Abstract
Starvation poses a fundamental challenge to cell survival. How autophagy promotes energy homeostasis in this setting has been extensively characterized 1, but how other mechanisms may be similarly critical is less understood. Here, we initially find that glyceraldehyde 3-phosphate dehydrogenase (GAPDH) inhibits Coat Protein I (COPI) transport by targeting a GTPase-activating protein (GAP) against ADP-Ribosylation Factor 1 (ARF1) to suppress COPI vesicle fission. We then find that GAPDH inhibits multiple other transport pathways also by targeting ARF GAPs. Defining a physiologic role for this broad inhibition, our results suggest that it is activated by the cell during starvation to reduce energy consumption in promoting energy homeostasis. These findings reveal a previously unappreciated level of coordination among the intracellular transport pathways, with this process underlying a new critical mechanism of cellular energy homeostasis.
The activation of ARF small GTPases initiates vesicular transport by recruiting coat proteins to intracellular membrane compartments for vesicle formation 2. In the case of COPI transport, the GAP that de-activates ARF1, known as ARFGAP1, also acts as an ARF effector by being a coat component 3, 4. To gain further insight into how ARFGAP1 acts in COPI transport, we sought to identify new interacting proteins. When ARFGAP1 was incubated with cytosol in a pulldown experiment, we identified GAPDH as one such protein (Extended Data, Table 1). Although well known to act in glycolysis, GAPDH is also known to have multiple non-glycolytic roles 5. Thus, to query whether it acts in COPI transport, we initially performed a COPI transport assay as previously described 6–8. In cells treated with small interfering RNA (siRNA) against GAPDH, we observed enhanced COPI transport (Fig 1a and Extended Data, ED Fig 1a). Targeting specificity was confirmed by a rescue experiment (Fig 1a and ED Fig 1b), and by treatment with another siRNA sequence (ED Fig 1c). The siRNA treatments were limited to two days to maintain cell viability (ED Fig 1d). Consistent with the effect of reducing GAPDH level, GAPDH overexpression had the opposite effect of inhibiting COPI transport (Fig 1b and ED Fig 1e). The expression levels of GAPDH in the different treatment conditions were also documented (ED Fig 1f). These initial results suggested that GAPDH acts as a negative regulator of COPI transport.
Fig 1. GAPDH inhibits COPI vesicle fission by targeting the GAP activity of ARFGAP1.
a,b, COPI transport in HeLa cells, n=10 fields of cells examined in a representative experiment (out of 3), mean +/− SD, two-tailed t-test: (a) *p=9.8E-07, **p=9.2E-09, (b) *p=6.8E-06. c, Vesicle reconstitution system, n=3, GDH (glutamate dehydrogenase), LDH (lactate dehydrogenase), GPDH (glycerol-3-phosphate dehydrogenase). d, Vesicle reconstitution system: EM image of Golgi membrane (left), bar = 50 nm; vesicle quantitation (right), n=10 EM meshes examined from a representative experiment (out of 3), *p=8.9E-07, e, GAP assay using ARF1 and ARFGAP1, and also with metabolic enzymes as indicated, n=3. f, Vesicle reconstitution system: EM image of Golgi membrane (left), bar = 50 nm; vesicle quantitation (right), n=10 EM meshes examined from a representative experiment (out of 3), *p=6.2E-06.
We next pursued the reconstitution of COPI vesicles from Golgi membrane, which has been instrumental in dissecting out the mechanistic details of COPI vesicle formation 6–8. The addition of GAPDH as another purified component inhibited this process (Fig 1c). As specificity control, multiple other metabolic enzymes did not have a similar effect (Fig 1c). We also performed electron microscopy (EM), and found that GAPDH induced the accumulation of buds with constricted necks on the Golgi membrane (Fig 1d). Thus, these results suggested that GAPDH inhibits COPI transport by targeting the fission stage of vesicle formation.
We then found that GAPDH could bind directly to ARFGAP1 (ED Fig 1g), and GAPDH interacts with ARFGAP1 in cells (ED Fig 1h). Thus, we next performed a GAP assay, which revealed that GAPDH, but not other metabolic enzymes, inhibited the catalytic activity of ARFGAP1 (Fig 1e). Complementing this finding, we found that a mutant ARFGAP1 deficient in catalytic activity could not promote COPI vesicle fission (Fig 1f). ARFGAP1 also acts as a coat component by promoting coat polymerization and cargo sorting 3, 4. However, GAPDH did not affect the interaction of ARFGAP1 with coatomer (ED Fig 1i) or with COPI cargo proteins (ED Fig 1j). Thus, we concluded that GAPDH inhibits COPI vesicle fission by targeting the GAP activity of ARFGAP1.
We also found that this role of GAPDH did not require its catalytic activity (Supplementary Information, and ED Fig 1k-l). Next, we sought to understand how GAPDH could be recruited to the Golgi to inhibit COPI transport. Led by a previous observation that starvation redistributes GAPDH from the cytosol to the nucleus 9, we initially performed a subcellular fractionation experiment and found that starvation also redistributed GAPDH to cytoplasmic membranes (ED Fig 2a). Confocal microscopy revealed that this redistribution involved the Golgi (ED Fig 2b), the trans Golgi network (TGN) (ED Fig 2c), the early endosomes (ED Fig 2d), and the late endocytic compartments (ED Fig 2e). We also detected a pool of GAPDH at the endoplasmic reticulum (ER), but the level of this ER pool was not affected by starvation (ED Fig 2f).
In light of the above results, we next examined whether GAPDH also inhibits other intracellular pathways. Performing a quantitative screen of the major pathways, as previously described 10, we found that siRNA against GAPDH did not affect transport from the ER to the Golgi (Fig 2a and ED Fig 2g), but enhanced transport from the Golgi to the plasma membrane (PM) (Fig 2b and ED Fig 2h). All three major endocytic routes were also enhanced. These included endocytic recycling (Fig 2c and ED Fig 2i), endocytic transport to the Golgi (Fig 2d and ED Fig 2j), and endocytic transport to the lysosome (Fig 2e and ED Fig 2k). Fluid-phase uptake was also enhanced by the siRNA treatment (Fig 2f and ED Fig 2l). However, clathrin-mediated endocytosis (Fig 2g), caveolae-dependent endocytosis (ED Fig 2m), and another clathrin-independent endocytosis (ED Fig 2n) were not affected. Moreover, similar to the case of COPI transport, we found that GAPDH overexpression had the opposite effect of inhibiting the affected pathways (Fig 2a-g). The specificity of our detection of GAPDH and organelle markers were also confirmed (Supplementary Information, and ED Figs 2o-r and 3a-l).
Fig 2. GAPDH inhibits other intracellular pathways by also targeting ARF GAPs. a-g.
Transport assays in HeLa cells, n=10 fields of cells examined in a representative experiment (out of 3), mean +/− SD, two-tailed t-test. (a) Transport of vesicular stomatitis virus G protein (VSVG) from the ER to the Golgi, NS1 (not significant)=0.625, NS2=0.438. (b) Transport of VSVG from the Golgi to the PM, *p=7.6E-08, **p=5.4E-06. (c) Endocytic recycling of transferrin (Tf) from the early endosome (EE) to the PM, *p=1.05E-07, **p=1.05E-07. (d) Endocytic transport of cholera toxin (CT) from the PM to the Golgi, *p=2.7E-09, **p=4.5E-04. (e) Endocytic transport of epidermal growth factor (EGF) from the PM to the lysosome, *p=1.6E-08, **p=7.2E-07. (f) Dextran endocytosis, *p=1.1E-10, **p=1.8E-06. (g) EGF endocytosis, NS1=0.9997, NS2=0.418. h-j, GAP assays using ARF6 and ACAP1 (h), using ARF1 and AGAP1 (i), or using Sar1p and Sec23p (j), n= 3.
We then examined whether GAPDH inhibits these additional pathways also by targeting ARF GAPs. ARF6 and its cognate GAP, ACAP1, act in endocytic recycling 11. We found that GAPDH inhibited the GAP activity of ACAP1 toward ARF6 (Fig 2h). ARF1 and its cognate GAP, AGAP1, act in endocytic transport to the lysosome 12. We found that GAPDH also inhibited the GAP activity of AGAP1 toward ARF1 (Fig 2i). Moreover, other metabolic enzymes did not have a similar effect (ED Fig 3m,n). In the case of exit from the ER, Sec23p is the cognate GAP for the ARF-like small GTPase, Sar1p 13. Consistent with our finding above that GAPDH does not inhibit this pathway, we found that GAPDH also did not inhibit the GAP activity of Sec23p toward Sar1p (Fig 2j). We then found that GAPDH bound directly to ACAP1 (ED Fig 3o) and AGAP1 (ED Fig 3p), but not Sec23p (ED Fig 3q). Thus, these results suggested that GAPDH inhibits the transport pathways by targeting ARF GAPs, which was further supported by our examination of transferrin (Tf) endocytosis (Supplementary Information, and ED Fig 3s-v). Also, led by the consideration that the catalytic domain in ARFGAP1 is conserved across many ARF GAPs, but not for Sec23p, we found that GAPDH bound to the catalytic domain of ARFGAP1 (ED Fig 3r), which suggested why GAPDH did not interact with Sec23p, and thus did not inhibit exit from the ER.
We then examined the effect of starvation on the transport pathways. Cells were incubated with a general starvation medium (Figs 3a-h), which lacks glucose and amino acids, Hank’s medium (ED Fig 4a-h), which lacks amino acids and serum, or media that lack either glucose (ED Fig 4i-p) or amino acids (ED Fig 4q-x). In all cases, the identical pathways were inhibited, and notably, these effects were reversed by siRNA against GAPDH (Fig 3a-h and ED Figs 4a-x).
Fig 3. AMPK phosphorylates GAPDH to inhibit the transport pathways for energy homeostasis.
a-h, Transport assays in HeLa cells, n=10 fields of cells examined in a representative experiment (out of 3), mean +/− SD, two-tailed t-test. (a) ER to Golgi, NS1=0.7084, NS2=0.175. (b) Golgi to ER, *p=5.7E-06, **p=6.3E-15. (c) Golgi to PM, *p=5.4E-06, **p=4.3E-11. (d) EE to the PM, *p=5.5E-09, **p=1.2E-12. (e) PM to Golgi, *p=5.4E-04, **p=6.2E-06. (f) PM to lysosome, *p= 1.7E-03, **p=1.9E-08. (g) Dextran endocytosis, *p=7.8E-08, **p=2.1E-12. (h) EGF endocytosis, NS1= 0.9994, NS2=0.7437. i, Total ATP level, n=3. j, Cell death, n=3. k, AMPK phosphorylation of GAPDH assessed by an in vitro kinase assay, n= 3. l,m, Distribution of GAPDH assessed by subcellular fraction, membranes (M), cytosol (C), n=2. n,p, Total ATP level, n=3. (o,q) Cell death, n=3.
We then considered that vesicular transport consumes substantial cellular energy. Thus, we hypothesized that the GAPDH-mediated transport inhibition enables the cell to conserve energy during starvation. In support of this hypothesis, we found that siRNA against GAPDH exacerbated the starvation-induced reduction in total ATP level, while GAPDH overexpression had the opposite effect (Fig 3i and ED Fig 5a-c). Moreover, siRNA against GAPDH enhanced starvation-induced cell death, while GAPDH overexpression had the opposite effect (Fig 3j). We ruled out that these effects could be attributed to the conventional role of GAPDH in glycolysis (Supplementary Information, and ED Fig 5d-f). We also confirmed the key findings on how GAPDH acts in another cell type (Supplementary Information, and ED Fig 5g-x).
Next, we pursued another line of investigation that further supported our hypothesis. AMPK plays a central role in energy homeostasis by sensing an energy deficit and then coordinating a wide range of cellular processes to restore energy homeostasis 14. Treating cells with siRNA against AMPKα, we found that starvation could no longer inhibit the transport pathways (ED Fig 6a-h), which was confirmed with a rescue experiment (ED Fig 6i,j), and with treatment using another siRNA sequence (ED Figs 6k-s). Complementing these findings, we found that siRNA against AMPKα1 prevented starvation from redistributing GAPDH to membrane compartments (ED Figs 7a-e).
We also activated AMPK in the normal (non-starved) condition by treating cells with 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), and found that it reproduced the effect of starvation in suppressing the identical transport pathways, and this effect was also reversed by siRNA against GAPDH (ED Fig 7f-m). This result was further confirmed using A769662, a more specific activator of AMPK (ED Fig 7n-u). Moreover, AICAR treatment in the normal condition was sufficient to redistribute GAPDH to membrane compartments (ED Fig 8a-e). Thus, the collective results suggested that starvation acts through AMPK and then GAPDH to inhibit the transport pathways.
Next, to examine how AMPK affects GAPDH, we were initially led by a previous observation that AMPK phosphorylates GAPDH at serine 122 (S122) 9. Performing an in vitro kinase assay, we confirmed this finding, as mutating this residue to alanine (S122A) abolishes the phosphorylation (Fig 3k and ED Fig 8f,g). However, when compared to acetyl-coA carboxylase (ACC), a well-established substrate of AMPK, GAPDH was phosphorylated less efficiently (ED Fig 8h-k), which could be explained by the sequence surrounding the S122 residue not representing an optimal consensus site for AMPK phosphorylation 15. We also pursued cell-based studies. Initially, we confirmed that starvation activates AMPK in cells (ED Fig 8l). We then found that starvation increased the phosphorylation of the S122 residue in GAPDH, and this effect was prevented by the S122A mutation (ED Fig 8m). We also found that this phosphorylation was prevented by siRNA against AMPKα1 (ED Fig 8n). Similar results were also seen in HEK293 cells (ED Fig 8o-q). We then replaced the endogenous GAPDH with the S122A mutant and found that starvation could no longer redistribute cytosolic GAPDH to membranes (Fig 3l). Accordingly, the S122A mutant also prevented starvation from inhibiting the transport pathways (ED Fig 9a-h). We also mutated the S122 residue in GAPDH to aspartate (S122D), which mimics constitutive phosphorylation at this site 9, and found that simply expressing the S122D mutant in the normal condition was sufficient to redistribute cytosolic GAPDH to membranes (Fig 3m). The expression of this mutant in the normal condition was also sufficient in inhibiting the transport pathways (ED Fig 9i-p).
The effects of mutating the S122 residue in GAPDH suggested yet another way of confirming that the GAPDH-mediated transport inhibition acts in energy homeostasis. Replacing the endogenous GAPDH with the S122A mutant in cells, we found that the decline in the total ATP level induced by starvation was exacerbated (Fig 3n). Cell viability was similarly exacerbated in this setting (Fig 3o). In contrast, replacing the endogenous GAPDH with the S122D mutant had the opposite effect of promoting the ATP level (Fig 3p) and cell viability (Fig 3q) during starvation.
We next noted that AMPK phosphorylation of the S122 residue in GAPDH has been found previously to promote a different non-glycolytic role of GAPDH. This involves GAPDH being translocated to the nucleus to stimulate Sirtuin1 (Sirt1) activity, which then activates a transcription program to promote autophagy 9. We first confirmed that our starvation medium (which lacks glucose and amino acids) activated autophagy (Figs 4a,b). We next found that the recruitment of GAPDH to the Golgi, which is needed to inhibit COPI transport, started immediately upon starvation (Fig 4c). In contrast, LC3 puncta formation, which tracks autophagosome formation, started more slowly (Fig 4d). Further distinguishing between the roles of GAPDH in transport inhibition and autophagy, we found that siRNA against Sirt1 did not have an appreciable effect on starvation-induced inhibition of COPI transport (Fig 4e). In contrast, this siRNA treatment prevented starvation from inducing autophagy (Fig 4f).
Fig 4. The roles of GAPDH in transport and autophagy are distinct.
a,b, Autophagy in HeLa cells assessed by (a) LC3 puncta formation, n=10 fields of cells examined in a representative experiment (out of 2), mean +/− SD, two-tailed t-test, *p=0.000152, or (b) LC3 gel shift, n=2. c, GAPDH recruitment to the Golgi in HeLa cells, n=6 fields of cells examined in one experiment. d, LC3 puncta formation in HeLa cells, n=10 fields of cells examined in one experiment. e, COPI transport assay in HeLa cells, n=10 fields of cells examined in a representative experiment (out of 3), mean +/− SD, two-tailed t-test, NS=0.8490. f, LC3 puncta formation in HeLa cells, n=10 fields of cells examined in a representative experiment (out of 2), mean +/− SD, two-tailed t-test, *p=0.00174. g-n, Transport assays in Atg5-deficient MEFs, n=10 fields of cells examined in a representative experiment (out of 3), mean +/− SD, two-tailed t-test, (g) ER to Golgi, NS=0.25. (h) Golgi to ER, *p=9.9E-08. (i) Golgi to PM, *p=5.9E-07 (j) EE to PM, *p=6.4E-05. (k) PM to Golgi, *p=7.6E-07. (l) PM to lysosome, *p=3.2E-07. (m) Dextran endocytosis, *p=1.2E-09, (n) EGF endocytosis, NS=0.9994. o, Total ATP level, n=3. p, Cell death, n=3. q, Summarizing how transport inhibition by GAPDH promotes cellular energy homeostasis.
We also examined mouse embryonic fibroblasts (MEFs) derived from Atg5-deficient mice, which cannot undergo autophagy 16, and found that GAPDH could still inhibit the identical transport pathways as those seen in autophagy-competent cells (Figs 4g-n). Moreover, siRNA against GAPDH could still induce a further exacerbation in the decline of total ATP level (Fig 4o) and cell viability (Fig 4p) during starvation. In another approach, we modified GAPDH so that its redistribution to the nucleus was prevented, but its recruitment to cytoplasmic membrane compartments was preserved. This form of GAPDH could still inhibit the transport pathways, and also promote ATP level and cell viability during starvation (Supplementary Information, and ED Fig 10a-u). Thus, multiple lines of evidence all point to the roles of GAPDH in transport inhibition and autophagy being distinct.
In summary, we have uncovered a new mechanism of energy homeostasis that is critical for the cell to survive during starvation. Mechanistically, this involves the activation of AMPK to induce the redistribution of cytosolic GAPDH to multiple membrane compartments, which then targets ARF GAPs to exert inhibition on different transport pathways (Fig 4q). As the cell encounters starvation in many settings, considerable effort has been devoted to elucidating mechanisms of cell survival during this daunting situation. Autophagy has emerged as a major mechanism 1. In the overall energy equation, autophagy acts on the supply side to increase energy availability. In contrast, the transport inhibition by GAPDH that we have uncovered acts on the demand side to reduce energy consumption. Despite being fundamentally distinct, the two mechanisms nevertheless funnel into a common goal of promoting energy homeostasis during starvation. Another notable distinction between the two mechanisms is that the transport inhibition by GAPDH occurs more rapidly than autophagy. This more rapid mechanism likely allows the cell to stave off the lethal consequences of starvation while longer-term solutions, such as autophagy, are being mobilized.
METHODS
Chemicals, proteins, and cells.
AICAR, chloroquine diphosphate, methyl-pyruvate, protease inhibitor cocktail, and phosphatase inhibitor cocktail were obtained (Sigma). AMPK activator A-769662 was obtained (Cayman). Alexa 546-conjugated transferrin, and Alexa 555-conjugated forms of EGF, dextran and CT were also obtained (Invitrogen).
Purification of coatomer, ARF1, ARFGAP1, BARS, Golgi-enriched membrane, and cytosol have been described 3, 17. Purification of ARF6, ACAP1, and AGAP1 have also been described 11, 12. Purified mammalian Sar1p and Sec23p were obtained (J. Goldberg, Memorial Sloan-Kettering Cancer Center, USA). Purified forms of GAPDH, glutamate dehydrogenase (GDH), glycerol-3-phosphate dehydrogenase (GPDH), and lactate dehydrogenase (LDH) were also obtained (Sigma), as well as purified activated AMPK and the SAMS peptide (HMRSAMSGLHLVKRR) (Promega).
For the purification of recombinant GAPDH, the pET30–2-GAPDH plasmid was transformed in BL21 competent cell. Bacteria were then resuspended in 5ml of lysis buffer (20mM Tris pH 8.0, 100 mM NaCl, 10mM imidazole, 1mg/ml lysozyme, and protease inhibitors cocktail). After gentle agitation at 4°C for 30minutes, the suspension was sonicated on ice 6 times for 10 seconds each, and then centrifuged at 12,000 × g for 20 minutes. The supernatant was recovered and then added to 0.5ml Ni-bead (50% slurry), followed by incubation at 4°C for 1 hour. Beads were then washed with 10ml washing buffer (20mM Tris pH 8.0, 100 mM NaCl, 10mM imidazole). Bound proteins were then eluted with elution buffer (20mM Tris pH 8.0, 100 mM NaCl, 250mM imidazole).
HeLa, BSC-1, COS-7, CHO, and HEK293 cells have been described previously 10, 18, 19. Wide-type and Atg5-deficient MEF cells were obtained (N. Mizushima, University of Tokyo, Japan). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum and supplemented with glutamine. Unless stated otherwise, starvation involves incubating cells in a medium consisted of phosphate-buffered saline (PBS) with 10% dialyzed bovine serum.
Mass Spectrometry.
Cytosol was purified from rat livers by homogenization in lysis buffer (1mM Tris pH 7.4, 800mM sucrose, 5mM EDTA and protease inhibitors) at 4°C, followed by centrifugation at 80,000 × g for 90 minutes. The resulting supernatant was dialyzed against dialysis buffer (25mM Tris at pH 8.0, 50mM KCl and 1mM DTT), followed by centrifugation at 150,000 × g for 90 minutes at 4°C. Aliquots were stored at −80°C. Pulldown experiment was performed by incubating GST or GST-ARFGAP1 on beads (5 ug) with 50μg rat liver cytosol in traffic buffer (25mM HEPES pH 7.2, 50mM KCl and 2.5mM Mg(OAc)2) for 1 hour followed by extensive washing. Samples were separated by SDS-PAGE and then stained with Coomassie blue, followed by gel excision for protein identification by mass spectrometry (Taplin Mass Spectrometry Facility, Harvard Medical School, USA). Those proteins (having at least five identified peptides) that interacted with GST-ARFGAP1 but not GST were considered specific.
Plasmids and antibodies.
Mammalian expression vectors that contain temperature-sensitive mutants (ts-045) of VSVG and VSVG-KDELR have been described 7. GAPDH for mammalian expression was subcloned into pcDNA3.1 and pEGFP-N1. GAPDH for bacterial expression (pET30–2-GAPDH) was obtained (Addgene, 83910). Mutant forms of GAPDH were generated using QuikChange Site-Directed-Mutagenesis (Stratagene). The sequences of oligonucleotides used for mutagenesis are listed in SI Table 1. The pEGFP-NES-GAPDH construct was obtained (W. Liu, Zhejiang University, China). IL2R-β tagged with HA and Venus (HA-Venus-IL2R-β) was also obtained (Thomas Waldman and Olga Anton Hurtaldo, NIH, USA). GST-ARFGAP1, GST-Wbp1, and GST-KDELR have been described 3, 17.
Antibodies against ARF1, ARFGAP1, β-COP, calnexin, cellubrevin, coatomer, giantin, GM130, Lamp1, Sec61p, TGN46 VSVG, TGN46, HA tag, and Myc tag have been described previously 10, 17, 19. The following antibodies were obtained: AMPKα (Cell Signaling, 2603S), β-actin (Ambion, AM4302), GFP (Invitrogen, GF28R), LC3 (Sigma, L7543), p62 (Santa Cruz, 28359), p172-AMPKα (Cell Signaling, 2531S), Rab11 (BD Biosciences, 610656), Sirt1 (Cell Signaling, 2493S), and Flag tag M2 (Sigma, F1804). Antibodies against GAPDH were obtained from three sources: Ambion (AM4300), Santa Cruz (FL335), and Sigma (G9545). Anti-pS/pT was obtained from Abcam (117253). Conjugated antibodies were also obtained (Jackson ImmunoResearch), which include Cy2- or Cy3-conjugated donkey antibodies against mouse or rabbit IgG, and horseradish peroxidase-conjugated donkey antibodies against mouse or rabbit IgG.
Transfections and siRNA.
Transfection of DNA plasmids was performed using FuGene6 (Roche). Transfection of siRNA was performed using Lipofectamine RNAiMAX (Invitrogen). Sequences used for siRNA against GAPDH and AMPKα1, and primers used to generate siRNA resistant forms of GAPDH and AMPKα1 are listed in Supplementary Information (SI Table1). Rescue plasmids were generated using QuikChange Site-Directed-Mutagenesis (Stratagene).
We found that siRNA against GAPDH for 2 days was optimal in reducing the endogenous level while maintaining cell viability. Rescue of siRNA against GAPDH involved the transfection of siRNA-resistant GAPDH for 12 hours to achieve limited GAPDH expression. GAPDH overexpression involved the transfection of GAPDH for 48 hours.
In vivo transport assays.
A quantitative microscopy-based approach, which involves the colocalization of model cargoes with organelle markers and coupled with kinetic analysis, was performed as previously described 8, 10.
For anterograde transport from ER to Golgi, cells were transfected with pROSE-VSVG-ts045-Myc for 1 day, and then incubate at 39°C for 4 hours to accumulate VSVG in the ER. Cells were then shifted to 32°C for different times as indicated in figures. Cells were then stained for giantin, followed by confocal microscopy to assess the arrival of VSVG to the Golgi.
For retrograde transport from the Golgi to the ER, cells were transfected with pROSE-VSVG-ts045-KDELR-Myc for 1 day, and then incubated at 32°C for 8 hours to achieve steady-state distribution at the Golgi. Cells were then shifted to 39°C for different times as indicated in the figures. Cells were then stained for giantin, followed by confocal microscopy to assess the exit of VSVG-KDELR from the Golgi.
For anterograde transport from the Golgi to the PM, cells were transfected with pROSE-VSVG-ts045-Myc for 1 day, and then incubated at 20°C for 2 hours to accumulate VSVG at the TGN. Cells were then shifted to 32°C for different times as indicated in the figures. Cells were then stained for TGN46, followed by confocal microscopy to assess the exit of VSVG from the Golgi.
For the recycling of Tf from the early recycling endosome to the PM, cells were incubated with Alexa 546-conjugated Tf (5μg/ml in DMEM) at 37°C for 2 hours to allow the steady-state accumulation of Tf in endosomes. Subsequently, cells were incubated with medium without Tf for different times as indicated in the figures. Cells were then stained for Rab11, followed by confocal microscopy to assess the exit of Tf from the early recycling endosome.
For the retrograde transport of CT from the PM to the Golgi, cells were incubated with Alexa 555-conjugated CT for 30 minutes at 4°C (0.5μg/ml in DMEM). After washing to clear unbound CT, cells were shifted to 37°C for different times as indicated in the figures. Cells were then stained for TGN46, followed by confocal microscopy to assess the arrival of CT to the Golgi.
For endocytic transport of EGF to the lysosome, cells were incubated with Alexa 555-conjugated EGF (1μg/ml in DMEM) for 1 hour at 4°C. Cells were then washed to clear unbound EGF, followed by shifting to 37°C for times indicated in the figures. Cells were stained for Lamp1, followed by confocal microscopy to assess the arrival of EGF to the lysosome.
For the fluid-phase uptake of dextran, cells were incubated with Alexa 555-conjugated dextran (0.2mg/ml) at 37°C for different times as indicated in the figures. Cells were then stained for EEA1, followed by confocal microscopy to assess the arrival of dextran to the early endosome.
For the endocytosis of EGF, cells were incubated with Alexa 555-conjugated EGF (1μg/ml in DMEM) for 1 hour at 4°C. Cells were then washed to clear unbound EGF, followed by shifting to 37°C for times indicated in the figures. Cells were stained for EEA1, followed by confocal microscopy to assess the arrival of EGF to the early endosome.
For the endocytosis of Tf, cells were incubated with Alexa 546-conjugated Tf (5μg/ml in DMEM) for 1 hour at 4°C. Cells were then washed to clear unbound Tf, followed by shifting to 37°C for times indicated in the figures. Cells were stained for EEA1, followed by confocal microscopy to assess the arrival of Tf to the early endosome.
For the endocytosis of CT, cells were incubated with Alexa 555-conjugated CT for 30 minutes at 4°C (0.5μg/ml in DMEM). Cells were then washed to clear unbound CT, followed by shifting to 37°C for different times as indicated in the figures. Cells were stained for EEA1, followed by confocal microscopy to assess the arrival of CT to the early endosome.
For the endocytosis of IL2R-β, cells were transfected with pCDH-HA-Venus-IL2R-β for 1 day at 37°C. Cells were then incubated with anti-HA antibody at 4°C for 1 hour to label the surface pool of the receptor. Cells were washed to clear unbound antibody, followed by shifting to 37°C for different times as indicated in the figures. Cells were then stained for EEA1, followed by confocal microscopy to assess the arrival of IL2R-β to the early endosome.
Confocal microscopy.
Colocalization studies were performed with either a Nikon or a Zeiss system. The Nikon system is equipped with the Nikon Eclipse TE2000U Inverted Microscope having a Plan Apo 60x/1.40 oil objective, Nikon D-Eclipse C1 confocal package with a 488 Laser (having 515/30 emission filter) and a 543 Laser (having 590/50 emission filter), and Nikon EZ-C1 version 3.90 acquisition software. The Zeiss system is equipped with the Zeiss Axio Observer Z1 Inverted Microscope having a Plan-Apochromat 63x objective, the Zeiss LSM 800 with Airyscan confocal package with Zeiss URGB (488 and 561 nm) laser lines, and Zen 2.3 blue edition confocal acquisition software.
For quantitation of colocalization, ten fields of cells were examined, with each field typically containing about 5 cells. Images were imported into the NIH Image J version 1.50e software, and then analyzed through a plugin software (https://imagej.net/Coloc_2). Under the “Image” tab, the “Split Channels” option was selected. Under the “Plugins” tab, “Colocalization Analysis” option was selected, and within this option, the “Colocalization Threshold” option was selected. Colocalization values were then calculated by the software, and expressed as the fraction of protein of interest (cargo or GAPDH) colocalized with an organelle marker.
In vitro reconstitution of COPI vesicle formation.
The reconstitution system was performed essentially as previously described 17. Briefly, Golgi membrane (0.2 mg/ml) was washed with 3M KCl, and then incubated with ARF1 (6 μg/ml) and coatomer (6 μg/ml) for the first-stage incubation that reproduces the ARF-dependent recruitment of coatomer onto Golgi membrane. The Golgi membrane was re-isolated and then incubated with ARFGAP1 (2 μg/ml), BARS (2 μg/ml) for the second stage that results in vesicle formation. GAPDH (1 μg/ml) was added at the second stage. EM examination of Golgi membrane using the whole-mount technique has been described previously 3. Quantitation involves the examination of 10 meshes per condition.
Fractionation of the cytoplasm to obtain total membrane versus cytosol.
Cells were washed with PBS, resuspended in homogenization buffer (0.25 M sucrose, 1 mM EDTA, and 20 mM HEPES-KOH, pH 7.4 and protease inhibitor cocktail) and then disrupted by passing through 28- gauge needles. After low-speed centrifugation (800 x g for 6 min) to spin out nuclei and unbroken cells, the resulting post-nuclear supernatant was centrifuged at 100,000 x g for 1 hour to obtain cytosol and total membrane fractions.
Other in vitro assays.
For pulldown assays, GST fusion proteins were bound to glutathione beads followed by incubation with purified proteins. For the GAP assay, recombinant forms of ARF small GTPase were first loaded by GTP and then incubated with different ARF GAPs in the presence of liposomes. GTP hydrolysis was then quantified by detecting the generation of free phosphate (ATPase/GTPase Activity Assay Kit, Sigma). The in vitro kinase assay was performed by incubating AMPK (0.1 ug) with the SAMS peptide (a sequence derived from ACC) (3 ug) or different GAPDH proteins (40 ug) in 50 ul of reaction buffer (25mM MOPS, pH 7.2, 12.5mM β-glycerol-phosphate, 25mM MgCl2, 5mM EGTA, 2mM EDTA, 0.25mM DTT, 100uM AMP and 50uM ATP) at 30°C for times as indicated. Phosphorylation of the substrate was then quantified using the ADP-Glo Kinase assay (Promega), which detects ADP generated from ATP in the kinase reaction.
Other in vivo assays.
Cellular ATP level was detected using ATPlite Luminescence Detection Assay System (Perkin Elmer), and was performed according to manufacturer’s instructions, with final values normalized for cell number. Cell death was assessed by incubating cells with propidium iodide (1 ng/μl), and then quantifying for positively stained cells by flow cytometry. Lactate level was measured using a colorimetric assay kit (Biovision). Oxygen consumption rate (OCR) was measured using an XFe24 Extracellular Flux Analyzer (Seahorse Biosciences), as previously described 20. Sirt1 activity was measured using Sirt1 Direct Fluorescent Screening Assay Kit (Cayman), as previously described 9. Autophagy was assessed through LC3 lipidation, either using confocal microscopy to quantify LC3 puncta formation, or western blotting to detect LC3 mobility shift, as previously described 9.
Experimental replicates.
All colocalization studies are shown as 10 technical replicates from a representative experiment, with 3 independent experiments having been performed. Metabolic studies involving OCR are done as 5 technical replicates, with 2 independent experiments having been performed. The result from a representative replicate is shown. All other studies show a representative experiment with the number of independent experiments indicated in the figure legend.
Statistical analysis.
Sample size is noted in the figure legends. No statistical methods were used to predetermine sample size. Statistical significance was determined using Excel or Prism software for the two-tailed Student’s t-test. No inclusion/exclusion criteria were pre-established. The experiments were not randomized. The investigators were not blinded to the group allocation during experiments and in outcome assessment.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The following figures have associated raw data: Figures 1a,b, 2a-g, 3a-j, 3n-q, 4c-e, 4g-p; Extended Data Figures 1c, 1k, 2m,n, 2s-v, 3s-v, 4a-x, 5a-c, 5g-x, 6a-h, 6j, 6l-s, 7f-u, 9a-p, 10h-u. For gel source data, see Supplementary Figure 1.
Extended Data
ED Fig 1. Further characterizing how GAPDH inhibits COPI transport.
a,b, HeLa cells were treated as indicated, and then the COPI transport assay was performed. A confocal image from a representative experiment (out of three) is shown, VSVG-KDELR (green), giantin (red), bar = 10 um. Line-scan analysis for the image is also shown. c, HeLa cells were treated as indicated, and then the COPI transport assay was performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test, *p=2.8E-07, **p=8.2E-07. d, HeLa cells were treated as indicated, and then cell death was quantified by flow cytometry, n=10, mean +/− SD, two-tailed t-test, NS=0.7314. e, HeLa cells were treated as indicated, and then the COPI transport assay was performed. A confocal image from a representative experiment (out of three) is shown, VSVG-KDELR (green), giantin (red), bar = 10 um. Line-scan analysis for the image is also shown. f, HeLa cells were treated as indicated, and then whole cell lysates were immunoblotted for proteins as indicated, n=3. g, GST fusion proteins were incubated with purified GAPDH in a pulldown experiment, followed by immunoblotting for proteins as indicated, n=3. h, HeLa cells were transfected with constructs as indicated, followed by immunoprecipitation for the Myc tag and then immunoblotting for GAPDH, n=2. i, GST fusion proteins as indicated were bound to beads and then incubated with purified coatomer in a pulldown experiment, followed by immunoblotting to detect β-COP or Coomassie-staining to detect GST fusion proteins, n=3. j, Cytoplasmic tails of cargoes as indicated were fused to GST, bound to beads, and then incubated with ARFGAP1, followed by immunoblotting with antibody against ARFGAP1 or Coomassie staining to detect GST fusion proteins, n=2. k, HeLa cells were treated as indicated, and then COPI transport was assessed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test, *p=1.4E-06. l, HeLa cells were transfected with GAPDH constructs as indicated, followed by immunoprecipitation of ARFGAP1 and then immunoblotting for the different forms of GAPDH as indicated, n=2.
ED Fig 2. Further characterizing how GAPDH affects other pathways.
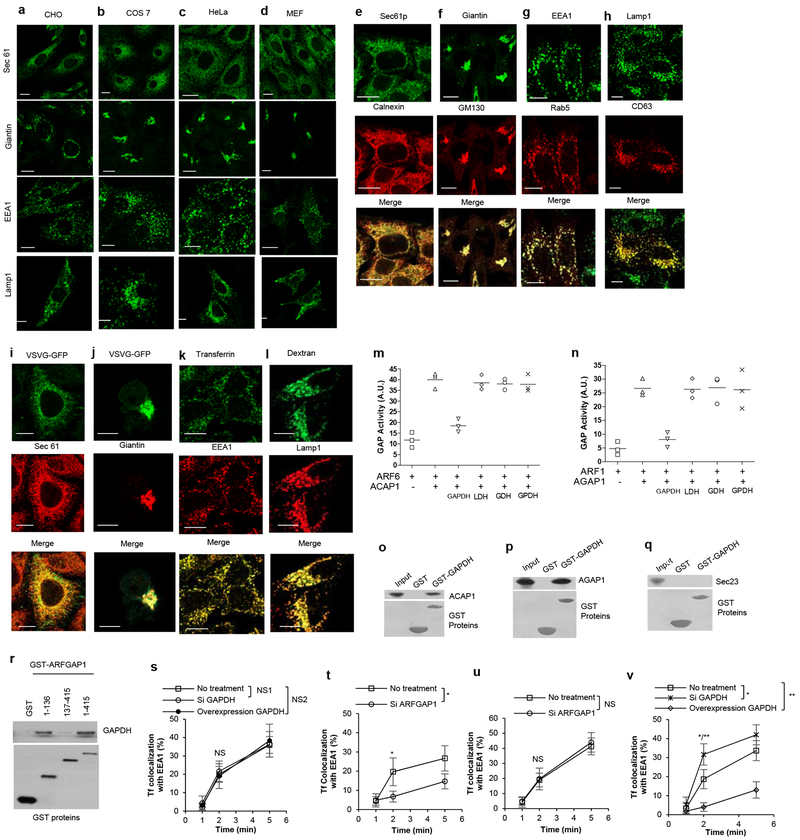
a, HeLa cells were treated as indicated, and then subjected to fractionation into cytosol (C) versus total membrane (M), followed by immunoblotting for proteins as indicated, n=2. b-f, HeLa cells were treated as indicated, and then examined for the colocalization between GAPDH and different organelle markers. A confocal image from a representative experiment (out of three) is shown, bar, 10 um (left); quantitation is also shown (right), n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (b) Colocalization of GAPDH (red) with giantin (green), *p=2.9E-04. (c) Colocalization of GAPDH (red) with TGN46 (green), *p=1.5E-04. (d) Colocalization of GAPDH (red) with EEA1 (green), *p=1.9E-03. (e) Colocalization of GAPDH (red) with Lamp1 (green), *p=7.2E-04. (f) Colocalization of GAPDH (red) with Sec61p (green), NS=0.8531. g-l, Transport assays were performed in HeLa cells. A confocal image from a representative experiment (out of three) is shown, bar = 10 um. Line-scan analysis for the representative image is also shown. (g) Transport from the ER to the Golgi, VSVG (green) and giantin (red), (h) Transport from the Golgi to the PM, VSVG (green) and TGN46 (red), (i) Transport from the RE to the PM, Tf (red), Rab11 (green), (j) Transport from the PM to the Golgi, CT (red), TGN46 (green), (k) Transport from the PM to the lysosome, EGF (red), Lamp1 (green), (l) Transport from the PM to the EE, dextran (red), EEA1 (green). m,n, Transport assays in HeLa cells, a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (m) CT endocytosis, NS1=1, NS2=0.3205. (n) interleukin-2 receptor beta subunit (IL2R-β) endocytosis, NS1=0.063, NS2=0.9264. o-r, HeLa cells were treated as indicated followed by immunofluorescence microscopy using different antibodies against GAPDH (o,q) or imaging for GFP-tagged GAPDH (p,r), n=2. Image from a representative experiment is shown, bar, 10 um.
ED Fig 3. Additional characterizations on GAPDH and its effects on the transport pathways.
a-d, The distribution of different organelle markers in CHO (a), COS-7 (b), HeLa (c), or MEF (d) cells were assessed by immunofluorescence microscopy, n=2. Image from a representative experiment is shown, bar, 10 um. e-h, Comparing the distribution of two markers against the same intracellular compartment using confocal microscopy, n=2. Image from a representative experiment is shown, bar, 10 um. (e) ER markers, Sec61p (green), calnexin (red). (f) Golgi markers, giantin (green), GM130 (red). (g) EE markers, EEA1 (green), Rab5 (red). (h) Lysosome markers, lamp1 (green), CD63 (red). i-l, Confirming the staining specificity of organelle markers using model cargoes that reside at specific intracellular compartments through confocal microcopy, n=2. Image from a representative experiment is shown, bar, 10 um. (i) GFP-tagged VSVG at the ER (green), Sec61p (red). (j) GFP-tagged VSVG at the Golgi (green), giantin (red), (k) fluorescently labeled Tf (green), EEA (red), (l) fluorescently labeled dextran (green), lamp1 (red). m-n, The GAP assay was performed using ARF6 and ACAP1 (m), or using ARF1 and AGAP1 (n), in the presence of different metabolic enzymes as indicated, n= 3. o-r, Pulldown studies to detect GAPDH binding directly to: ACAP1 (o), AGAP1 (p), Sec23p (q), or different portions of ARFGAP1 as indicated (r), n=3. s-v, HeLa (s,u) or BSC-1 (t,v) cells were treated as indicated, followed by transport assay for Tf endocytosis. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test. (s) NS1=0.4646, NS2=0.6973. (t) *p=1.8E-04. (u) NS=0.8073. (v) *p=3.6E-05. **p=1.7E-06.
ED Fig 4. Different ways of starving cells lead to identical pathways being inhibited, and these inhibitions require GAPDH.
a-h, HeLa cells were incubated in Hank’s medium, and then transport assays were performed. Quantitation of an experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (a) Transport of VSVG from the ER to the Golgi, NS1=0.1944, NS2=0.05. (b) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=2.4E-06, **p=4.2E-10. (c) Transport of VSVG from the Golgi to the PM, *p=1.7E-05, **p=6.7E-08. (d) Transport of Tf from the early endosome (EE) to the PM, *p=2.9E-05, **p=6.9E-09. (e) Transport of CT from the PM to the Golgi, *p=2.4E-03, **p=7.8E-07. (f) Transport of EGF from the PM to the lysosome, *p=1.9E-06, **p=1.5E-02. (g) Transport of dextran from the PM to the EE. *p=7.8E-03, **p=3.4E-07. (h) Transport of EGF from the PM to the EE, NS1=0.1485, NS2=0.6378. i-p, HeLa cells were incubated in medium without glucose, and then transport assays were performed. Quantitation of an experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (i) Transport of VSVG from the ER to the Golgi, NS1=0.6921, NS2=0.5648. (j) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=4.4E-04, **p=6.5E-07. (k) Transport of VSVG from the Golgi to the PM, *p=1.6E-02, **p= 6.7E-05. (l) Transport of Tf from the early endosome (EE) to the PM, *p=1.9E-04, **p=2.8E-09. (m) transport of CT from the PM to the Golgi, *p=7.4E-04, **p=4.2E-05. (n) Transport of EGF from the PM to the lysosome, *p=7.6E-03, **p=3.2E-09. (o) Transport of dextran from the PM to the EE, *p=2.2E-03, **p=1.6E-05. (p) Transport of EGF from the PM to the EE, NS1=0.8648, NS2=0.8946. q-x, HeLa cells were incubated in medium without amino acids, and then transport assays were performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (q) Transport of VSVG from the ER to the Golgi, NS1=0.1419, NS2=0.3379. (r) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=1.9E-05, **p=1.1E-06. (s) Transport of VSVG from the Golgi to the PM, *p=1.1E-03, **p=6.6E-07. (t) Transport of Tf from the early endosome (EE) to the PM, *p=7.2E-05, **p=8.4E-07. (u) transport of CT from the PM to the Golgi, *p=1.1E-05, **p=1.8E-06. (v) Transport of EGF from the PM to the lysosome, *p=1.1E-02, **p=9.3E-08. (w) Transport of dextran from the PM to the EE, *p=7.8E-03, **p=1.2E-08. (x) Transport of EGF from the PM to the EE, NS1=0.9731, NS2=0.8159.
ED Fig 5. Further characterizing the effects of starvation and GAPDH.
a-c, HeLa cells were starved using Hank’s medium (a), medium that lacks glucose (b), or medium that lacks amino acids (c), and then total ATP level was measured, n=3. d, HeLa cells were treated as indicated, followed measurement of lactate production, n=3. e, HeLa cells were treated as indicated, followed measurement of oxygen consumption rate, n=3. f, The relative abundance of various glycolytic enzymes in HeLa cells. Data is derived from a public database (https://pax-db.org/protein). g-n, HEK293 cells were treated as indicated. Transport assays were then performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (g) Transport of VSVG from the ER to the Golgi, NS1=0.6468, NS2=0.2133. (h) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=4.3E-07, **p=1.3E-03. (i) Transport of VSVG from the Golgi to the PM, *p=2.1E-03, **p=1.9E-05. (j) Transport of Tf from the early endosome (EE) to the PM, *p=6.3E-05, **p=4.7E-04. (k) transport of CT from the PM to the Golgi, *p=6.5E-04, **p=1.5E-02. (l) Transport of EGF from the PM to the lysosome, *p=7.8E-07, **p=6.5E-08. (m) Transport of dextran from the PM to the EE, *p=3.6E-06, **p=3.4E-02. (n) Transport of EGF from the PM to the EE, NS1=0.2375, NS2=0.5291. o-v, HEK293 cells were treated as indicated. Transport assays were then performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (o) Transport of VSVG from the ER to the Golgi, NS1=0.6649, NS2=0.84. (p) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=4.6E-03, **p=2.5E-06. (q) Transport of VSVG from the Golgi to the PM, *p=7.1E-05, **p=1.1E-06. (r) Transport of Tf from the early endosome (EE) to the PM, *p=7.2E-06, **p=6.5E-09. (s) transport of CT from the PM to the Golgi, *p=5.6E-03, **p=8.8E-09. (t) Transport of EGF from the PM to the lysosome, *p=1.1E-02, **p=2.9E-08. (u) Transport of dextran from the PM to the EE, *p=2.4E-04, **p=2.4E-06. (v) Transport of EGF from the PM to the EE, NS1=0.7924, NS2=0.4675. w,x, HEK293 cells were treated as indicated, followed by starvation, and then quantitation of total ATP level (w) or cell death (x), n=3, mean with SD are shown.
ED Fig 6. Inhibition of transport pathways by starvation and AMPK.
a-h, HeLa cells were starved using a general starvation medium (lacking glucose and amino acids), and then transport assays were performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (a) Transport of VSVG from the ER to the Golgi, NS1=0.3506, NS2=0.9126. (b) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=8.6E-04, **p=5.8E-07. (c) Transport of VSVG from the Golgi to the PM. *p=1.0E-07, **p=4.4E-09. (d) Transport of Tf from the early endosome (EE) to the PM, *p=4.2E-05, **p=4.4E-10. (e) transport of CT from the PM to the Golgi, *p=2.6E-04, **p=9.5E-07. (f) Transport of EGF from the PM to the lysosome, *p=2.0E-04, **p=2.2E-13. (g) Transport of dextran from the PM to the EE, *p=1.4E-04, **p=1.6E-06. (h) Transport of EGF from the PM to the EE, NS1=0.1085, NS2=0.1408. i, HeLa cells were treated as indicated and then the whole cell lysate was immunoblotted for proteins as indicated, n=2 experiments. j, HeLa cells were starved, and then the COPI transport assay was performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test, *p=1.1E-04, **p=3.5E-06. k, HeLa cells were treated as indicated and then the whole cell lysate was immunoblotted for proteins as indicated, n=2 experiments. l-s, HeLa cells were starved, and then transport assays were performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (l) Transport of VSVG from the ER to the Golgi, NS1=0.6077, NS2=0.5535. (m) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=3.5E-06, **p=3.6E-10. (n) Transport of VSVG from the Golgi to the PM, *p=4.1E-03, **p=4.4E-06. (o) Transport of Tf from the early endosome (EE) to the PM, *p=3.7E-06, **p=1.1E-09. (p) transport of CT from the PM to the Golgi, *p=7.6E-04, **p=8.7E-11. (q) Transport of EGF from the PM to the lysosome, *p=3.1E-08, **p=1.3E-02. (r) Transport of dextran from the PM to the EE, *p=1.1E-06, **p=3.7E-10. (s) Transport of EGF from the PM to the EE, NS1=0.228, NS2=0.1738.
ED Fig 7. Effects of AMPK on GAPDH distribution and transport pathways.
a-e, HeLa cells were treated as indicated, and then examined for the colocalization between GAPDH and different organelle markers. A confocal image from a representative experiment (out of three) is shown, bar, 10 um (left); quantitation is also shown (right), n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (a) Colocalization of GAPDH (red) with giantin (green), *p=2.9E-03, NS=0.6397. (b) Colocalization of GAPDH (red) with TGN46 (green), *p=6.5E-03, NS=0.6413. (c) Colocalization of GAPDH (red) with EEA1 (green), *p=3.5E-03, NS=0.8793. (d) Colocalization of GAPDH (red) with Lamp1 (green), *p=9.4E-04, NS=0.551. (e) Colocalization of GAPDH (red) with Sec61p (green), NS1=0.5361, NS2=0.4243. f-m, HeLa cells were treated as indicated, and then transport assays were performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (f) Transport of VSVG from the ER to the Golgi, NS1=0.5250, NS2=0.8291. (g) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=4.6E-05, **p=2.9E-08. (h) Transport of VSVG from the Golgi to the PM, *p=1.4E-04, **p=6.9E-10. (i) Transport of Tf from the early endosome (EE) to the PM, *p=2.0E-07, **p=1.0E-11. (j) transport of CT from the PM to the Golgi, *p=4.0E-04, **p=3.9E-08. (k) Transport of EGF from the PM to the lysosome, *p=7.1E-04, **p=4.2E-08. (l) Transport of dextran from the PM to the EE, *p=1.4E-07, **p=1.9E-09. (m) Transport of EGF from the PM to the EE, NS1=0.6088, NS2=0.4061. n-u, HeLa cells were treated as indicated, and then transport assays were performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (n) Transport of VSVG from the ER to the Golgi, NS1=0.5559, NS2=0.8576. (o) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=4.1E-05, **p=2.3E-09. (p) Transport of VSVG from the Golgi to the PM, *p=4.2E-03, **p=6.3E-05. (q) Transport of Tf from the early endosome (EE) to the PM, *p=7.2E-04, **p=3.7E-11. (r) transport of CT from the PM to the Golgi, *p=2.3E-04, **p=3.0E-08. (s) Transport of EGF from the PM to the lysosome, *p=2.9E-03, **p=5.8E-06. (t) Transport of dextran from the PM to the EE, *p=2.9E-02, **p=1.6E-08. (u) Transport of EGF from the PM to the EE, NS1=0.6856, NS2=0.9248.
ED Fig 8. Further characterizing how AMPK acts on GAPDH.
a-e, HeLa cells were treated as indicated, and then examined for the colocalization between GAPDH and different organelle markers. A confocal image from a representative experiment (out of three) is shown, bar, 10 um (left); quantitation is also shown (right), n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (a) Colocalization of GAPDH (red) with giantin (green), *p=1.5E-04. (b) Colocalization of GAPDH (red) with TGN46 (green), *p=2.2E-04. (c) Colocalization of GAPDH (red) with EEA1 (green), *p=2.1E-03. (d) Colocalization of GAPDH (red) with Lamp1 (green), *p=7.1E-05. (e) Colocalization of GAPDH (red) with Sec61p (green), NS=0.9933. f, Purity of GAPDH forms assessed by Coomassie gel staining, n=2. g, Purity of AMPK complex assessed by Coomassie gel staining, n=2. h-j, AMPK was incubated with wild-type GAPDH (h), S122A mutant GAPDH (i), or a peptide derived from ACC known as SAMS (j) in the in vitro kinase assay, followed by quantitation of phosphorylation over time, n=3. k, calculation of the stoichiometry of phosphorylation from the results shown in (h-j). l-q, Whole cell lysates, derived from HeLa (l-n) or HEK293 (o-q) cells, were treated as indicated and then immunoblotted for proteins as indicated, n=2.
ED Fig 9. Further characterizing how GAPDH mediates the ability of starvation to inhibit the transport pathways.
a-h, HeLa cells were transfected with the S122A mutant GAPDH, followed by starvation. Transport assays were then performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test done for the following transport assays: (a) Transport of VSVG from the ER to the Golgi, NS1=0.4144, NS2=0.7463 (b) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=3.8E-05, **p=1.9E-09. (c) Transport of VSVG from the Golgi to the PM. *p=3.6E-03, **p=1.4E-06. (d) Transport of Tf from the early endosome (EE) to the PM, *p=1.4E-03, **p=1.4E-05. (e) transport of CT from the PM to the Golgi, *p=7.9E-05, **p=1.5E-07. (f) Transport of EGF from the PM to the lysosome, *p=4.0E-03, **p=1.3E-09. (g) Transport of dextran from the PM to the EE, *p=3.1E-05, **p=1.2E-08. (h) Transport of EGF from the PM to the EE, NS1=0.7549, NS2=0.2279. i-p, HeLa cells were transfected with the S122D mutant GAPDH. Transport assays were then performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (i) Transport of VSVG from the ER to the Golgi, NS=0.8289. (j) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=2.2E-05. (k) Transport of VSVG from the Golgi to the PM, *p=6.3E-04. (l) Transport of Tf from the early endosome (EE) to the PM, *p=6.9E-06. (m) transport of CT from the PM to the Golgi, *p=1.3E-04. (n) Transport of EGF from the PM to the lysosome, *p=1.6E-03. (o) Transport of dextran from the PM to the EE, *p=7.4E-04. (p) Transport of EGF from the PM to the EE, NS=0.5473.
ED Fig 10. Further confirmation that the roles of GAPDH in transport and autophagy are distinct.
a, HeLa cells were transfected with GFP-tagged forms of GAPDH as indicated followed by immunofluorescence microscopy, n=2. Representative image from an experiment is shown, bar, 10 um. b, HeLa cells were transfected with GFP-tagged forms of GAPDH as indicated followed assessment of Sirt1 activity, n=3. c, HeLa cells were transfected with GFP-tagged forms of GAPDH as indicated, and then LC3 puncta formation was quantified. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test, *p=2.3E-05, NS=0.5598. d, HeLa cells were transfected with GFP-tagged forms of GAPDH as indicated, and then LC3 lipidation was assessed by immunoblotting, n=2. e, HeLa cells were treated as indicated, and then LC3 puncta formation was quantified. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test, *p=1.7E-05. f, HeLa cells were treated as indicated, and then LC3 lipidation was assessed by immunoblotting, n=2. g, HeLa cells were treated as indicated, and then p62 level was assessed by immunoblotting, n=2. h-o, HeLa cells were treated as indicated. Transport assays were then performed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test: (h) Transport of VSVG from the ER to the Golgi, NS=0.8155. (i) COPI transport of VSVG-KDELR from the Golgi to the ER, *p=1.4E-05. (j) Transport of VSVG from the Golgi to the PM, *p=5.5E-04. (k) Transport of Tf from the early endosome (EE) to the PM, *p=3.3E-07. (l) transport of CT from the PM to the Golgi, *p=2.0E-03. (m) Transport of EGF from the PM to the lysosome, *p=2.4E-06. (n) Transport of dextran from the PM to the EE, *p=3.9E-05. (o) Transport of EGF from the PM to the EE, NS=0.7725. p,q, HeLa cells were treated as indicated, followed by quantitation of total ATP level (p) or cell death (q), n=3, mean +/− SD is shown. r, HeLa cells were treated as indicated, followed by quantitation of total ATP level, n=3, mean +/− SD is shown. s,t HeLa cells were treated as indicated, and then COPI transport was assessed. Quantitation of a representative experiment (out of three) is shown, n=10 fields of cells examined, mean +/− SD, two-tailed t-test, *p=3.5E-05 (s), NS=0.7907 (t). (u) HeLa cells were treated as indicated, followed by quantitation of total ATP level, n=3, mean +/− SD is shown.
ED Table 1. Proteins that interact with ARFGAP1.
ARFGAP1 as a GST fusion protein was incubated with cytosol for a pulldown experiment, followed by mass spectrometry to identify interacting proteins.
| Abbreviation | Protein Name | MWT(kDa) |
|---|---|---|
| Tln1 | Protein Tln1 | 270 |
| Copa | Coatomer subunit alpha | 138 |
| Ap2b1 | AP-2 complex subunit beta | 105 |
| Ap2a2 | AP-2 complex subunit alpha-2 | 104 |
| Copb2 | Coatomer subunit beta' | 102 |
| Copg1 | Coatomer subunit gamma-1 | 98 |
| Arcn1 | Coatomer subunit delta | 57 |
| Aldh2 | Aldehyde dehydrogenase, mitochondrial | 56 |
| Fh | Fumarate hydratase, mitochondrial | 54 |
| Ctbp1 | C-terminal-binding protein 1 | 47 |
| Cth | Cystathionine gamma-lyase | 44 |
| Akrx7a2 | Aflatoxin B1 aldehyde reductase member 2 | 41 |
| Taldo1 | Transaldolase | 37 |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | 36 |
| Apoe | Apolipoprotein E | 36 |
| Mdh2 | Malate dehydrogenase | 36 |
| Grhpr | Grhpr protein | 36 |
| Eif2s1x | Eukaryotic translation initiation factor 2 subunit 1 | 36 |
| Hadh | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 34 |
| Haao | 3-hydroxyanthranilate 3,4-dioxygenase | 33 |
| Gnmt | Glycine N-methyltransferase | 33 |
| Qprt | Nicotinate-nucleotide pyrophosphorylase | 31 |
| Psma3 | Proteasome subunit alpha type-3 | 28 |
| Psme2 | Proteasome activator complex subunit 2 | 27 |
Supplementary Material
Acknowledgements
This work was supported by grants from the US National Institutes of Health to VWH (GM058615 and GM115683), J. Loscalzo (HL61795, HL119145, HG007690, and GM107618), and WMO (HL128802), and a grant from the American Heart Association to J. Loscalzo (D700382).
Footnotes
Competing Interests The authors declare no competing interests.
REFERENCES
- 1.Singh R & Cuervo AM Autophagy in the cellular energetic balance. Cell Metab 13, 495–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donaldson JG & Jackson CL ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 12, 362–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang JS et al. ARFGAP1 promotes the formation of COPI vesicles, suggesting function as a component of the coat. J Cell Biol 159, 69–78 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SY, Yang JS, Hong W, Premont RT & Hsu VW ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J Cell Biol 168, 281–290 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sirover MA On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta 1810, 741–751 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Yang JS et al. A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance. Nat Cell Biol 10, 1146–1153 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang JS et al. COPI acts in both vesicular and tubular transport. Nat Cell Biol 13, 996–1003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park SY, Yang JS, Schmider AB, Soberman RJ & Hsu VW Coordinated regulation of bidirectional COPI transport at the Golgi by CDC42. Nature 521, 529–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C et al. AMPK-Dependent Phosphorylation of GAPDH Triggers Sirt1 Activation and Is Necessary for Autophagy upon Glucose Starvation. Mol Cell 60, 930–940 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Bai M et al. ARFGAP1 promotes AP-2 dependent endocytosis. Nat Cell Biol 13, 559–567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J et al. An ACAP1-Containing Clathrin Coat Complex for Endocytic Recycling. J Cell Biol 178, 453–464 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nie Z et al. Specific regulation of the adaptor protein complex AP-3 by the Arf GAP AGAP1. Dev Cell 5, 513–521 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Barlowe C et al. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Hardie DG, Ross FA & Hawley SA AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13, 251–262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaffer BE et al. Identification of AMPK Phosphorylation Sites Reveals a Network of Proteins Involved in Cell Invasion and Facilitates Large-Scale Substrate Prediction. Cell Metab 22, 907–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuma A et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Yang JS et al. A role for BARS at the fission step of COPI vesicle formation from Golgi membrane. EMBO J 24, 4133–4143 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoe T et al. The KDEL receptor, ERD2, regulates intracellular traffic by recruiting a GTPase-activating protein for ARF1. Embo J 16, 7305–7316 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai J et al. ACAP1 Promotes Endocytic Recycling by Recognizing Recycling Sorting Signals. Dev Cell 7, 771–776 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Oldham WM, Clish CB, Yang Y & Loscalzo J Hypoxia-Mediated Increases in l-2-hydroxyglutarate Coordinate the Metabolic Response to Reductive Stress. Cell Metab 22, 291–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The following figures have associated raw data: Figures 1a,b, 2a-g, 3a-j, 3n-q, 4c-e, 4g-p; Extended Data Figures 1c, 1k, 2m,n, 2s-v, 3s-v, 4a-x, 5a-c, 5g-x, 6a-h, 6j, 6l-s, 7f-u, 9a-p, 10h-u. For gel source data, see Supplementary Figure 1.