Abstract
Early diagnosis of diseases (before they become advanced and incurable) is essential to reduce morbidity and mortality rates. With the advent of novel technologies in clinical laboratory diagnosis, microbead-based arrays have come to be recognized as an efficient approach, that demonstrates useful advantages over traditional assay methods for multiple disease-related biomarkers. Multiplexed microbead assays provide a robust, rapid, specific, and cost-effective approach for high-throughput and simultaneous screening of many different targets. Biomolecular binding interactions occur after applying a biological sample (such as blood plasma, saliva, cerebrospinal fluid etc.) containing the target analyte(s) to a set of microbeads with different ligand-specificities that have been coded in planar or suspension arrays. The ligand-receptor binding activity is tracked by optical signals generated by means of flow cytometry analysis in the case of suspension arrays, or by image processing devices in the case of planar arrays. In this review paper, we discuss diagnosis of cancer, neurological and infectious diseases by using optically-encoded microbead-based arrays (both multiplexed and single-analyte assays) as a reliable tool for detection and quantification of various analytes.
Keywords: Early diagnosis, Microbead array applications, Multiplexing, Biomarker, Cancer, Infectious disease, Neurological disease
Graphical abstract
1. Introduction
Many life-threatening diseases start out at clinically undetectable levels, and steadily increase in severity with time, until symptoms eventually appear. The sooner that the disease is diagnosed, the more successful therapies will be in curing, treating and reversing the progress of the disease. Early detection of disease has therefore assumed an essential role in modern medical therapy. Surveillance and monitoring of the progression and/or the management of the disease, and individually assessing the response to treatment are becoming the hallmarks of personalized medicine [1,2].
Microbead-based arrays are an emerging technology used for early diagnosis, and in simultaneous detection, quantification and profiling of a range of targets of interest relevant to a particular disease. Preliminary work in this field was carried out as early as 1926 when various particulate materials were used in biological investigations. The first systematic study of the development of well-defined albumin microspheres for diagnostic applications was performed in the late 1960s by Rhodes, Scheffel, Wagner, Zolle and their colleagues [3]. Attempts towards optimizing and developing antibody-based multiplexed assays (and commercial instruments and kits) date back to over 20 years ago [4]. By use of a multiplex detection-based system, scientists can predict the possibility of disease occurrence before the appearance of the first clinical symptoms at the very early stages. Biomarkers may be related to genetic information such as a mutation or change in amino acid positions in a double-stranded DNA or RNA structure, alterations in a complex protein or gene structure, or the appearance of a single specific (or multiple) antigens that correlate with the presence of a disease. This technology also has significant applications in the analysis of protein/gene/DNA profiles, experiments for drug discovery, research, and optimization of clinical laboratory diagnosis [5–9]. Certain particular biomarkers have been shown to be characteristic of many specific disease states, or other physiological disturbances of an organism that can be used as an indicator to diagnosis or predict disease [10–13].
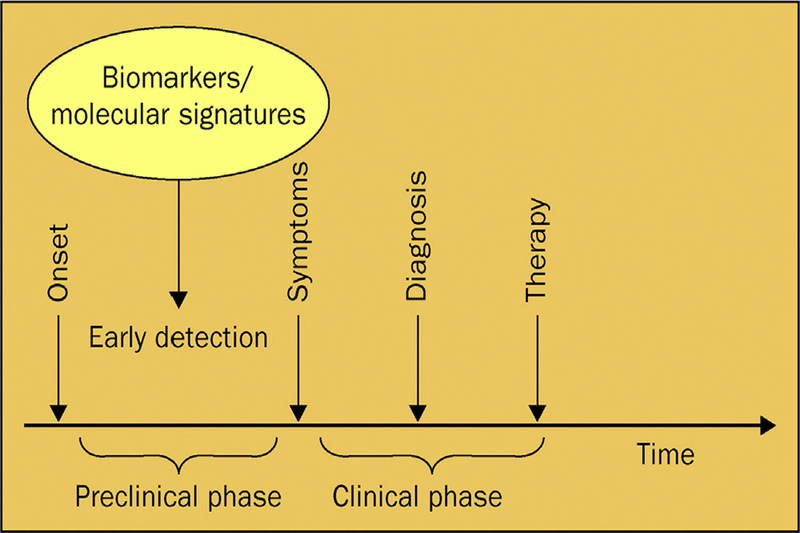
Cancer progression can be broadly divided into two phases (Fig. 1). The first preclinical phase starts at the initial point when enough irreversible mutations in the cells have taken place, until the first noticeable symptoms of disease are detected. The more observable clinical phase comprises the period between symptoms appearing and commencement of therapy. Early detection is defined as taking place in the preclinical stage, and the finding of prognostic or diagnostic biomarkers in this phase may allow effective interventions in order to prevent any symptoms even appearing [3].
Fig. 1. An epidemiological perspective of cancer progression.
Adapted from Ref. [11] With permission.
Microbeads are defined as spherical polymeric particles in the size range from 0.5 to 500 mm diameter. Reactions take place on the surface of these microbeads that function as a solid-support surface to capture analytes (molecular targets in the sample) of interest.
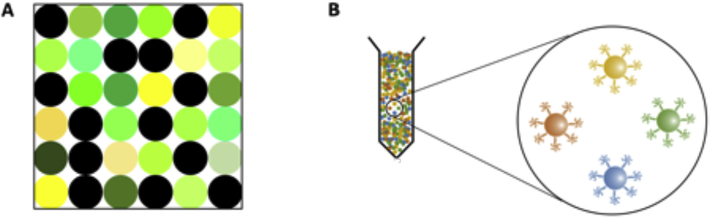
There are two basic different types of microbead-based technologies: solid-state planar bead arrays, and liquid-state suspension bead arrays, which both have extensive multiplexing capability (Fig. 2). In the microbead planar array format (Fig. 2A), microspheres are attached in place at known locations onto a solid surface by various means (such as creating microholes and micro-machined cavities etc.). The solid surfaces can be made of polymers or glass [14–16]. Binding reactions happen in the same way as for suspension arrays, and ultimately there is a two-dimensional array consisting of false or true reacted spots. The identity of each spot is known from its location in the grid [17]. In order to analyze the interactions occurring on the solid support a fluorescent image with a CCD camera is captured. In the next steps, image processing and annotation are carried out by software designated for that imaging system [7,18–20]. The chief benefit of using a planar microarray is the fact that thousands of separate tests can be implemented in parallel, making it suitable for powerful applications in genomics, proteomics, and drug discovery [8]. “MesoScale discovery” (MULTIARRAY/MULTI-SPOT) and FastQuant (FAST Quant System) with an ability to measure 10 analytes, and “Searchlight” which is a 24-plex assay are examples of commercial multiplexed planar format microarrays [21].
Fig. 2.
Schematic representation of A) planar array and B) suspension array.
The multiplexed suspension arrays (Fig. 2B), are composed of a mixture of optically encoded polymeric microspheres each conjugated to a specific antibody or other ligand designed to recognize and capture a different specific analyte.
High-throughput screening of biomarkers in clinical analysis can be achieved by a multiplexing approach, which means simultaneous quantification of many different biomarkers whose interrelated expression levels combined together, may be indicative of an individual disease, or show the presence of one particular type of disease such as cancer, neurological or infectious disease. Multiplexed bioassays increase the sensitivity and provide a more efficient, fast and accurate diagnosis using a single relatively small sample volume [8,22,23].
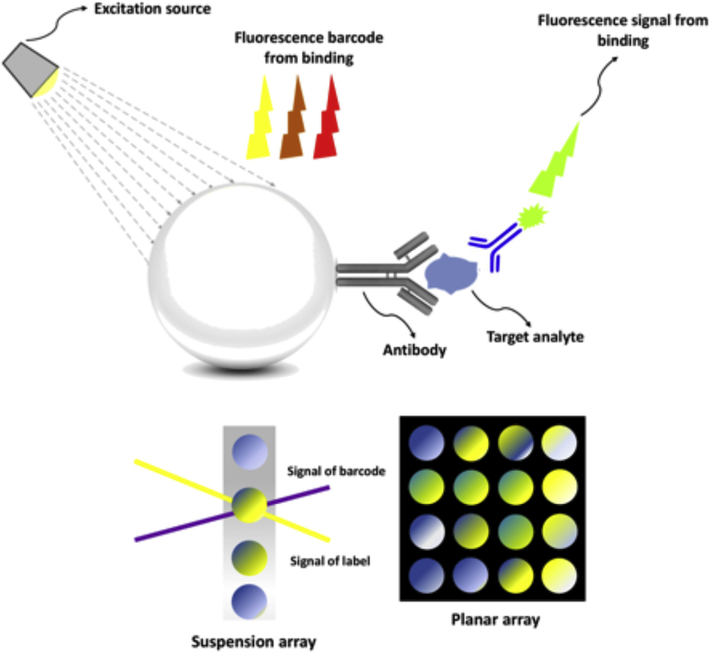
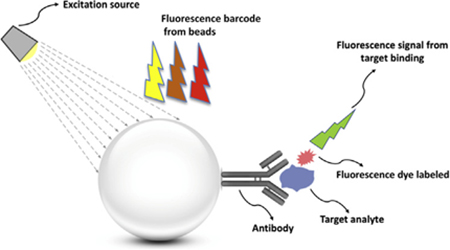
In suspension bead-based assays, suspended beads are mixed with a sample containing different antigens or analytes [17,22,24,25]. The detection principles of suspension bead based assays can be categorized into three subtypes; protein determination (immunoassays), nucleic acid detection, and receptor-ligand assays [26]. Firstly, the microbead-based immunoassay detection principle mainly includes sandwich-type assays and competitive type assays [26]. In a competitive assay, the interaction of unlabeled analyte (usually the antigen in the sample) with the labeled antibody on the surface of microspheres is the goal. A reduction in the binding of the labeled antigen then signifies the presence of the unlabeled form. In this system, the detectable signal declines with an increase in analyte concentration, which creates a high background signal. On the other hand, in non-competitive immuno-assay formats (usually sandwich-type formats), the antigens are captured by bead-conjugated primary antibodies and then sandwiched between the primary antibody and a matching fluorescently labeled secondary detector antibody as is used in a traditional sandwich immunoassay complex [17,27]. This assay type can achieve the highest level of sensitivity because of the use of two matching antibodies (Fig. 3). The amount of labeled secondary antibody bound to the bead-bound antigen is proportional to the amount of antigen present in the original sample. For this reason the sandwich assay is one of the most sensitive and popular techniques in early diagnosis [28]. In addition, microbead assays can be applied for specific nucleic acid sequence detection, in which the analyte sequence hybridizes with oligonucleotide probes coated on the microspheres. This assay makes the identification of single nucleotide polymorphisms (SNPs) or point mutations in the examined sample, feasible [26]. Microparticles can also be a good substrate for fluorescently labeled enzyme tests and for ligand immobilization.
Fig. 3.
Schematic illustration of principle of a bead-based assay.
The molecular interactions that occur on the microspheres can be read out by means of flow cytometry. The application of flow cytometry for screening of microbeads and molecular interactions, started in the 1970s [29]. Flow cytometry provides the ability to measure the fluorescence from a wide range of optically encoded microspheres and from the labeled secondary antibody at high speeds (50 million events per day) and with high sensitivity [7,8,17,30]. In suspension array technology, interactions occur in all dimensions on the microbead surface, hence they show faster binding kinetics, and attain better hybridization rates between capture ligand and target analytes [6,8,31]. Overall, both approaches provide a high degree of multiplexing, good sensitivity and specificity, decreased labor requirements, and low cost. However, suspension bead arrays have higher binding rates, faster decoding speed, more flexibility, better quality, faster binding kinetics and lower analysis time compared to planar bead arrays [6–8,23].
Although this approach is beneficial, it suffers from some limitations which are discussed below. The level of multiplexing in suspension bead based assays is limited by factors such as availability of reagents (antibodies, oligonucleotides, etc.) and the utilized assay chemistries (antibody sandwich assay, hybridization, PCR etc), which can cause significant cross reactivity and loss of measurable response at very high multiplexing levels [29]. From the immunological aspect, the availability of matched antibody pairs that do not cross-react can be limiting [32].
Bead-based optical immunoassays are based on the interaction between an antibody and an antigen, where the bead has an intrinsic fluorescent barcode and the analyte or secondary antibody has a different fluorescent label [33–35]. Although fluorescence is the commonest optical signal, different read-outs such as chemiluminescence [36], electrochemiluminescence [37], and surface-plasmon resonance (SPR) [38] have been used.
The basic concept of multiplexing is to develop microspheres that have optical barcodes on their surface or embedded inside them, so the specific binding ligand that has been attached to each bead can be identified and retrieved. One of the widely used methods for multiplexing is barcoding the microbeads with different amounts and different wavelengths of a set of fluorescent dyes that are introduced into the interior or attached on the surface of the microbeads. Bead-based assays for analytes is an attractive strategy for rapidly determining large numbers of concentrations [21]. Bead based assays are a user-friendly technique; i.e., the measurable parameters can be adjusted by the user. The wide variation in types of analytes increases its applications (nucleic acids, antigens, antibodies, receptors). The assay is repeatable due to the fact that it is flow cytometry based. Analysis of up to 100 reactions or assays per well is feasible by this assay [21]. This outstanding property provides the ability to combine a large number of different assays, like DNA, receptor-ligand, immuno-assay and enzyme to solve a tricky problem in life science research, drug discovery as well as diagnostic areas. However, still there are some limitation. For instance, in [39] the authors reported that the 100-plex Luminex multiplex method was unable to detect many cytokines in a high percentage of patient plasma samples. They reported that interference by biomolecules such as heterophilic antibodies, present in complex sample matrices such as plasma, is the leading cause of the incapability of the multiplex method to distinguish these analytes.
After applying the barcode to a microsphere, attaching the specific binding ligand, a labeling step is usually required to monitor whether the binding event of the analyte has occurred. Direct conjugation of a fluorescent label to the sample itself is the most prevalent method for labeling. Indirect labeling using a labeled secondary antibody in a sandwich format is also used. An important factor, which should be considered is that the overlap in emission spectra between the barcode and the label should be prevented or minimized [8,28,31,40]. Fluorescence is the most useful method for encoding microbeads, and labeling due to its sensitivity ease of detection with readily available laboratory instruments. There are different kinds of fluorescent particles with application in early detection sensors, including organic compounds [40], quantum dots [35,41–45], fluorescent proteins [28,46] etc. Among them, QDs can provide better discrimination and identification of barcodes based on their narrower spectral line width and better photostability respectively [47]. Due to the size and shape-dependent optical properties of QDs, using a diverse mixtures of QDs, and altering the intensity levels of several of them can theoretically produce over 1 million barcodes, which paves the way for achieving highly multiplexed assays [47]. The next sections provide an overview on surveys that have been performed on microbead-based detection systems for early detection of different diseases.
2. Neurological diseases
2.1. Multiple sclerosis
Multiple sclerosis (MS) is a common disease that leads to chronic neurological disability in young adults, it affects approximately 400,000 people in the United States, and worldwide, 2.5 million individuals [48–50]. It is an inflammatory neurodegenerative disorder of the central nervous system (CNS), with four major clinical subtypes. These are: remitting MS (RRMS), secondary progressive MS (SPMS), primary progressive MS (PPMS), and progressive relapsing MS (PRMS). MS results in the formation of multifocal demyelinating lesions in the white matter and also gray matter lesions [51]. Early diagnosis is crucial in MS treatment, since if treatment is not commenced until the MS displays its first clinical symptoms, irreparable damage may occur At present the earliest possible stage that patients can be identified and given treatment is after a first clinical demyelinating event [49].
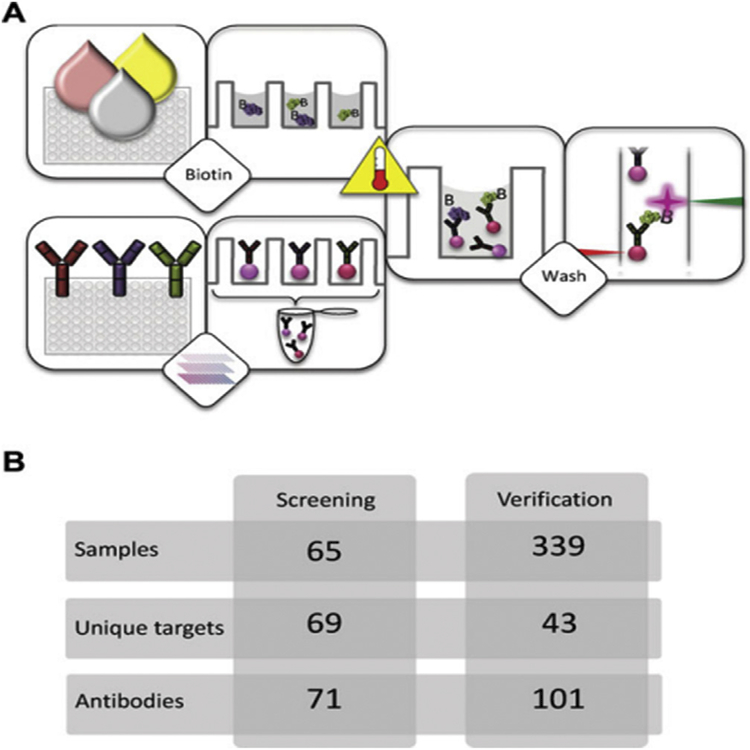
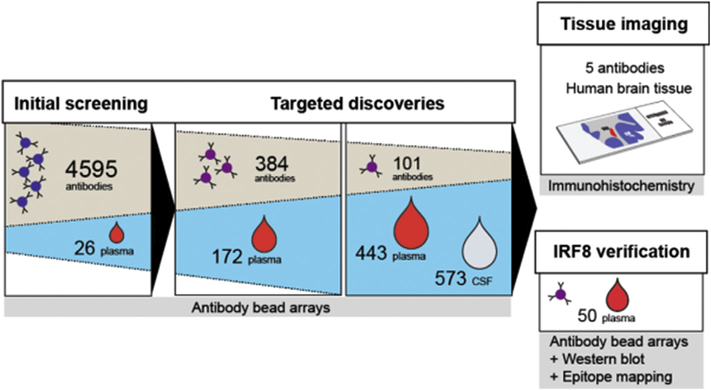
MS clinical detecting is challenging, usually involving repeated neuroimaging over time. The lack of unequivocal early diagnostic tests with associated delays in the commencement of treatment increases risk of significant brain and spinal cord damage. Likewise, the limited number of validated biomarkers that accurately reflect disease activity, complicates treatment of clinically-defined MS and trials of disease-modifying interventions. To address this issue, researchers have applied microbead-based array technologies in MS screening and evaluation. Haggmark € et al. used antibody suspension bead arrays for protein profiling of cerebrospinal fluid (CSF) within the brain in multiple sclerosis patients [52]. To successfully reduce systematic intensity differences between sample groups, they supplemented the labeling reaction with bovine serum albumin (BSA) and Immunoglobulin G (IgG). They also evaluated the effect of heat treatment of CSF proteins on the analysis, and the entire set of 339 CSF samples were heat treated at both 56 C and 72 C. Two proteins, growth associated protein 43 (GAP43) and alpha-1-antichymotrypsin precursor which is coded by SerpinA3 gene were found to have different intensity levels between sample groups. GAP43 was found at lower levels in secondary progressive MS compared to early stages of MS, and lower than the control group of samples from other neurological diseases. SERPINA3 instead was observed at higher levels in all MS patients compared to controls. They also suggested that since SERPINA3 is an acute phase protein that is released early in the course of other diseases, the level of the protein GAP43 in CSF would be more reliable for MS detection. In Fig. 4, the schematic overview of this workflow is shown.
Fig. 4. Overview of workflow and study setup for MS detection in CSF.
(A) The suspension bead array technology enables the analysis of directly labeled samples for multiplex protein profiling. Crude samples are distributed in a randomized fashion in 96-well plates before they are diluted and labeled with biotin. In parallel, antibodies are immobilized on magnetic, color-coded beads and all bead identities are subsequently pooled to create an antibody array in suspension. The labeled samples are heat treated before their incubation with the bead array. After this, unbound proteins are washed away and a streptavidin-conjugated fluorophore added for target detection. In the used instrumentation, one laser is for the identity of beads and the other laser for the fluorophore used for detection of captured, biotinylated target proteins.
(B) In this study, a small set of samples (n ¼ 65) was utilized for assay development and an initial screening with 71 antibodies targeting 69 unique proteins. At a later stage, a subset of 43 targets selected based on results from the screening were additionally analyzed in the set of 339 CSF samples. At this point, multiple antibodies per target were included, resulting in a 101-plex bead array. Adapted from Ref. [52]. With permission. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In another study, Bystrom et al. applied affinity proteomic assays to obtain protein profiles related to MS in body fluids for subsequent tissue analysis [51] (Fig. 5). Starting from an initial screening, follow-up and targeted assays were applied on suspension bead arrays for multiplexed screening of plasma and CSF of MS patients. All antibodies were conjugated to carboxylated color-coded magnetic beads (MagPlex-C, Luminex [53]) arranged in bead arrays in 384-well plates. From these investigations, antibodies targeting interferon regulatory factor 8 (IRF8), interleukin 7 (IL7), solute carrier family 30 member 7 (SLC30A7), methyltransferase like 14 (METTL14), and GAP43 were most indicative of disease state and progression, and were consequently chosen for immunofluorescence analysis of post-mortem brain tissue sections from MS patients.
Fig. 5. Study overview of MS detection in brain tissue.
Application of antibody suspension bead array technology to discover target protein candidate and evaluating them by immunofluorescence analysis of post-mortem brain tissue sections from MS patients. Adapted from Ref. [51] with permission.
2.2. Alzheimer’s disease
Alzheimer’s disease (AD) is a progressive neurodegenerative pathology associated with dementia in humans [54,55] With the aging of the population, the incidence of AD has increased gradually, now It affects more than 35 million people worldwide [56].
AD is characterized by protein-misfolding and deposition (beta-amyloid plaques and tau tangles) in the brain, which result in neuronal cell death, inflammation and brain damage, and is thought to commence years before the neurodegeneration that causes dementia symptoms. Surveys assessing cerebrospinal fluid biomarkers of AD disease have shown that 3 possible biomarkers; Ab42 peptide, total tau (t-tau) and phosphorylated tau (p-tau) have good correlation with pathologic hallmarks. The combination of low CSF Ab42, and high CSF tau have been accurately linked with pathological characteristics of post-mortem AD [57]. Extracellular amyloid plaques and intracellular neurofibrillary tangles are considered as the chief pathological findings in AD. Amyloid b peptides (Ab) occur with the number of amino-acids varying between 36 and 43, and Ab40 and Ab42 are the main constituents of senile plaques. and detecting in body fluids can be employed in predicting the severity and progression at early or preclinical stages of AD [58]. Among all existing biomarkers; Aβ(1–42) is most promising for AD detection, since Ab42 CSF levels fall by about 50% in AD patients relative to healthy individuals of the same age (presumably because the Ab42 is deposited in plaques). The sensitivity and specificity of low Aβ42 levels have been proved to be as high as 80e90% [59,60]. Biomarker measurement not only provides an opportunity for early diagnosis of AD, but also can aid in evaluating treatment effects [61]. Biomarkers can help in studying therapies, since they allow researchers to detect which individuals should take part in new treatment trials, based on the treatment target [62]. While biological markers like magnetic resonance imaging (MRI), positron emission tomography (PET) scans and CSF have paved the way for AD detection, the collection of biomarkers from CSF can be invasive and painful, and expensive imaging systems not be readily available in rural areas, underserved communities, underinsured individuals or developing countries [57,63,64]. Therefore, there is a need to develop a simple and less costly approach to test blood or cerebrospinal fluid, in order to detect and monitor AD biomarkers. Therefore, early diagnosis of cognitive impairment is important for affected people to get treatments and health care services, as soon as possible.
In 2012, Kang et al. developed an xMAP-Luminex multiplex flow cytometric method for early detection of AD biomarkers in cognitively normal individuals to predict the progression to AD dementia in subjects with MCIusing Innogenetics AlzBio3 immunoassay re-agents [65]. The INNO-BIA AlzBio3 is a fluorimetric immunoassay in which the different ligands are absorbed selectively on encoded beads covalently conjugated to a first monoclonal antibody (AT270 for P-tau (181 P), AT120 for tau, 4D7A3 for phosphorylated ta). The assay has the following features: (1) a wide dynamic range of ready-to-use calibrators, (2) time savings for the simultaneous analyses of three biomarkers in one analytical run, (3) reduction of human error, potential of reduced cost of reagents, and (4) a modest reduction of sample volume as compared to conventional enzyme-linked immunosorbent assay (ELISA) methodology [65,66]. Factors that influenced inter-laboratory and intra-laboratory variability were also well analyzed in this paper [65]. Variability in analytical results could be caused by pre-analytical, analytical, and post-analytical factors, and sources of variability attributable to the kit itself.
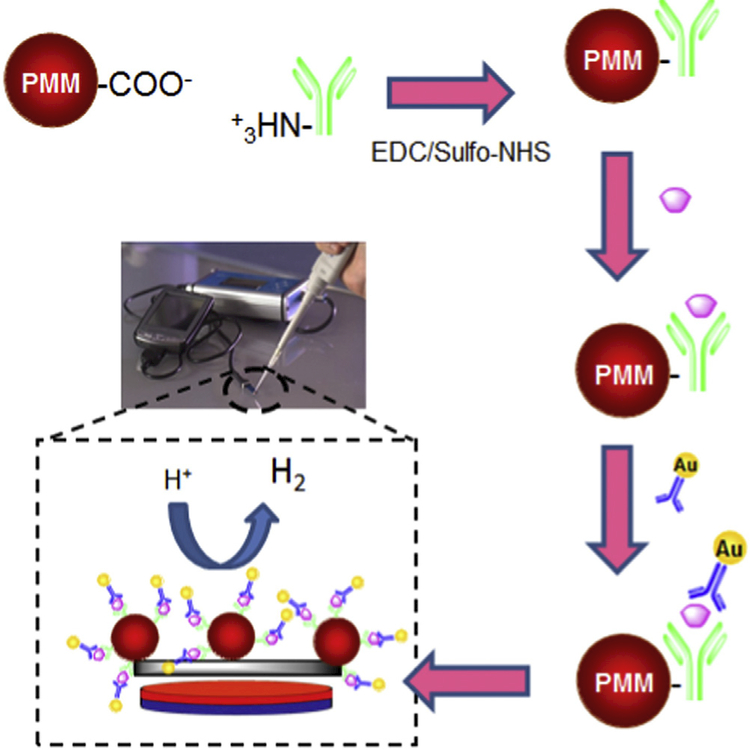
de la Escosura-Muniz~ et al. described a magneto-sandwich immunoassay based on antibody-modified porous magnetic microspheres (PMMs) to detect beta amyloid and ApoE in clinical CSF, serum and plasma samples of patients suffering from AD (Fig. 6) [67].
Fig. 6. Scheme of the magneto-sandwich immunoassay.
PMMs were modified with antibodies, AD biomarker capture, labeling with AuNPs and electrocatalytic recognition based on the hydrogen evolution reaction (HER) on screen-printed carbon electrodes (SPCEs) and a portable potentiostat. Adopted from Ref. [67] with permission.
ApoE4 is a specific isoform of apolipoprotein E, which is an important genetic risk factor for AD as it causes an excess formation of amyloid in the brain. The concentration of ApoE4 in the CSF, plasma and blood of AD patients is in the range of mg.ml−1. Also, the required detection limit of beta amyloid for early detection of AD in patients samples is in the range of pg. ml−1 [67].
Au nanoparticle (AuNP) tags and electrocatalytic hydrogen evolution was also used as detection signals. PMMs were chosen due to their advantageous properties; possession of carboxyl groups and high porosity which allowed and increased antibody immobilization. This immunoassay platform showed high sensitivity with a detection limit of about 7.1 ng ml−1.
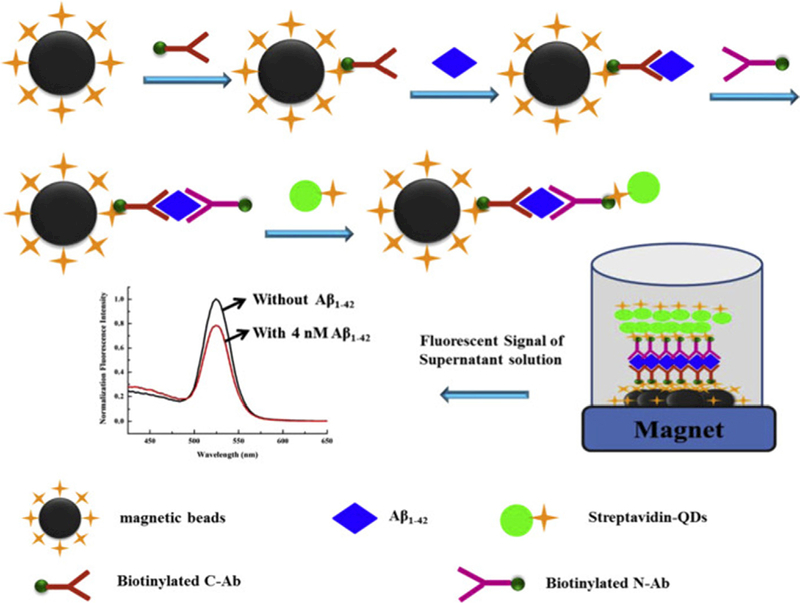
Pi et al. used streptavidin-modified QDs as signal reporters to develop a new sandwich immunoassay based on a suspension magnetic bead array for detection of Ab (1–42) AD biomarker) in human CSF. It has proved that in the early stage of AD, the beta amyloid concentration in human cerebrospinal fluid (CSF) start to drop. The levels of Aβ (1–42) in healthy individuals and AD patients are 0.31 ng ml −1 and 0.16 ng ml −1 respectively [68].
In their assay, in the presence of the analyte Aβ (1–42), strepta- vidin modified QDs bound to magnetic beads through the formation of C-Ab-Aβ1–42-N-Ab sandwich immunocomplexes, which were removable by an external magnetic field. As the Aβ (1–42) concentration increased, more QDs bound to magnetic beads (MBs) so the fluorescence intensity of supernatant decreased gradually as shown in Fig. 7. They also evaluated the fluorescence intensity of MB after magnetic separation [69]. The fluorescence was observed to be very weak due to the quenching effect of MB on the QDs. From the relationship between intensity and added Aβ(1–42) concentrations, the concentration of Aβ(1–42) in the CSF sample was estimated to be about 0.64 nM (3.1 ng ml−1).
Fig. 7. Schematic diagram of sandwich immunoassay to detect Aβ1–42.
Adapted from Ref. [69] With permission.
3. Inflammatory diseases
Inflammation is a defense reaction that an organism mount against harmful stimuli such as tissue injury or infectious agents. The defense mechanisms that protect an organism against infections can be divided into; cellular defenses, and humoral defenses. The correct balance in the inflammatory response is crucial to homeostasis: an insufficient response may result in immunodeficiency, which can lead to infection and cancer; an excessive response might result in morbidity and autoimmune diseases such as rheumatoid arthritis, and Crohn’s disease. Moreover other diseases including atherosclerosis, diabetes, Alzheimer’s disease, multiple sclerosis, and cerebral and myocardial ischemia have been linked to excessive systemic inflammation [70].
Cytokines are small soluble proteins that are released by immune cells. These molecules can modify the behavior and properties of a wide variety of different cell types, and cytokine profiles have been correlated with different parameters of an immune responses. As cytokines have multiple and overlapping functions, the evaluation of the whole set of cytokines secreted in any microenvironment (e.g., a site of inflammation) is more useful than the measurement of a single isolated cytokine, in understanding the complete mechanism of immunological alterations observed in patients suffering, for instance, from inflammatory and autoimmune pathologies [71,72]. In addition, it is essential to have a good measure of the physiological range of cytokines in healthy subjects [56]. In recent years, sandwich microarray-based assays have been widely used in the analysis of human cytokines, for two reasons. Firstly, high affinity antibodies and high specificity antibodies are already available in diagnostic kits based on sandwich ELISAs. Secondly, as cytokines are released from cells, culture supernatants are relatively simple to process compared to cellular lysates [73]. Thus, assays can make use of hundreds of specially prepared magnetic beads, or microspheres labeled with cytokine-specific antibodies. As a proof of concept, Khan et al. compared cytokine detection kits from different manufacturer (multiplex kits from LINCO Research, Bio-Rad Laboratories, R&D Systems, and BioSource International) and showed that the bead arrays (the Luminex 100) system was comparable to ELISA kits, with the benefit of time and cost savings, due to the capability of multi-analyte profiling [74]. The cytokine concentrations obtained from the four kits revealed similar patterns, but the absolute levels were different [74]. De Jager and co-workers successfully monitored the differences between cytokine profiles from activated lymphocyte from healthy individuals and from patients with chronic inflammatory diseases in culture supernatant of human peripheral blood mononuclear cells stimulated with antigen and mitogen by the use of a fluorescent-bead-based detection assay [32]. The early detection of the cytokine profile, could indicate and predict the onset of sepsis after trauma, and may improve management of sepsis by preventing a late or misleading diagnosis [75]. For this, Hsu et al. applied a multiplex sandwich immunoassay based on suspension microarrays to evaluate changes in plasma protein levels, such as cytokines, chemokines, soluble receptors, and matrix metal-loproteinases [76]. As a result, patient samples could be divided into a trauma group and a sepsis group. Four soluble receptors: sFas ligand, intercellular cellular adhesion molecule (sICAM-1), tumor necrosis factor receptor I (sTNF-RI), and tumor necrosis factor receptor II (sTNF-RII) were observed to be significantly higher in patients with sepsis than in those with trauma. Visentin et al. utilized class I and class II anti-human leukocyte antigen (HLA) single-antigen flow beads with sera from four patients with high titers of antibodies to analyze the mechanism of the complement interference phenomenon [77]. Kellar et al. demonstrated that multiplexed fluorescent bead assays were applicable to simultaneously quantify at least eight human cytokines in human serum. The major constraint of multiplexing is the number of beads that can be merged in a single limited volume. To address this issue, they utilized conventional blocking protocols, these assays can be applied with human serum [78]. More recently Chen et al. successfully designed a high-throughput screening system based on bead-based suspension gene-array technology for detection of 20 pathogens associated with acute respiratory tract infections [79]. The lowest concentrations in the range of 100/ml to 104/ml could be readily measured, and detection efficiency was influenced by various experimental parameters, including the amplification efficiency of primers, binding of the probe and its target sequence, hybridization temperature, etc. [79].
4. Viral infectious disease; hepatitis B
Hepatitis B liver infection accounts for 800,000 deaths per year mostly from liver cancer and cirrhosis, and ranks as the 15 t h cause of death worldwide [80]. Hepatitis B represented a breakthrough in the overall understanding of viral infections, their distribution and their consequences. Current diagnostics for HBV encompass standard serological tests followed by HBV DNA quantification by PCR. DNA detection is a crucial part of HBV detection since it determines the need for treatment and is used to monitor patients during antiviral therapy [81].
In one study [47] the authors clinically validated Quantum Dot (QD) barcode technology for diagnosing patients infected with HBV. In summary, the viral DNA was first extracted from patient sera using magnetic microbeads, various regions of the extracted genome were amplified by recombinant polymerase amplification (RPA), amplified products were sensed by a multiplexed QD bar-code assay, and finally fluorescent signals were measured by flow cytometry. They proved that analysis of multiple regions of the viral genome improved clinical sensitivity by an average 31% [47]. Wang et al. used near-infrared (NIR) emitting CdSeTe/CdS/ZnS core/shell/shell QD-encoded microbeads combined with flow cytometry using a single laser for multiplexed detection of HBV (hepatitis B virus) [82]. They conducted a 2-plex hybridization assay for antigen of the hepatitis B virus (HBsAg) and hepatitis B viral protein (HBcAg) and a 3-plex hybridization assay for antibody of the hepatitis B virus (HBsAb), hepatitis B e antibody (HBeAb), and hepatitis B core antibody (HBcAb). The synthesis of zinc-modified CdSeTe-core QDs synthesis is described in the paper. They proved that CdSeTe/CdS/ZnS QD-encoded microbeads were a flexible and reliable platform for multiplexed immunoassay of protein samples. A benefit of this assay is that for all six assays a single 488 nm laser was used, while other systems require two or more lasers [82].
5. Cancer
Cancer is one of the most prevalent fatal diseases in the world, typified by uncontrolled cell growth that invades adjacent tissues and metastasizes to distant organs. Early and accurate detection of cancer significantly decreases the mortality rate and can lead to successful treatment and long-term survival.
Many traditional detection methods such as polymerase chain reaction (PCR), enzyme-linked immunosorbent assay (ELISA), electrophoresis, surface plasmon resonance (SPR), surface enhanced Raman spectroscopy (SERS), microcantilevers, colori-metric assays, electrochemical assays, fluorescence methods, etc., have been developed but they all have limits such as lower accuracy, specificity and sensitivity for clinical diagnostic applications [83,84]. The required limit for detection of cancer biomarkers in patient samples such as serum, is in the range of femtomolar (pg. mL−1) concentrations or lower [85]. As most key cancer biomarkers are present at only very low levels at the early stages of cancer, and not very different from normal levels, achieving low detection limits is vital for early diagnosis of cancer [85]. Also, multiplexing is important for simultaneous and early detection of multiple tumor markers (TMs), accurate diagnosis achieved by quantitative analysis of biomarkers of interest, hence, it is expected to become a significant part of clinical laboratory investigation in the future [8,86].
5.1. Breast and ovarian cancer
Breast cancer is the most common malignancy and the second cause of cancer-related death in American females [87]. The five-year survival for breast cancer depends on tumor stage; 98% of patients whose tumor is detected at very early stages (stages 0 and I) can expect to survive 5 years. Five-year survival for stage II tumors is approximately 85%, stage III 60%, and stage IV only 20%. Totally, breast cancer has an approximate 80% 5-year survival [87].
Hence, early detection allows surgical resection at an early stage when survival is maximal. with a more successful patient response. As breast cancer is a heterogeneous disease a panel of numerous biomarkers could be more useful for early detection of it [88].
Some studies have demonstrated that cytokine profiles may be useful for the diagnosis and monitoring of several types of cancers especially breast and ovarian cancers. Gorelik et al. applied multiplex LabMAP system (Luminex) to identify markers of female reproductive cancer, and distinguish them from benign pelvic tumors. A sandwich immunoassay was used according to the protocol provided by Luminex Corporation for simultaneous analysis of multiple markers. The captured biotinylated antibodies were covalently conjugated to carboxylated polystyrene microbeads and phycoerythrin-conjugated goat anti-mouse Immunogolubin G used as a fluorescent label to determine the binding intensity. Various cytokines (cytokines/chemokines, growth factors and antigens) were explored as a newclass of tumor markers and showed more accurate diagnosis capability in combination with the traditional marker, cancer antigen-125 (CA-125) than using just CA-125 alone. Microbeads were analyzed by Bioplex suspension array [89]. In another study, Dehqanzada et al. used the same procedure for analysis of a panel of serum cytokines in breast cancer patients and also evaluated cancer vaccine trials (before and after vaccination) using Luminex technology. Levels of 22 cytokines consisting of interleukin (IL)-1α, −1β, −2, −4, −5, −6, −7, −8, −10, −12, −13, −15, −17, interferon gamma (IFN-γ), granulocyte -colony stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), tumor necrosis factor (TNF-α), inter-feron gamma-induced protein 10 (IP-10), macrophage inflammatory protein (MIP-1a), chemokine (C-C motif) ligand 5 (RANTES), exotoxin and monocyte chemotactic protein-1 (MCP-1) were measured. Specific differences were found in cytokine levels in sera of breast cancer patients compared to healthy controls and in vaccinated patients. Levels of each cytokine were detected as mean fluorescence intensity (MFI) and analyzed by flow cytometry [90].
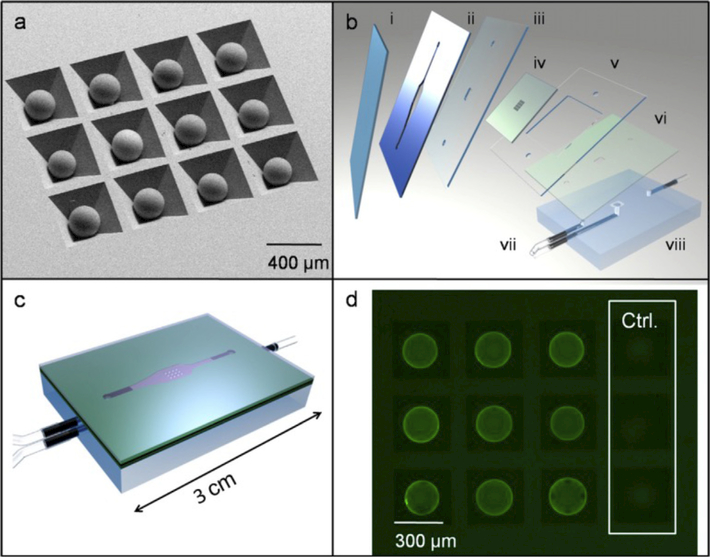
In 2009, Jokerst et al. described a microfluidic biosensor for multiplexed measurement of three important cancer markers, carcinoembryonic antigen (CEA), cancer antigen 125 (CA125), and Her-2/Neu (C-erbB-2) in both serum and saliva samples [91]. These “nano-bio-chips” (NBC) used a sandwich immunoassay; primary antibodies were covalently immobilized on microporous agarose beads to capture antigens; a fluorescence signal was generated after coupling of a QD-labeled antibody to the antigens. The application of QD probes, amplified the signal 30 times relative to that of standard organic fluorophores, and also reduced the limits of detection by nearly 2 orders of magnitude (0.02 ng mL−1 CEA; 0.11pM CEA) in comparison with ELISA [91]. A summary of the work is shown in Fig. 8.
Fig. 8. Schematic diagram of nano-bio chips for cytokines.
(a) SEM photomicrograph of beads in anisotropically etched silicon chip.
(b) Chip (iv) is fitted between double-sided adhesive layer (ii) and cover slip (i) with laminate layers (ii, v, vi) included to direct fluid flow through PMMA base (vii) and inlet and outlet ports (vii).
(c) Sealed LOC assembly.
(d) Fluorescent image of beads after immunoassay including negative controls as imaged with 1 s of CCD camera integration (exposure) time. Adapted from Ref. [91] With permission. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In the majority of studies, human cancer antigens have been used as biomarkers for early diagnosis of breast and ovarian cancer, especially CA 125, a cancer-specific antigen. It can be used in combination with other biomarkers to provide robust and accurate detection of cancer at different stages. Kim et al. developed a multiplexed immunobead-based system for combined detection of early stage ovarian cancer serum markers. The Luminex system was used to analyze several markers on the surface of microbeads in a sandwich immunoassay format. Primary antibodies were linked to the bead surface and binding was visualized by dye-tagged secondary antibodies. Detection antibodies labeled with biotin and streptavidin-R-phycoerythrin to identify antigen-antibody coupling allowed simultaneous determination and quantification of a panel of biomarkers in the liquid phase. The use of additional markers plus CA125 would enhance detection efficiency. Multiple biomarkers including transthyretin and apolipoprotein A1, together with CA125 showed better prediction of ovarian cancer. For precise assessment, serum samples from healthy donors and patients with benign tumors were used as controls, and results demonstrated higher sensitivity and specificity of diagnosis and improved rate of detection when CA125 was used in combination with the two afore mentioned markers compared with single biomarkers [92].
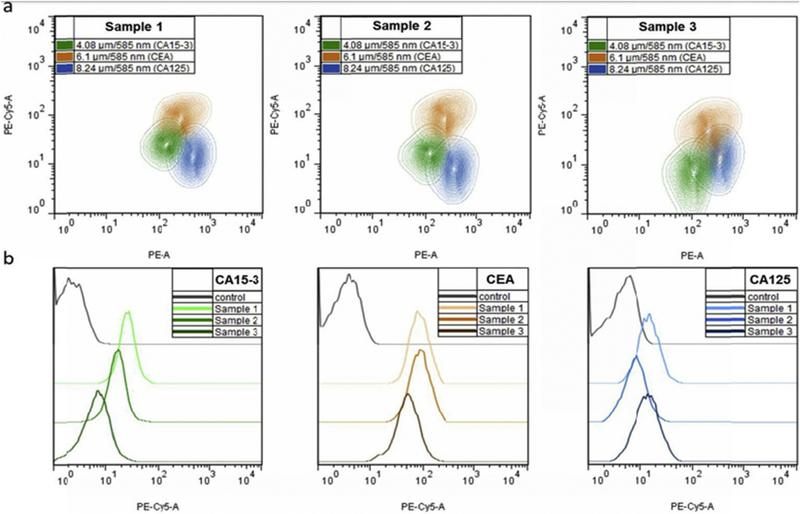
Brazhnik et al. presented a novel bead-based (suspension) array for detection of breast and ovarian human serum tumor markers, each individual antigen coupled to its specific QD-encoded microbead, and detected in serum samples and by flow cytometry. Polymer coated melamine resin microbeads were prepared by layer-by-layer assembly onto the charged surface of polystyrene latex beads, and tagged by water soluble CdSe/ZnS QDs emitting at 585 nm (orange range). Biotinylated monoclonal capture antibodies were conjugated to the surface of QD-tagged microbeads, that were then incubated in patient serum samples. Each bead-bound antibody bound the particular antigen of interest. Binding was detected by biotinylated antibodies and a streptavidin linked fluorophore complex (Tri-COLOR label, 670 nm). Serum samples from healthy donors were compared by ELISA as a “gold standard” to determine the exact amount of antigen levels. Flow cytometry analysis of multiplexed QDebead microarrays used for simultaneous detection of CA 15–3, CE A, and CA 125, markers of female reproductive-system tumors, in clinical serum samples is displayed in Fig. 9a.
Fig. 9. Flow cytometry analysis of multiplexed QDebead microarrays used for simultaneous detection of CA 15e3, CE A, and CA 125, markers of female reproductive system tumors, in clinical serum samples.
(a) Simultaneous detection of three cancer markers in three individual serum samples from patients with different stages of breast cancer.
(b) Comparative histograms indicating different levels of each target marker in three analyzed serum samples in comparison with the control sample. FSC-A, bead size; PE-A bead optical code (QD 585 nm fluorescence); PE Cy5-A, the amount of the cancer marker detected (fluorescence of the streptavidin-Tri-COLOR visualization label). Reprinted from [93] With permission. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Comparison of the results of flow cytometry analysis between serum samples from cancer-positive patients with healthy donors (cancer-negative patients), demonstrated a clear distinction in antigen levels between cancer antigen 15–3 (CA 15–3), carcinoembryonic antigen (CEA), and cancer antigen 125 (CA 125) markers, as shown in Fig. 9b [93].
5.2. Prostate cancer
Prostate cancer ranked as the third most common cancer in men accounting for almost 1 in 5 new diagnoses. The recognition of Prostate Specific Antigen (PSA) as a hallmark of this disease, and the rise in aging male population in many countries, triggered a movement towards early detection and treatment in the United States, followed by Europe [94]. Serum level of PSA is suggested by the American Cancer Society as a potential protein biomarker for early detection of prostate cancer. PSA serum concentration signifying the probability of early stage prostate cancer is 4–10 ng mL−1, while in healthy individuals it is 0.5–2 ng mL−1 [85].
Brazhnik et al. proposed an efficient and comprehensive method for simultaneous monitoring and early diagnosis of several human prostate specific antigens (PSA) by means of a multiplex QD-encoded microbead suspension immunoassay, and proved it to be a reliable, flexible and ultrasensitive approach compared to conventional single-analyte ELISA assay. Green fluorescent QD-tagged microbeads were prepared by a layer-by-layer deposition procedure. The detection system was based on a sandwich immunoassay consisting of capture monoclonal antibodies, biotinylated secondary detection monoclonal antibodies and a Tri-COLOR fluorescent streptavidin complex (Fig. 10a). The microbead complex was interrogated using flow cytometry for simultaneous determination of a panel comprised of different prostate specific antigens (Fig. 10b) [95].
Fig. 10. QD-encoded microbeads for multiplexed cancer diagnosis.
(a) Schematic of the designed lab-on-a-bead system for detection of prostate-specific antigen (PSA).
(b) Flow cytometry dot plots of the suspension array based on two microbead populations carrying different fluorescent nanocrystal codes for simultaneous detection of free PSA and total PSA in clinical serum samples. A green (PE-A-negative, FITC-A-positive) microbead population was used for free PSA detection, and an orange (PE-A-positive, FITC-A-negative) microbead population was used for the total PSA detection Panel A Detection of two PSA forms in the serum of a healthy female donor (the PSA-negative control). Panel B. Detection of two PSA forms in the serum of a prostate cancer-positive male patient (the PSA-positive control). Red fluorescence shifts of different intensities in the PE-Cy5-A channel indicate binding of different amounts of PSA. Reprinted from [95] With permission. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
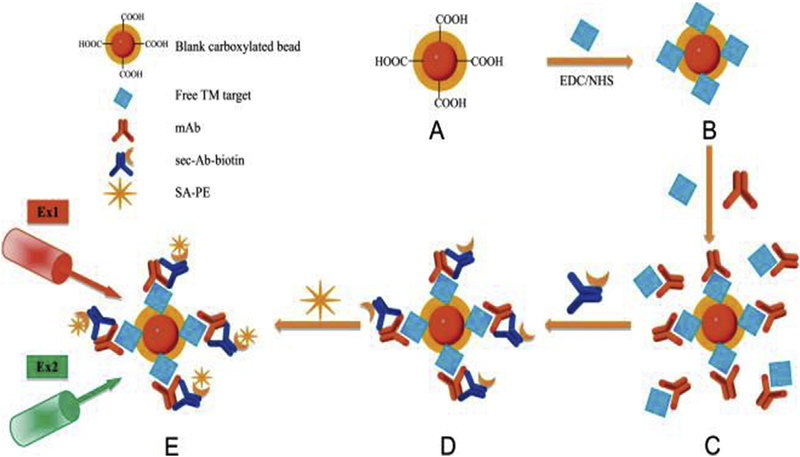
Since PSA is not sufficient on its own for detection and profiling of prostate cancer, it is necessary to choose multiple biomarkers to ensure a simultaneous, fast and more accurate monitoring. Liu et al. developed an optimized suspension array for multiplex quantitative diagnosis of multiple prostate cancer biomarkers including PSA, prostate-specific membrane antigen (PSMA), prostate stem cell antigen (PSCA) and prostatic acid phosphatase [28] in serum samples. This lab-on-a-bead system consisted of competitive immunoassay format. Distinct microbeads labeled with red and orange fluorescent dyes within the beads (to recognize their unique codes) were coated by pre-selected prostate biomarkers that then competed with the free biomarkers in human serum to interact with various dilutions of antigen specific monoclonal antibodies. Binding was visualized by a biotin-conjugated secondary antibody and streptavidin-R-phycoerythrin (SA-PE) as a fluorescent green dye. The suspension array standard curves correlated well with biomarker concentrations. The median fluorescent intensity (MFI) of the bead population was read by xPONENT™ 3.0 software, based on Luminex xMAP® technology. Better results were obtained when most of the carboxyl sites on the microbead surface were saturated by tumor markers. Fig. 11 [96].
Fig. 11. Schematic diagram of the detection procedures for simultaneous and combined detection of multiple tumor markers (TMs) for prostate cancer (PC by suspension array.
(A) The carboxylated blank red and orange fluorescent beads were activated.
(B) The free TM target was coupled onto activated carboxylated beads by EDC/NHS to prepare bead probe set as the beadeTM compound.
(C)The TM on the bead and the free TM were allowed to compete for their corresponding mAb in solutions; then, the beadeTMemAb compound was formed.
(D)Sec-Ab- biotin was added to capture them Ab in solutions and to result in the beadeTMemAbesec-Ab-biotin compound.
(E) Streptavidin-R-phycoerythrin (SA-PE) was added into the solutions to couple with the compound by biotin-avidin interaction. The beadeTMemAbesec-Ab-biotineSA-PE compound was ready for laser beaming. The red laser (Ex1) was used to excite the red and orange fluorescent dyes within the beads to recognize their unique coded numbers. Simultaneously, a green laser (Ex2) was used to excite SA-PE that combined on the bead surface. The available fluorescent signalsdMFIs that only combined on the beads were recorded and analyzed by xPONENT™ 3.0 software. Reprinted from [96] With permission. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
5.3. Lung cancer
According to American Cancer Society, lung cancer accounts for about 25% of all cancer deaths among men and women. The number of people that die of lung cancer is more than other types of cancer such as colon, breast, and prostate. 57% of lung cancers are identified at an advanced stage, due to the fact that lung cancer is often a silent disease in the early stages, while a small minority of cases (16%) are diagnosed at an early local stage [97]. Therefore, microbead array technology could help with lung cancer detection at asymptomatic stages. Ye et al. detected p53, p16, retinoblastoma (Rb), and epidermal growth factor receptor (EGFR) mutations simultaneously by means of suspension arrays for early and rapid diagnosis of lung cancer. Hot-spot mutations in p53, p16, Rb, and EGFR genomic DNA was detected by oligonucleotide specific probes covalently linked onto the bead surface by a carbodiimide coupling method. Bound sequences were hybridized with biotinylated polymerase chain reaction (PCR) amplicons, tagged with SA-PE as a label in a single liquid phase reaction. Genomic DNA extracted from both lung tumor specimens and healthy tissues with PCR amplifications was used to amplify each hot spot mutation region. Coupling reactions were visualized with flow cytometry and analyzed using the Luminex100. The sensitivity and specificity were higher using a panel of four markers than using a single marker. The mutation rate of the panel of four markers was higher indicating its potential in early detection of lung cancer at early stages. Moreover, p16 mutations occurred mostly in non-small cell lung cancer; while the mutation rate of Rb was higher in small cell lung cancer [98].
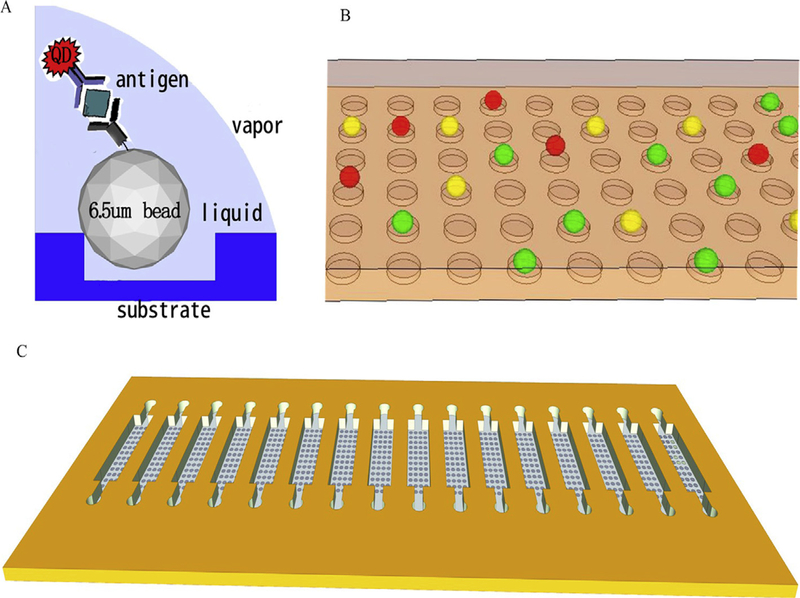
Carcinoembryonic antigen (CEA), fragments of cytokeratin 19 (CYRFA21–1) and neuron-specific enolase (NSE) are the most three common serum biomarkers for lung cancer that have often been used in clinical diagnosis [99]. Liu et al. fabricated a bead-based on-chip assay for multiplex diagnosis of the three afore-mentioned lung cancer biomarkers in human serum using planar and suspension arrays. As illustrated in Fig. 12, the microbead array was constructed by an array of microwells within a PDMS layer using soft lithography technology, each magnetic microbead was physically trapped in microwells and the target biomarkers were detected by sandwich immunoassay linked to their specific antibodies. Each bead carried a unique spectral signal, the antigen-antibody interactions was visualized by three different color QDs used as labeling agents specific for detection of the three individual biomarkers. The advantage of this approach was the separation of beads on a chip in an ordered microarray, avoiding the aggregation of beads and signal interference compared to other microfluidic systems, but it also had limitations on the number of beads that could be used in each microwell because more beads led to reduced binding of antigens and weaker QD signals. Therefore, the number of beads in each microwell and the QD labels were optimized to achieve higher sensitivity. They achieved a low detection limit in the ng/ml range [100].
Fig. 12. Schematic illustrations of bead-based assay system for sensitive detection of three biomarkers using QDs.
(A) Reaction principle for the bead-based sandwich assay;
(B) Three colors of beads after reaction in the chip;
(C) The structure of the chip. Reprinted from [100] With permission. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In another study, Wu et al. developed a fluorescent immuno-assay analysis by simultaneously measuring these three lung cancer biomarkers and visualizing antibody-antigen interactions in a sandwich immunoassay format by multicolor quantum dots (QDs) with different emission spectra as codes for each individual antigen. They utilized magnetic microbeads as array elements in a suspension array and achieved multiplex monitoring of target proteins in a single serum sample. Detection limits were lower (pg/ml range), compared to previous studies (ng/ml range) for all these cancer markers, because of the higher detection sensitivity of suspension arrays compared with planar arrays. Flow cytometry based data analysis is expensive and depends on a fluidic system which limits its wide application in clinical practice, so they developed a procedure to detect biomarkers by applying a micro-image analysis methodology. This software converts images from RGB (red, green, blue) to an HSI (hue, saturation, intensity) color model and measured mean fluorescent intensity produced by QDs which determines target concentration on each bead [101].
5.4. Colorectal cancer
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in both genders according to the American Cancer Society. A recent study on early detection of colorectal cancer was carried out in 2016 b y a group of researchers from Madrid, Spain. Villar-V azquez et al. presented a bead-based bioassay based on Luminex magnetic microbeads for early diagnosis and identification of 16 tumor-associated antigens (TAAs) of CRC and control sera samples in a fast and accurate multiplex analysis. A set of antigens including Transcription factor II B (GTF2B), epidermal growth factor like repeats and discoidin domains 3 (EDIL3), Tyrosine-protein kinase HCK (HCK), Proto-oncogene serine/threonine-protein kinase (PIM1), Serine/Threonine Kinase 4 (STK4), and tumpr protein 53 (p53) were chosen as the optimum combination of TAAs for CRC diagnosis giving high specificity and sensitivity. Since humoral responses emerge several months before clinical symptoms, auto-antibodies against TAAs can be used as specific biomarkers. Due to the fact that autoantibodies may be developed against different TAAs, there is a need to be able to detect hundreds of targets by spectrally distinct magnetic beads simultaneously instead of single-analyte ELISA assays stages [102].
5.5. Melanoma
According to American Cancer Society, skin cancer is by far the most common type of cancer but it is usually not serious. Melanoma accounts for only about 1% of skin cancers, but it is a major cause of a cause of skin cancer deaths. In order to assess multi markers associated with this malignancy, Yurkovetsky and coworkers, investigated several cytokines, chemokines, proangiogenic factors as well as growth factors, and soluble receptors in the sera of patients suffering from melanoma and healthy sera as control samples by multiplex microbead immuno-assay analysis (Luminex). The results demonstrated high sensitivity and specificity [103].
5.6. Non-specific tumor markers
Malignant tumors cause 7.6 million deaths every year, however many deaths can be avoided by early screening and detection. As mentioned above, many specific tumor types have various bio-markers that have been identified as more or less specific for that particular tumor type. However, a major goal of medical research is to answer the long-held question “Can there be a blood test for cancer?” A single tumor marker would not be sufficient to detect cancer because of its limited specificity. As a result, analysis of a complete set of tumor markers could increase the accuracy of disease diagnosis. On the other hand, multi analyte detection can reduce sample and time consumption. Thus, for the simultaneous detection of various molecules multiplex immunoassay of a set of tumor markers has become a highly attractive goal to meet the growing demand for diagnostic application. Multiplex suspension bead arrays have been tested to detect the amount and status of tumor markers within a biological sample [104–107].
In a recent study (2016) concerning multiplexed detection of tumor markers, Leng et al. reported magnetic-fluorescent barcodes based on cadmium-free NIR-emitting QDs, which were combined with flow cytometry using a single laser for 5-plex detection of five tumor markers consisting of alpha-fetoprotein (AFP), carcinoembryonic antigen (CEA), cancer antigen 19e9 (CA199), cancer antigen 125 (CA125), and cancer antigen 242 (CA242) [108]. Bifunctional barcodes were made by coupling cadmium-free NIR-emitting CuInS2/ZnS QDs with superparamagnetic Fe3O4 nano-particles in microspheres through SPG (Shirasu Porous Glass) membrane emulsification technique, and cadmium-free NIR CuInS2/ZnS were applied to reduce fluorescence quenching and lower intrinsic toxicity [108]. They claimed that the magnetic separation-based multiplex assay displayed enhanced sensitivity (about twofold) for all the tumor markers in comparison with the non-magnetic multiplex assay, which may have been linked to reduced non-specific biofouling.
A novel suspension array based on fluorescence-encoded polystyrene beads (FFPBs) with a coreeshell structure, was introduced by Long et al. for simultaneous detection of the tumor markers AFP, carcinoembryonic antigen (CEA), and prostate specific antigen (PSA) which associated with hepatocellular cancer, colorectal cancer, and prostate cancer. The suspension immunoassay was carried in a sandwich format where antibodies were immobilized on the FFPB surface and then clinical sera containing the antigen proteins were added to the beads. fluorescence-encoded polystyrene beads (FFPBs) had the following advantages: (1) The FFPBs provided an adequate amount of carboxyl functional groups for efficient coupling of capture probes; (2) the quantity of the impregnated fluorophore had a negligible impact on the coupling efficiency; and (3) the FFPBs exhibited precise fabrication ability and good detection reproducibility. The results of applying the proposed suspension array to 46 clinical serum samples demonstrated close correlation with the reference electrochemiliminescence immunoassay method [107]. In a study [105] the authors investigated a multiplex suspension bead assay based on Luminex (Austin,TX, USA) for detection of tumor markers. They simultaneously determined the concentration of nine tumor markers in 1114 human serum samples (546 patients with tumors, 158 patients with non-tumor inflammatory diseases, and 410 normal controls). The nine TMs were a-fetoprotein (AFP), carcinoembryonic antigen (CEA), cancer antigen 125 (CA125), cytokeratin 19 fragment (CYFRA 21e1), cancer antigen 424 (CA242), free-prostate specific antigen (f-PSA), total-prostate specific antigen (t-PSA), neuron-specific enolase (NSE) and b-human chorionic gonadotropin (free b-hCG). They compared multiplex suspension bead assays with conventional assays used in clinical laboratories to see if the results from suspension bead arrays were comparable to conventional methodologies. The serum samples were tested with both methods. Nine tumor markers were tested simultaneously with the Luminex assay and individually with conventional methods. As shown in Table 1, the Luminex assay and the conventional methods displayed the same levels of sensitivity.
Table 1.
Accuracy rate in tumor prediction by the detection of tumor markers.
| TM | Method | TP/+a | TN/-b | Sensitivity (%) | Specificity (%) | Youden’s indexc | Conformityd | Accuracy rate (%) |
|---|---|---|---|---|---|---|---|---|
| AFP (liver cancer) | Luminex-100 | 36/40 | 313/324 | 90.00 | 96.60 | 0.87 | 0.86 | 95.88 |
| Compared method | 36/40 | 311/324 | 90.00 | 95.99 | 0.86 | 95.33 | ||
| CA125 (ovarian cancer) | Luminex-100 | 26/30 | 309/324 | 86.67 | 95.37 | 0.82 | 0.82 | 94.83 |
| Compared method | 26/30 | 311/324 | 96.67 | 95.99 | 0.93 | 96.05 | ||
| CYFRA21–1 (lung cancer) | Luminex-100 | 27/39 | 317/324 | 69.23 | 97.84 | 0.67 | 0.61 | 94.77 |
| Compared method | 27/39 | 296/324 | 69.23 | 91.36 | 0.61 | 88.98 | ||
| CA242 (Pancreatic, gastric, rectum cancer and cholangiocarcinoma) | Luminex-100 | 36/41 | 246/324 | 87.80 | 75.93 | 0.64 | 0.61 | 77.26 |
| Compared method | 37/41 | 236/324 | 90.24 | 72.84 | 0.63 | 74.79 | ||
| Free β-Hcg (ovarian cancer) | Luminex-100 | 4/30 | 322/324 | 13.33 | 99.38 | 0.13 | 0.13 | 92.09 |
| Compared method | 4/30 | 324/324 | 13.33 | 100.00 | 0.13 | 92.66 | ||
| NSE (lung cancer) | Luminex-100 | 4/39 | 319/324 | 10.36 | 98.46 | 0.09 | 0.08 | 88.98 |
| Compared method | 6/39 | 317/324 | 15.38 | 97.84 | 0.13 | 88.98 | ||
| Free PSA (prostate cancer) | Luminex-100 | 5/10 | 316/324 | 50.00 | 97.53 | 0.48 | 0.48 | 96.11 |
| Compared method | 8/10 | 317/324 | 80.00 | 97.84 | 0.78 | 97.31 | ||
| Total PSA (prostate cancer) | Luminex-100 | 7/10 | 333/324 | 70.00 | 87.96 | 0.58 | 0.57 | 87.43 |
| Compared method | 8/10 | 330/324 | 80.00 | 87.35 | 0.67 | 87.13 |
TP: true positive. TP/+: true positive/tumor patients.
TN: true negative. TN/−: true negative/normal subjects.
Youden’s index is a single statistic that captures the performance of a diagnostic test. It is equal to the risk difference for a dichotomous test. Youden’s index = Sensitivity + Specificity−1. Youden’s index has minimum and maximum values of −1 and + 1, respectively, with a value of +1 representing the optimal value for analgorithm.
Conformity index is in analogy to Youden’s index and describes the accordance between 9TM and the comparison test: (TP with both assays/+) +(TN with both assays/−) −1. reprinted from [105] With permission.
They reported that the Luminex system not only offered all of the benefits of conventional methods, but also facilitated evaluation of multiple markers with the advantages of automation, increased throughput, flexibility, and reduced sample usage. Thus, the Luminex-100 technology is a promising method for clinical diagnosis [105]. Microsphere-based flow cytometry assays (MFCA) established by Luminex involves a capture antibody coupled covalently to internally dyed polystyrene microspheres. The fluorophores which are red and orange can identify presence of tumor markers, a reporter molecule labeled with a fluorescent marker binds to analyte captured on the bead for detection [106].
Sun et al. reported on a microsphere-based flow cytometric assay (MFCA) to quantify tumor markers. MFCA were set up as a sandwich immunoassay and each microsphere represented an assay by itself which decreased the risk of cross recognition and increased specificity. They chose 5 common abdominal tumor markers in serum samples: human AFP, CEA, CA19–9, CA24–2, and cancer antigen 72e4 (CA72–4). Table 2 highlights the comparison between reproducibility, upper and lower detection limits, simultaneous multi-analyte detection, sensitivity and accuracy of 5 tumor markers. The results showed that their multiplexed assay was reproducible, sensitive, accurate and compared well with ELISA as a standard assay [106].
Table 2. Working detection ranges of standard curves for 5 tumor markers by MFCA and ELISA.
(E: ELISA; Mm: MFCA multiplexed; Ms: MFCA single. 50% value was the halfway point between the lower and upper ranges).
| Ms | Lower Mm | E | Ms | 50% Mm | E | Ms | Upper Mm | E | Ms | Total Mm | E | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFP | 0.016 | 0.016 | 0.004 | 0.30 | 0.36 | 0.02 | 16 | 16 | 4 | 1024× | 1024× | 256× |
| CEA | 0.004 | 0.004 | 0.004 | 0.16 | 0.20 | 0.02 | 64 | 64 | 4 | 4096× | 4096× | 256× |
| CA19–9 | 0.063 | 0.063 | 0.016 | 2.1 | 2.0 | 0.22 | 64 | 64 | 4 | 1024× | 1024× | 256× |
| CA24–2 | 0.016 | 0.016 | 0.001 | 0.46 | 0.40 | 0.009 | 64 | 64 | 0.25 | 1024× | 1024× | 256× |
| CA72–4 | 0.004 | 0.004 | 0.001 | 0.22 | 0.16 | 0.055 | 64 | 64 | 4 | 4096× | 4096× | 256× |
A more recent study was conducted by Zhao and co-workers in 2010. They designed a photonic suspension array based on silica colloidal crystal beads (SCCBs) using a sandwich format for multiplex detection of tumor makers. The aim of this study was evaluation of tumor markers such as AFP, CEA, CA 125 and CA 19e9 which are used in diagnosis of hepatocellular cancer, yolk-sac cancer, colorectal cancer, gastric cancer, and lung cancer. SCCBs were formed by the assembly of monodisperse colloidal nano-particles and displayed ordered hexagonal symmetry. As a result, the SCCBs provided a higher surface area which led to more dye molecules participating in photon absorption, more space for biomolecule immobilization and more rapid immunoreactions. These properties provided high sensitivity, acceptable resolution, good detection reproducibility, and the results were in acceptable agreement with those from parallel single-analyte tests of clinical serum samples [104].
6. The present and future outlook for microbead arrays
Microbead array technology promises a wide range of applications in medical, environmental, and industrial analysis due to its versatility. In regard to medical applications, multiplex microbead arrays can be utilized in biosensors and immunoassay diagnostic and screening applications. Moreover, there are applications in cell separation, study of phagocytosis and blood flow, study of cell motility, hemoperfusion and extracorporeal therapy, targeted drug delivery, and much more besides [109]. In particular the diagnosis of multi-causal/multi-gene diseases, allowing comprehensive disease management, and investigating complex cellular functions are all possible with this methodology [4]. Table 3 compares the merits and demerits of traditional and conventional methods used for biomarker validation with microbead array technology and the chief characteristics of antibody-based array systems are presented in Table 4. Nevertheless, until now most commercial multiple assays have only been employed in research laboratories and nonclinical testing such as The Zeus Scientific, Inc. AtheNA Multi-Lyte® EBV IgG system which obtained premarket FDA clearance in 2012 (Error! Hyperlink reference not valid.www.zeusscientific.com/products/athena-multi-lyte/athena-ebv-igg-plus-test-system2). Only a limited number of these assays have been approved by FDA for clinical testing such as the EraGen Biosciences Inc MultiCode®-RTx HSV 1&2 Kit (http://www.phenomenelleangels.com/images/HSV-clearance-pressrelease.pdf) and the Immucor PreciseType™ Human Erythrocyte Antigen Molecular Bead Chip test [86]. Some of the FDA approved systems largely consisting of lateral flow planar protein multiplex arrays and immunoassays used for point-of care diagnosis are covered in the following section.
Table 3.
A comparison between advantages and disadvantages of the conventional methods used for antibody validation technology.
| Detection method | Advantages | Disadvantages | Ref |
|---|---|---|---|
| ELISA | Specificity, sensitivity, sample does not need purification prior to analysis; availability; no radiation hazards; long shelf life | Can measure only one analyte at a time; time-consuming; requiring trained personnel and some specific equipment; requiring relatively large sample volumes; costly | [30,115] |
| Western blotting (Immunoblotting) | Sensitivity (detect as little as 0.1 ng of protein in a sample); Specificity | Non-specific bond (due to high antibody concentration); Inaccuracy (generating false-positive and false-negative results; High costs and technical demand | [73,96,116] |
| Microarray chip-based screening (planar array) | Printing tens of thousands of features on a single microscope slide; High throughput; and low cost per assay; required small amount (5 μl) of sample | Relatively poor productivity and detection sensitivity; limited binding rates; lower decoding speed in compared with suspension array; and overall low flexibility | [5,8] |
| Bead-based microarrays (suspension arrays) | Multiplexing capabilities; Faster binding kinetics; lower sample consumption; reproducibility; specificity and sensitivity; Flexibility in target selection; fast and robust analysis; High-throughput; Easy fabrication and cost –effectiveness | Limitation related to some antibodies cross-reacting with serum proteins; rely on recognized available antibodies; Matrix Effecta | [8,117] |
| Immunohistochemistry (IHC) | in situ detection of antigens in fixed tissues; convenience of sample submission; safe handling of potential human pathogens; retrospective studies of stored specimens; rapidity; and the detection of nonviable organisms | Difficult standardization (antigen retrieval); challenging quantitation; the need for well-trained personnel (for standardization and interpretation); success depends on antibody (mono- vs. polyclonal) | [118,119] |
| Electrochemiluminescence immunoassay | Sensitivity; selectivity; short incubation times; decreased dilutions and repeats; versatility | Toxic; volatile; and requires high concentrations of sample (usually up to 100 mM) | [120,121] |
| Sandwich based assayb | does not require the proteins to be labeled; simple sample preparation; vastly increases throughput; high specificity | two noncompeting affinity reagents are required for each protein. | [73] |
| Antigen capture methodc | Useful for discovery rather than quantitative analysis | labeling of antigenic epitopes of proteins that tend to lose their affinity; non-specific bond formation | [73] |
Matrix Effect is caused by components except analyte of interest which affects the accuracy which require multiple sample dilutions to decrease matrix effect.
In a sandwich immunoassay, capture antibodies are immobilized on the solid support, and bound proteins are detected using a second, labeled detection antibody.
In this assay, proteins are similarly captured by immobilized antibodies, but the captured proteins are detected directly. This is usually accomplished by chemically labeling the complex mixture of antigens before applying them to the array. In the two-color version of this assay, two samples are labeled independently with distinguishable fluorophores, and the samples are mixed before applying them to the array.
Table 4.
Main characteristics of antibody-based array systems. Adapted from Ref. [4] With permission.
| Characteristics | Antibody-based |
|---|---|
| Principle | Antibody-antigen interaction |
| Required reagents/information | antibody pairs |
| Quantification | Flow cytometry and others |
| Multiplexicity | 1–100 |
| Clinical diagnostic test | Yes |
| Sample enrichment | No |
| Sensitivity | >sub ng/mL–pg/mL |
| Reproducibility | Excellent |
| Assay development | Time and resource demanding |
| Consumable | High demand |
| Robustness | Yes |
| Throughput | High |
| Matrix effect | Yes |
| Sample manipulation | No |
| Automation | Yes |
-
(1)
Triage® Cardio ProfilER® 4-plex system uses a portable lateral flow platform to measure troponin-I, creatine kinase-MB, myoglobin, and brain natriuretic peptide (BNP) to allow further evaluation of chest pain [21].
-
(2)
The BioPlex®@ 2200 antinuclear antibodies (ANA) screen was FDA cleared on June 19, 2012, and permitted to proceed to the market. BioPlex® and its supported software are intended for the qualitative screening of specific antinuclear antibodies (ANA), the quantitative detection of antibodies to double standard DNA (dsDNA), and the semi-quantitative detection of ten (10) separate antibody assays in human serum and/or ethylenediaminetetraacetic acid (EDTA) treated or heparinized plasma (Error! Hyperlink reference not valid.www.accessdata.fda.gov/cdrh_docs/pdf11/k113610.pdf).
-
(3)
Bead Block™ microspheres are used for the embolization of hypervascular tumors, including uterine fibroids and arteriovenous malformations (AVMs). Their safety and effectiveness were confirmed by non-clinical, animal testing on March 7, 2016 (https://www.fda.gov/cdrh/510k/K094018.pdf).
At present suspension immunoassays are the predominant technology for FDA-cleared multiplexed protein detection and quantitation, for analyzing antigens or antibodies in the serum of patients with allergies, autoimmune or infectious diseases in clinical laboratories [4]. Clinical applications of this methodology require establishment of internationally accepted calibration standards, performance criteria, and quality control programs. To the best of our knowledge, this approach has not yet been widely applied to early diagnosis. The most important limitation in microbead-based multiplex assays lies in the need to obtain a large collection of specific antibodies for a wide range of analytes. Despite the availability of many monoclonal and polyclonal antibodies, standardization and calibration of several hundreds of these reagents is not practicable to produce a reliable assay in a multiplex format while, meeting the need for sensitivity and specificity. Similarly, production of a large set of proteins for protein micro-array technology remains a challenging task. To tackle this problem for antibody microarrays, several studies have been carried out to substitute costly antibody products [110,111]; with the introduction of aptamers from nucleic acid libraries to function as ligands for the specific detection of proteins [111–113]. Krishnan et al. suggested that future studies in this methodology should aim at developing four major areas; the overall technology, improvement in antibodies, more rapid assays, and automation [30]. Within the next few years, bead chemistry, instrumentation, and software are destined to undergo some important advances, as many companies have their own computational platforms like Luminex. Antibodies are critical ingredients of protein microbeads arrays, since the entire results depend on them. Considering antibodies may cross-react with serum proteins and immunoglobulins, Krishnan et al. argued that we should reduce this dependency on antibodies by considering alternatives approaches such as aptamers [30]. The third area that should be improved, is reducing the assay time. This can be achieved by omitting the washing step, ‘no wash’ multiplex assays can be performed in 1–2 h but with lower sensitivity. These authors [30] also predicted that the next decade is likely to witness a considerable increase in the number of diagnostic clinical laboratories. In line with modern trends, these will likely be largely automated, since all steps of the multiplex microbead assay system, from sample preparation to instrument operation, are capable of being carried out automatically. Bio-Rad has developed a multiplex instrument to enable high-throughput analysis of clinical samples. Additionally, it is expected that within the next few years, substantial number of new markers, proteins, and RNAs that are specifically related to the both normal physiological state and different disease states will emerge. These advances can hopefully pave the way for advancement of bead-based multiplexed assays. However, microbead diagnostic technology continues to advance at a swift pace with the state-of-the art researching possessing limits of detection down to the zeptomole. Microbeads with the unique ability to multiplex in a simple mode and competitive sensitivity, are becoming a versatile platform that could challenge the gold standard diagnostic methods still used today [114].
7. Conclusion
Accurate and real-time medical diagnosis can greatly improve the treatment efficacy and survival rate. Tracking the specific bio-markers, which are indicative of disease initiation and progression, is a mean to reach this end; early diagnosis. Over several years, microbeads have become a potential substrate for probe biomolecule immobilization due to their inherent high surface-to-volume ratio, sensitivity, simplicity, speed, multiplexing capability and cost effectiveness. This article, reviews a range of previous applications that utilized microbead-based array in the detection and early detection of a variety of serious diseases: Neurological disease; Multiple Sclerosis, Alzheimer’s disease; Inflammatory disease; Viral infectious disease; Cancers; Breast and Ovarian cancer, Prostate cancer, Lung cancer, Melanoma and Non-specific tumor markers. In addition, a comparison between the proposed methodology with other conventional detecting methods and issues regarding microbeads-based detection systems are discussed in the last section. Microsphere synthesis, biomarker selection and conjugation and the optical features optimization are the most pivotal researches in this field. Future surveys on the present topic are therefore required with the intention of meeting the needs of clinical detection and prospective in the growing commercialization of detection kits for all disease.
highlights.
Multiplexed immunoassays are a step forward in early medical diagnosis.
Up to 100 fluorescent barcodes in the microbeads can be distinguished.
Both suspension arrays and planar arrays can be used.
Cancer diagnosis using tumor markers in biosamples such as serum.
Neurological, inflammatory and infectious diseases are also candidates.
Acknowledgments
Michael R Hamblin was supported by US NIH grants R01AI050875 and R21AI121700.
Biography

Michael R Hamblin Ph.D. is a Principal Investigator at Wellman Center for Photomedicine, Massachusetts General Hospital, and an Associate Professor Harvard Medical School. He has interests in photodynamic therapy and photobiomodulation. He has published 386 peer-reviewed articles, is Editor/Associate Editor for 10 journals and serves on NIH Study-Sections. He has an h-factor 83 and > 27,000 citations. He has authored/edited 23 textbooks on PDT and photomedicine including SPIE proceedings. Dr Hamblin was elected as a Fellow of SPIE in 2011, received 1st Endre Mester Lifetime Achievement Award Photomedicine from NAALT in 2017, and Outstanding Career Award from Dose Response Society in 2018.
References
- [1].Dudley JT, et al. , Personalized medicine: from genotypes, molecular phenotypes and the quantified self, towards improved medicine, Pac Symp Biocomput (2015) 342–346. [PMC free article] [PubMed] [Google Scholar]
- [2].Chen R, Snyder M, Systems biology: personalized medicine for the future? Curr. Opin. Pharmacol 12 (5) (2012) 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Srinivas PR, Kramer BS, Srivastava S, Trends in biomarker research for cancer detection, Lancet Oncol 2 (11) (2001) 698–704. [DOI] [PubMed] [Google Scholar]
- [4].Yigitbası T, Abuelzein E, Multiplex immunoassay and bead based multiplex. Trends in immunolabelled and related techniques, InTech, Rijeka (2012) 351–360. [Google Scholar]
- [5].Fritzler MJ, Fritzler ML, Microbead-based technologies in diagnostic auto-antibody detection, Expert Opinion on Medical Diagnostics 3 (1) (2009) 81e89. [DOI] [PubMed] [Google Scholar]
- [6].Rödiger S, et al. , Nucleic acid detection based on the use of microbeads: a review, Microchimica Acta 181 (11e12) (2014) 1151–1168. [Google Scholar]
- [7].Nolan JP, Sklar LA, Suspension array technology: evolution of the flat-array paradigm, Trends Biotechnol. 20 (1) (2002) 9–12. [DOI] [PubMed] [Google Scholar]
- [8].Leng Y, et al. , Suspension arrays based on nanoparticle-encoded micro-spheres for high-throughput multiplexed detection, Chem. Soc. Rev 44 (15) (2015) 5552–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fraser LA, et al. , Oligonucleotide functionalised microbeads: indispensable tools for high-throughput aptamer selection, Molecules 20 (12) (2015) 21298–21312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Golubnitschaja O, Flammer J, What are the biomarkers for glaucoma? Surv. Ophthalmol 52 (6) (2007) S155–S161. [DOI] [PubMed] [Google Scholar]
- [11].Barber RC, Biomarkers for early detection of Alzheimer disease, J. Am. Osteopath. Assoc 110 (9) (2010) S10. [PubMed] [Google Scholar]
- [12].Pepe MS, et al. , Phases of biomarker development for early detection of cancer, J. Natl. Cancer Inst. (Bethesda) 93 (14) (2001) 1054–1061. [DOI] [PubMed] [Google Scholar]
- [13].Luo X, Davis JJ, Electrical biosensors and the label free detection of protein disease biomarkers, Chem. Soc. Rev 42 (13) (2013) 5944–5962. [DOI] [PubMed] [Google Scholar]
- [14].Thompson JA, Microbead-based Biosensing in Microfluidic Devices, 2011.
- [15].Vostrý M, Multiplex immunoassays: CHIPS and beads, Electronic Journal of the IFCC 20 (4) (2010) 162. [PMC free article] [PubMed] [Google Scholar]
- [16].Kojima T, et al. , Immobilization of proteins onto microbeads using a DNA binding tag for enzymatic assays, J. Biosci. Bioeng 121 (2) (2016) 147–153. [DOI] [PubMed] [Google Scholar]
- [17].Wilson R, Cossins AR, Spiller DG, Encoded microcarriers for high-throughput multiplexed detection, Angew. Chem. Int. Ed 45 (37) (2006) 6104–6117. [DOI] [PubMed] [Google Scholar]
- [18].Breitling R, Biological microarray interpretation: the rules of engagement, Biochim. Biophys. Acta Gene Struct. Expr 1759 (7) (2006) 319–327. [DOI] [PubMed] [Google Scholar]
- [19].Berrar DP, Dubitzky W, Granzow M, A Practical Approach to Microarray Data Analysis, Springer, 2003. [Google Scholar]
- [20].Freudenberg JM, Comparison of background correction and normalization procedures for high-density oligonucleotide microarrays, Institut fur Informatik (11) (2005) 120. [Google Scholar]
- [21].Yigitbası T, Multiplex immunoassay and bead based multiplex, in: Trends in Immunolabelled and Related Techniques, 2012. (InTech). [Google Scholar]
- [22].Hsu HY, Joos TO, Koga H, Multiplex microsphere-based flow cytometric platforms for protein analysis and their application in clinical proteomicsefrom assays to results, Electrophoresis 30 (23) (2009) 4008–4019. [DOI] [PubMed] [Google Scholar]
- [23].Sukhanova A, Nabiev I, Fluorescent nanocrystal-encoded microbeads for multiplexed cancer imaging and diagnosis, Crit. Rev. Oncol.-Hematol 68 (1) (2008) 39–59. [DOI] [PubMed] [Google Scholar]
- [24].Braeckmans K, et al. , Encoding microcarriers: present and future technologies, Nat. Rev. Drug Discov 1 (6) (2002) 447–456. [DOI] [PubMed] [Google Scholar]
- [25].Saralidze K, Koole LH, Knetsch ML, Polymeric microspheres for medical applications, Materials 3 (6) (2010) 3537–3564. [Google Scholar]
- [26].Antal-Szalmas P, et al. , Measurement of soluble biomarkers by flow cytometry, EJIFCC 23 (4) (2013) 135. [PMC free article] [PubMed] [Google Scholar]
- [27].Kingsmore SF, Multiplexed protein measurement: technologies and applications of protein and antibody arrays, Nat. Rev. Drug Discov 5 (4) (2006) 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Herb ath M, et al. , Exploiting fluorescence for multiplex immunoassays on protein microarrays, Methods Appl. Fluoresc 2 (3) (2014) 032001. [DOI] [PubMed] [Google Scholar]
- [29].Nolan JP, Mandy F, Multiplexed and microparticle-based analyses: quantitative tools for the large-scale analysis of biological systems, Cytometry 69 (5) (2006) 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Krishhan V, Khan IH, Luciw PA, Multiplexed microbead immunoassays by flow cytometry for molecular profiling: basic concepts and proteomics applications, Crit. Rev. Biotechnol 29 (1) (2009) 29–43. [DOI] [PubMed] [Google Scholar]
- [31].Jun B-H, et al. , Fluorescence-based multiplex protein detection using optically encoded microbeads, Molecules 17 (3) (2012) 2474–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].De Jager W, et al. , Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells, Clin. Diagn. Lab. Immunol 10 (1) (2003) 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Abdorahim M, et al. , Nanomaterials-based electrochemical immunosensors for cardiac troponin recognition: an illustrated review, Trac. Trends Anal. Chem 82 (2016) 337–347. [Google Scholar]
- [34].Abdolrahim M, et al. , Development of optical biosensor technologies for cardiac troponin recognition, Anal. Biochem 485 (2015) 1–10. [DOI] [PubMed] [Google Scholar]
- [35].Mohagheghpour E, et al. , Controllable synthesis, characterization and optical properties of ZnS: Mn nanoparticles as a novel biosensor, Mater. Sci. Eng. C 29 (6) (2009) 1842–1848. [Google Scholar]
- [36].Wolter A, Niessner R, Seidel M, Detection of Escherichia coli O157: H7, Salmonella typhimurium, and Legionella pneumophila in water using a flow-through chemiluminescence microarray readout system, Anal. Chem 80 (15) (2008) 5854–5863. [DOI] [PubMed] [Google Scholar]
- [37].Zhang Y, et al. , Goldesilver nanocomposite-functionalized graphene sensing platform for an electrochemiluminescent immunoassay of a tumor marker, RSC Adv 3 (34) (2013) 14701–14709. [Google Scholar]
- [38].Karlsson R, Applications of Surface Plasmon Resonance for Detection of Bispecific Antibody Activity, 2015.
- [39].Liu MY, et al. , Multiplexed analysis of biomarkers related to obesity and the metabolic syndrome in human plasma, using the Luminex-100 system, Clin. Chem 51 (7) (2005) 1102–1109. [DOI] [PubMed] [Google Scholar]
- [40].Resch-Genger U, et al. , Quantum dots versus organic dyes as fluorescent labels, Br. J. Pharmacol 5 (9) (2008) 763–775. [DOI] [PubMed] [Google Scholar]
- [41].Kairdolf BA, et al. , Semiconductor quantum dots for bioimaging and bio-diagnostic applications, Annu. Rev. Anal. Chem 6 (1) (2013) 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tayebi M, et al. , Synthesis, surface modification and optical properties of thioglycolic acid-capped ZnS quantum dots for starch recognition at ultralow concentration, J. Electron. Mater 45 (11) (2016) 5671–5678. [Google Scholar]
- [43].Tayebi M, et al. , Thioglycolic acid-capped CdS quantum dots conjugated to a-amylase as a fluorescence probe for determination of starch at low concentration, J. Fluoresc 26 (5) (2016) 1787–1794. [DOI] [PubMed] [Google Scholar]
- [44].Tayebi M, et al. , Determination of total aflatoxin using cysteamine-capped CdS quantum dots as a fluorescence probe, Colloid Polym. Sci 294 (9) (2016) 1453–1462. [Google Scholar]
- [45].Yaraki MT, et al. , Synthesis and optical properties of cysteamine-capped ZnS quantum dots for aflatoxin quantification, J. Alloy. Comp 690 (2017) 749–758. [Google Scholar]
- [46].Jun B-H, et al. , Multilayer fluorescence optically encoded beads for protein detection, Anal. Biochem 396 (2) (2010) 313–315. [DOI] [PubMed] [Google Scholar]
- [47].Kim J, et al. , Clinical validation of quantum dot barcode diagnostic technology, ACS Nano 10 (4) (2016) 4742–4753. [DOI] [PubMed] [Google Scholar]
- [48].Ziemssen T, Ziemssen F, The role of the humoral immune system in multiple sclerosis (MS) and its animal model experimental autoimmune encephalomyelitis (EAE), Autoimmun. Rev 4 (7) (2005) 460–467. [DOI] [PubMed] [Google Scholar]
- [49].Miller JR, The importance of early diagnosis of multiple sclerosis, J. Manag. Care Pharm 10 (3) (2004). S4. [PubMed] [Google Scholar]
- [50].Owens GM, Economic burden of multiple sclerosis and the role of managed sare organizations in multiple sclerosis management, Am. J. Manag. Care 22 (6 Suppl) (2016) p. s151. [PubMed] [Google Scholar]
- [51].Byström S, et al. , Affinity proteomic profiling of plasma, cerebrospinal fluid, and brain tissue within multiple sclerosis, J. Proteome Res 13 (11) (2014) 4607–4619. [DOI] [PubMed] [Google Scholar]
- [52].Häggmark A, et al. , Antibody-based profiling of cerebrospinal fluid within multiple sclerosis, Proteomics 13 (15) (2013) 2256–2267. [DOI] [PubMed] [Google Scholar]
- [53].Dunbar SA, Applications of Luminex® xMAP™ technology for rapid, high-throughput multiplexed nucleic acid detection, Clin. Chim. Acta 363 (1) (2006) 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Choi I, Lee LP, Rapid detection of Ab aggregation and inhibition by dual functions of gold nanoplasmic particles: catalytic activator and optical reporter, ACS Nano 7 (7) (2013) 6268–6277. [DOI] [PubMed] [Google Scholar]
- [55].A.s. Association, 2014 Alzheimer’s disease facts and figures, Alzheimer’s Dementia 10 (2) (2014) e47–e92. [DOI] [PubMed] [Google Scholar]
- [56].Anstey KJ, Cherbuin N, Herath PM, Development of a new method for assessing global risk of Alzheimer’s disease for use in population health approaches to prevention, Prev. Sci 14 (4) (2013) 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Biagioni MC, Galvin JE, Using Biomarkers to Improve Detection of Alzheimer’s Disease, 2011. [DOI] [PMC free article] [PubMed]
- [58].Zhou Y, et al. , Detection of Ab monomers and oligomers: early diagnosis of Alzheimer’s disease, Chem Asian J 11 (6) (2016. March 18) 805–817. [DOI] [PubMed] [Google Scholar]
- [59].Blennow K, Hampel H, CSF markers for incipient Alzheimer’s disease, Lancet Neurol 2 (10) (2003) 605–613. [DOI] [PubMed] [Google Scholar]
- [60].Formichi P, et al. , Cerebrospinal fluid tau, Ass, and phosphorylated tau protein for the diagnosis of Alzheimer’s disease, J. Cell. Physiol 208 (1) (2006) 39–46. [DOI] [PubMed] [Google Scholar]
- [61].McKhann GM, Albert MS, Sperling RA, Changing diagnostic concepts of Alzheimer’s Disease, in: Alzheimer’s Disease-modernizing Concept, Biological Diagnosis and Therapy, Karger Publishers, 2012, pp. 115–121. [Google Scholar]
- [62].A.s. Association, 2017 Alzheimer’s disease facts and figures, Alzheimer’s Dementia 13 (4) (2017) 325–373. [Google Scholar]
- [63].Berti V, et al. , Early detection of Alzheimer’s disease with PET imaging, Neurodegener. Dis 7 (1–3) (2010) 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Vemuri P, et al. , Serial MRI and CSF biomarkers in normal aging, MCI, and AD, Neurology 75 (2) (2010) 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kang J-H, et al. , Simultaneous analysis of cerebrospinal fluid biomarkers using microsphere-based xMAP multiplex technology for early detection of Alzheimer’s disease, Methods 56 (4) (2012) 484–493. [DOI] [PubMed] [Google Scholar]
- [66].Elshal MF, McCoy JP, Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA, Methods 38 (4) (2006) 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].de la Escosura-Muniz A, et al. , Alzheimer’ s disease biomarkers detection in human samples by efficient capturing through porous magnetic micro-spheres and labelling with electrocatalytic gold nanoparticles, Biosens. Bio-electron 67 (2015) 162–169. [DOI] [PubMed] [Google Scholar]
- [68].Pi J, et al. , A sandwich immunoassay for detection of Ab1–42 based on quantum dots, Talanta 146 (2016) 10–15. [DOI] [PubMed] [Google Scholar]
- [69].Pi J, et al. , A sandwich immunoassay for detection of Ab 1–42 based on quantum dots, Talanta 146 (2016) 10–15. [DOI] [PubMed] [Google Scholar]
- [70].Tracey KJ, The inflammatory reflex, Nature 420 (6917) (2002) 853–859. [DOI] [PubMed] [Google Scholar]
- [71].Kleiner G, et al. , Cytokine levels in the serum of healthy subjects, Mediat. Inflamm (2013) 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Elsabahy M, Wooley KL, Cytokines as biomarkers of nanoparticle immunotoxicity, Chem. Soc. Rev 42 (12) (2013) 5552–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].MacBeath G, Protein microarrays and proteomics, Nat. Genet 32 (2002) 526–532. [DOI] [PubMed] [Google Scholar]
- [74].Khan SS, et al. , Multiplex bead array assays for detection of soluble cytokines: comparisons of sensitivity and quantitative values among kits from multiple manufacturers, Cytometry B Clin. Cytometry 61 (1) (2004) 35–39. [DOI] [PubMed] [Google Scholar]
- [75].Carrigan SD, Scott G, Tabrizian M, Toward resolving the challenges of sepsis diagnosis, Clin. Chem 50 (8) (2004) 1301–1314. [DOI] [PubMed] [Google Scholar]
- [76].Hsu H-Y, et al. , Suspension microarrays for the identification of the response patterns in hyperinflammatory diseases, Med. Eng. Phys 30 (8) (2008) 976–983. [DOI] [PubMed] [Google Scholar]
- [77].Visentin J, et al. , Deciphering complement interference in antiehuman leukocyte antigen antibody detection with flow beads assays, Transplantation 98 (6) (2014) 625–631. [DOI] [PubMed] [Google Scholar]
- [78].Kellar KL, et al. , Multiplexed fluorescent bead-based immunoassays for quantitation of human cytokines in serum and culture supernatants, Cytometry 45 (1) (2001) 27–36. [DOI] [PubMed] [Google Scholar]
- [79].Chen Y-S, et al. , Development of a bead-based suspension array for the detection of pathogens in acute respiratory tract infections, Exp. Biol. Med (2016), 1535370216647128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lavanchy D, Kane M, Global epidemiology of hepatitis B virus infection, in: Hepatitis B Virus in Human Diseases, Springer, 2016, pp. 187–203. [Google Scholar]
- [81].E.A.F.T.S.O.T. Liver, EASL clinical practice guidelines: management of chronic hepatitis B virus infection, J. Hepatol. (Amst.) 57 (1) (2012) 167–185. [DOI] [PubMed] [Google Scholar]
- [82].Wang X, et al. , NIR-emitting quantum dot-encoded microbeads through membrane emulsification for multiplexed immunoassays, Small 9 (19) (2013) 3327–3335. [DOI] [PubMed] [Google Scholar]
- [83].Wu L, Qu X, Cancer biomarker detection: recent achievements and challenges, Chem. Soc. Rev 44 (10) (2015) 2963–2997. [DOI] [PubMed] [Google Scholar]
- [84].Siegel R, Naishadham D, Jemal A, Cancer statistics, 2013, CA A Cancer J. Clin 63 (1) (2013) 11–30. [DOI] [PubMed] [Google Scholar]
- [85].Rusling JF, et al. , Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer, Analyst (Cambridge, U.K.) 135 (10) (2010) 2496–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ling MM, Ricks C, Lea P, Multiplexing molecular diagnostics and immunoassays using emerging microarray technologies, Expert Review of Molecular Diagnostics 7 (1) (2007) 87–98. [DOI] [PubMed] [Google Scholar]
- [87].Misek DE, Kim EH, Protein biomarkers for the early detection of breast cancer, Nternational Journal of Proteomics (2011) 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Opstal-van Winden AW, et al. , A bead-based multiplexed immunoassay to evaluate breast cancer biomarkers for early detection in pre-diagnostic serum, Int. J. Mol. Sci 13 (10) (2012) 13587–13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Gorelik E, et al. , Multiplexed immunobead-based cytokine profiling for early detection of ovarian cancer, Cancer Epidemiol. Biomark. Prev 14 (4) (2005) 981–987. [DOI] [PubMed] [Google Scholar]
- [90].Dehqanzada ZA, et al. , Assessing serum cytokine profiles in breast cancer patients receiving a HER2/neu vaccine using Luminex® technology, Oncol. Rep 17 (3) (2007) 687–694. [PubMed] [Google Scholar]
- [91].Jokerst JV, et al. , Nano-bio-chips for high performance multiplexed protein detection: determinations of cancer biomarkers in serum and saliva using quantum dot bioconjugate labels, Biosens. Bioelectron 24 (12) (2009) 3622–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kim Y-W, et al. , Development of multiplexed bead-based immunoassays for the detection of early stage ovarian cancer using a combination of serum biomarkers, PLoS One 7 (9) (2012), e44960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Brazhnik K, et al. , Multiplexed analysis of serum breast and ovarian cancer markers by means of suspension beadequantum dot microarrays, Physics Procedia 73 (2015) 235–240. [Google Scholar]
- [94].Adhyam M, Gupta AK, A review on the clinical utility of PSA in cancer prostate, Indian journal of surgical oncology 3 (2) (2012) 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Brazhnik K, et al. , Quantum dot-based lab-on-a-bead system for multiplexed detection of free and total prostate-specific antigens in clinical human serum samples, Nanomed. Nanotechnol. Biol. Med 11 (5) (2015) 1065–1075. [DOI] [PubMed] [Google Scholar]
- [96].Liu N, et al. , Simultaneous and combined detection of multiple tumor bio-markers for prostate cancer in human serum by suspension array technology, Biosens. Bioelectron 47 (2013) 92–98. [DOI] [PubMed] [Google Scholar]
- [97].Howlader N, Noone A, Krapcho M, SEER Cancer Statistics Review, 1975–2012 [seer. Cancer. Gov/csr. 1975_2012/, Based on the November 2014 SEER Data Submission, Posted to the SEER Website April 2015], National Cancer Institute, Bethesda, MD, 2015. [Google Scholar]
- [98].Ye Y, et al. , Combined detection of p53, p16, Rb, and EGFR mutations in lung cancer by suspension microarray, Genet. Mol. Res 8 (4) (2009) 1509–1518. [DOI] [PubMed] [Google Scholar]
- [99].Stieber P, et al. , CYFRA 21–1: a new marker in lung cancer, Cancer 72 (3) (1993) 707–713. [DOI] [PubMed] [Google Scholar]
- [100].Liu L, et al. , Bead-based microarray immunoassay for lung cancer biomarkers using quantum dots as labels, Biosens. Bioelectron 80 (2016) 300–306. [DOI] [PubMed] [Google Scholar]
- [101].Wu S, et al. , Multiplexed detection of lung cancer biomarkers based on quantum dots and microbeads, Talanta 156 (2016) 48–54. [DOI] [PubMed] [Google Scholar]
- [102].Villar-Vazquez R, et al. , Development of a novel multiplex beads-based assay for autoantibody detection for colorectal cancer diagnosis, Proteomics 16 (8) (2016. April) 1280–1290. [DOI] [PubMed] [Google Scholar]
- [103].Yurkovetsky ZR, et al. , Multiplex analysis of serum cytokines in melanoma patients treated with interferon-a2b, Clin. Canc. Res 13 (8) (2007) 2422–2428. [DOI] [PubMed] [Google Scholar]
- [104].Zhao Y, et al. , Multiplex detection of tumor markers with photonic suspension array, Anal. Chim. Acta 633 (1) (2009) 103–108. [DOI] [PubMed] [Google Scholar]
- [105].Wang Y, et al. , Evaluation of a method for the simultaneous detection of multiple tumor markers using a multiplex suspension bead array, Clin. Biochem 45 (16) (2012) 1394–1398. [DOI] [PubMed] [Google Scholar]
- [106].Sun K, et al. , Establishment of multiplexed, microsphere-based flow cyto-metric assay for multiple human tumor markers1, Acta Pharmacol. Sin 28 (12) (2007) 2011–2018. [DOI] [PubMed] [Google Scholar]
- [107].Long Y, et al. , Multiplex immunodetection of tumor markers with a suspension array built upon coreeshell structured functional fluorescence-encoded microspheres, Anal. Chim. Acta 665 (1) (2010) 63–68. [DOI] [PubMed] [Google Scholar]
- [108].Leng Y, et al. , Magnetic/fluorescent barcodes based on cadmium-free near-infrared-emitting quantum dots for multiplexed detection, Adv. Funct. Mater 26 (42) (2016) 7581–7589. [Google Scholar]
- [109].Arshady R, Microspheres for biomedical applications: preparation of reactive and labelled microspheres, Biomaterials 14 (1) (1993) 5–15. [DOI] [PubMed] [Google Scholar]
- [110].Hanes J, et al. , Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display, Nat. Biotechnol 18 (12) (2000) 1287–1292. [DOI] [PubMed] [Google Scholar]
- [111].Hallborn J, Carlsson R, Automated screening procedure for high-throughput generation of antibody fragments, Biotechniques 33 (2002) 30–37. [PubMed] [Google Scholar]
- [112].Porschewski P, et al. , Using aptamers as capture reagents in bead-based assay systems for diagnostics and hit identification, J. Biomol. Screen 11 (7) (2006) 773–781. [DOI] [PubMed] [Google Scholar]
- [113].Smith D, et al. , Sensitivity and specificity of photoaptamer probes, Mol. Cell. Proteomics 2 (1) (2003) 11–18 [DOI] [PubMed] [Google Scholar]
- [114].Lam A, Automation of a multiplexed microbead-based DNA assay platform, in: Institute of Biomaterials and Biomedical Engineering, University of Toronto, 2015. [Google Scholar]
- [115].Spielberg F, et al. , Field testing and comparative evaluation of rapid, visually read screening assays for antibody to human immunodeficiency virus, Lancet 333 (8638) (1989) 580–584. [DOI] [PubMed] [Google Scholar]
- [116].Mahmood T, Yang P-C, Western blot: technique, theory, and trouble shooting, N. Am. J. Med. Sci 4 (9) (2012) 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ellington AA, et al. , Antibody-based protein multiplex platforms: technical and operational challenges, Clin. Chem 56 (2) (2010) 186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].de Matos LL, et al. , Immunohistochemistry as an important tool in bio-markers detection and clinical practice, Biomark. Insights 5 (2010) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Gorham J, et al. , Biotechnology in the diagnosis of infectious diseases and vaccine development, Manual of standards for diagnostic tests and vaccines for terrestrial animals (2004) 69–90. [Google Scholar]
- [120].Mathew B, Biju R, Thapalia N, An Overview of Electrochemiluminescent (ECL) Technology in Laboratory Investigations, 2005. [PubMed]
- [121].Hu L, Xu G, Applications and trends in electrochemiluminescence, Chem. Soc. Rev 39 (8) (2010) 3275–3304. [DOI] [PubMed] [Google Scholar]