Abstract
Background:
Clostridium difficile infection (CDI) is the leading cause of antibiotic-associated diarrhea worldwide. As a result, the US Centers for Disease Control and Prevention have designated C. difficile as an urgent threat. Despite the global public health risk posed by CDI, little is known about its epidemiology on the African continent. This article describes the common occurrence of CDI from a cross-section of consecutively seen, randomly enrolled patients presenting with diarrhea at two major hospitals in Kenya.
Methods:
Patients presenting with diarrhea at two major hospitals in Kenya from May to July 2017 were enrolled. After signing the informed consent, stool samples, demographic data, medical history, prior antibiotic use, and HIV status were obtained from the patients. C. difficile was detected and validated by toxigenic culture and PCR.
Results:
The average age of the patients was 35.5 years (range 3–86 years); 59% were male and 41% were female. Out of 105 patient s tools tested, 98 (93.3%) were positive for C. difficile by culture. PCR analysis confirmed C. Difficile-specific genes, tcdA, tcdB, and tcdC, in the strains isolated from the stools. Further, 82.5% of the stools had C. difficile isolates bearing the frame-shift delection associated with hypervirulent strains. Remarkably, 91.9% of the stools that tested positive for C. difficile came from patients under 60 years old, with 64.3% being less than 40 years of age.The majorityof the patients (85%) reported over-the-counter antibiotic use in the last 30 days before the hospital visit.
Conclusions:
Together, the results revealed an unusually high incidence of C. difficile in the stools analyzed, especially among young adults who are thought to be less vulnerable. Comprehensive research is urgently needed to examine the epidemiology, risk factors, pathogenesis, comorbidities, clinical outcomes, antibiotic susceptibility, and genetic makeup of C. difficile strains circulating on the African continent.
Keywords: Clostridium difficile infections in Africa, C. difficile pathogenesis, C. difficile epidemiology, CDI in young adults, Antibiotics and CDI
Introduction
Clostridium (Clostridioides) difficile is a leading cause of antibiotic-associated diarrhea worldwide. Morbidity and mortality associated with C. difficile infections (CDI) have increased significantly over the last decade. The annual cost of treatment in the USA ranges between $1.9 and $7.0 billion (Zhang et al., 2016). The US Centers for Disease Control and Prevention has designated C. difficile as an urgent threat. The risk of CDI increases with broadspectrum antibiotic use (Owens et al., 2008). Other CDI-associated risk factors include old age, the use of gastric acid-suppressing drugs, comorbidities, immunodeficiency, and inflammatory bowel disease (Khanna and Pardi, 2012).
Toxigenic C. difficile strains produce and release toxins A and B, which cause disease (Geric et al., 2004; Kuehne et al., 2010; Lyerly et al., 1985; Rupnik et al., 2001; Voth and Ballard, 2005). During infection, the toxins are internalized by the host cells, leading to monoglucosylation of GTPases of the Rho family in the cytosol (Just et al., 1995). The infiltrated toxins cause the release of various immunomodulatory mediators, resulting in massive inflammation and the accumulation of neutrophils in the colon. The downstream effect of the toxins is mild to severe diarrhea and in more complicated cases, pseudomembranous colitis. Other CDI-associated symptoms include abdominal cramping, profuse diarrhea, and abdominal pain (Borriello, 1998).
Despite the public health risks posed by CDI, little is known about the epidemiology of this life-threatening pathogen in Africa. Such knowledge is critical, because most African countries are popular tourist destinations and may serve as direct routes for disseminating CDI. Given the practice of unregulated antibiotic use on the African continent, it was hypothesized that the incidence of CDI would be high among diarrhea patients. To investigate this hypothesis, the presence of C. difficile was determined in the stools of patients reporting diarrhea at two regional hospitals in Kenya, a popular tourist destination. The results revealed an alarming frequency of C. difficile in the patients with diarrhea, especially in young adults originally thought to be less vulnerable.
Materials and methods
Stool sample collection
This study was approved by the ethics review boards of Kenyatta National Hospital/University of Nairobi, Kisii Teaching and Referral Hospital, and the Kenyan Ministry of Health. The Kenyatta National Hospital is located in an urban area of Nairobi and serves patients from Nairobi and surrounding rural towns, as well as patients needing specialized care from other parts of the country. The Kisii Teaching and Referral Hospital is located in a rural area and serves people living in the surrounding rural and peri-urban areas. Patients who visit this hospital either are referred by doctors or are self-referred.
All patients that reported to these hospitals with diarrhea from May to July 2017 were sequentially enrolled in the study. To limit bias, no diarrhea patient was excluded from the study, except a few patients who refused to participate. After signing a consent form, the patients were administered questionnaires to capture data on demographics, previous antibiotic use, and medical history including HIV status (if known). Each patient was asked to collect a stool sample for the study. The samples were carefully aliquoted into sterile 1-ml tubes with screw caps using a sterile spatula and stored at −80 °C until shipped on cold packs to the University of Texas Health Science Center, Houston, Texas, USA for analysis.
Detection of C. difficile in stool by culture
The presence of C. difficile in the diarrheal stools was determined and validated using toxigenic culture (Darkoh et al., 2011a) and PCR. An anaerobic condition was maintained in a Bactron 600 anaerobic chamber (Sheldon Manufacturing, Cornelius, OR, USA) using 5% CO2, 10% H2, and 85% N2. A loopful of the frozen stools was carefully spread on the C. difficile-specific culture plates (Darkoh et al., 2011a) using sterile single-use loops; the plates were then incubated anaerobically at 37 °C for 48 h. This medium contains 250μg/ml D-cycloserine, 8μg/ ml cefoxitin, and 0.025% p-cresol, which specifically select for C. difficile and eliminate other non-C. difficile anaerobes on the plate. The numbers of stools that grew colonies were enumerated. To obtain C. difficile isolates for further analysis, all of the colonies on each plate were pooled and enriched by culturing for 24 h anaerobically at 37 C in brain heart infusion (BHI) broth containing 300 mg/ml D-cycloserine and 8μg/ml cefoxitin. Freezer stocks (1 ml) of each culture were made in 10% dimethylsulfoxide (DMSO) and stored at –80 C. Cell pellets from the remaining culture were stored at −20° for PCR analysis.
PCR analysis
For PCR analysis, DNA was isolated from each of the bacterial pellets using the Gene Reagent Pack on the Corbett Life Science X-tractor platform (Qiagen, Valencia, CA, USA). The concentration of the extracted DNA was determined using NanoDrop (ThermoScientific, Wilmington, DE, USA). PCR was performed using primers specific for toxins A and B (TcdA2), TcdC (TNC), and the 16S ribosomal RNA gene (16S rRNA) as control (Fiedoruk et al., 2015; Fry et al., 2012; Griffiths et al., 2010; Lemee et al., 2004; Li et al., 2017; Liu et al., 2018; Murray et al., 2009). The sequences of the primers used are: TcdA2 F-50AGATTCCTATATTTACATGACAATAT30, R- 5´GTATCAGGCATAAAGT AATATACTTT3´; TNC F- 5´GAGCACAAAGGGTATTGCTCTACTGGC3´, R- 5´CCAGACACAGCTAATCTTATTTGCACCT3´; 16S rRNA F- 5´ACACG GTCCAAACTCCTACG3´, R-5´AGGCGAGTTTCAGCCTACAA3´. The PCR amplification was done using OneTaq Quick-Load 2 Master Mix (New England Biolabs, Ipswich, MA, USA) with an initial denaturationtemperatureof94 Cfor 30 sand 36cyclesof94 Cfor30 s,55 C for 30 s, and 68 C for 30 s, with a final extension of 68 C for 5 min. The PCR products were analyzed using 1% agarose gel electrophoresis, stained with ethidium bromide, and imaged using BioDoc-It Imaging System (UVP, Upland, CA, USA).
Toxin assays
Toxins A and B in the culture supernatants of the isolates were detected using the Cdifftox activity assay for toxin activity (Darkoh et al., 2011b) and C. difficile TOX A/B II ELISA test (TechLab, Blacksburg, VA, USA) for toxin production. For the Cdifftox activity assay, the culture was centrifuged for 10 min at 10 000 xg and the supernatant (250 μl) was added to 30 μl of 0.2-μ filtered 30 mM p-nitrophenyl-β-D-glucopyranoside (Sigma-Aldrich, St. Louis, MO, USA) in a sterile 96-well plate. The sample was incubated aerobically at 37 C for 4–24 h and absorbance at 410 nm was measured using a SpectraMax I3 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA).
For the ELISA test, 200 μl of the supernatant was used and the manufacturer’s instructions were followed.
Statistical analysis
The data were analyzed using GraphPad Prism (GraphPad Software, San Diego, CA, USA). The non-parametric one-way Kruskal–Wallis test followed by Mann–Whitney U-test was used to determine differences in toxin production and toxin activity. Pearson’s Chi-square test wae used to determine the relationship between history of antibiotic use and CDI positivity. Statistical significance was defined as p ≤ 0.05.
Results
A total of 105 stools collected from non-biased, sequentially enrolled diarrhea patients were examined for the presence of C. difficile; 92 were collected from Kisii Teaching and Referral Hospital and 13 from Kenyatta National Hospital. Out of the 105 stools, 98 (93.3%) grew C. difficile colonies on the culture plates, whereas seven (6.7%) showed no growth (Table 1). To validate the culture results, PCR was performed to detect C. difficile-specific genes (tcdA, tcdB, and tcdC). Of the 98 samples analyzed by PCR, 97 (98.9%) were positive for tcdA, tcdB, or tcdC and only one (1.1%) was negative. The tcdA and tcdB genes were amplified in 95 (97.9%) of the 97 samples. Further, the tcdC gene was amplified in all of the 97 samples, of which 80 (82.5%) had isolates bearing the frame-shift deletion associated with hypervirulent strains (Carter et al., 2011; Curry et al., 2007; Hundsberger et al., 1997; Matamouros et al., 2007; Warny et al., 2005). C. difficile isolates from all of the 95 stools that had colonies bearing the tcdA and tcdB genes produced active toxins. However, neither colonies from the single stool sample that tested negative for the tcdA, tcdB, and tcdC genes nor the two samples that tested negative for the tcdA and tcdB genes produced toxins.
Table 1.
Detection and confirmation of Clostridium difficile in 105 diarrheal stool samples by culture and PCR.a
| Number of patients | Percentage (%) | |
|---|---|---|
| Total number of patients | 105 | |
| C. difficile detection by culture | ||
| Positive | 98 | 93.3 |
| Negative | 7 | 6.7 |
| PCR analysis of pooled C. difficile colonies | ||
| Presence of tcdA/tcdB or tcdC genes | ||
| Positive | 97 | 98.9 |
| Negative | 1 | 1.1 |
| tcdA+/tcdB+ | ||
| Positive | 95 | 97.9 |
| Negative | 2 | 2.1 |
| tcdA+/tcdB+ | ||
| Positive | 92 | 96.8 |
| Negative | 3 | 3.2 |
| tcdA+/tcdB− | ||
| Positive | 2 | 2.1 |
| Negative | 95 | 97.9 |
| tcdC+ | ||
| Positive | 97 | 100 |
| Negative | 0 | 0 |
| tcdC deletion | ||
| Wild-type | 17 | 17.5 |
| Deletion | 80 | 82.5 |
Stools were streaked on C. difficile plates containing 250 g/ml D-cycloserine and 8 μg/ml cefoxitin and incubated anaerobically at 37° C for 48 h. To validate the culture results, PCR was performed using primers specific for C. difficile genes (tcdA, tcdB, or tcdC) on the colonies isolated from the plates.
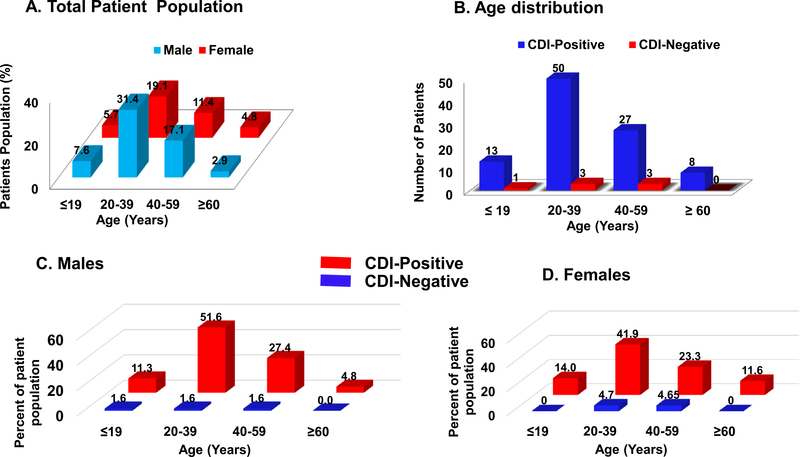
Of the patients enrolled, 59% were male (average age 34 years) and 41% were female (average age 37 years). Male and female patients had a similar prevalence of C. difficile and there was no statistically significant correlation (p = 0.456) between the age groups (Figure 1). The average age of all of the patients was 35.5 years (range 3–86 years): 13.3% were <20 years of age, 50.5% were 20–39 years old, 28.6% were 40–59 years old, and 7.6% were 60–86 years old. Strikingly, 91.9% of the stools that tested positive for C. difficile came from patients under 60 years of age, with 64.3% being less than 40 years old (Figure 1B). About 90.5% of the patients reported having diarrhea for less than 1 month, whereas 9.5% indicated prolonged diarrhea lasting 1–3 months prior to the hospital visit. Moreover, 4.8% of the patients reported being HIVpositive, 28.6% reported being HIV-negative, and 66.7% did not know their HIV status. All of the HIV-positive patients were C. difficile-positive.
Figure 1.
Distribution of the total patient population based on sex and age (A) and by the presence of Clostridium difficile in stool based on age (B) and sex (C and D). All patients presenting diarrhea at Kenyatta National Hospital and Kisii Teaching and Referral Hospital in Kenya from May to July 2017 were sequentially enrolled in the study. To limit bias, no diarrhea patient was excluded, except a few patients who refused to participate. The population was 59% male and 41% female.
About 85% of the patients reported over-the-counter antibiotic use 30 days before the hospital visit, and no significant difference was found between the age groups (p = 0.485) or sexes (p = 0.760) (Table 2). Pearson’s Chi-square test demonstrated a significant relationship between history of antibiotics use and CDI positivity (p = 0.035). The most widely used over-the-counter antibiotics reported by the patients were amoxicillin, metronidazole, cephalosporins, ciprofloxacin, and azithromycin.
Table 2.
Distribution of antibiotics taken by the patients in the last 30 days before the hospital visit.
| Number | % | Age (years) |
|||
|---|---|---|---|---|---|
| Mean | Range | ||||
| Antibiotic use in last 30 days | |||||
| Yes | 89 | 84.8 | 35.1 | (3–86) | |
| No | 16 | 15.2 | 37.9 | (13–61) | |
| Antibiotic class | Generic name | ||||
| Unreported antibiotic | 16 | 15.2 | 37.9 | (13–61) | |
| Penicillins | |||||
| Amoxicillin | 31 | 29.5 | 36.8 | (5–86) | |
| Nitroimidazoles | Ampicillin | 2 | 1.9 | 34.5 | (29–40) |
| Cephalosporins | Metronidazole | 20 | 19.1 | 31.4 | (3–58) |
| Cephalosporin | 10 | 9.5 | 31.5 | (19–60) | |
| Cephalexin | 1 | 1 | 35 | – | |
| Fluoroquinolones | Ceftriaxone | 9 | 8.6 | 34.3 | (17–61) |
| Ciprofloxacin | 6 | 5.7 | 31.8 | (18–42) | |
| Macrolides | Levofloxacin | 2 | 1.9 | 56.5 | (56–57) |
| Sulfonamides | Azithromycin | 4 | 3.8 | 31 | (22–39) |
| Glycopeptides | TMP–SMX | 2 | 1.9 | 45.5 | (43–48) |
| Vancomycin | 1 | 1 | 71 | – | |
TMP–SMX, trimethroprim–sulfamethoxazole.
Discussion
Most CDI studies have been reported from industrialized regions and little is known about the infection on the African continent. Moreover, C. difficile has traditionally been neglected as an important diarrhea-causing pathogen in Africa. Since Kenya is a popular destination for tourists around the world, it was sought to establish the presence of C. difficile by examining a cross-section of unbiased, sequentially enrolled diarrhea patients in two hospitals. Astoundingly, the study results demonstrated that a large number of stools from young adults presenting with diarrhea at the two hospitals had C. difficile. This rate is unusually higher than that reported in industrialized countries and the few studies conducted on the African continent (Onwueme et al., 2011; Seugendo et al., 2015), but the patient population and demographics are different. Moreover, the methods used in the current study (culture with PCR validation) are more sensitive than those used in the previous studies conducted in Africa.
The higher CDI frequency in Kenya undoubtedly relates to the overuse of antibiotics, which are widely available over-the-counter with no strict regulation governing access to the drugs. The patients in this study were unique because they went to the hospital following unsuccessful treatment of their diarrhea by self-medication. This practice is common in African countries and may potentially exacerbate CDI rates and antibiotic resistance more than previously thought. While beta-lactam antibiotics are well known to predispose people to CDI, 19% of the CDI patients in this study had taken metronidazole before presenting to the hospital. Of importance, metronidazole was not associated with cure when used as self-treatment. Thus, the current study adds to the growing evidence that metronidazole is less effective in treating CDI (Johnson et al., 2014).
Comorbidities such as HIV infection may have contributed to the high prevalence of CDI in this population, a known association (Keeley et al., 2016). HIV infection in the study population was invariably associated with CDI, although only a small number of HIV-positive patients were enrolled. However, other diarrhea-causing pathogens such as norovirus and Escherichia coli were not tested for in this study. The finding that 64.3% of the CDI patients were less than 40 years of age and 27.6% were 40–59 years old was unexpected. Such a high frequency of CDI in young adults with diarrhea has not been observed elsewhere, and this may highlight the changing epidemiology and virulence of C. difficile strains.
A limitation of this study was the lack of data on the symptoms exhibited by the patients and clinical outcomes, as this was an initial study to establish the presence of C. difficile among diarrheal patients in Kenya. Another limitation is that other diarrhea-causing pathogens could not be excluded in this study and therefore C. difficile may not have been the sole pathogen associated with diarrhea in these patients. Nonetheless, with such an unusually high level of CDI among the diarrhea patients, a larger study has been planned to comprehensively examine the epidemiology, risk factors, co-infections, pathogenesis, comorbidities, clinical outcomes, antibiotic resistance, and the genetic makeup of C. difficile strains circulating in this population.
Acknowledgements
This work was supported in part, by NIH R01 grant number R01AI116914, a Molecular Basis of Infectious Diseases Training Grant from the NIH Institute of Allergy and Infectious Diseases (T32AI055449), and the Gillson-Longenbaugh Foundation.
Footnotes
Ethical approval
This study was approved by the ethics review boards of Kenyatta National Hospital/University of Nairobi, Kisii Teaching and Referral Hospital, and the Kenyan Ministry of Health.
None of the authors have any conflict to report.
References
- Borriello SP. Pathogenesis of Clostridium difficile infection. J Antimicrob Chemother 1998;41(Suppl C):13–9. [DOI] [PubMed] [Google Scholar]
- Carter GP, Douce GR, Govind R, Howarth PM, Mackin KE, Spencer J, et al. The antisigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLoS Pathog 2011;7(10)e1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry SR, Marsh JW, Muto CA, O‘Leary MM, Pasculle AW, Harrison LH. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol 2007;45(1):215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkoh C, DuPont HL, Kaplan HB. Novel one-step method for detection and isolation of active-toxin-producing Clostridium difficile strains directly from stool samples. J Clin Microbiol 2011a;49(12):4219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkoh C, Kaplan HB, DuPont HL. Harnessing the glucosyltransferase activities of Clostridium difficile for functional studies of toxins A and B. J Clin Microbiol 2011b;49(8):2933–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedoruk K, Daniluk T, Rozkiewicz D, Zaremba ML, Oldak E, Sciepuk M, et al. Conventional and molecular methods in the diagnosis of community-acquired diarrhoea in children under 5 years of age from the north-eastern region of Poland. Int J Infect Dis 2015;37:145–51. [DOI] [PubMed] [Google Scholar]
- Fry PR, Thakur S, Abley M, Gebreyes WA. Antimicrobial resistance, toxinotype, and genotypic profiling of Clostridium difficile isolates of swine origin. J Clin Microbiol 2012;50(7):2366–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geric B, Rupnik M, Gerding DN, Grabnar M, Johnson S. Distribution of Clostridium difficile variant toxinotypes and strains with binary toxin genes among clinical isolates in an American hospital. J Med Microbiol 2004;53(Pt 9):887–94. [DOI] [PubMed] [Google Scholar]
- Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol 2010;48 (3):770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, von Eichel-Streiber C. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur J Biochem 1997;244(3):735–42. [DOI] [PubMed] [Google Scholar]
- Johnson S, Louie TJ, Gerding DN, Cornely OA, Chasan-Taber S, Fitts D, et al. Vancomycin, metronidazole, or tolevamer for Clostridium difficile infection: results from two multinational, randomized, controlled trials. Clin Infect Dis 2014;59(3):345–54. [DOI] [PubMed] [Google Scholar]
- Just I, Selzer J, Wilm M, von Eichel-Streiber C, Mann M, Aktories K. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 1995;375(6531):500–3. [DOI] [PubMed] [Google Scholar]
- Keeley AJ, Beeching NJ, Stott KE, Roberts P, Watson AJ, Beadsworth MB. Clostridium difficile: a healthcare-associated infection of unknown significance in adults in sub-Saharan Africa. Malawi Med J 2016;28(2):66–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S, Pardi DS. Clostridium difficile infection: new insights into management. Mayo Clin Proc 2012;87(11):1106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature 2010;467(7316):711–3. [DOI] [PubMed] [Google Scholar]
- Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, et al. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol 2004;42 (12):5710–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Jia H, Yang H, Ji B, Liu Y, Peng X, et al. Preliminary screening of type IV secretion system in divergent geographic sources of Clostridium difficile. Exp Ther Med 2017;14(5):4405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Li WG, Zhang WZ, Wu Y, Lu JX. Molecular characterization of Clostridium difficile Isolates in China from 2010 to 2015. Front Microbiol 2018;9:845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun 1985;47(2):349–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamouros S, England P, Dupuy B. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol 2007;64(5):1274–88. [DOI] [PubMed] [Google Scholar]
- Murray R, Boyd D, Levett PN, Mulvey MR, Alfa MJ. Truncation in the tcdC region of the Clostridium difficile PathLoc of clinical isolates does not predict increased biological activity of Toxin B or Toxin A. BMC Infect Dis 2009;9:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwueme K, Fadairo Y, Idoko L, Onuh J, Alao O, Agaba P, et al. High prevalence of toxinogenic Clostridium difficile in Nigerian adult HIV patients. Trans R Soc Trop Med Hyg 2011;105(11):667–9. [DOI] [PubMed] [Google Scholar]
- Owens RC Jr., Donskey CJ, Gaynes RP, Loo VG, Muto CA. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin Infect Dis 2008;46(Suppl 1):S19–31. [DOI] [PubMed] [Google Scholar]
- Rupnik M, Brazier JS, Duerden BI, Grabnar M, Stubbs SL. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 2001;147(Pt 2):439–47. [DOI] [PubMed] [Google Scholar]
- Seugendo M, Mshana SE, Hokororo A, Okamo B, Mirambo MM, von Muller L, et al. Clostridium difficile infections among adults and children in Mwanza/Tanzania: is it an underappreciated pathogen among immunocompromised patients in sub-Saharan Africa?. New Microbes New Infect 2015;8:99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 2005;18(2):247–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 2005;366(9491):1079–84. [DOI] [PubMed] [Google Scholar]
- Zhang S, Palazuelos-Munoz S, Balsells EM, Nair H, Chit A, Kyaw MH. Cost of hospital management of Clostridium difficile infection in United States-a meta-analysis and modelling study. BMC Infect Dis 2016;16(1):447. [DOI] [PMC free article] [PubMed] [Google Scholar]