from the significantly increased genomic resolution of WGS/WES, emphasises that large sub-clones after the MRCA remain spatially restricted in CRCs and consequently that the previously observed, widespread clonal ‘intermixing’ in some CRCs10 may reflect the shape, size, and boundary location of discrete sub-clones as well as outwards radiation of low frequency clones. Furthermore, whilst the “Big Bang” was broadly consistent with ITH measured in CRAs, our present study finds that sub-clonal driver mutations in the absence of selective sweeps occur commonly in these tumours.
In a study analogous to ours, Stachler et al26 exome-sequenced 5-11 samples of oesophageal carcinoma and its precursor, Barrett’s oesophagus (BE) from 5 patients. Comparing the two studies reveals both similarities and differences. BE is not a discrete tumour and is generally a highly polyclonal lesion, reflected in multiple “initiating” deletion mutations in CDKN2A and a series of clonal expansions without selective sweeps. By comparison, CRAs are discrete and probably have monoclonal origins usually caused by bi-allelic APC mutation, followed either by selective sweeps, or by polyclonal expansions reminiscent of BE. We note that in both BE27,28 and CRA, data are consistent with malignant progression sometimes occurring from a sub-clone that does not have the largest driver mutation burden.
In summary, we have used measurements of intra-tumour heterogeneity to reveal the evolutionary trajectories of colorectal tumour cell populations across what appears to be a rather flat fitness landscape for adenomas, with a higher, sharper peak occupied by cancers. Our data refine the Fearon and Vogelstein model4 of CRC progression by showing that driver mutations do not necessarily lead to hard selective sweeps and that progression to CRC can involve punctuated evolution.
Methods
Sample acquisition and processing
Oxfordshire Research Ethics Committee B gave permission for the study (protocol 05/Q1605/66), and all samples were collected with informed patient consent, that was obtained by the local clinicians prior to tissue collection. Fresh-frozen biopsies from 24 colorectal adenomas or carcinomas were obtained from the John Radcliffe Hospital, Oxford or from University Hospital, Basel. In the case of CRAs, the biopsies were obtained from endoscopic resections, whereas the CRC biopsies were obtained from surgical resections. Any residual bulk cancer surplus to diagnostic requirements was also acquired. Paired normal biopsies were also taken from regions clearly separate from the tumour.
Library preparation and sequencing
DNA was extracted from tumour regions and morphologically normal tissue using the Qiagen DNeasy® kit. The sequencing library preparations were performed using either the NEBNext® DNA kit or in the case of exome sequencing the Illumina TruSeq® exome kit. Sequencing of these biopsies was carried out using standard protocols on the Illumina HiSeq 2000 by the Genomics Core at the Wellcome Trust Centre for Human Genetics, Oxford. The FastQC program29 was used to assess raw sequencing quality and coverage and depth were assessed using the GATK package (specifically the DepthOfCoverage module)30.
Pre-processing and nucleotide variant calling
Reads in FastQ format were aligned to version 19 of the human genome reference using BWA version 0.7.531. The Picard package32 was used to identify duplicate reads and the Samtools package33 was used to count the number of reads in the binary alignment map (.bam) files. Single nucleotide variant (SNA) calling was performed using the Platypus tool34. Variant call format (.vcf) files were annotated with AnnoVar35 and converted to tab-delimited file format using snpSift36. SNAs were categorised as somatic if they were present in at least one tumour sample and either the normal sample had <40X coverage and zero mutant reads or ≥40X depth and ≤1 mutant read. To obtain high confidence and consistent variants for phylogenetic analysis, only variants called with depths consistently ≥10X and allele frequency ≥1% in one or more regions were retained.
Calling small insertions and deletions
Indel calling remains problematic and highly inaccurate37. For this reason we focussed mainly on potential driver events. We identified a set of reliable indels by performing a first pass using the Scalpel tool38 then verifying the presence in the Platypus call sets and also by visual inspection using the IGV browser39. On visual inspection, we found that many ubiquitous indels were actually called incompletely across the biopsy sets by Platypus and sometimes missed all together by Scalpel, highlighting the inconsistency of the currently available tools.
Driver mutation identification
We used two classifications of driver genes (Table S3). First, we identified CRC drivers using the IntOGen database 2016.5 (https://www.intogen.org/). Driver mutations were then classified using a two-tier system. Tier 1 driver mutations were considered to be very likely involved in colorectal carcinogenesis and included canonical mutations such as most protein-truncating APC SNAs, BRAF V600E and KRAS codon 12, 13, 61, 117 and 146 changes. For non-canonical mutations, tier 1 status was assigned to protein-truncating mutations in tumour suppressor genes or recurrently occurring mutations in CRCs in the COSMIC database (http://cancer.sanger.ac.uk/cosmic). Tier 2 mutations, considered to be of lower confidence as drivers, comprised all other coding or splice site changes in the same set of genes. These genes are defined in Table S3a. Second, we used a more restrictive definition of the top 5% significantly mutated genes in the TCGA publication exclusively (non-MSI+ carinomas). These genes are defined in Table S3b.
Coverage normalisation
We normalised for differences in sequencing coverage to avoid bias in mutation calling and intra-tumour heterogeneity measurements due to unequal coverage between samples. To do this, we identified a subset of 110,533 exonic regions consistently sequenced at ≥10X in all biopsies across all tumours. To normalise coverage between samples, we individually sub-sampled each .bam file such that each contained a roughly equal number of reads. This equated to around 17,000,000 reads per biopsy, covering in total 25Mb of exonic regions.
We then generated 100 sets of ‘mini-bams’ (one bam from each sample) where each bam contained the same 10,000 regions randomly selected regions from the original 110,533 well-covered regions. For each set of mini-bams we repeated the joint Platypus calling procedure (as per the above). The mutation frequency was calculated by simply taking the resulting number of variants and the total length of the 10,000 regions in each iteration.
Deep sequencing
In order to validate the exome and genome sequencing, and to search for additional low allele frequency somatic mutations, we sequenced a panel of 50 CRC driver genes in selected tumour regions. These genes encompassed all well established driver events including the top 15 genes from the IntOGen list (see Table S3) and included both tier 1 and 2 driver mutations. Coding regions were captured using molecular inversion probes (MiPs) and sequenced using the Illumina NextSeq. Molecule tagging was used to ensure that reads were derived from unique tumour DNA molecules. Further details of the gene panel and the sequencing protocol are available on request.
All of the subset of 44 driver mutations identified by WES/WGS from three carcinomas and five adenomas were validated. We also were able to identified two additional ubiquitous APC mutations that had not been called in the WGS/WES data.
CNA calling
We used CloneHD40 to call absolute copy number and LOH profiles for each sample. We first collected raw read depth data across the genomes using Samtools. For WGS samples, we used 1kb non-overlapping windows (excluding difficult regions such as centromeric). For WES samples, bed files corresponding to the exome capture kit were used and the data were further placed onto 20kb bins. For each set we utilised the corresponding normal sample to identify outlier bins, which we removed. Furthermore, variant call files for each sample set were used to identify germline heterogeneous loci to collect B-allele data, informative for LOH and/or unbalanced aberrations in the tumour samples.
We then ran filterHD (part of the CloneHD tool) to identify changes in the read depth and B-allele tracks that go beyond the noise resulting from finite sequencing depth for each set40. Such “jump” locations were used as input for cloneHD copy number calling for the tumour samples. For WES samples we also used the corresponding normal samples to correct for platform related bias in the read depth tracks. The bias correction is important for WES samples and not using it would result in a large number of loci with high jump probability, i.e., over-segmentation (see discussion about bias correction40).
We validated the general patterns of large CNA to calls from SNP arrays (Illumina Global Screening Array) using the OncoSNP program on the majority of tumours. Overall, 93% of the larger CNAs were in congruence.
For this analysis, we did not report structural variants or chromosomal hypermutation events such as chromothripsis and chromoplexy.
Exclusion of tumours from analysis of heterogeneity
The ability to accurately measure ITH depends on the number of biopsies available from the tumour41. We assessed the relationship between biopsy number and genetic divergence and found no correlation (Fig S1B). A subsampling bootstrap analysis of three neoplasms each with a high number of biopsies showed that at least four biopsies per neoplasm provided a suitably accurate measurement of ITH (Fig S1C); hence neoplasms with less than four biopsies available were excluded from ITH analysis.
Analysis on sub-clonal populations
Since larger sub-clones might disturb the overall phylogenetic analysis, we investigated the existence of such in the samples with whole genome sequencing using the Battenberg algorithm42. Briefly, the algorithm phases heterozygous SNPs with use of the 1000 genomes genotypes as a reference panel. The resulting haplotypes are corrected for occasional errors in phasing in regions with low linkage disequilibrium. After segmentation of the resulting b-allele frequency (BAF) values, t-tests are performed on the BAFs of each copy number segment to identify whether they correspond to the value resulting from a fully clonal copy number change. If not, the copy number segment is represented as a mixture of 2 different copy number states, with the fraction of cells bearing each copy number state estimated from the average BAF of the heterozygous SNPs in that segment.
Clusters of subclonal substitutions were identified in whole genome sequencing data using a Bayesian Dirichlet process (DP) in n dimensions, where n is the number of related samples as previously described43. For each mutation, the allele frequency was converted to a cancer cell fraction (CCF) prior to clustering, allowing for purity and copy number estimates obtained from the Battenberg algorithm, as described previously43. Clusters were identified as local peaks in the posterior mutation density obtained from the DP. For each cluster, a region representing a ‘basin of attraction’ was defined by a set of planes running through the point of minimum density between each pair of cluster positions. Mutations were assigned to the cluster in whose basin of attraction they were most likely to fall, using posterior probabilities from the DP.
We investigated the geographical spread of subclones by analysing the presence of each subclone across samples from each patient. Subclones may be: clonal (defined as CCF ≥ 0.9) in all samples; clonal in some samples and absent (defined as CCF ≤ 0.1) from others; present subclonally in a single sample; present in multiple samples and subclonal in at least one. The first three categories represent clones/subclones that are in defined geographical regions of various sizes, whereas the last category represents subclones that are more diffuse, indicative of subclone mixing across regions covering multiple biopsies. The percentage of mutations in this last category was low (median 12%, range 5-26%), indicating that where subclones were present they were generally in defined geographical regions.
Phylogenetic analysis
We built phylogenetic trees from the SNAs sets for each tumour using PAUP* software. We first converted each variant set into a binary matrix, where the rows related to a particular biopsy or the normal sample and the columns related to a specific variant. The binary encoding (0/1) designated absence or presence of a variant. A nexus file was used to specify the parsimony parameters needed for the tree construction along with the variant matrix. The following functions and parameters were used: (i) the outgroup function was used to root all resulting trees to the normal sample - effectively a column on the mutation matrix containing only zeros; (ii) the hsearch function was used to perform a heuristic search of 10,000,000 trees from the given tree space, with 1000 of the shortest trees output for the main analysis; (iii) the bootstrap function was used to perform a sub-sampling procedure 10,000 times that involved randomly selecting a set of mutations from the binary matrix (with replacement), with the proportion of each branch instance was reported in a log file; and (iv) the alltrees function was used in the cases where less than 10 biopsies were present. This made it possible to perform an extended 'brute-force' run to acquire the definitely shortest tree(s) from the total search space, at the expense of computational time. The resulting .tre files were visualised and converted to .pdf format using FigTree software44. The homoplasy indexes for the most parsimonious tree in a given set was automatically calculated and output to the PAUP* log file.
To obtain the shortest and thus most parsimonious tree, an Rscript using the ape package45 was used to input the .tre file. In all cases expect carcinoma 6 only one tree was the shortest. In the case of carcinoma 6 the 8 shortest trees were visually compared revealing that a specific clade was unresolved across these trees though the main topology was the same. For this case we build a consensus tree using the ape package, consensus function.
We also tested for phylogenetic consistency across the variant types. We produced subsets of the SNAs using only non-synonymous or synonymous mutations or if the whole genome was available, the exome and then compared the resulting phylogenies with the original total data trees. Comparisons were performed in three ways: (i) topology structure, where identical topology matches were noted; (ii) the number of consistent terminal clades represented across each tree; and (iii) statistical comparison of tree topologies using the Penny and Hendy (P&H) symmetrical distance method. The P&H method was implemented using an R script and the apTreeshape package46. To obtain a p-value for the P&H index we build a distribution of random trees of the same length and number of taxa for a given set, thus enabling a probability to be determined for obtaining a given P&H index.
To perform tree balance analysis (which was only appropriate for carcinoma 6 that possessed > 15 samples), we used the Colless’s test function as implemented in the R apTreeShape package. We tested the topology of carcinoma 6 against a balanced ‘Yule’ tree, hence the p-value represents the likelihood of imbalance given this branching process.
Diversity analysis: adenoma versus cancer
To compare the SNA diversity of adenomas and carcinomas, we performed two main analyses. Firstly, we wanted to compare the proportion of truncal SNAs from the total SNAs called in adenomas and carcinomas, but since certain tumours had many more biopsies, we performed a biopsy-wise down-sampling procedure. Here, in tumours with more than four biopsies (this was considered a reasonable minimum) we randomly selected various four-biopsy combinations and calculated the proportion of variants classified as trunk, branch and leaf. The distributions of these proportions was then plotted with the quartiles for the four-biopsy samples.
Second, the downsampled call sets were used to assess the SNA diversity by measuring the SNA divergence of a pair of randomly selected biopsies from a given tumour. The diverging (non-shared) somatic variants were given as a number per Mb of the genome across the 100 down-sampled iterations.
Mutational signatures with EMu
For each tumour set, we classified all SNAs based on their flanking trinucleotide sequence context47 and whether they fell onto the trunk (shared) or branches (including trunk and leaves) of the phylogenetic tree. Inference of mutational processes and their activities was then determined using the EMu algorithm48. Through this analysis we identified 4 signatures (Signatures A, B, C and D). The resemblance to the COSMIC signatures was determined by comparing 96-channel mutation frequencies and by visual inspection.
Comparison of spatial and genetic distances
To obtain estimates of the physical distances between each biopsy from any tumour, we produced normalised measurements of the number of pixels on a straight-line drawn between the centre points of the biopsy locations. The photographs of the tumour and biopsy locations were used as reference. We then used the ape R package to obtain the inter-taxa distances for each of the phylogenetic trees and performed a linear regression of the physical on the phylogenetic distance for each tumour.
Digital Pathology
Patient material was fixed in 10% buffered formalin and paraffin-embedded. Tumour blocks were sectioned at 4 μm. Ki67 and β-catenin proteins were analysed by immunohistochemistry using a Leica autostainer with haematoxylin counterstaining according to standard protocols (ki-67 antibody, DAKO MIB-1 cat no M7240, 1:150; β-catenin antibody, BD biosciences cat no 610154, 1:50; DAB chromogen). All slides were scanned at 200x magnification on a 3D Histech Pannoramic Scanner and stored in MIRAX (.mrxs) format.
Digital image analysis was performed by a board-certified pathologist (VHK) using the HALO™ image analysis software, v.2.0.1145.19 (Indica Labs, Corrales, NM 87048, USA). Briefly, invasive cancer regions were identified on each scanned tissue section and manually annotated. Normal mucosa and regions of necrosis were excluded. Next, the HALO ™ classifier machine learning algorithm was trained to categorize tumour tissue, stroma and background regions for each stain. The goodness of classification was visually controlled on all cases. Marker-positive cancer cells were identified using the cytonuclear algorithm. Cellular recognition was trained and optimized on invasive cancer tissue. Colour segmentation was performed. The H&E nuclear stain was set as R=0.644, G=0.716, B=0.267, and the DAB positive stain as R=0.268, G=0.570, B=0.776. Algorithm settings are provided in Table S9. ki-67 and β-catenin stains were calibrated using normal, non-proliferative mucosal cells outside of the crypt bases. The total counts and percentage of marker-positive cells in the tumour cell compartment were recorded.
General statistical analyses
All analyses were performed in R or STATA. Unless otherwise stated, all statistical comparisons of two distributions used the Wilcoxon test (Wilcox.test function in R). Unless otherwise stated, 95% confidence intervals of medians were determined using a bootstrapping analysis of the median values (1000 repeats of 20 samples [10 adenoma, 10 carcinoma] with replacement). Data in contingency tables were analysed using Fisher’s exact test. Researchers were not blinded to sample type.
Power calculation for detecting a difference in mutation burden
We calculated the power to detect a difference in the mutation burden of 9 adenomas versus 10 carcinomas. In accordance with the measured burdens, we assumed 90 mutations per adenoma (std. dev. 35 mutations) and considered the power of a t-test to detect a difference between the carcinomas with a variety of higher burdens, requiring a (one tailed) significance level of p<0.05. Standard formulae were used to calculate power. Our data had very good power to detect a 50% increase in burden in carcinomas and good/fair power to detect a 33% increase (Figure S10).
CNA timing model
Full details of the model are presented as a Supplementary Note. Matlab code to calculate the CNA timing is available from the GitHub project page: https://github.com/daniel-temko/CNVTiming.
We modelled the last common ancestor of WGS tumour samples by obtaining the SNAs (together with their associated variant allele frequencies) and informative CNAs (asymmetric copy number gains/amplifications or cnLOH) that were observed in all biopsies of a given tumour. Using an assumption of constant SNA mutation rate over time, we sought to time individual clonal CNAs along the history of the ancestral cell lineage, by partitioning the SNAs present in the same region of the CNA into those accumulated before and after the CNA.
The premise of the timing model is to use the information from all CNAs under consideration in a joint likelihood maximisation of their respective timing. The individual timings are estimated using the assumption that the SNAs in a CNA region will accumulate following a Poisson process, based itself on the assumption of a given mutation rate. The SNVs in a given CNA region were partitioned into those on the non-amplified and amplified DNA strands (referred to as alpha and beta variants respectively in the supplementary note) using a Gaussian mixture model, implemented using the R package ‘mixtools’ and the ‘normalmixEM’ function. Here the parameter mu (the starting cluster means) was inferred from the cellularity and copy state of the CNA. For example: in a biopsy with 100% tumour content, the expected cluster means of a trisomy region are 0.33 and 0.67, representing, the non-amplified and amplified chromosomes. We performed a heuristic search (using 10,000 iterations of the normalmixEM function) to assess the confidence in the inferred cluster centres. Only cases where clustering was deemed successful, determined by the final means of the two clusters being located within one standard deviation of the expected cluster means, were passed forward for timing analysis. Note that ‘complex CNAs’ – those CNAs where there was reason to suspect multiple alterations occurred at the same locus (for example focal gain after an arm gain) - were excluded from the analysis.
The statistical assessment of the distribution of the CNA timings was performed by comparing the distribution of timings for each tumour against a uniform distribution using the Kolmogorov–Smirnov test. The duration of the comparative interval was set to the interval between the inferred initiation of the adenoma (timing of the 5q LOH event) and the time most recent common ancestor (MRCA). In the cases where no 5q event was detected, the time of initiation of adenoma growth was taken as the average time of 5q LOH in the two cases where this event could be timed (adenoma 4 and carcinoma 5).
Supplementary Material
Acknowledgements
SL, TAG (A19771) and IT (A27327) are funded by Cancer Research UK. We acknowledge core funding to the Wellcome Trust Centre for Human Genetics from the Wellcome Trust (090532/Z/09/Z). TG and SL were also supported by Bowel and Cancer Research Charity small grant scheme. VM was supported in part by funding from The Wellcome Trust (098051). DCW is supported by the Li Ka Shing foundation. XJ and IT are supported by an ERC advanced grant (EVOCAN-340560). The S:CORT study is funded by MRC and Cancer Research UK.
Footnotes
Data availability
Raw data (bam and vcf files) are available via the EGA accession code: EGAS00001003066.
Individual contributions
IT, TG and SL conceived and designed the study. RG, JE, LMW, KH SL and IT provided samples. HD, AMB, SB, LC performed experiments. WC, MK, VM, PM, RA and DW performed bioinformatics analysis. WC and DT performed mathematical analysis. CG, AA and VK performed image analysis. MJ, MRJ and LMW performed pathology assessment. ED, TM, and the S:CORT consortium provided reference data. WC, ML, VM, DT, RA, VK, XJ, DW, YF, MK, SA, DW, AS, SL, TG and IT analysed data. WC, AS, SL, TG and IT performed evolutionary analysis. WC, TG and IT wrote the manuscript with input from all authors.
The authors have no competing interests to declare.
References
- 1.Morson BC. Evolution of cancer of the colon and rectum. Cancer. 1974;34:845–849. doi: 10.1002/1097-0142(197409)34:3+<845::aid-cncr2820340710>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 2.Ashton-Rickardt PG, et al. High frequency of APC loss in sporadic colorectal carcinoma due to breaks clustered in 5q21-22. Oncogene. 1989;4:1169–1174. [PubMed] [Google Scholar]
- 3.Powell SM, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith G, et al. Mutations in APC, Kirsten-ras, and p53—alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433–9438. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muzny DM, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proc Sixth Int Congr Genet. 1932;1:356–366. [Google Scholar]
- 11.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127ps10–127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 12.Blum MGB, François O. On statistical tests of phylogenetic tree imbalance: The Sackin and other indices revisited. Math Biosci. 2005;195:141–153. doi: 10.1016/j.mbs.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Fischer A, Illingworth CJ, Campbell PJ, Mustonen V. EMu: probabilistic inference of mutational processes and their localization in the cancer genome. Genome Biol. 2013;14:R39. doi: 10.1186/gb-2013-14-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katainen R, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 16.Quirke P, et al. DNA aneuploidy in colorectal adenomas. Br J Cancer. 1986;53:477. doi: 10.1038/bjc.1986.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones AM, et al. Analysis of copy number changes suggests chromosomal instability in a minority of large colorectal adenomas. J Pathol. 2007;213:249–256. doi: 10.1002/path.2234. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Liang L, Fang J-Y, Xu J. Somatic gene copy number alterations in colorectal cancer: new quest for cancer drivers and biomarkers. Oncogene. 2016;35:2011–2019. doi: 10.1038/onc.2015.304. [DOI] [PubMed] [Google Scholar]
- 19.Durinck S, et al. Temporal Dissection of Tumorigenesis in Primary Cancers. Cancer Discov. 2011;1:137–143. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman S, et al. The relative timing of mutations in a breast cancer genome. PloS One. 2013;8:e64991. doi: 10.1371/journal.pone.0064991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim T-M, et al. Subclonal Genomic Architectures of Primary and Metastatic Colorectal Cancer Based on Intratumoral Genetic Heterogeneity. Clin Cancer Res Off J Am Assoc Cancer Res. 2015;21:4461–4472. doi: 10.1158/1078-0432.CCR-14-2413. [DOI] [PubMed] [Google Scholar]
- 23.Uchi R, et al. Integrated Multiregional Analysis Proposing a New Model of Colorectal Cancer Evolution. PLOS Genet. 2016;12:e1005778. doi: 10.1371/journal.pgen.1005778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki Y, et al. Multiregion ultra-deep sequencing reveals early intermixing and variable levels of intratumoral heterogeneity in colorectal cancer. Mol Oncol. 2017;11:124–139. doi: 10.1002/1878-0261.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim T-M, et al. Clonal origins and parallel evolution of regionally synchronous colorectal adenoma and carcinoma. Oncotarget. 2015;6:27725–27735. doi: 10.18632/oncotarget.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stachler MD, et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet. 2015;47:1047–1055. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maley CC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2016;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 28.Ross-Innes CS, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47:1038–1046. doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babraham Bioinformatics. FastQC A Quality Control tool for High Throughput Sequence Data. [cited 2013 Apr 6]; [Internet]. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 30.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010 Jul 19;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. [cited 2015 Oct 12];2013 Available from: http://arxiv.org/abs/1303.3997.
- 32.broadinstitute/picard. [cited 2016 Oct 1];GitHub. 2016 [Internet]. Available from: https://github.com/broadinstitute/picard. [Google Scholar]
- 33.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009 Aug 15;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, WGS500 Consortium et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014 Aug;46(8):912–8. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010 Sep;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 2012 Apr 1;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasan MS, Wu X, Zhang L. Performance evaluation of indel calling tools using real short-read data. [cited 2016 Aug 4];Hum Genomics. 2015 Dec;9(1) doi: 10.1186/s40246-015-0042-2. [Internet]. Available from: http://www.humgenomics.com/content/9/1/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narzisi G, O’Rawe JA, Iossifov I, Fang H, Lee Y, Wang Z, et al. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat Methods. 2014 Aug 17;11(10):1033–6. doi: 10.1038/nmeth.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013 Mar 1;14(2):178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer A, Vázquez-García I, Illingworth CJR, Mustonen V. High-Definition Reconstruction of Clonal Composition in Cancer. Cell Rep. 2014 Jun;7(5):1740–52. doi: 10.1016/j.celrep.2014.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werner B, Traulsen A, Sottoriva A, Dingli D. Detecting truly clonal alterations from multi-region profiling of tumours. Sci Rep. 2017 Mar 27;7 doi: 10.1038/srep44991. srep44991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The Life History of 21 Breast Cancers. Cell. 2012 May 25;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swofford DL. PAUP* 4.0. [cited 2015 Aug 2];2003 [Internet]. Available from: http://paup.csit.fsu.edu/
- 44.Rambaut A. FigTree. [cited 2017 Mar 28]; [Internet]. Available from: http://tree.bio.ed.ac.uk/software/figtree/
- 45.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004 Jan 22;20(2):289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 46.Bortolussi N, Durand E, Blum M, Francois O. apTreeshape: statistical analysis of phylogenetic tree shape. Bioinformatics. 2005 Dec 1;22(3):363–4. doi: 10.1093/bioinformatics/bti798. [DOI] [PubMed] [Google Scholar]
- 47.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013 Aug 14;500(7463):415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer A, Illingworth CJ, Campbell PJ, Mustonen V. EMu: probabilistic inference of mutational processes and their localization in the cancer genome. Genome Biol. 2013;14:R39. doi: 10.1186/gb-2013-14-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.