Summary
Protein complexes are assemblies of subunits that have co-evolved to execute one or many coordinated functions in the cellular environment. Functional annotation of mammalian protein complexes is critical to understanding biological processes as well as disease mechanisms. Here, we used genetic co-essentiality derived from genome-scale RNAi- and CRISPR-Cas9- based fitness screens performed across hundreds of human cancer cell lines to assign measures of functional similarity. From these measures, we systematically built and characterized functional similarity networks which recapitulate known structural and functional features of well-studied protein complexes and resolve novel functional modules within complexes lacking structural resolution, such as the mammalian SWI/SNF complex. Finally, by integrating functional networks with large protein-protein interaction networks, we discovered novel protein complexes involving recently-evolved genes of unknown function. Taken together, these findings demonstrate the utility of genetic perturbation screens alone and in combination with large-scale biophysical data to enhance our understanding of mammalian protein complexes in normal and disease states.
Introduction
The derivation of gene-gene relationships is a central goal of systems genetics (Baliga et al., 2017). Several gene properties including co-temporal expression, co-evolution, and physical interaction between protein products have proven to be informative in the identification of functionally related genes such as those coding for subunits of protein complexes (de Juan et al., 2013; Gingras et al., 2007; Jansen et al., 2002; Ramani et al., 2008). One particularly powerful approach is to define genetic interactions by probing for epistatic relationships between genes, in which the phenotypic readout of a genetic perturbation depends on the status of a second gene (Baryshnikova et al., 2013).
Genetic interaction mapping has been most extensively pursued in S. cerevisiae, in which crosses between gene knockout strains coupled with cellular fitness readouts enabled systematic measurements of genetic interactions (Pan et al., 2004; Schuldiner et al., 2005; Tong et al., 2004). Studies have indicated that genes functioning within similar biological processes tend to share genetic interaction partners (Collins et al., 2007; Kelley and Ideker, 2005; Schuldiner et al., 2005), which motivated the construction of genome-scale functional similarity networks for yeast (Costanzo et al., 2010; Costanzo et al., 2016). In these networks, functionally related genes share an edge based on the similarity of their genetic interaction profiles, ultimately yielding a modular, hierarchical model of the cell, in which genes with coordinated functions, such as members of the same protein complex, cluster in to functional modules.
Given the utility of this approach for inferring gene function, a major interest in the field lies in deriving a global functional similarity maps for human cells. However, the generation of such datasets faces major limitations. In addition to the roughly 16-fold increase in combinatorial space of all possible double knockouts in human cells compared to yeast (due to the four-fold increase in genes), human screening libraries for simultaneously knocking out multiple genes of interest are relatively new and still face unsolved technical challenges (Boettcher et al., 2018; Du et al., 2017; Han et al., 2017; Najm et al., 2018; Shen et al., 2017). In contrast, genome-scale single-gene knockout libraries are technically more advanced and have been used extensively in pooled fitness screens (Doench, 2018). In theory, performing an equivalent double-knockout experiment with a single-gene knockout library would require screening for fitness effects across a massive cell line collection, in which each cell line contains a precise genetic knockout stably derived from an isogenic background. While such a cell line collection does not currently exist, we hypothesized that an analogous approach may be feasible using a collection of cancer cell lines with sufficient diversity in fitness responses, in which diversity does not arise from single knockouts but rather through genomic and transcriptomic variation en masse.
We and others have observed that cancer cell lines exhibit highly variable genetic dependencies for cellular fitness (Bertomeu et al., 2018; Blomen et al., 2015; Hart et al., 2015; Hart et al., 2017a; McDonald et al., 2017; Meyers et al., 2017; Tsherniak et al., 2017; Wang et al., 2015; Wang et al., 2017b) in a manner that reflects the diverse genomic and transcriptomic alterations a cell may accumulate during tumorigenesis. Project Achilles (Broad Institute of MIT and Harvard) seeks to systematically map genetic vulnerabilities across large collections of cancer cell lines, including the Cancer Cell Line Encyclopedia (CCLE) (Barretina et al., 2012), and has recently performed genome-scale perturbation screens in 501 cancer cell lines using RNA interference (RNAi) (Tsherniak et al., 2017) and in 342 cancer cell lines using CRISPR-Cas9 (Meyers et al., 2017). Because the genomic and transcriptomic state of each cancer cell line gives rise to a unique overall fitness response upon perturbation of each gene, these datasets may provide an opportunity derive gene-gene functional relationships and to construct a modular network of cell function.
In this study, we evaluate the use of large-scale RNAi and CRISPR-Cas9 genetic perturbation datasets for the construction of a human functional similarity network. We focused on protein complexes because of their modular composition, coordinated function, and involvement in many biological processes (Pereira-Leal et al., 2006). Following validation that correlated fitness profiles between gene pairs in both RNAi and CRISPR-Cas9-based screens represent informative measures of known functional relationships, such as physical interactions, we then developed a network permutation approach to evaluate whether gene modules coding for protein complex subunits are significantly correlated in their fitness profiles. After benchmarking against a gold standard protein complex dataset (Core CORUM), we find that ~40% of gold standard protein complexes form significantly connected functional modules in our networks. We find that these functional similarity networks reproduced structural features of known protein complexes, as well as identified intra-complex functional modularity in complexes with no known structure. Finally, we applied this approach to a set of computationally-predicted but unvalidated protein complexes (hu.MAP), and describe a pair of functionally related but uncharacterized genes whose protein products form a novel protein complex. Taken together, these findings establish the utility of large-scale fitness screening in cancer cell lines to reveal functional and structural features of protein complexes, and prime the field for the global derivation of a human functional similarity network from large-scale genetic perturbation screens.
Results
Interacting proteins exhibit coordinated fitness effects upon genetic depletion in cancer cell lines
Protein complexes execute specific molecular functions that require the proper assembly and activity of their interacting subunits (Figure 1A, left). Depletion of individual subunits required for complex function would be predicted to produce similar phenotypic effects on fitness (Figure 1A, right). To assess this premise systematically in human cells, we analyzed recently-generated datasets from large-scale RNAi- and CRISPR-Cas9- based fitness screening efforts of hundreds of cancer cell lines via Project Achilles (https://portals.broadinstitute.org/achilles) (Cowley et al., 2014; Meyers et al., 2017; Tsherniak et al., 2017). Over 600 cancer cell lines representing 23 different lineages were screened across the RNAi and CRISPR-Cas9 datasets, representing an extremely diverse set of cellular contexts (Figure S1A, Table S1).
Figure 1. Genes encoding protein complex subunits display coordinated fitness variation across genetic screens performed in human cancer cell lines.
(A) Schematic of normal and perturbed protein complex biogenesis.
(B) Fitness profiles for genes encoding subunits of five different protein complexes screened in the CRIPSR-Cas9 fitness dataset, annotated by their gene name abbreviations and cellular localization. Both rows (genes) and columns (cell lines) are hierarchically clustered.
(C) Graphical representation of RNAi- and CRISPR-Cas9-based screening datasets and analysis pipelines (n=501 and n=342 cell lines, respectively; Project Achilles, Broad Institute).
As an illustration of this premise, the fitness effect upon CRIPSR-Cas9 knockout of subunits of various protein complexes across 342 cell lines are shown (Figure 1B). The fitness effects upon gene knockout varied widely across cell lines, and analysis of both RNAi and CRISPR-Cas9 datasets demonstrated that genes coding for Core CORUM protein complex subunits exhibited significantly greater fitness variation than genes that do not (Wilcoxon rank sum test, p < 1E-3) (Figure S1B, Table S2). Additionally, fitness profiles for genes encoding subunits of the same protein complexes are strikingly concordant, and correlation clustering by fitness profile resulting in a genes being grouped by protein complex membership (Figure 1B). Across both datasets, pairs of genes sharing Core CORUM protein complex membership exhibit significantly greater correlations of fitness profiles than protein subunits from different complexes (Figure S1C, Kolmogorov–Smirnov test, p = 2.2e-16 for both RNAi and CRISPR-Cas9). This result suggests the potential to use correlated fitness effects across cancer cell lines as a measure of functional similarity and therefore identify and resolve functional relationships within and between protein complexes.
In order to test this observation for interacting proteins in general, we collated all large-scale, annotated protein-protein interaction (PPI) datasets generated by individual or joint research groups over the past five years (Drew et al., 2017; Hein et al., 2015; Marsh et al., 2013; Rolland et al., 2014; Thul et al., 2017; Wan et al., 2015), including Core CORUM as the gold-standard literature reference (Ruepp et al., 2010). We again found that gene pairs with an annotated PPI consistently exhibited greater fitness profile correlations across all datasets than gene pairs that did not have an annotated interaction (Figure S1D). Furthermore, after binning gene pairs by the strength of their fitness correlations in the RNAi and CRISPR datasets (as performed in (Hart et al., 2017a)using the KEGG Pathway Database), we found that the most strongly correlated gene pairs were the most enriched for interacting protein products across PPI datasets (Figure S1E).
Taken together, these results demonstrate that interacting proteins tend to exhibit correlated fitness profiles in large-scale genetic perturbation screens, and that top- ranked fitness correlations were highly enriched for interacting proteins, suggesting that many protein complexes may exist as correlated modules within these fitness screening datasets. In order to further test this, we analyzed both the RNAi and CRISPR-Cas9 screening data in parallel. From these screening data, all collected at one screening facility and subjected to statistical models that eliminate off target effects, we filtered for genes whose depletion had significant fitness effects to obtain 6300 genes in the RNAi dataset and 8997 genes in the CRISPR-Cas9 dataset (Figure 1C). We then performed Pearson correlations on fitness profiles for all pairwise combination of genes, rank-order normalized the vector of correlations for each gene, symmetrized by taking the best rank across gene pairs, and used these data as the basis for all networks generated below.
Using RNAi and CRISPR-Cas9 screening data to generate functional similarity networks for hundreds of human protein complexes
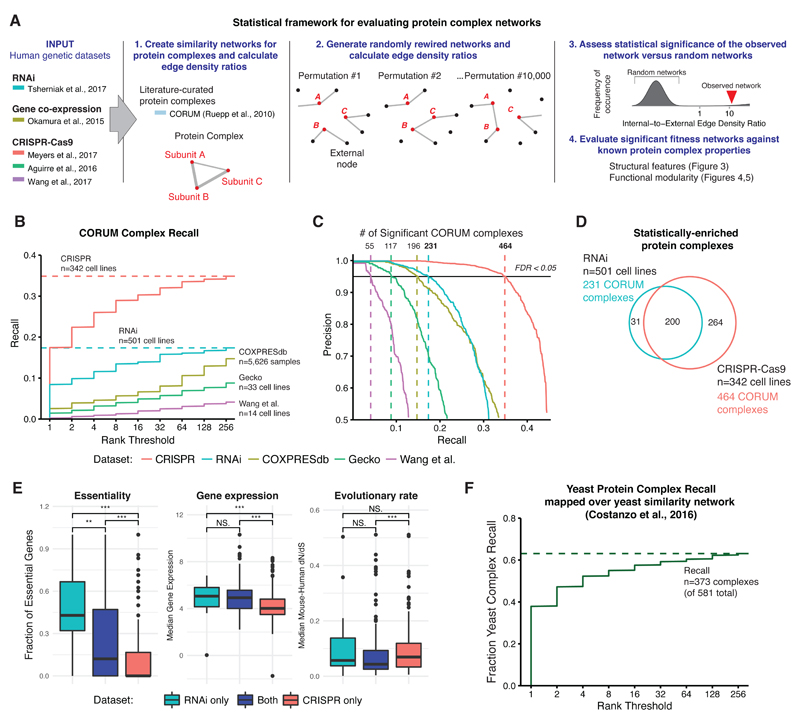
To systematically assess the extent to which fitness screening data can inform the biology of protein complexes, we developed a network-based approach for analyzing both RNAi and CRISPR fitness datasets (Figure 2A (summary), Figure S2 (detail), see Methods). We represent protein complexes as a functional similarity network consisting of nodes for each protein subunit and edges when the measure of functional similarity between two subunits exceeds some threshold. We define functional similarity using the symmetric rank-normalized correlations as above. At any given rank threshold, edges may connect two subunits within a protein complex of interest (internal edges), or they may connect subunits between complexes (external edges). To determine the set of protein complexes whose subgraphs are statistically enriched for edges in this network, we calculated the ratio of internal edge density versus external edge density for each protein complex across several rank thresholds (Figure S2A-C). We then determined the statistical significance of this ratio using an empirical null distribution generated from 10,000 randomly rewired networks while preserving node degree. Finally, we visualized the functional similarity network for protein complexes that exceed statistical significance (False Discovery Rate (FDR) < 0.05) (Figure S2D) and analyzed their functional and structural features.
Figure 2. A statistical framework for nominating significant protein complex fitness correlation networks.
(A) Overview of the statistical framework for identifying significant protein complex fitness correlation networks (see Figure S2).
(B) Fraction of human protein complexes recalled at FDR < 0.05 in fitness correlation datasets (RNAi, CRISPR, Gecko and Wang et al.) and a gene expression correlation dataset (COXPRESdb), plotted against a log range of rank correlation thresholds. Fraction of CORUM complex recall is defined as the fraction of CORUM protein complexes (n=1286) that exhibit correlations at or below that rank threshold.
(C) Precision-recall curve for the protein complexes in each dataset.
(D) Venn diagram depicting overlap between CORUM protein complexes statistically enriched with top-ranked correlations in CRISPR and RNAi datasets.
(E) Biologic properties of protein complexes with significant correlations in RNAi, CRISPR, or both datasets (Wilcoxon rank sum test, ** p < 1e-2, *** p < 1e-3, N.S. = not significant).
(F) Statistical framework in (A) applied to a yeast correlation dataset derived from a genome-scale pairwise interaction map (Costanzo et al., 2016). A cumulative total of 373 yeast protein complexes with statistically significant fitness networks were recalled at rank 256, representing 64.2% of total yeast protein complexes.
To compare the performance of our methodology on the RNAi and CRISPR fitness datasets described above, we included two additional recently published CRIPSR-Cas9 screening datasets: one consisting of 14 CRISPR-Cas9-based screens performed in acute myeloid lymphoma cancer lines (Wang et al., 2017b), and the other consisting of 33 cancer cell lines of diverse lineage screened with the GeCKOv2 sgRNA library (Aguirre et al., 2016) (Figure 2A, S3A-B). Additionally, to draw comparisons with previous studies of correlated gene expression profiles of protein complexes, we included a large-scale mRNA co-expression dataset, COXPRESdb (Okamura et al., 2015), containing pairwise correlations across 5000+ publically available RNA-seq datasets (Figure 2A). We generated the gene network for each of these five datasets and determined the set of Core CORUM protein complexes whose subgraphs are significantly enriched for internal edges at various rank thresholds.
The CRISPR-Cas9 and RNAi fitness datasets both captured a greater fraction of human protein complexes than the smaller GeCKOv2 or Wang et al. datasets - likely due to the scale of cancer cell lines screened - and also outperformed the COXPRESdb network across top rank thresholds (Figure 2B-C). Specifically, by rank 256, 17% of all Core CORUM protein complexes were captured at statistical significance in the RNAi dataset and 35% of all complexes in the CRISPR dataset. The distribution of the cellular localization of protein complexes enriched in both RNAi or CRISPR datasets was similar to the localization distribution for all human protein complexes (Core CORUM), indicating that protein complexes from a variety of processes and pathways exhibit significant functional similarity networks (Figure S3C).
In total, we found that 495 out of 1331 Core CORUM complexes had statistically significant fitness networks in either RNAi or CRISPR dataset, with 231 complexes from RNAi, 464 from CRISPR, and 200 overlapping between the two (Figure 2D, Table S3). We identified features of protein complexes that underpin differences in recall between the two genetic perturbation datasets. Protein complexes recalled only in the CRISPR dataset were significantly depleted of core essential genes (Hart et al., 2015; Hart et al., 2017b), lower median gene expression levels, and had lower sequence conservation (Wilcoxon rank sum test, p < 1e-3) (Figure 2E, Table S3).
To further benchmark this performance against a large-scale functional mapping effort in a model organism, we generated the functional similarity network from a dataset generated by Costanzo et al. in their effort to map all pairwise genetic interactions in S. cerevisiae (Costanzo et al., 2016). Applying our methodology to this functional similarity network, which is derived by calculating pairwise correlations of genetic interaction profiles, we captured 64% (373/581) of all yeast complexes as significantly correlated subgraphs (Figure 2F).
We visualized individual functional similarity networks for each of the 495 Core CORUM protein complexes recalled in either dataset, first by determining the dataset in which that protein complex exhibits the greatest enrichment for internal edges (Figure S3D), and second by choosing a rank threshold specifically for the complex that optimizes the ratio of internal to external edges (see Methods). We found that these functional similarity networks vary greatly in their network topology (Figure S3E) and included many well-studied protein complexes (26S Proteasome, Mediator, RNA Pol II, STAGA, mammalian SWI/SNF, and others) with structural or functional features that we sought to examine in greater depth.
Functional similarity networks recapitulate structural and functional modules of protein complexes
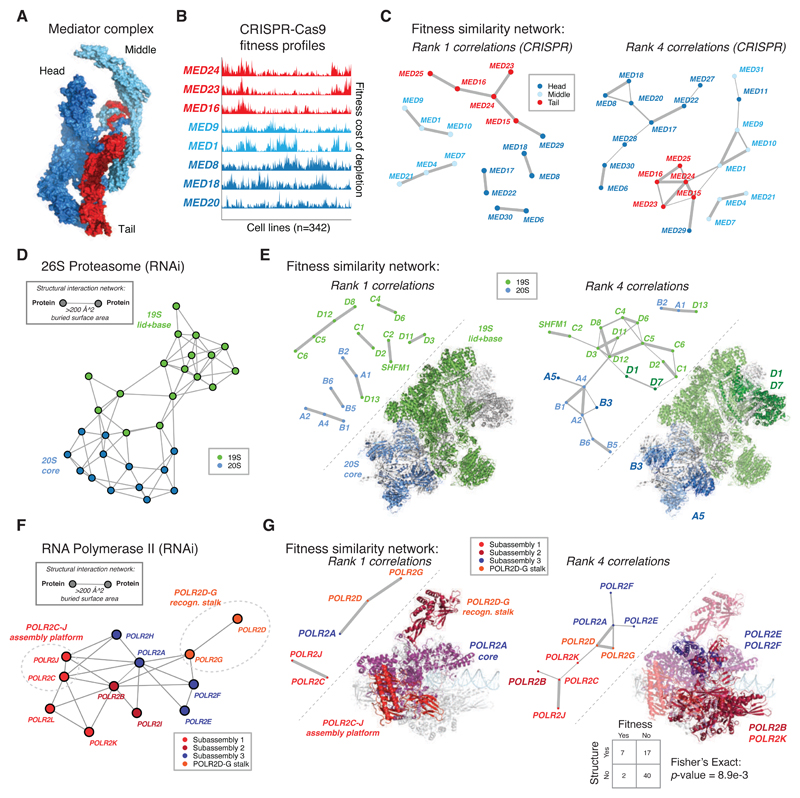
Large protein complexes are often hierarchically assembled from smaller sub-assemblies that have specific modular functions. We hypothesized that for protein complexes with a sufficient number of subunits and sufficiently distinct functional componentry, this modular structure may be reflected in the functional similarity networks. The Mediator complex is an evolutionarily conserved transcriptional activator composed of three stable modules – the Head, Middle and Tail (Figure 3A) – and one detachable, cell cycle specific module (Cyclin Kinase Module, not crystallized) (Nozawa et al., 2017; Tsai et al., 2017). Recent biochemical and genetic studies have demonstrated that the Head, Middle and Tail modules exhibit differential genomic targeting and have specialized roles in the context of Mediator complex global function (Jeronimo et al., 2016; Petrenko et al., 2016). Depletion of subunits belonging to the Head, Middle and Tail modules results in distinct cell fitness effects across cancer cell lines (Figure 3B). Correspondingly, the CRISPR functional similarity network for the Mediator complex largely contains edges between subunits belonging to the same structural module at various rank thresholds (Figure 3C). Comparatively, a gene expression similarity network for the Mediator complex derived from the COXPRESdb dataset captured few correlations between the Head or Middle modules, even at a lenient rank threshold of 50 (Figure S4A).
Figure 3. Fitness correlation networks highlight functional modules of protein complexes with solved structures.
(A) The Mediator complex (PDB 5U0P) is a modular complex composed of functionally distinct sub-assemblies (Head, Middle and Tail modules).
(B) Fitness profiles from the CRISPR-Cas9 dataset of representative subunits of the Mediator complex Head, Middle and Tail modules, colored as in (A). Both rows (genes) and columns (cell lines) are hierarchically clustered.
(C) CRISPR-Cas9 fitness correlation network for Mediator complex, with subunits colored by module membership, and edges between nodes thresholded either at rank one (left) or rank four (right).
(D) The 26S proteasome is composed of the 20S core and 19S regulatory particles, shown here as modules in a structural interaction network, in which each node represents a subunit and each edge represents a physical interaction (buried surface area, Å^2) between subunits in the solved structure (PDB 5GJR).
(E) Fitness correlation networks in the RNAi dataset at different fitness rank thresholds reflect the sub-complex structural organization of the proteasome. Sequentially including edges across rank levels reveals edges preferentially linking genes within the same sub-complex. Proteasome subunit names are abbreviated to their shortest identifying sequence (ex: PSMA1 -> A1).
(F) The RNA polymerase II complex (PDB 5FLM), represented as a structural interaction network. The protein complex is composed of four distinct subassemblies, in particular, two functionally obligate heterodimeric subunits: the assembly core (POLR2C-J) and the detachable recognition stalk (POLR2D-G).
(G) The fitness correlation network for RNA Pol II in the RNAi dataset at different rank thresholds. The overlap between structural edges and functional edges present between protein complex subunits is statistically significant (Fisher’s Exact Test, p-value = 8.9e-3).
We sought to understand whether or not there were additional protein complexes that displayed overlapping physical and functional modularity. After restricting our analysis to large protein complexes (10 or more subunits) that displayed significant fitness networks, we compared fitness network communities with those inferred from PPI networks, using the Bioplex 2.0 dataset and a Protein Data Bank (PDB) structural interaction network (Figure S4B-C, Table S4). In comparison to the 36 protein complexes displaying overlapping modularity between these structural interaction networks and hu.MAP (another protein interaction network), we found that 10 protein complexes showed significantly overlapping fitness network modules versus their structural counterparts, and only two displayed overlapping structural and mRNA co-abundance network modularity (Figure S4D, Table S5). Several of these protein complex assemblies, including the 26s proteasome, RNA polymerase holoenzyme, and the COP9 signalosome (Figure S4D, inset), showed overlapping functional and physical modularity over recognizable subcomplexes and assembly modules.
The 26S proteasome is composed of the 19S regulatory and 20S core sub-assemblies joined by a common interface (Figure 3D). The RNAi functional similarity network clearly distinguished the subunits of the 19S and 20S particles; of the 31 genes encoding proteasome subunits included in this dataset, 20 were involved in a top-ranked correlation with another proteasome subunit, largely within the same particle (Figure 3E, left). This is consistent with the fact that depletion of the 19S and 20S particles was previously shown to have differing effects on cancer cells, despite both particles being part of the same macromolecular assembly (Dambacher et al., 2016). While co-expression networks of 26S proteasome subunits are strongly connected, they fail to reach significance using a module overlap test for structural networks (Figure S4E).
Heteromeric complexes follow energetically favorable ordered assembly pathways, forming intermediate configurations during this process (Ahnert et al., 2015). The RNA Polymerase II subunits POLR2C and POLR2J form a heterodimer that acts as an assembly platform to nucleate the remainder of the assembly pathway, which occurs via three subassembly intermediates (Figure 3F) (Wild and Cramer, 2012), while the POLR2D-G detachable heterodimeric recognition stalk selectively associates with the complex via POLR2A during transcription elongation (Werner and Grohmann, 2011). The RNAi functional similarity network of the RNA Pol II complex identified the POLR2C-J and POLR2D-G heterodimers as Rank 1 pairwise correlations (Figure 3G, left). The network further highlighted functional relationships between the Subassembly 3 components that anchor the recognition stalk to the complex(Tan et al., 2003) and the assembly platform component POLR2K and its binding partner POLR2B, which assemble together on the POLR2C-J heterodimer (Figure 3G, right) (Wild and Cramer, 2012). The observed overlap between edges in the functional similarity network and the structural interaction network is statistically significant (Two-sided Fisher’s exact test, p-value = 8.9e-3). The co-expression network for RNA Pol II does not resolve this functional modularity (Figure S4F).
To test the extent to which such heterodimers exhibit correlation in fitness profiles globally, we curated a set of 271 heterodimeric interactions from the full PDB structural interaction dataset (Table S4) and found significantly higher ranked correlations in both the RNAi and CRISPR datasets than in COXPRESdb dataset (Figure S4G). Taken together, these results suggest that stably interacting, functionally obligate subunits, such as those in heterodimers or structural submodules of protein complexes, exhibit highly ranked fitness correlations in our fitness datasets.
Functional similarity networks resolve shared subunits with functionally diverged protein complexes
Novel complexes can form over evolutionary time via partial duplication and divergence of specific subunits, which can adopt new function while maintaining stable interactions with shared subunits from the original complex (Pereira-Leal et al., 2006). The eukaryotic RNA polymerases I, II and III arose according to this paradigm, with select subunits duplicating and diverging between Archaea and Eukaryotes (Carter and Drouin, 2009) (Figure S5A). Hierarchical clustering performed on fitness profiles of RNA polymerase I, II, and III subunits in the RNAi dataset functionally distinguish RNA Pol III from RNA Pol I and II (Figure S5B). Interestingly, components of the heterodimeric POLR1C-D assembly platform (homologous to POLR2C-J) show functional similarity to both Pol I and Pol III in the functional similarity network for the three polymerases (Figure S5C). This heterodimer is shared between both complexes, and blocking their dimerization precludes assembly of either complex in S. cerevisiae (Mann et al., 1987).
The functional similarity network of the STAGA/ATAC family of complexes, which are known to deposit acetylation marks genome-wide (Spedale et al., 2012), reveals structural modularity and suggests distinct functional characteristics (Figure S5D). Although members of the STAGA complexes cluster distinctly from members of the ATAC complex in the CRISPR dataset (Figure S5E), three subunits shared between both complexes – KAT2A, SGF29 and TADA3 – appear centrally in the functional similarity network, bridging the two complexes (Figure S5F). The gene co-expression network did not recapitulate this modularity or shared subunit membership for the RNA Polymerases and the STAGA/ATAC family of complexes (Figure S5G-H). These results collectively demonstrate the resolution with which RNAi- and CRISPR- fitness networks report on the modularity and assembly of protein complexes within defined complex families.
Functional similarity identifies a novel functional module of the mammalian SWI/SNF complex
We next turned to a complex of unknown structure and incompletely defined subunit composition, the mammalian SWI/SNF (mSWI/SNF or BAF) ATP-dependent chromatin remodeling complex. mSWI/SNF complexes are combinatorially assembled into 12-15 subunit heteromorphic ~1.5-2MDa complexes (Figure 4A), which utilize the energy of ATP hydrolysis to remodel nucleosomal architecture and oppose Polycomb repressive complexes, thus facilitating DNA accessibility and gene expression activation (Kadoch et al., 2017). Recent human genetic studies have unmasked recurrent mutations in the genes encoding mSWI/SNF subunits in over 20% of human cancers and in neurodevelopmental disorders such as intellectual disability syndromes (Kadoch and Crabtree, 2015). The mechanistic interpretation of the mSWI/SNF mutational spectrum is complicated by the incomplete functional characterization of several recently identified subunits, as well as the combinatorial subunit configurations produced by several paralogous, even tissue-specific, subunits (Kadoch et al., 2013). Therefore, we sought to apply our methodology to study the mSWI/SNF complex. The mSWI/SNF functional similarity network from both RNAi and CRISPR datasets revealed three distinct functional modules (Figure 4B-C, S6A). The first corresponds to a core set of BAF complex components (ARID1A, SMARCB1, and SMARCE1 and the SMARCA4 ATPase subunit), while a second module is composed of distinguishing subunits of the PBAF variant of mSWI/SNF complexes (PBRM1, ARID2, BRD7). The third functional module that did not correspond to any known configuration of the mSWI/SNF complex and is composed of one established mSWI/SNF subunit, SMARCD1; one recently discovered subunit, BRD9 (Kadoch et al., 2013); and one putative subunit, GLTSCR1 (Ho et al., 2009), whose highest ranked fitness correlations were with SMARCD1 and BRD9 (Figure 4B, S6A).
Figure 4. Fitness correlation mapping identifies biochemically distinct modules of mammalian SWI/SNF complexes.
(A) Schematic depicting subunits of the mammalian SWI/SNF family of ATP-dependent chromatin remodeling complexes.
(B) Fitness correlation network (from RNAi dataset) between mSWI/SNF subunits resolves three functional modules: core BAF (SMARCA4, ARID1A, SMARCB1 and SMARCE1), PBAF (PBRM1, ARID2, BRD7, PHF10) and a novel functional module that contains two previously characterized subunits (SMARCD1, BRD9) and one putative subunit (GLTSCR1).
(C) Hierarchical clustering performed on fitness profile correlations from the RNAi dataset groups subunits into distinct modules.
(D) Density sedimentation experiments using 10-30% glycerol gradients performed on nuclear extracts from CCRF cells links two functional modules to known complexes, BAF (blue bar) and PBAF (red bar), and one to a novel assembly of distinct size and composition (green bar).
(E) Rare cancers characterized by mSWI/SNF perturbations exhibit mutually exclusive loss of one of the BAF core module genes or paralog families (containing SMARCA4, ARID1A, SMARCB1, SMARCE1). SCCOHT = small cell carcinoma of the ovary, hypercalcemic type. In addition, specific intellectual disability syndromes are caused by heterozygous mutations in BAF core module genes. i
To experimentally determine whether these three functional modules exist as distinct biochemical entities, we performed size fractionation followed by immunoblot on nuclear extracts isolated from CCRF cells. BAF-specific and PBAF-specific subunits separated into assemblies of different sizes, as shown by their migration in distinct fractions of 10-30% glycerol gradients (Figure 4D). BRD9 migrated in lower molecular weight fractions of the gradient, indicating an unexpected smaller subassembly with SMARCD1. Immunoprecipitation studies further confirmed that the novel module binds the catalytic subunit SMARCA4 but fails to bind subunits found exclusively in other sized-fractions, such as ARID1A, ARID2, and BRD7 (Figure S6B). Immunoprecipitation of the SMARCA2 mSWI/SNF ATPase subunit from cancer cell line nuclear extracts coupled with mass-spectrometry resolved high numbers of peptides of GLTSCR1 with minimal background signal (Figure S6C). These results are supported by large-scale published co-fractionation (Figure S6D) (Wan et al., 2015) and co-immunoprecipitation datasets (Figure S6E) (Huttlin et al., 2015), which suggest binding interactions but do not resolve mSWI/SNF modularity.
Together, the functional organization of mSWI/SNF subunits unveiled by functional similarity networks suggests the existence of three concurrently-expressed mSWI/SNF family complexes that have distinct function and are assembled on a common catalytic subunit or module. This has significant implications for recently published studies employing small molecule targeting of the BRD9 subunit (Hohmann et al., 2016), and advances understanding of mSWI/SNF complex combinatorial assembly, a major and unmet goal in the field. Mechanistic dissection of the functions of this novel mSWI/SNF functional module on chromatin will require further study (Michel et al., unpublished data). Importantly, functional modules of mSWI/SNF complexes were not explained by co-expression; for example, ARID1A and ARID2 assemble into mutually exclusive mSWI/SNF complexes (BAF and PBAF, respectively), are functionally distinct, and bind different sets of subunits, but exhibit one of the highest co-expression profiles among mSWI/SNF subunits in human normal tissue samples (Figure S6F, G). Finally, it is interesting to note that several diseases which are near-uniformly characterized by mSWI/SNF complex perturbations contain homozygous or heterozygous mutations in gene families related to the core BAF functional module (Figure S6A).
Discovery of novel subunits and protein complexes from a combined physical-functional network approach
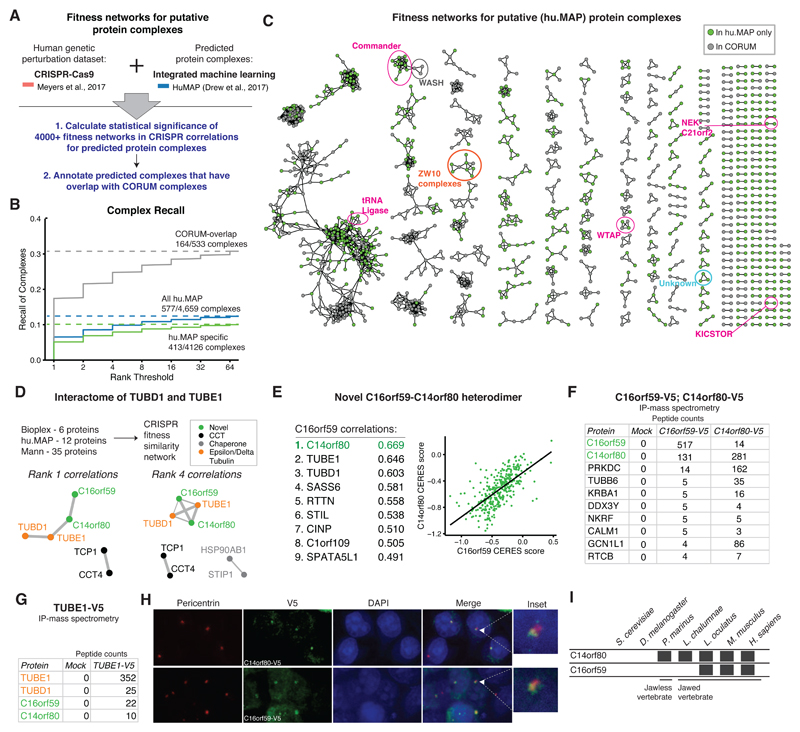
Given the degree to which functional similarity networks recapitulate CORUM protein complex features, we conducted a second analysis on a set of predicted, non-validated protein complexes to nominate targets for further discovery. Based on the highly enriched overlap between the hu.MAP predicted PPI network and the CRISPR functional similarity network at top ranks (Figure S1E), as well as the correlations in more lowly expressed, evolutionarily recent complexes (Figure 2E) captured in the CRISPR dataset, we used this dataset to identify significant fitness correlations among 4000+ predicted hu.MAP protein complexes (Figure 5A). Our methodology identified 577 complexes recapitulated in the functional similarity network representing a recall of 12.4% at an FDR of 0.05 (Figure 5B). Of the 533 hu.MAP protein complexes showing significant overlap with the CORUM dataset (defined as 80% or more of the subunits), 164 protein complexes (30.7%) were recalled in the functional similarity network, compared to 413 out of 4,126 protein complexes (10%) without CORUM overlap.
Figure 5. A combined physical-functional interaction map highlights validated and novel interactions.
(A) Strategy for the generation of fitness similarity networks for putative protein complexes. The statistical framework for identifying significant protein complex fitness correlation networks (Figure 2A) was applied to the hu.MAP complex dataset. Hu.MAP exhibits high level of complex enrichment within the CRISPR-Cas9 correlation dataset (Figure S1E).
(B) Fraction of hu.MAP protein complexes (interactions) recalled in the CRISPR fitness correlation datasets. Of the 4,659 predicted complexes, 577 exhibit significant fitness networks.
(C) Statistically significant fitness correlation networks for hu.MAP complexes. Recently discovered protein complexes consisting of genes of unknown function are highlighted in magenta, and complexes with novel components that were selected for validation are labeled in orange and blue. Proteins found in the Core CORUM set are marked in gray, while proteins unique to the hu.MAP complex list are marked in green.
(D) In order to discover novel elements of the epsilon- and delta-tubulin interactome, 53 putative TUBE1 and TUBD1 interactors from three different large-scale protein-protein interaction networks were assembled and used to generate a fitness similarity network from the CRISPR-Cas9 dataset. Out of all 53 putative interactors, only two proteins, C16orf59 and C14orf80, exhibited top ranked correlations with TUBE1 and TUBD1.
(E) Proteins exhibiting top ranked fitness correlations with C16orf59 are predominantly centrosomal. The top ranked correlation to C16orf59 is with another gene of unknown function, C14orf80. A scatterplot showing the correlation between CRISPR-Cas9 CERES scores of the C16orf59 and C14orf80 proteins across 300+ cell lines is shown.
(F) IP/mass-spectrometry results for V5-tagged C16orf59 and C14orf80 immunoprecipitations. Total peptide counts are indicated, ranked by overall abundance in the C16orf59 purification.
(G) IP/mass spectrometry of transiently transfected epsilon-tubulin (TUBE1) co-precipitates TUBD1 as well as the C16orf59-C14orf80 heterodimer.
(H) Immunofluorescence performed for pericentrin (centrosomal marker) and V5 (C14orf80 and C16orf59), with DAPI nuclear stain. Both proteins exhibit centrosomal localization. Panel magnification= 60X.
(I) Evolutionary history of the C14orf80 and C16orf59 genes. Both are evolutionarily recent, with C16orf80 present only after the jawless-jawed vertebrate transition, while C14orf80 is present from jawless vertebrates forward.
This set of 577 recalled complexes in the functional similarity network (Figure 5C, Table S6) is comprised of 7,323 total proteins, 5,668 of which do not appear in the Core CORUM dataset. Notably, we found that many of these fitness networks correspond to hu.MAP complexes that have been recently validated as novel protein complexes (i.e. C12orf66- SZT2 heteromer of the KICKSTOR complex (Wolfson et al., 2017), and C16orf62-COMMD (Phillips-Krawczak et al., 2015) (Figure S7A-B). The recently discovered Commander complex (Wan et al., 2015) correlates with the WASHC4 and WASHC5 components of its known interacting complex, the WASH complex, and the KICKSTOR components SZT2 and C12orf66 correlate strongly with its mTOR pathway interactor, GATOR1(Peng et al., 2017; Wolfson et al., 2017). The gene co-expression networks for the same complexes did not reveal these interactions (Figure S7C-F).
In order to identify novel interactions among these functional similarity networks for experimental validation, we scored each protein complex by taking the average product between the CRISPR correlation value and the hu.MAP probability weight over all gene pairs in the protein complex. We then ranked significant protein complexes by this score and identified complexes or subunits with no known literature annotation (Figure S8A, Table S6). Based on this strategy, we selected two unknown genes each with multiple interaction predictions for validation studies: C16orf59 (interaction pairs: TUBD1, TUBE1), and C19orf25 (interaction pairs: NRZ complex members).
C16orf59 and C14orf80 form a heterodimeric complex that binds delta and epsilon tubulins
Tubulins are major elements of the eukaryotic cytoskeleton and are critical for cellular processes such as cell division and motility (Turk et al., 2015). Of the many characterized isoforms within the tubulin protein superfamily, the delta and epsilon tubulin variants remain incompletely understood. Recent work has suggested that these isoforms exist as an evolutionarily conserved module (Turk et al., 2015) and may be involved in forming triplet microtubules that are critical for centriole assembly (Wang et al., 2017a).
We assembled a combined TUBD1 and TUBE1 interactome consisting of 53 purported interactors from three different mass spectrometry datasets (Figure 5D, S8B). The CRISPR functional similarity network for these genes identified only two of the 53 purported interactors, and both were genes of no annotated function: C16orf59 as annotated above, and additionally, C14orf80 (Figure 5E). The fitness profile of C16orf59 also correlated strongly with SASS6, a core centrosome component that is necessary for centrosomal duplication (Leidel et al., 2005) and the basal body component RTTN, suggesting that the function of C16orf59 is centrosome-related (Figure 5E). However, since the top fitness correlation of C16orf59 was with C14orf80, we hypothesized that these two unknown proteins may directly interact. To test this, we lentivirally introduced V5-tagged versions of C16orf59 and C14orf80 into HEK293T cells and performed immunoprecipitation with subsequent mass spectrometry-based proteomics. C16orf59 and C14orf80 reciprocally immunoprecipitated one another, suggesting that they form a heterodimeric protein complex (Figure 5F, S8C, Table S7). This interaction was also reported in the recently-published Bioplex 2.0 network (Marsh et al., 2013). Finally, both proteins precipitated with TUBE1 in a TUBE1-V5 immunoprecipitation (Figure 5G, S8C, Table S8), further supporting the evidence that the C16orf59 and C14orf80 heterodimer is a centrosomal and an interactor with delta and epsilon tubulin. To determine the subcellular localization of the C16orf59 and C14orf80 proteins, we performed immunofluorescence experiments in HEK293T cells containing V5-tagged C16orf59 and C14orf80 constructs. Consistent with the strong fitness correlation between C16orf59 and centrosome components, both C16orf59 and C14orf80 were found in the centrosomal components of nuclei marked by pericentrin as a centrosomal control (Figure 5H). Given the relatively recent evolutionary history of these two proteins— C14orf80 arose during jawless vertebrates, while C16orf59 arose in jawed vertebrates (Figure 5I) – our data suggest that the two uncharacterized proteins C16orf59 and C14orf80 form a vertebrate-specific centrosomal protein complex.
C19orf25 selectively binds the cytoplasmic module of the ZW10 protein complex family
The mammalian NRZ complex is composed of NBAS, ZW10, and RINT1 and is descended from its yeast predecessor, Dsl1. Both NRZ and Dsl1 complexes are involved in transport between the ER and the Golgi in their respective organisms (Tagaya et al., 2014). The ZW10 protein evolutionarily diverged to assemble into a second, nuclear-localized complex, RZZ, which facilitates dynein recruitment to the kinetochore (Vleugel et al., 2012). The RZZ complex is composed of four subunits: Rod (KNTC1 in humans), ZWILCH, and ZWINT and the shared ZW10 subunit (Figure S9A). Previous efforts to purify ZW10 precipitated a factor, C19orf25, that did not appear to have kinetochore localization, despite binding ZW10 (Kops et al., 2005). We hypothesized that the C19orf25 functional similarity network would discern which form of the ZW10-nucleated complexes it associates with. The top fitness correlations of C19orf25 all are members of either NRZ or the STX18 SNARE complex which transiently docks NRZ on the ER membrane (STX18 and BNIP1) (Figure S9B-C). The ‘moonlighting’ ZW10 protein is strongly correlated with both protein complex configurations, while C19orf25 forms specific correlations with the NRZ and STX18 complexes (Figure S9D-E). Consistent with the prediction that C19orf25 is functionally associated with NRZ and not RZZ, immunofluorescence experiments with V5-tagged C19orf25 confirmed cytoplasm-specific localization (Figure S9F). Finally, we were able to reproduce the previously observed interaction between C19orf25 and RINT1/ZW10 using IP/mass-spectrometry of V5-C19orf25 from cytoplasmic extract (Figure S9G, S8C, Table S8). Given that C19orf25 is an evolutionarily recent protein complex subunit (Figure S9H), present only in bony vertebrates, our data suggest that C19orf25 is an evolutionarily recent addition to the cytoplasmic NRZ complex that arose concurrently with members of the RZZ complex.
Discussion
The ability to functionalize individual subunits and modules of protein complexes remains a major challenge, especially for those complexes with incompletely resolved protein subunit membership and structural information. Here, we demonstrate that protein complex componentry as well as differential function between subunits can be elucidated using large-scale genetic perturbation screens across diverse cellular contexts— in this case, hundreds of cancer cell lines. This study, to the best of our knowledge, represents the largest and most comprehensive analysis of protein complexes using fitness screening in human cells to date. These studies provide a conceptual framework for further study of functional relationships between proteins in both normal and disease-associated states (associated with genetic mutations, gene variants, or gene expression changes) as increasing fitness datasets continue to emerge.
Future work to merge disease genetics with physical and functional interactions may help reveal the molecular basis of certain human diseases. Indeed, examining disease-associated alleles coding for interacting proteins may define convergent pathways and novel targets for therapeutic intervention. For instance, the putative hu.MAP interacting subunits C15orf41 and CDAN1 display strongly correlated CRISPR-Cas9 fitness profiles (Table S6), and the genes encoding these two proteins harbor mutually exclusive mutations in the majority of congenital dyserythropoietic anemias(Aguirre et al., 2016). Similarly, our findings with respect to the core BAF functional module (SMARCA4, SMARCB1, SMARCE1, ARID1A) are particularly timely and relevant to disease biology. Intriguingly, all rare cancer types known to be driven by mSWI/SNF complex perturbation (defined as ≥70% of tumors with protein-level loss of a single mSWI/SNF subunit) exhibit mutually exclusive and complete loss of one of the genes or paralog families in the core functional module of BAF complexes identified in our analysis (Figure S6A). The small percentage of these human tumor types not explained by their prevailing characteristic perturbation instead exhibit loss of one of the other members of the core BAF functional module we identified (Hasselblatt et al., 2011; Schneppenheim et al., 2010; van den Munckhof et al., 2012). Finally, both Coffin-Siris and Nicolaides-Baraitser intellectual disability syndromes are driven by germline heterozygous mutations of core BAF module genes in a mutually exclusive manner (Figure S6A). Understanding the convergent functional contributions of mSWI/SNF subunits mutated in the specific cancers and intellectual disability syndromes highlighted above has remained a major recent challenge in the field. Our findings suggest a synergistic function between these four mSWI/SNF subunits, informing future studies to address the structural basis underlying these convergent functional correlations and disease-associated mutational patterns.
Several challenges still remain with respect to expanding the degree of protein complex capture from these or similar datasets, particularly for those complexes that do not demonstrate variable essentiality for cellular fitness. Additional approaches and screening readouts, such as cellular morphology (Rohban et al., 2017), may be able to better classify complex subunits that predominantly have morphological effects rather than fitness effects. This is particularly relevant for the utility of these datasets in the context of emerging genes-to-variants studies, and for the functional characterization of other human disease-linked genetic mutations. In addition, as similar genome-scale genetic perturbation screens are performed across increasingly larger and diverse sets of cell lines (and normal cell types), commensurate bioinformatic approaches will be required to address normalization methods and to enable further integration with machine learning-based PPI classifiers and other ensemble approaches. We provide all fitness correlations from both RNAi and CRISPR-Cas9 datasets as well as the statistically significant fitness networks for both CORUM and hu.MAP complexes as resources for the larger research community.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Cigall Kadoch, Ph.D. (cigall_kadoch@dfci.harvard.edu).
Experimental model and subject details
293T (ATCC #CRL-3216) and A549 (ATCC #CCL-185, male) cell lines were passaged and grown in DMEM media, supplemented with Glutamax and pen/strep and 10% FBS. CCRF-CEM (ATCC #CCL-119, female) cells were grown in suspension in RPMI 1640 supplemented with 10% FBS, glutamine, and pen/strep.
Method details
Lentiviral packaging and Infection
pMD2.G and psPAX2 lentiviral packaging vectors was co-transfected with pLX317 vector containing the clone of interest into HEK-293T cells, using PEI as a transfection reagent. Cells were incubated for 72 hours, and the media was filtered with a 0.4 uM filter before being either concentrated with an ultracentrifuge (20,000 RPM for 2.5 hours) or added directly to cells plated at 70% confluence with 1:1000 Polybrene.
Nuclear extract isolation, immunoprecipitation, and mass-spectrometry of BAF complex subunits
Harvested cells were incubated in Buffer A (25 mM HEPES pH 7.6, 5 mM MgCl2, 25 mM KCl, 0.05 mM EDTA, 10% glycerol and 0.1% NP40 with protease inhibitor (Roche), 1 mM DTT and 1 mM phenylmethylsulfonyl fluoride (PMSF)) for 10 minutes and the pellets were resuspended in 600 μl of Buffer C (10 mM HEPES pH 7.6, 3 mM MgCl2, 100 mM KCl, 0.5 mM EDTA and 10% glycerol with protease inhibitor, 1 mM DTT and 1 mM PMSF) with 67 μl of 3 M (NH4)2SO4 for 20 minutes. The lysates were spun down using a tabletop ultracentrifuge at 100,000 rpm at 4°C for 10 minutes. Nuclear extracts were precipitated with 200 mg of (NH4)2SO4 on ice for 20 minutes and finally purified as pellets by ultracentrifugation at 100,000 rpm at 4°C for 10 minutes. The pellets were resuspended in IP Buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mM EDTA and 1% Triton-X100 with protease inhibitor, 1 mM DTT and 1 mM PMSF) for the subsequent experiments. Immunoprecipitation was performed with antibodies targeting BRD7 (Bethyl, A302-304A), BRD9 (Abcam, ab137245), ARID1A (Bethyl, A301-041), SMARCA4 (Abcam, [EPNCIR111A], ab110641) and SMARCA2 (Bethyl, A301-015A).
For proteomic analysis, antibodies were crosslinked with dimethyl pimelimidate (DMP) to Gammabind G Sepharose beads (GE) prior to immunoprecipitation from nuclear extract. Captured protein was eluted using 6M urea/600mM NaCl and digested with trypsin. Mass spectrometry was performed (using a Thermo Exactive Plus Orbitrap) by the Taplin Mass Spec Facility (Harvard Medical School). To analyze the results, peptides that were present in control precipitations (mock) were removed from the bait peptide list, and each protein was then ranked according to the number of total peptides captured.
Density Sedimentation (10-30% glycerol gradients)
Nuclear extract (800 μg, quantified by Bradford assay) was resuspended in 200 ul of 0% glycerol HEMG buffer (supplemented with protease inhibitors and DTT) and overlaid onto a 11 ml 10%–30% glycerol (in HEMG buffer) gradient prepared in a 14 x 89 mm polyallomer centrifuge tube (Beckman Coulter). Tubes were centrifuged in an SW40 rotor at 4°C for 16 hr at 40,000 rpm. Fractions (0.550 ml) were collected and used in immunoblot analyses. For BRD9 and PHF10 blots, fractions were first concentrated using 10 uL of Strataclean resin (Agilent) before loading.
Immunoblot
Protein was loaded onto Bis-Tris 4-12% gradient Novex gels and run for 150V for 90 minutes. A wet transfer was performed for 2.5 hours at 165 mA at 4 degrees Celsius onto PVDF membranes. After transfer, membranes were blocked in 5% milk for 1 hour at room temperature before applying primary antibody (see below) and fluorescently labeled secondary antibodies for visualization using the LI-COR Odyssey.
Antibodies used
| Subunit | Company | Catalog # |
|---|---|---|
| SMARCA4 | Santa Cruz | sc-17796 |
| ARID1A | Bethyl | A301-041 |
| SMARCE1 | Bethyl | A300-810A |
| SMARCB1 | Santa Cruz | sc-166165 |
| SS18 | Cell Signaling | #21792 |
| DPF2 | Abcam | ab134942 |
| SMARCD2 | Santa Cruz | sc-101162 |
| SMARCC1 | Santa Cruz | sc-10756 |
| PBRM1 | Bethyl | A301-591A |
| ARID2 | Santa Cruz | sc-166117 |
| BRD7 | Santa Cruz | sc-376180 |
| SMARCD1 | Santa Cruz | sc-135843 |
| BRD9 | Abcam | ab66443 |
| PHF10 | Invitrogen | PA5-30678 |
Large-scale purification of factors for mass spectrometry (C14orf80, C16orf59, C19orf25, TUBE1)
After lentiviral introduction and selection, 293T cells with V5 tagged bait proteins were expanded to 15-20 confluent 15 cm dishes, or about 1E9 cells per preparation. Cells were scraped off the dishes and pelleted at 3000 rpm for 5 minutes at 4 degrees C. Cells were then resuspended in complete hypotonic buffer (10mM Tris pH 7.5, 10 mM KCL, 1.5 mM MgCl2, 1M DTT, 100 mM PMSF, 1000x protease inhibitor cocktail) and lysed for 5 minutes on ice. Lysate was then pelleted at 3000 rpm for 5 minutes at 4 degrees C and the upper cytoplasmic layer was collected, to which 3M KCl was added to a final concentration of 150 mM KCl, and rotated at 4 degrees C for 1 hour. After rotation, the cytoplasmic extract was spun at 20,000 RPM for 1 hour at 4°C in SW28 tubes in an ultracentrifuge. The lipid phase was extracted with a P1000 tip and the remainder filtered through a 0.45 uM filter (Steriflip). This was then incubated with V5 beads and rotated overnight at 4 degrees C. Beads were then pelleted and washed 6 times with 12 mL of high salt buffer. After the last wash, beads were transferred into a 500 uL tube. For V5 elution, immunoprecipitations with V5 antibodies were eluted off of beads with 2.5 M glycine and quenched for pH normalization with Tris buffer.
Immunoflourescence (IF)
Cells with overexpression of a tagged bait were split to 30-60% confluency onto a 24-well plate with appropriately sized and sterilized coverslips. Cells were allowed to adhere overnight. In the case of C14orf80 and C16orf59 experiments, cells were treated with nocodazole overnight to promote cell cycle arrest. When cells were at the appropriate density and treatment time, cells were washed once with PBS in the plate and covered with either -20 degree Celsius methanol for 5 minutes (for C14orf80 and C16orf59 overexpression) or with 4% paraformaldehyde for 20 minutes (for C19orf25 overexpression), and then washed twice with IF wash buffer (0.1% NP40, 1 mM sodium azide, PBS 1X) and blocked overnight in blocking buffer (IF wash buffer + 10% FBS, filtered through 0.2um filter). After blocking was complete, antibodies were diluted into blocking buffer and this solution was placed on the coverslips for 3 hours at room temperature. The dilutions were performed as follows:
| Antibody | Manuf. | Cat. | Species | Dilution |
|---|---|---|---|---|
| Pericentrin | Abcam | ab4448 | Rabbit | 1:4000 |
| V5 | Thermo | R960-25 | Mouse | 1:1500 |
| V5 | Cell Signalling | #13202 | Rabbit | 1:3000 |
| Anti-KDEL | Abcam | ab50601 | Rat | 1:300 |
This was followed by 3 washes with IF buffer (rinse, 5 minute incubation, repeat). Secondary antibody was diluted in blocking buffer 1:1000 and the slides were incubated for 1 hour. This was followed by a second round of 3 IF wash buffer washes (rinse, 5 minute incubation, repeat). Slides were then removed from the 24 well plate and mounted onto coverslips with mounting media containing DAPI stain.
| Antibody | Manuf. | Cat. | Dilution |
|---|---|---|---|
| Goat anti-Mouse IgG Alexa Fluor Plus 555 |
Thermo | A32727 | 1:1000 |
| Goat anti-Rat IgG Alexa Fluor 647 |
Thermo | A-21247 | 1:1000 |
| Goat anti-Rabbit IgG Alexa Fluor 546 |
Cell Signaling | A-11010 | 1:1000 |
Imaging and Analysis
Images were captured at 60x magnification on a spinning disc confocal microscope. Images were taken as an 11 layer z-stack, which was then z-projected using the maximal value per pixel across stacks. ImageJ software was used for image processing and figure generation.
Quantification and Statistical Analysis
Using fitness thresholds to define genes for analysis
Fitness screening data from RNAi screens in 501 cell lines (Tsherniak et al., 2017) and from CRISPR screens in 342 cell lines (Meyers et al., 2017) were downloaded from the Project Achilles Data Portal (https://portals.broadinstitute.org/achilles). Copy-number corrected versions of two additional CRISPR-Cas9 screening datasets, Wang et al. (Wang et al., 2017b) and GeCKO (Aguirre et al., 2016), were also used as comparisons (Meyers et al., 2017). Genes for downstream analysis were filtered for the presence of fitness effects upon genetic depletion across cancer cell lines: for both of the main datasets used in the paper (Project Achilles RNAi and CRISPR), genes were only included if the cell line most dependent on that gene exceed a cutoff (-2 for the RNAi dataset, -0.3 for CRISPR dataset) and expresses that gene above a threshold of 0.5 RPKM. This resulted in a final set of 6300 genes in RNAi and 8997 genes in CRISPR. For parallel analyses involving gene expression (COXPRESdb Hsa3.c1-0, http://coxpresdb.jp/download.shtml), a union of the RNAi and CRISPR fitness genes was used.
Fitness profile plots
For each of the selected genes in Figure 1B and 3B, their CRISPR-Cas9 fitness profiles were scaled between 0 and 1, where 0 represents the minimum essentiality and 1 represents the maximum essentiality of that gene across cell lines. A soft threshold was applied (scores were taken to the 3rd power), and both genes and cell lines were hierarchically clustered using complete linkage and correlation as a distance measure. Profiles were plotted using the ggjoy R package.
Fitness variation
For each gene in the RNAi and CRISPR datasets, the variance of that gene’s fitness scores was calculated across all cell lines in which the gene was screened. For Figure S1B, genes were then binned by their membership or absence in the CORUM dataset (RNAi: No interaction, n = 4850. Interaction: 1455. CRISPR: No interaction, n = 7299. Interaction, n = 1698). Significance was assessed using the Wilcox rank sum test.
Enrichment of correlated gene pairs among protein-protein interactions
Pearson correlations between the fitness profiles of all gene pairs were calculated in both RNAi and CRISPR datasets, using pairwise complete observations only (i.e. only in cell lines where both genes were screened). For Figure 1C, genes present in the Core CORUM dataset of literature curated protein complexes were paired and broken into two bins: gene pairs from the same complex, and gene pairs from different complexes. The resulting distributions were compared using a two-sample KS test. In Figure 1D, for each protein-protein interaction dataset, each possible gene pair with was binned into one of two groups: interacting protein products or non-interacting protein products, and boxplots were then shown for these two groups across each of the available protein-protein interaction datasets. For Figure 1E, enrichment of protein-interactions among correlated gene pairs of varying strengths was calculated in the same was as previously done for pathways (Hart et al., 2017a). Correlated protein pairs were binned into 1000 bins based on their ranked correlation. The cumulative log likelihood of protein pairs to interact was calculated for all bins for all protein-protein interaction datasets. Log likelihoods for the first 100,000 ranked correlations are shown.
Similarity network significance for protein complexes
For each correlation dataset (RNAi, CRISPR, COEXPRESdb, GeCKO and Wang et al. and Costanzo et al. http://thecellmap.org/costanzo2016/) we performed a row-wise rank-transformation on the gene-gene correlation matrix, and symmetrized by taking the maximum rank between gene pairs.
For a given collection of genesets (Core CORUM, hu.MAP complexes, yeast complexes) and a similarity network at a given rank threshold, the significance of the observed internal-to-external edge density of each of the genesets in the collection is determined by calculating an empirical p-value using degree-preserved randomized networks. For the observed network and each of 10,000 randomized networks with preserved degree sequence, we calculated the ratio of the internal edge density to the external edge density (Figure S2) of each of the subgraphs of the genesets under consideration. We determined the empirical p-values representing the significance of each geneset as the number of times this ratio in the null models exceeds that of the ratio in the observed network. We then apply a false discovery rate (FDR) correction to the p-values per correlation dataset. We used a FDR cutoff of 0.05 for significance.
To generate protein complex dataset recall plots, the cumulative percentage of protein complexes with significant fitness networks was plotted as a function of the rank threshold. We also plot the fraction of complexes recalled as a function of FDR for each correlation dataset.
To further characterize protein complexes that scored as significant in RNAi only (n = 31), CRISPR only (n = 264), or both (n = 200) in Figure 2E, we assembled a set of gene features. Core essential genes were defined as the union of the CEG1 (Hart et al., 2015) and CEG2 (Hart et al., 2017b) datasets. Gene expression data was taken from the CCLE RNA-seq data for these cell lines (Barretina et al., 2012), and human-mouse dN/dS was obtained from BioMart (Smedley et al., 2015). Differences between the three groups of protein complexes were assessed with the Wilcoxon rank sum test.
For protein complexes that showed significant correlations in both RNAi and CRISPR datasets, we calculated the edge density ratio for that complex over all ranks in both datasets. Each protein complex was then assigned to either RNAi or CRISPR datasets as shown in Figure S2D, depending on which showed the largest edge density ratio for that complex over all rank thresholds.
Network visualizations
Global fitness network plots (Figures S3E and 5C)
Functional similarity networks were plotted in Cytoscape. In order to systematically choose rank thresholds for network visualizations for protein complex genesets in Figures S3E and 5C, we took the difference between the cumulative sum of internal edges (weighted by the inverse rank) and the cumulative sum of weighted external edges at each possible rank threshold, and chose the threshold that maximizes this difference.
Individual complex similarity plots
For individual complexes, network visualizations were generated using edges weighted by the inverse rank of the correlation. Functional similarity networks shown for individual complexes were shown with the top ranked correlation thresholds. For similarity networks highlighting shared subunits, we chose more lenient rank thresholds within the top 10% of ranked correlations. For coexpression networks, we chose a uniform rank 50 threshold, as many of the coexpresion networks did not have internal edges to be shown at more stringent rank thresholds.
Networks were plotted with ggraph and ggiraph using the Fruchterman and Reingold force-directed layout algorithm as implemented in igraph R package. Node colors were chosen based on literature curated annotations (protein complex composition, functional modules, assembly components).
Interface size dataset
Using the entire set of heteromeric protein complex structures in the PDB as of 2017-03-13, we identified polypeptide chains with >90% sequence identity to a human protein-coding gene. The sizes of all interfaces formed between pairs of subunits were calculated between all pairs of subunits using AREAIMOL as implemented in the CCP4 suite (Winn et al., 2011). For each pair of human protein-coding genes, the largest physical interface identified in the PDB was used for our analyses, and interactions with buried surface areas < 200 Å^2 were filtered from the dataset.
Structural heterodimers were defined to be protein pairs with at least 40% of each subunit’s total surface area involved in the buried surface interface between subunits. A KS test was performed on the scaled rank correlations for each of the 271 structural heterodimers across the three datasets compared (RNAi, CRISPR, COXPRESdb).
Modularity overlap enrichment
Protein complexes that had significant fitness networks in either RNAi or CRISPR and at least 10 total subunits were considered for modularity analysis. For each of these 118 protein complexes, we generated networks using the following structural edgelists (Bioplex 2.0, PDB, and huMAP), as well as genetic edgelists (RNAi or CRISPR, CoxpresDB) with optimal thresholds chosen as stated above.
For each protein complex, we performed Louvain community detection as implemented in the igraph R package across networks derived from these datasets. We then assessed overlap significance between module assignments using two-sided Fisher's exact tests. For each of the three datasets shown (Coexpression, Fitness, and huMAP), networks were compared to available PDB or Bioplex physical networks. FDR correction was applied across p-values in each of the three groupings.
Heatmaps and clustering
Hierarchical clustering was performed on fitness profile correlation matrices using the Euclidian distance metric and complete linkage clustering. Heatmaps were visualized using the heatmap.2 function in the gplots R package.
Scoring significant hu.MAP complexes
Predicted protein complexes from hu.MAP were obtained from their website (http://proteincomplexes.org/download). The hu.MAP interaction network is weighted by a predicted pairwise probability of interaction between each protein, ranging from 0.75 (the significance cutoff) to 1 (maximum probability). In order to score protein complexes by a combination of physical interaction evidence and correlation strength, we took the product of the average CRISPR correlations and average predicted interaction probability for all gene pairs in that complex, and ranked significant complexes by that score. We then selected protein complexes for validation that included one or more subunits of uncharacterized function. The top 300 protein complexes by this scoring paradigm were included as a supplement (Table S7).
Gene evolution tables
The following ENSMBL gene trees were used as references for Figures 5I and S9H:
C14orf80: ENSGT00390000011474
C16orf80: ENSGT00390000011149
RINT1: ENSGT00390000017006
NBAS: ENSGT00390000012474
ZW10: ENSGT00390000016427
C19orf25: ENSGT00390000007991
ZWILCH: ENSGT00390000013696
ZWINT: ENSGT00390000017639
KNTC1: ENSGT00390000007883
Supplementary Material
Acknowledgements
We thank members of the Kadoch Lab and the Genetics Perturbation Platform (GPP) at the Broad Institute for helpful discussions. This work was supported in part by the National Science Foundation Graduate Research Fellowship (2015185722), the NSF Training Grant in Genetics and Genomics (T32GM096911), and the Quantitative Cell Biology Network (NSF MCB-1411898) to J.P. C.K. is supported by the NIH DP2 Director’s New Innovator Award (1DP2CA195762-01), the American Cancer Society Research Scholar Award RSG-14-051-01-DMC, and the Pew- Stewart Scholars in Cancer Research Grant. J.M. is supported by a Medical Research Council Career Development Award (MR/M02122X/1). The GPP at the Broad Institute is supported by the Carlos Slim Foundation in Mexico through the Slim Initiative for Genomic Medicine and the NCI grant U01 CA176058 to W.C.H. We thank R. St. Pierre, B. Tye, M. Sonnett, R. Gopalakrishnan, Z. McKenzie, J. Otto, F. Winston, R.E. Kingston, and M. Meyerson for helpful discussions.
Footnotes
Author Contributions
J.P., R.M.M., A.T., and C.K. conceived of and designed the study. J.P., R.M.M., A.E.S, and A.T. performed all analyses. J.P., B.C.M., and N.M. performed all experimental validation studies. S.H.C. contributed novel insights and data. F.V., B.A.W., W.C.H. and A.T. performed and directed RNAi- and CRISPR-Cas9-based screening efforts as part of Project Achilles (Broad Institute), J.N.W. and J.A.M. provided structural data and performed structural analyses of protein complexes. J.P., R.M.M., and C.K. wrote the manuscript.
Data and Software Availability
All code used to generate figures in the manuscript are available through an online repository: https://github.com/robinmeyers/pan-meyers-et-al. All data from this manuscript are available at: https://figshare.com/s/87b9ba98da066d524ae7.
References
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discovery. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahnert SE, Marsh JA, Hernandez H, Robinson CV, Teichmann SA. Principles of assembly reveal a periodic table of protein complexes. Science. 2015;350:aaa2245. doi: 10.1126/science.aaa2245. [DOI] [PubMed] [Google Scholar]
- Baliga NS, Björkegren J, Boeke JD, Boutros M, Crawford N, Dudley AM, Farber CR, Jones A, Levey AI, Lusis AJ, et al. The State of Systems Genetics in 2017. Cell Systems. 2017;4:7–15. doi: 10.1016/j.cels.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Myers CL, Andrews B, Boone C. Genetic interaction networks: toward an understanding of heritability. Annual review of genomics and human genetics. 2013;14:111–133. doi: 10.1146/annurev-genom-082509-141730. [DOI] [PubMed] [Google Scholar]
- Bertomeu T, Coulombe-Huntington J, Chatr-Aryamontri A, Bourdages KG, Coyaud E, Raught B, Xia Y, Tyers M. A High-Resolution Genome-Wide CRISPR/Cas9 Viability Screen Reveals Structural Features and Contextual Diversity of the Human Cell-Essential Proteome. Molecular and cellular biology. 2018;38 doi: 10.1128/MCB.00302-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen VA, Májek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, Sacco R, van Diemen FR, Olk N, Stukalov A, et al. Gene essentiality and synthetic lethality in haploid human cells. Science (New York, NY) 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- Boettcher M, Tian R, Blau JA, Markegard E, Wagner RT, Wu D, Mo X, Biton A, Zaitlen N, Fu H, et al. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nature Biotechnology. 2018;36:170. doi: 10.1038/nbt.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R, Drouin G. The Increase in the Number of Subunits in Eukaryotic RNA Polymerase III Relative to RNA Polymerase II Is due to the Permanent Recruitment of General Transcription Factors. Molecular Biology and Evolution. 2009;27:1035–1043. doi: 10.1093/molbev/msp316. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science (New York, NY) 2016;353 doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, East-Seletsky A, Ali LD, Gerath WF, Pantel SE, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Scientific data. 2014;1 doi: 10.1038/sdata.2014.35. 140035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife. 2016;5 doi: 10.7554/eLife.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nature reviews Genetics. 2013;14:249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- Doench JG. Am I ready for CRISPR? A user's guide to genetic screens. Nature reviews Genetics. 2018;19:67–80. doi: 10.1038/nrg.2017.97. [DOI] [PubMed] [Google Scholar]
- Drew K, Lee C, Huizar RL, Tu F, Borgeson B, McWhite CD, Ma Y, Wallingford JB, Marcotte EM. Integration of over 9,000 mass spectrometry experiments builds a global map of human protein complexes. Molecular systems biology. 2017;13:932. doi: 10.15252/msb.20167490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D, Roguev A, Gordon DE, Chen M, Chen S-HH, Shales M, Shen JP, Ideker T, Mali P, Qi LS, et al. Genetic interaction mapping in mammalian cells using CRISPR interference. Nature methods. 2017;14:577–580. doi: 10.1038/nmeth.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A-C, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nature Reviews Molecular Cell Biology. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, Bassik MC. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nature biotechnology. 2017;35:463–474. doi: 10.1038/nbt.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown Kevin R, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Hart T, Koh C, Moffat J. Coessentiality And Cofunctionality: A Network Approach To Learning Genetic Vulnerabilities From Cancer Cell Line Fitness Screens. bioRxiv. 2017a [Google Scholar]
- Hart T, Tong AHYHY, Chan K, Van Leeuwen J, Seetharaman A, Aregger M, Chandrashekhar M, Hustedt N, Seth S, Noonan A, et al. Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens. G3 (Bethesda, Md) 2017b doi: 10.1534/g3.117.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselblatt M, Gesk S, Oyen F, Rossi S, Viscardi E, Giangaspero F, Giannini C, Judkins AR, Frühwald MC, Obser T, et al. Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. The American journal of surgical pathology. 2011;35:933–935. doi: 10.1097/PAS.0b013e3182196a39. [DOI] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann AF, Martin LJ, Minder JL, Roe J-SS, Shi J, Steurer S, Bader G, McConnell D, Pearson M, Gerstberger T, et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nature chemical biology. 2016;12:672–679. doi: 10.1038/nchembio.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome research. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Langelier M-FF, Bataille AR, Pascal JM, Pugh BF, Robert F. Tail and Kinase Modules Differently Regulate Core Mediator Recruitment and Function In Vivo. Molecular cell. 2016;64:455–466. doi: 10.1016/j.molcel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Science advances. 2015;1 doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Williams RT, Calarco JP, Miller EL, Weber CM, Braun SM, Pulice JL, Chory EJ, Crabtree GR. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nature genetics. 2017;49:213–222. doi: 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nature biotechnology. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G, Kim Y, Weaver BAA, Mao Y, McLeod I, Yates JR, Tagaya M, Cleveland DW. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gönczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nature Cell Biology. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Mann C, Buhler JM, Treich I, Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987;48:627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Hernández H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell. 2013;153:461–470. doi: 10.1016/j.cell.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald ER, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, Belur D, Castelletti D, Frias E, Gampa K, et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell. 2017;170:577. doi: 10.1016/j.cell.2017.07.005. 1535066112. [DOI] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. Computational correction of copy-number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nature genetics. 2017 doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Strand C, Donovan KF, Hegde M, Sanson KR, Vaimberg EW, Sullender ME, Hartenian E, Kalani Z, Fusi N, et al. Orthologous CRISPR-Cas9 enzymes for combinatorial genetic screens. Nature biotechnology. 2018;36:179–189. doi: 10.1038/nbt.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa K, Schneider TR, Cramer P. Core Mediator structure at 3.4 Å extends model of transcription initiation complex. Nature. 2017;545:248–251. doi: 10.1038/nature22328. [DOI] [PubMed] [Google Scholar]
- Okamura Y, Aoki Y, Obayashi T, Tadaka S, Ito S, Narise T, Kinoshita K. COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic acids research. 2015;43:6. doi: 10.1093/nar/gku1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Molecular cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Peng M, Yin N, Li MO. SZT2 dictates GATOR control of mTORC1 signalling. Nature. 2017;543:433–437. doi: 10.1038/nature21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB, Levy ED, Teichmann SA. The origins and evolution of functional modules: lessons from protein complexes. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2006;361:507–517. doi: 10.1098/rstb.2005.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko N, Jin Y, Wong KH, Struhl K. Mediator Undergoes a Compositional Change during Transcriptional Activation. Molecular cell. 2016;64:443–454. doi: 10.1016/j.molcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Krawczak CA, Singla A, Starokadomskyy P, Deng Z, Osborne DG, Li H, Dick CJ, Gomez TS, Koenecke M, Zhang J-SS, et al. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Molecular biology of the cell. 2015;26:91–103. doi: 10.1091/mbc.E14-06-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Li Z, Hart GT, Carlson MW, Boutz DR, Marcotte EM. A map of human protein interactions derived from co-expression of human mRNAs and their orthologs. Molecular systems biology. 2008;4:180. doi: 10.1038/msb.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohban MH, Singh S, Wu X, Berthet JB, Bray M-AA, Shrestha Y, Varelas X, Boehm JS, Carpenter AE. Systematic morphological profiling of human gene and allele function via Cell Painting. eLife. 2017;6 doi: 10.7554/eLife.24060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Research. 2010;38 doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneppenheim R, Frühwald MC, Gesk S, Hasselblatt M, Jeibmann A, Kordes U, Kreuz M, Leuschner I, Martin Subero JI, Obser T, et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. American journal of human genetics. 2010;86:279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nature methods. 2017;14:573–576. doi: 10.1038/nmeth.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh M, Baldock R, Barbiera G, et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Research. 2015;43 doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedale G, Timmers HT, Pijnappel WW. ATAC-king the complexity of SAGA during evolution. Genes & development. 2012;26:527–541. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagaya M, Arasaki K, Inoue H, Kimura H. Moonlighting functions of the NRZ (mammalian Dsl1) complex. Frontiers in cell and developmental biology. 2014;2:25. doi: 10.3389/fcell.2014.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Prysak MH, Woychik NA. Loss of the Rpb4/Rpb7 Subcomplex in a Mutant Form of the Rpb6 Subunit Shared by RNA Polymerases I, II, and III. Molecular and Cellular Biology. 2003;23:3329–3338. doi: 10.1128/MCB.23.9.3329-3338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, et al. A subcellular map of the human proteome. Science (New York, NY) 2017;356 doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- Tong A, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global Mapping of the Yeast Genetic Interaction Network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tsai K-LL, Yu X, Gopalan S, Chao T-CC, Zhang Y, Florens L, Washburn MP, Murakami K, Conaway RC, Conaway JW, et al. Mediator structure and rearrangements required for holoenzyme formation. Nature. 2017;544:196–201. doi: 10.1038/nature21393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564. doi: 10.1016/j.cell.2017.06.010. 1916796928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk E, Wills AA, Kwon T, Sedzinski J, Wallingford JB, Stearns T. Zeta-Tubulin Is a Member of a Conserved Tubulin Module and Is a Component of the Centriolar Basal Foot in Multiciliated Cells. Current biology : CB. 2015;25:2177–2183. doi: 10.1016/j.cub.2015.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Munckhof P, Christiaans I, Kenter SB, Baas F, Hulsebos TJ. Germline SMARCB1 mutation predisposes to multiple meningiomas and schwannomas with preferential location of cranial meningiomas at the falx cerebri. Neurogenetics. 2012;13:1–7. doi: 10.1007/s10048-011-0300-y. [DOI] [PubMed] [Google Scholar]
- Vleugel M, Hoogendoorn E, Snel B, Kops GJ. Evolution and function of the mitotic checkpoint. Developmental cell. 2012;23:239–250. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Bezginov A, et al. Panorama of ancient metazoan macromolecular complexes. Nature. 2015;525:339–344. doi: 10.1038/nature14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Kong D, Hoerner CR, Loncarek J, Stearns T. Centriole triplet microtubules are required for stable centriole formation and inheritance in human cells. bioRxiv. 2017a doi: 10.7554/eLife.29061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science (New York, NY) 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, Sabatini DM. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 2017b;168:890. doi: 10.1016/j.cell.2017.01.013. 942505984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nature Reviews Microbiology. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- Wild T, Cramer P. Biogenesis of multisubunit RNA polymerases. Trends in biochemical sciences. 2012;37:99–105. doi: 10.1016/j.tibs.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D: Biological Crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Wyant GA, Gu X, Orozco JM, Shen K, Condon KJ, Petri S, Kedir J, Scaria SM, et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017 doi: 10.1038/nature21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.