Abstract
Floral induction that initiates bolting and flowering is crucial for reproductive fitness in radishes. CONSTANS-like (CO-like, COL) genes play an important role in the circadian clock, which ensures regular development through complicated time-keeping mechanisms. However, the specific biological and functional roles of each COL transcription factor gene in the radish remain unknown. In this study, we performed a genome-wide identification of COL genes in the radish genome of three cultivars including ‘Aokubi’, ‘kazusa’ and ‘WK10039’, and we analyzed their exon-intron structure, gene phylogeny and synteny, and expression levels in different tissues. The bioinformatics analysis identified 20 COL transcription factors in the radish genome, which were divided into three subgroups (Group I to Group III). RsaCOL-09 and RsaCOL-12 might be tandem duplicated genes, whereas the others may have resulted from segmental duplication. The Ka/Ks ratio indicated that all the COL genes in radish, Arabidopsis, Brassica rapa, Brassica oleracea, Capsella rubella and rice were under purifying selection. We identified 6 orthologous and 19 co-orthologous COL gene pairs between the radish and Arabidopsis, and we constructed an interaction network among these gene pairs. The expression values for each COL gene during vegetable and flower development showed that the majority of Group I members had similar expression patterns. In general, the expression of radish COL genes in Groups I and III decreased during development, whereas the expression of radish COL genes in Group II first increased and then decreased. Substantial numbers of radish COL genes were differentially expressed after vernalization treatment. The expression levels of RsaCOL-02 and RsaCOL-04 were significantly increased during vernalization treatment, while the expression of RsaCOL-10 was significantly decreased. These outcomes provide insights for improving the genetic control of bolting and flowering in radish and other root vegetable crops, and they facilitate genetic improvements to radish yields and quality.
Introduction
The transition from vegetative development to bolting and flowering is critical for reproductive success in the plant life cycle. The initiation of flowering is controlled by complex genetic networks and could be effected by diverse plant hormones and environmental factors such as light, temperature, and length of day [1–3]. In Arabidopsis thaliana, approximately 180 genes participate in flowering-time control, and they are involved in six interactive regulatory pathways including the vernalization pathway, autonomous pathway, photoperiod pathway, gibberellin (GA) pathway, ambient temperature pathway, and age pathway [1,3,4]. These genes interact within different pathways to ensure that flowering occurs under appropriate conditions. For example, the primary flowering repressor FLOWERING LOCUS C (FLC) encodes a MADS-box transcription factor, and it integrates both the autonomous and the vernalization pathways [5]. Other flowering pathway integrators such as FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and LEAFY (LFY) were also confirmed as convergence points for different flowering pathways [6–8].
CONSTANS-like (CO-like, COL) genes play a vital role in the regulation of plant flowering through the photoperiod pathway, integrating the circadian clock, light signals and meristem genes identified as flowering time controls [9–11]. As a phloem-specific transcription activator of FT, CO promotes flowering by up-regulating the transcription of the FT and TWIN SISTER OF FT (TSF) genes [12]. At the posttranscriptional level, CO is degraded by the ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) in the dark, and, in the morning, it is degraded by a pathway activated by the photoreceptor PHYTOCHROME B (PHYB). These transcriptional and posttranscriptional regulations ensure that CO activates FT and TSF transcription only during long days. CO belongs to an Arabidopsis gene family containing 16 other genes that encode putative transcription factors [13]. Structurally, typical COL genes contain two conserved domains, a plant-specific C-terminal CCT (also termed CO, CO-like, TOC1) domain and an N-terminal zinc finger B-box domain, which is also found in animals [14]. The COL genes belong to a larger transcription factor gene family named the B-BOX (BBX) family, which can be divided into five groups based on the presence of one or two B-BOX motifs and the presence or absence of the CCT domain [15]. Previous studies showed that most COL genes containing a CCT domain are involved in controlling the flowering time in some plant species [16,17].
The availability of the Arabidopsis genome sequence and annotation provides a useful tool for a comparative analysis of COL transcriptional regulators. In addition, many tools and databases such as the PlantTFDB database (version 3.0, http://planttfdb.cbi.pku.edu.cn) were developed for identifying, clustering, aligning and functionally analyzing plant transcription factors [18]. The CO-like genes in Arabidopsis are classified into three major groups; group I included AtCO and AtCOL1 to AtCOL5 with two B-boxes; group II contained AtCOL6 to AtCOL8 and AtCOL16 with one B-box; and group III consisted of COL9-COL15 with one B-box and another diverged zinc finger domain [13,19]. An analysis identified 17 COL genes in Oryza sativa that belonged to 30 rice BBX genes, and they were divided into four phylogenetic groups [17]. Genome-wide analyses have also been performed for COL transcription factors in the Brassicaceae family. Song et al. (2015) found 25 Brassica rapa COL genes, and they investigated the evolutionary pattern of COL genes in 34 Angiospermae (27 eudicots, six monocots and one basal angiosperm), three Gymnospermae, one Pteridophyta, one Bryophyta and six Chlorophyta species [20].
The radish (Raphanus sativus L., 2n = 2x = 18) is an annual diploid species of the Brassicaceae family, and it is also an economically important vegetable crop that is produced worldwide. The primary edible part of the radish is the tuberous root, and it contains various nutrients and medicinal compounds [21]. The optimum timing of bolting and flowering are vital for economic organ production and for preventing premature bolting and flowering in radishes. Recently, one draft genome assembly of the radish Raphanus sativus var. hortensis was published, resulting in a total of 383 Mb of sequences and 54,357 genes [22]. In 2016, another genome assembly of the radish R. sativus cv. WK10039 was published, which containing 344.0 Mb of sequences could be integrated into nine chromosome pseudomolecules out of 426.2 Mb total sequences. In total, 46,514 protein-coding genes were predicted and annotated [23]. The transcriptomes of various tissues in radish including the roots and leaves, have been assembled and analyzed [24–27]. These databases provide powerful tools for the genome-wide characterization of bolting and flowering genes in radish. Previous studies have reported a list of functional genes and microRNAs (miRNAs) related to radish bolting and flowering based on transcriptomic sequencing, expression profiling and transgenic approaches [2,28,29]. However, the specific biological and functional roles of the CO-like transcription factor genes in radish remain unknown.
This study is the first report on the genome-wide identification of CO-like genes in the radish. In the present study, we identified the COL genes in the radish genome and classified them into three groups based on the analysis of their exon-intron structure, gene phylogeny and synteny, as well as their expression in specific tissues. The results provide insights into the genetic networks regulating bolting and flowering in radish and other vegetable crops in Brassicaceae, and they will also facilitate the genetic improvement of radish yield, nutritional value and commercial quality.
Materials and methods
Sequence retrieval and COL members identification in radish
The genome, genes and corresponding protein sequences of the radish were downloaded from the NODAI Radish Genome Database (http://www.nodai-genome-d.org, cultivar ‘Aokubi’), Radish Genome Database (http://radish-genome.org) and Raphanus sativus Genome Database (http://radish.kazusa.or.jp/, cultivar ‘kazusa’). The Markov models of two identical domains for the CO-like transcription factors, PF06203.13 (CCT) and PF00643.23 (zinc finger B-box), were downloaded from the Pfam database (http://pfam.xfam.org/) [30,31]. All radish proteins were aligned with these two Markov models using the HMMER program with a cut-off E-value of 1e-4 separately. Only the proteins that were aligned with both models were selected as CO-like protein candidates. The COL genes that were located at a distance of 10 kb on the same chromosome or scaffold were considered tandemly duplicated genes.
COL genes identification in representative plants
The genome and annotated proteins of 19 representative species in the primary lineages of the plant kingdom were also collected. The annotated proteins of algae (Chlamydomonas reinhardtii), Coccomyxa subellipsoidea, Dunaliella salina, moss (Physcomitrella patens), lycophyte (Selaginella moellendorffii), Amborella trichopoda, Aquilegia coerulea, Arabidopsis thaliana, Capsella rubella, Eutrema salsugineum, grape (Vitis vinifera), poplar (Populus trichocarpa), tomato (Solanum lycopersicum), maize (Zea mays) and rice (Oryza sativa) were downloaded from the Pfam database (v11) [32]. The genomic information on the gymnosperm plant Norway spruce (Picea abies) [33] was collected from the Congenie Website (http://congenie.org/). The sequences of the sacred lotus genes and proteins were downloaded from Lotus-DB (http://lotus-db.wbgcas.cn/, v1.0) [34]. The proteins of Brassica rapa were obtained from the BRAD (Brassica database, http://brassicadb.org/brad/), those of B. napus from the GENOSCOPE database (http://www.genoscope. cns.fr/brassicanapus/data/) and those of B. oleracea from BolBase (http://www.ocri-genomics. org/bolbase/, v1.0). The candidate proteins that only contain the fragmental CCT or zinc finger B-box domains were eliminated manually. Protparam (http://web.expasy.org/protparam/) [35] was employed in the physical and chemical characteristic analysis of the COL proteins in those analyzed species, including the molecular weight, theoretical isoelectric point (pI), atomic composition formula, instability index, aliphatic index and grand average of hydropathicity (GRAVY).
Phylogenetic analysis of the COL genes
The collection of protein sequences from multiple species was used for a phylogenetic analysis in Planta. First, a pairwise alignment and a multiple alignment were performed using the ClustalX2 program with the Gonnet protein weight matrix [36], and then a maximum likelihood phylogenetic tree was built with the MEGA program (v6.06) using the Jones-Taylor-Thornton (JTT) model with 1000 bootstrap replicates using the full CDS sequence for a partial 70% length [37]. Uniform rates and homogeneous lineages were adopted, and the partial deletion with a site coverage cutoff of 70% were used for gaps/missing data treatment. The frequency of each divergent branch was displayed when it was higher than 50%. The figure was beautified with information from the group using Adobe Illustrator software.
Gene structure and motif analysis
The gene structure was analyzed using the Gene Structure Display Server tool (http://gsds.cbi.pku.edu.cn/, v2.0) [38]. MEME software (http://meme.nbcr.net/meme/, v4.12.0) was used to search for motifs among the proteins [39]. The searching motif window length was set from 10 bp to 100 bp. Only widely distributed motifs that occurred in at least 3 protein sequences were retained. These motifs were drawn in two separate figures, in accordance with the phylogenetic trees. The top 10 motifs with the lowest E-values were reported, which were displayed according to a pattern in which the more statistically significant (lower E-value) motifs came first.
Identification of orthologous and paralogous genes
OrthoMCL software (v2.0.3) [40] was employed in searching for orthologous, co-orthologous and paralogous genes in radish, Arabidopsis, Brassica rapa, Brassica oleracea, Capsella rubella and rice using entire COL protein sequences. The E-value cut-off of an all-against-all BLASTP alignment process was set at 1e-5, and the alignment with a match cut-off value lower than 50 was eliminated. To evaluate the divergence of duplicated radish COL genes, the synonymous rate (Ks), nonsynonymous rate (Ka), and evolutionary constraint (Ka/Ks) between paralogous pairs of genes were calculated with the KaKs_calculator tool and paraAT software using the method developed by Nei and Gojobori (http://cbb.big.ac.cn/software). The divergence time was calculated for homologous pairs among five dicotyledonous plants using the formula T = Ks/2R, where R is the rate of divergence for nuclear genes from plants, and it was considered equal to 1.5 × 10−8 synonymous substitutions per site per year for dicotyledonous plants [41]. The selected homologous pairs of four species including radish, Arabidopsis, Brassica rapa, and rice were gathered and displayed using Cytoscape software (http://www.cytoscape.org, v2.8.3) [42].
Gene expression analysis in radish tissues and cytoplasmic male sterility and in response to vernalization
The gene expression data of the COL genes were gathered from the NODAI genome database, which was used in a previous report [22]. The RNA-seq data from the 7 d root, 7 d leaf, 14 d root, 14 d leaf, 20 d root, 20 d leaf, 40 d cortex, 40 d cambium, 40 d xylem, 40 d root tip, 40 d leaf, 60 d cortex, 60 d cambium, 60 d xylem, 60 d root tip, 60 d leaf, 90 d cortex, 90 d cambium, 90 d xylem, 90 d root tip, and 90 d leaf of Japanese radish cultivar ‘Aokubi’ were used. The gene expression profile in each sample was standardized using the RPKM (reads per kilobase per million measure) method. The expression profile of the COL genes from each sample was analyzed using the HemI program (http://hemi.biocuckoo.org/) with the average hierarchical clustering method after being normalized by a logarithmic base (log2). The hierarchy was obtained by clustering the gene expression in both the horizontal axis and the vertical axis using a Pearson distance similarity metric. A discrete bar containing 21 colors was used to represent the gene expression values.
Furthermore, high-throughput RNA sequencing was performed to characterize the transcriptome of radish buds with a length that was approximately 1.5 mm from two CMS lines (HYBP-B and YH-B) possessing the CMS-inducing orf138 gene and corresponding to near-isogenic maintainer lines (HYBP-A and YH-A) [43]. The sequencing data of 4 radish lines were downloaded from the NCBI SRA database under bioproject PRJNA273265, with two replicates sequenced for each line. The TopHat program (https://ccb.jhu.edu/software/tophat/ index.shtml, v2.1.0) was used to map the reads to the NODAI radish genome; the expression profile of all the spinach genes was then obtained with the FPKM (Fragments Per Kilobase of exon per million fragments Mapped) value using Cufflinks software (http://cole-trapnell-lab.github.io/cufflinks, v2.2.1) under the guidance of annotated gene models with a GFF file. The radish COL gene expression profiles from each sample were analyzed using the HemI program.
The gene expression of the radish seedlings at three different time points during vernalization in another report was also used in this study. The samples were labeled RT1, RT2, and RT3 for the room temperature treatment (RT), VE1, VE2, and VE3 for the early stage of vernalization (VE), and VL1, VL2, and VL3 for the late stage of vernalization (VL) [44]. The gene expression value obtained by the FPKM method was then drawn using the HemI program.
Prediction of the regulating network of radish COL genes
The interaction network of radish COL genes was constructed using information in Arabidopsis Interactions Viewer (http://bar.utoronto.ca/interactions/cgi-bin/arabidopsis_interactions_viewer.cgi) to build an Arabidopsis COL regulating network, and then each Arabidopsis gene was replaced by the corresponding radish orthologs and co-orthologs. This approach is based on the theory that homologs are structurally and functionally similar. In addition, the gene expression of radish organs during different developmental stages were also used to calculate the Pearson correlation coefficient. First, the radish genes with low expression, including the highest expression lower than 2 or an accumulated expression lower than 5 in 21 sequencing samples, were eliminated. Second, the Pearson correlation coefficient was calculated for any combination of two radish gene expression values using R language. Finally, the gene expression values of the radish COL genes were only retained if the P-value was lower than 0.1 and if the absolute Pearson correlation coefficient (PCC) value was higher than 0.9. WEGO software (http://wego.genomics.org.cn/) was then used to plot a GO functional figure for these genes with GO term hits to view the distribution of gene functions [45].
Results and discussion
Genome-wide identification of COL genes in radish
The bioinformatics analysis identified 20 COL transcription factors in the radish genome among 54,357 coding genes. All of the radish COL genes were well conserved, and they were designated ‘RsaCOL’ with a serial number and sorted by the E-value of the CCT domain (S1 Table). The molecular weights of the COL proteins in the radishes ranged from 33134.20 to 46404.79, which was very similar to those in Arabidopsis (Table 1). The theoretical pI of the COL proteins in radish varied from 5.09 to 7.57, including one alkaline protein and 19 weakly acidic proteins. In contrast, all 17 COL proteins in Arabidopsis were acidic. In addition, the number of acidic proteins in other plants was less than 3. The aliphatic index of COL proteins ranged from 48.46 to 71.21, and the GRAVY index ranged from -0.704 to -0.318. The position of COL genes RsaCOL-09 and RsaCOL-12 were Rs_scaf1+: 669331, 670395 and Rs_scaf1+: 673375, 674690, which was located in close proximity. This finding indicated that these genes might be tandemly duplicated, whereas the others may have resulted from segmental duplication.
Table 1. Classification and chemical characterization of radish COL genes.
| ID | Gene | CDS Length | Whole Gene Length | Protein Length | Molecular Weight | pI | Formula | Instability index | Aliphatic index | GRAVY | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RsaCOL-01 | RSG31377 | 1206 | 1448 | 401 | 45258.99 | 5.80 | C1949H3080N574O615S27 | 50.10 | unstable | 61.10 | -0.711 |
| RsaCOL-02 | RSG42550 | 1209 | 1439 | 402 | 45545.01 | 5.43 | C1963H3086N574O628S24 | 56.76 | unstable | 62.59 | -0.755 |
| RsaCOL-03 | RSG21446 | 1041 | 1134 | 346 | 37997.54 | 6.08 | C1659H2573N471O521S17 | 46.24 | unstable | 67.98 | -0.316 |
| RsaCOL-04 | RSG07010 | 1245 | 1503 | 414 | 46404.79 | 5.19 | C2005H3147N579O648S21 | 52.75 | unstable | 65.97 | -0.711 |
| RsaCOL-05 | RSG35888 | 1029 | 1139 | 342 | 37407.90 | 5.59 | C1641H2542N450O520S16 | 49.33 | unstable | 66.78 | -0.248 |
| RsaCOL-06 | RSG11754 | 1047 | 1616 | 348 | 37611.95 | 6.02 | C1620H2534N474O522S19 | 40.38 | unstable | 61.41 | -0.451 |
| RsaCOL-07 | RSG34470 | 1059 | 1575 | 352 | 38036.25 | 5.68 | C1639H2554N474O535S18 | 35.84 | stable | 61.28 | -0.464 |
| RsaCOL-08 | RSG03422 | 1275 | 1604 | 424 | 47004.50 | 5.77 | C2001H3184N594O661S27 | 44.00 | unstable | 60.02 | -0.711 |
| RsaCOL-09 | RSG01422 | 1065 | 1065 | 354 | 39437.70 | 6.79 | C1674H2630N510O552S22 | 60.02 | unstable | 57.34 | -0.720 |
| RsaCOL-10 | RSG31262 | 924 | 1000 | 307 | 33134.20 | 6.60 | C1436H2271N419O455S14 | 46.79 | unstable | 71.21 | -0.318 |
| RsaCOL-11 | RSG03335 | 1113 | 1452 | 370 | 41063.03 | 6.36 | C1766H2803N521O568S20 | 49.30 | unstable | 49.30 | -0.696 |
| RsaCOL-12 | RSG01423 | 1128 | 1316 | 375 | 42777.35 | 7.57 | C1820H2846N562O592S22 | 60.57 | unstable | 52.80 | -0.970 |
| RsaCOL-13 | RSG01107 | 1092 | 1641 | 363 | 40845.23 | 6.57 | C1736H2748N526O578S19 | 63.36 | unstable | 61.24 | -0.825 |
| RsaCOL-14 | RSG23074 | 1134 | 1403 | 377 | 41112.13 | 5.47 | C1722H2677N521O592S30 | 60.69 | unstable | 48.46 | -0.704 |
| RsaCOL-15 | RSG09942 | 1062 | 1588 | 353 | 38890.86 | 5.75 | C1639H2544N490O555S28 | 49.65 | unstable | 51.98 | -0.663 |
| RsaCOL-16 | RSG40602 | 1161 | 1417 | 386 | 43064.44 | 5.74 | C1863H2920N522O598S27 | 58.52 | unstable | 70.70 | -0.541 |
| RsaCOL-17 | RSG03894 | 1074 | 1234 | 357 | 39387.21 | 5.09 | C1651H2564N492O572S29 | 60.82 | unstable | 49.75 | -0.746 |
| RsaCOL-18 | RSG13348 | 1050 | 1411 | 349 | 38859.07 | 5.71 | C1625H2552N490O553S33 | 57.97 | unstable | 48.97 | -0.705 |
| RsaCOL-19 | RSG14263 | 1203 | 1377 | 400 | 44361.63 | 5.91 | C1895H2964N552O616S32 | 51.26 | unstable | 59.97 | -0.508 |
| RsaCOL-20 | RSG28563 | 1002 | 2399 | 333 | 38743.45 | 6.13 | C1677H2636N502O523S17 | 51.79 | unstable | 62.31 | -0.925 |
Several research centers have been interested in the whole sequencing of the radish genome and transcriptome, resulting in several versions of the radish genome and genetic information [22,46,47]. Till now, only one radish genome assembly contained gene location information in chromosome level, whereas other assemblies in scaffold level. In the present study, we primarily used the radish genome assembly in the NODAI Radish Genome Database to identify the COL genes in radish. In addition, we also ran HMMER software on other radish genome versions for a better comparison. Almost every COL gene in the ‘Aokubi daikon’ cultivar were matched with the corresponding COL gene in the ‘kazusa’ cultivar, except RsaCOL-20 (S2 Table). A total of 20 radish COL genes were also searched in the cultivar ‘WK10039’ (S2 Table), and their location in radish chromosomes were shown in S1 Fig. The availability of genome sequences from various species accelerates the genome-wide identification of gene families in plants. In radish, entire MADS-box genes and WRKY genes have been identified through genome-wide analysis [48,49]. The identification of RsaCOL genes in the present study provides additional knowledge about the gene family in radish and a foundation for further investigating the flowering regulatory networks in radish and other Brassicaceae vegetables.
Phylogenetic relationship and evolutionary divergence of radish COL proteins
To provide insight into the evolution of COL genes in different species of the plant kingdom, we performed a comparative analysis using the genes from a total of 71 whole-genome-sequenced plant species. These species covered the primary lineages in Planta, including 7 algae, one lycophyte, one gymnosperm, a monocot, Eusasterids II, Euasterids I, Eurosids I, Eurosids II, Malpighiales, and some representative species, including Amborella, columbine, and sacred lotus.
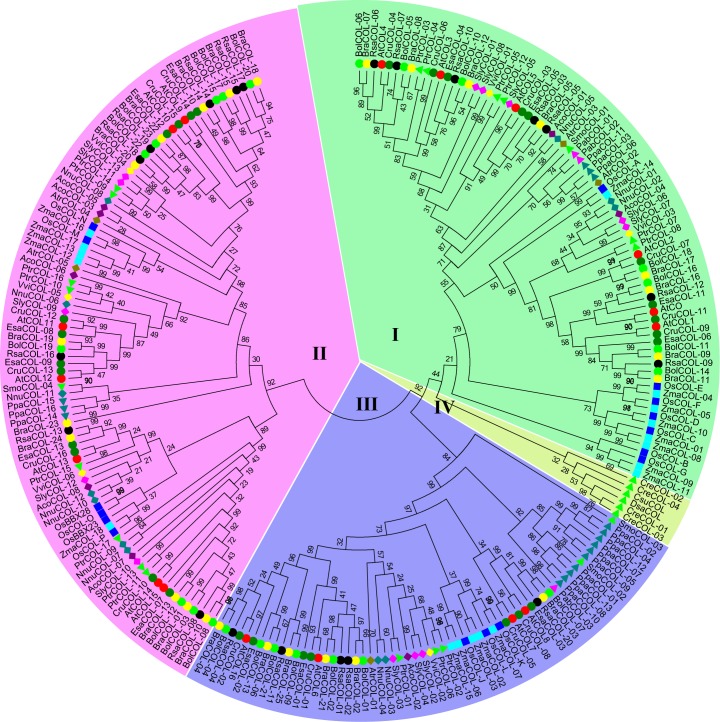
We identified 5 A. trichopoda COL genes, 8 A. coerulea COL genes, 12 sacred lotus COL genes, 16 rice COL genes, 3 Norway spruce COL genes, 17 Populus COL genes, 6 grape COL genes and 18 maize COL genes (S3 Table). C. subellipsoidea and D. salina had the lowest number of COL genes, at one gene for each species. C. reinhardtii and S. moellendorffii had 4 COL genes each. The number of COL genes in E. salsugineum and tomato were also the same (13 COL genes for each species), whereas in the case of C. rubella and P. patens, the number of COL genes was 16 COL genes for each species. A previous report showed that P. patens contained 17 COL genes [50], whereas only 16 COL genes were identified in this report. Compared with other genera in Brassicaceae, those two Brassica species contained large numbers of COL genes, specifically, 23 genes in B. oleracea and 25 genes in B. rapa. This result was consistent with a previous report, and it was likely due to a genome triplication event [20]. To gain insight into the evolution of COL genes, we constructed a phylogenetic tree to characterize the families of COL genes (Fig 1). The phylogenetic tree was generally consistent with previous reports [19,20].
Fig 1. Phylogenetic tree of the COL proteins from 16 plant species.
The phylogenetic tree was constructed based on the 90% shared amino acid sites using the maximum likelihood method. The abbreviations represent the species as follows: At, Arabidopsis thaliana; Atr, Amborella trichopoda; Bra, Brassica rapa; Bol, Brassica oleracea; Cru, Capsella rubella; Esa, Eutrema salsugineum; Nnu, Nelumbo nucifera; Pab, Picea abies; Ppa, Physcomitrella patens; Ptr, Populus trichocarpa; Rsa, Raphanus sativus; Sly, Solanum lycopersicum; Sme, Selaginella moellendorffii; Tca, Theobroma cacao; Vvi, Vitis vinifera; Os, Oryza sativa; and Zma, Zea mays.
A previous analysis of CO-like genes in Arabidopsis classified the family into three broad groups based on the number and divergence of the B-box zinc finger domain [13]. In another study, the genes were referred to as Group I to III genes [19]. Song et al. classified the CO-like genes in A. trichopoda, P. taeda, P. sitchensis, P. abies, S. moellendorffii and P. patens into groups A to C according to the distribution of the B-box and zinc finger domains [20]. We grouped the COL genes into Groups I to III; the Group I members had two zinc finger B-boxes, whereas only one B-box was present in Groups II and III. Group III members had an additional diverged zinc finger.
In the present study, the COL genes in alga were grouped into a diverged branch; thus, the branches that contained C. reinhardtii, C. subellipsoidea and D. salina were named Group IV. The amount of a large number of transcription factor families in P. abies was several fold higher than that in Arabidopsis or Physcomitrella, which could be due to the polyploidy and complex nature of the P. abies genome. However, only two COL genes in Group I and one COL gene in Group III were identified. Similarly, one COL gene in Group I, one COL gene in Group II and two COL genes in Group III were identified in S. moellendorffii. Three, three and ten COL genes in P. patens were classified into Groups I, II and III, respectively. These genes were grouped tightly, indicating that these four clades were relatively more original than the other clades. The COL genes in monocots were closely grouped, and the rice COL genes had similar COL genes corresponding to maize COL genes. For example, every rice COL gene had a corresponding gene in maize in Group I, whereas in Group II, OsCOL-K, OsCOL-L and OsCOL-J had one, two and two corresponding COL genes in maize, respectively. In Group II, OsCOL-N had one corresponding gene in maize, and OsCOL-M had three corresponding genes in a branch, whereas four rice COL genes (OsBBX28, OsCOL-O, OsBBX23 and OsCOL-O) corresponded to one maize gene. Genome duplication was also found in the Brassicaceae family. In several branches, one or two radish COL genes corresponded to the Arabidopsis COL gene. The phylogenetic tree showed the possibility of a COL gene that evolved divergently before the origin of Lycopodium, and it divided into three groups and evolved independently.
Gene structure of RsaCOL genes
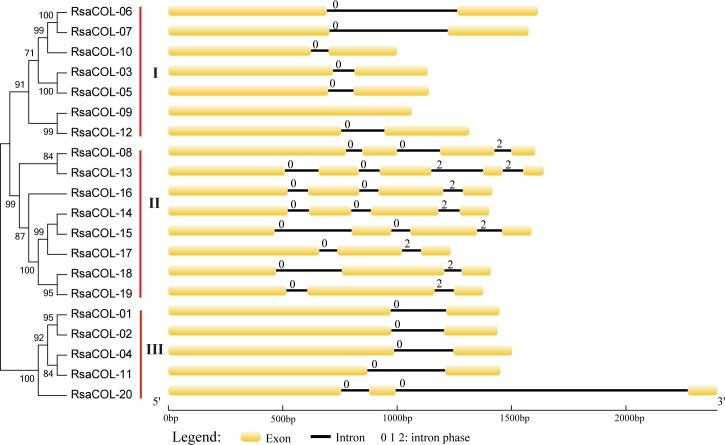
To compare the radish COL genes, their exon-intron structures were predicted, and the results are shown in Fig 2. The phylogenetic trees of the RsaCOL genes were constructed by Maximum-Likelihood method with 1000 bootstraps using full CDS sequences. The COL genes in radish were divided into 3 groups, Group I, Group II and Group III, which is similar to the grouping in Arabidopsis and rice [19]. In general, genes in the same group have similar numbers of exons and introns and even intron phases, indicating that these genes shared a conserved splicing pattern. The RsaCOL genes in Group I contained two exons, except for RsaCOL-12, which contained one exon. In Group II, five RsaCOL genes contained four exons, whereas three RsaCOL genes contained three exons. Only RsaCOL-20 contained two introns, whereas the other members in Group III contained one intron. The intron phase of all the intron-containing RsaCOL genes in Group I and Group III were 0, whereas the first intron phase and the last intron phase of all the intron-containing RsaCOL genes in Group II were 0 and 2, respectively. The length of the last intron in RsaCOL-20 was over 1000 bp, while the other introns in all the RsaCOL genes were short. Because most of the genes shared conserved splicing patterns, we hypothesized that the intron increased event that occurred in RsaCOL-20 might have resulted from a fragment insert based on the sequence analysis. In contrast, the intron loss event that occurred in RsaCOL-09 resulted from a fragment deletion, and it contained a full intron and parts of exons.
Fig 2. Gene structure of the radish COL genes.
Red boxes represent exons, and black and blue lines represent introns and UTRs, respectively. The lengths of the exons and introns were drawn according to the lengths of the sequences.
Conserved motifs located in RsaCOL protein sequences
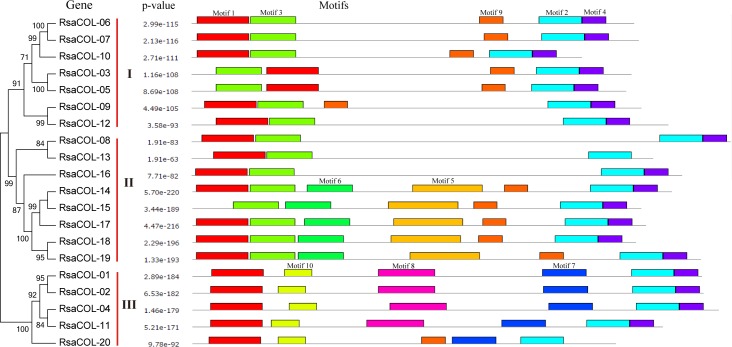
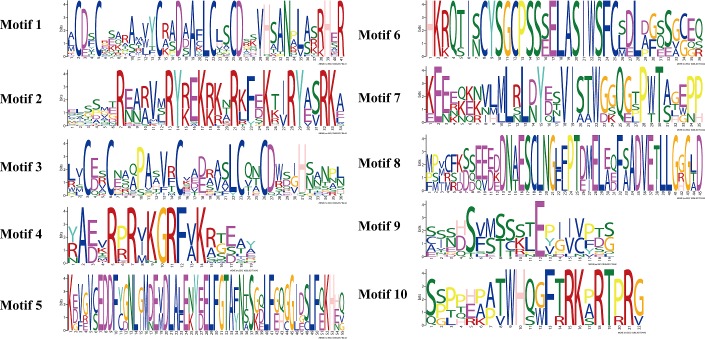
To explore the conserved domains and motifs, MEME software was employed to analyze the sequence alignment of the COL proteins in radish. The motifs were listed using serial numbers for Motif 1 to Motif 15 according to the ascending E-value of the alignment (Fig 3 and Fig 4). Motif 1 and Motif 2 were the most conserved motifs that could be found in all the COL proteins, followed by Motif 3 and Motif 4, which were conserved in 18 and 15 radish COL proteins, respectively, possibly because Motif 1 and Motif 2 were zinc finger domains and because Motif 3 was a CCT domain. Motif 3 is usually located nearly after Motif 1 in the protein sequences in Group I and Group II, except that Motif 1 was lost in RsaCOL-15 and the inversion events in RsaCOL-03 and RsaCOL-05. Motif 4 and Motif 9 were located in partial members of the COL genes in these three groups. Moreover, some motifs were only present in one unique group and were shared by all the members within the group. This was the case for Motif 7, Motif 8 and Motif 10 in Group III. In addition, Motif 6 was only found in 5 COL proteins in Group II; thus, it was the representative motif of Group II. For these motifs in radish COL proteins, the longest motif was 45 amino acids (aa), whereas the shortest motif was 19 aa.
Fig 3. Conserved motifs embedded in the radish COL proteins.
The phylogenetic tree, the number of conserved motifs and their distribution in each protein with their relative combined P-values.
Fig 4. Amino acid sequences of each conserved motif in the radish COL proteins.
The font size represents the frequency of the respective amino acid.
The orthologous, co-orthologous and paralogous COL genes in radish, Arabidopsis, rice and P. patens
To shed light on the vertical descent from a single ancestral gene and on duplication, a comparative analysis was performed to identify the orthologous, co-orthologous and paralogous gene pairs in radish, Arabidopsis, Brassica rapa, Brassica oleracea, Capsella rubella and rice. Orthologs are genes derived from a single gene (vertical descent) [51]. The meaning of co-orthologs is slightly different, in that they consist of two or more genes in one lineage that are collectively orthologous to one or more genes in another lineage due to lineage-specific duplication(s). Paralogous genes result from a lineage-specific duplication(s) that occurs subsequent to a given speciation event, and they are defined only relative to a speciation event, with no absolute meaning [52].
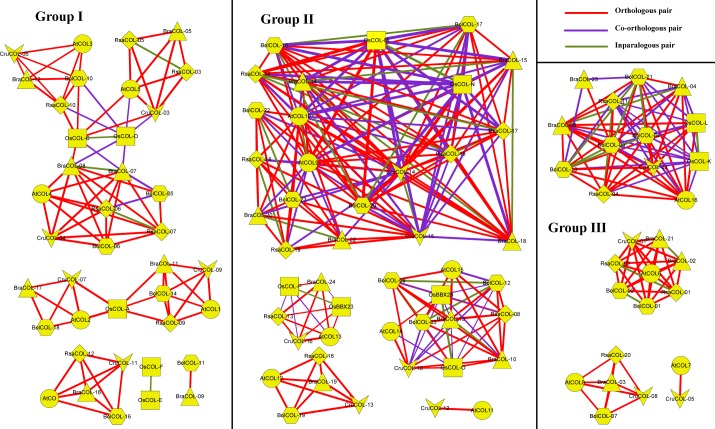
We identified 273 orthologous, 103 co-orthologous and 37 paralogous COL gene pairs among radish, Arabidopsis, Brassica rapa, Brassica oleracea, Capsella rubella and rice according to OrthoMCL software (S4 Table). In the network of homologous gene pairs, all the COL genes of radish, Arabidopsis, Brassica oleracea and Capsella rubella were included. A majority of COL genes in Brassica rapa and rice were included, except for two Brassica rapa COL genes and three rice COL genes. This homologous gene pair network was highly consistent with the phylogenetic tree, which could be clearly divided into three parts.
For orthologous gene pairs, 19, 34, 33, 19, and 6 orthologous gene pairs were identified in groups of radish-Arabidopsis, radish-Brassica rapa, radish-Brassica oleracea, radish-Capsella rubella and radish-rice COL genes (Fig 5). In addition, 6, 1, 4, 6 and 11 co-orthologous gene pairs were identified in those groups, providing comparative information on COL genes among lower plants, eudicots and monocots. Generally, the ratio of orthologous pairs of closer plants with the radish in the phylogenetic tree exceeded the ratio of co-orthologous pairs. The RsaCOL-03 and RsaCOL-05 were two orthologs to AtCOL5, and their divergence time was 11.95 million years ago, illustrating that they were duplicated from a common ancestor with AtCOL5 and that they diverged recently. Eight paralogous pairs were found in radish, with more than one pair in Arabidopsis and six in rice. There were no orthologous/co-orthologous genes for AtCOL7 and AtCOL11 in the Brassica genus and radish, whereas there was one counterpart for each gene in Capsella rubella, which indicated that there was COL gene loss in the Brassica-Raphanus lineage. This approach provides a better understanding of the general trends in the evolution of radish COL genes and of the reconstruction of the evolutionary history of each gene in its entirety.
Fig 5. Orthologous, co-orthologous and paralogous COL gene pairs.
The ellipse, diamond and rectangle shapes indicate genes belonging to radish, Arabidopsis, Brassica rapa, Brassica oleracea, Capsella rubella and rice. The red, purple and green lines indicate orthologous, co-orthologous and paralogous relationships, and the width of the line is associated with the relationship index as produced by OrthoMCL software.
Selection and divergence time
Nonsynonymous (amino acid-replacing, Ka) and synonymous (Ks) substitution rates among protein-coding sequences were used to reveal the DNA sequence evolution mechanisms. The Ka/Ks ratio was used to estimate the selective strength for DNA sequence evolution, with a Ka/Ks > 1 indicating positive selection, Ka/Ks < 1 indicating purifying (negative) selection, and a Ka/Ks close to 1 indicating a neutral mutation [53]. ParaAT software (http://cbb.big.ac.cn/software) is capable of constructing multiple protein-coding DNA alignments in parallel for a large number of homologs [54]. To investigate the selection mechanisms of COL genes during evolution, we calculated the Ka, Ks and divergence time among the paralogous gene pairs obtained here (S4 Table). Notably, the homologous gene pairs were identified by OrthoMCL software using the ML method, whereas the Ka, Ks and divergence time were calculated based on the Nei and Gojobori method. This universally used method for alignment and Ks calculation was stricter in identifying the homologous gene pairs; therefore, some homologous gene pairs identified in OrthoMCL software were not regarded as homologous pairs by this method, and neither their Ks nor their divergence time were available. For example, the AtCOL16 and OsCOL-L were considered as orthologs by OrthoMCL software, whereas the Ks was not available by ParaAT and KaKs_Calculator software.
In Brassicaceae, three rounds of whole-genome duplication (WGD) occurred after its lineage diverged from the monocot lineage. The most recent WGD event occurred 50 to 65 MYA [55,56], prior to the divergence of the species in the Brassicaceae family. Several reports have estimated that the Arabidopsis-Brassica split took place 33 to 43 MYA. Notably, a further hexaploidization event (α’ whole-genome triplication [WGT] event occurred recently in the common ancestor of Brassica and Raphanus) occurred 22 to 29 MYA [55,57,58]. Subsequently, the α’ duplicates of the Brassica and Raphanus species shared ancestry over the following 5 to 16 million years ago (MYA), followed by 13 to 19 MYA of independent evolution in the Brassica and Raphanus genera [59]. Studies on the Brassica species suggested that after α’ WGT, >50% of the Brassica duplicate genes may have been lost via deletion and pseudogenization; part of them might have been lost in a biased fashion [60], and Raphanus species might have evolved along a similar path. There were 17 COL genes in Arabidopsis, which were assumed to be 54 duplicated genes immediately after a WGT event. However, only 37% (20 genes) were retained. The case of the COL genes would provide a better understanding of duplicate evolution post-WGT regarding the rate of pseudogenization in duplicate genes and the patterns of expression divergence in Raphanus.
Almost all calculated homologous COL gene pairs in the selected six plants had a Ka/Ks ratio of less than 1, indicating the purifying selection of these genes. The exception was the BolCOL-09 and BraCOL-25 gene pair, with a Ka/Ks ratio of 1.05, suggesting that the gene pair might go through neutral mutation. The Ka/Ks ratio of the only existing paralogous gene pair (AtCOL9-AtCOL10) in Arabidopsis was 0.19, and the ratio of the radish paralogous gene pair fell into a range from 0.12 to 0.32. The divergence times of the paralogous gene pairs in Arabidopsis and Capsella rubella were 31.03 and 35.99 MYA, respectively. The divergence times of the radish COL paralogous gene pairs ranged from 11.95 to 19.54 MYA, which occurred during a WGT event. The divergence time of the paralogous gene pairs in Brassica rapa was 9.05 to 18.50 MYA, while the divergence time of paralogous gene pairs in Brassica oleracea was 1.92 to 16.62 MYA.
The divergence times of a large portion of the orthologous gene pairs between radish-Arabidopsis, radish-Capsella rubella and radish-Brassica species were 10.97 to 28.39 MYA, 15.13 to 32.51 MYA, and 3.63 to 19.98 MYA, respectively. These findings suggested those orthologs with Arabidopsis and Capsella rubella duplications occurred during an α’ WGT event (22–29 MYA), the shared ancestry era of Brassica and Raphanus as well as an independent evolution era (13–19 MYA), whereas those orthologs with Brassica species occurred during an independent evolution era.
Predicted regulating network of radish COL proteins
To better understand the regulating network in which radish COL genes are involved, an interaction network of radish COL genes was constructed using a computational approach. The Pearson correlation coefficient values of over one hundred gene pairs were larger than zero, whereas the PCC of four genes were less than zero, which indicates that COL proteins primarily have a positive interaction with other proteins in radish. Ten gene pairs were not calculated, and they are thus shown in green lines, indicating that their regulation patterns were unclear. The results showed that the radish COL genes played significantly different roles in the interaction-regulating network (Fig 6). For example, RsaCOL-12 and RsaCOL-09 interacted with 32 and 23 proteins, respectively, suggesting that the two genes have significant regulation effects at the transcriptional level. The RSG26538 gene was homologous to the AtBBX32 in Arabidopsis, which was a B-box domain transcription factor. It was predicted to be involved in the blue light signaling pathway, the negative regulation of transcription, the DNA-templated photomorphogenesis, the red or far-red light signaling pathway, the regulation of flower development, the regulation of transcription, the DNA-templated response to chitin, transcription, and being DNA-templated by GO enrichment analysis. In Arabidopsis, the AtCOL3 could target FT in the presence of AtBBX32 to regulate the flowering pathway, thus playing a role at the interface between the light and the clock to modulate the timing and duration of specific reproductive stages [61]. Therefore, we hypothesized that RsaCOL-03-RSG26538 was a corresponding module of BBX32-COL3, and it might participate in the regulatory mechanism affecting reproductive development in radish. Additionally, the transgenic of the Arabidopsis AtBBX32 and overexpression in soybean significantly increased the grain yield in soybeans, which were potentially caused by the timing of reproductive development in transgenic soybeans, leading to the increased duration of the pod and seed development period. At the molecular level, AtBBX32 influenced the transcript levels of the soybean clock genes GmTOC1 and LHY-CCA1-like2 (GmLCL2) and the clock-regulating gene CO [62]. Because these clock genes played vital roles in regulating many key agronomic traits, it would be worthwhile to use this approach as an efficient strategy for genetic manipulation in this regulatory pathway for increased radish agricultural productivity.
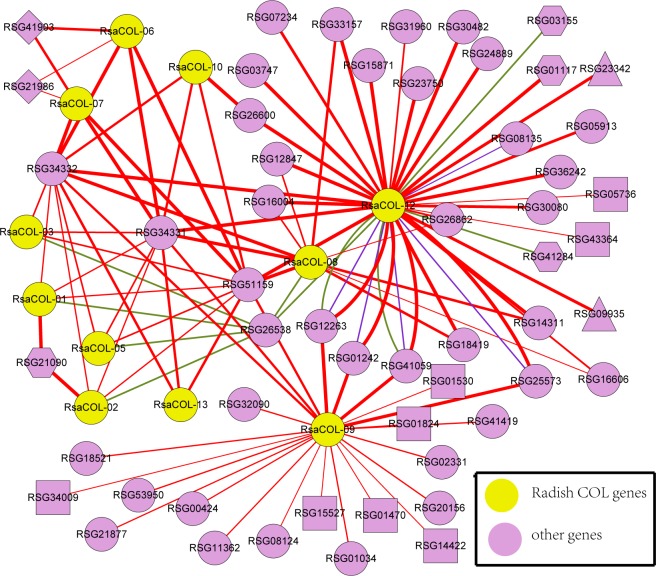
Fig 6. The interaction network of COL genes in radish according to the orthologs in Arabidopsis.
Red and blue indicate that the Pearson correlation coefficient (PCC) index is above or below 0, respectively; green indicates that the PCC index of the interaction was unclear. The ellipse, diamond, triangle, rectangle and hexagon shapes indicate that this protein might be detected in the nucleus, vacuole, plastid, cytosol or unclear, respectively. If a protein could be found in several cell parts, including the nucleus, it is only represented by an ellipse.
Both RSG34331 and RSG51159 interacted with 11 radish COL proteins, including RsaCOL-01 to RsaCOL-03, RsaCOL-05 to RsaCOL-10, RsaCOL-12, and RsaCOL-13. The annotation showed that RSG34331 and RSG51159 were both LOV kelch protein 2 (LKP2). Previous reports have demonstrated that the overexpression of LKP2 causes arrhythmia under both constant light and constant darkness, hypocotyl elongation, and postponed flowering under long days [63]. Therefore, we predicted that the radish LKP2s (RSG34331 and RSG51159) were potentially close to the circadian oscillator, and there was a positive interaction among radish homologs and AtCO, AtCO1, AtCO3, AtCO4, AtCO5, AtCO6, AtCO15. The RSG26538 had a positive interaction with RsaCOL-09 and RSG51159, and an unclear interaction with RsaCOL-01, RsaCOL-02, RsaCOL-03, RsaCOL-05, RsaCOL-08 and RsaCOL-12.
Expression pattern of the radish COL genes during development
We investigated the expression of each COL gene using published RNA-seq data for different radish tissues during vegetative and reproductive development. The expression level of each gene was calculated using the RPKM method. We also analyzed the average and median expression values of all the genes in a full radish gene dataset that contained 64,657 Augustus gene models for comparison, which was stable in each sample and available for comparison across samples. The average expression value of all the radish genes was approximately 15, whereas the median expression value of all the radish coding genes ranged from 0.1 to 0.2 (S5 Table). According to the RPKM values, all the RsaCOL genes were expressed in the leaf, and 19 of these genes were expressed at relatively high levels (RPKM>1) in at least one tissue (Fig 7). The expression values of RsaCOL-16 were lower in all the samples (lower than 0.34), suggesting that it may not act as a functional gene.
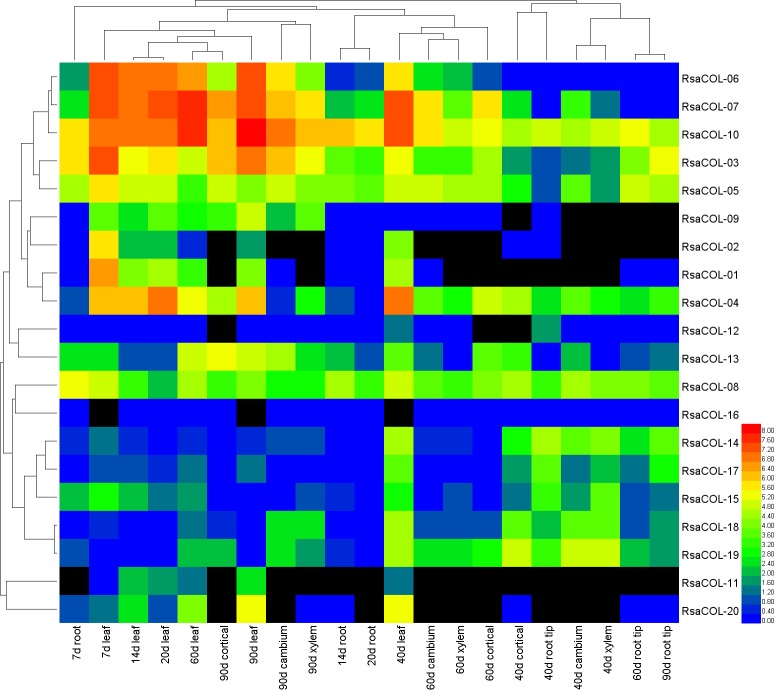
Fig 7. The heat map shows the expression profile of the radish COL genes over multiple development periods.
Clustering was performed using both horizontal and vertical axes. Interestingly, the clustering of the vertical axis including 20 radish COL genes was consistent with the phylogenetic tree. The genes RsaCOL-03, RsaCOL-05, RsaCOL-06, RsaCOL-07, RsaCOL-09, and RsaCOL-10 in Group I were clustered closely in both the expression clustering and the phylogenetic tree; RsaCOL-12 was also clustered relatively close to those genes. RsaCOL-01, RsaCOL-02 and RsaCOL-04 in Group III also illustrated a similar expression pattern, while RsaCOL-11 and RsaCOL-20 were clustered closely. The members in Group II were also significantly expressed following a similar pattern except for RsaCOL-13.
In general, the expression of radish COL genes in Group I and Group III decreased during development, whereas the expression of Group II COL genes in radish first increased and then decreased during development. In the leaves, the expression of RsaCOL-06 was highest at 7 d (191.17), gradually decreased at 14 d (120.91) and 20 d (140.57), and reached the lowest at 40 d (61.98), then finally increased at 60 d and 90 d. Moreover, at the same developmental stage, the expression of COL genes in the leaf was higher than it was in the cortex, cambium and xylem, and thus higher than it was in the root. This finding could be a result of circadian clock receptor and coordination regulation. The identification and characterization of RsaCOL genes that regulate the circadian clock and flower development at the molecular level could contribute to an understanding of the genetic mechanisms underlying bolting and flowering in root vegetables and facilitate radish cultivar improvement.
The Pearson correlation coefficient of gene expression showed that nine COL radish proteins were highly co-expressed with 898 other radish proteins. RsaCOL-07 and RsaCOL-06 were observed to be co-expressed with 298 and 288 proteins, respectively. The RSG09940 was annotated as a set domain-containing protein, which was co-expressed with RsaCOL-06, RsaCOL-07, RsaCOL-10 and RsaCOL-11. RSG02061, RSG05030, RSG08140, RSG10915, RSG11552, RSG13476, RSG15344, RSG16356, RSG20934, RSG29339, RSG33164, RSG45245, and RSG51054 were co-expressed with 3 radish COL genes (S6 Table).
To provide insight into the functions of newly identified radish genes, all radish genes and COL co-expressed genes were annotated using GO databases. A Gene Ontology (GO) enrichment analysis was performed to provide a dynamic, controlled vocabulary and hierarchical relationships for the information on molecular functions, cellular components and biological processes [64,65]. This approach provided important access to the underlying fundamental functions of new genes. In total, 42,347 out of 64,655 (65.50%) radish-coding genes were assigned with GO terms, whereas 867 out of 898 (96.55%) co-expressed genes were annotated (Fig 8, S7 Table). This result indicated that a large portion of those co-expressed COL genes were well-studied, and the ratio of co-expressed genes was higher than that of all the radish coding genes in substantial GO level 2 terms. The three most common categories for biological processes were cellular processes (679), metabolic processes (673) and biological regulation (393). For molecular functions, binding (463) and catalytic activity (426) were the two most abundant categories. For cellular components, the annotated genes involved in cell parts (840), cells (840) and organelles (782) were the most abundant entries.
Fig 8. The GO annotation for all the radish coding genes and COL co-expressed genes.
Expression pattern of the radish COL genes during development
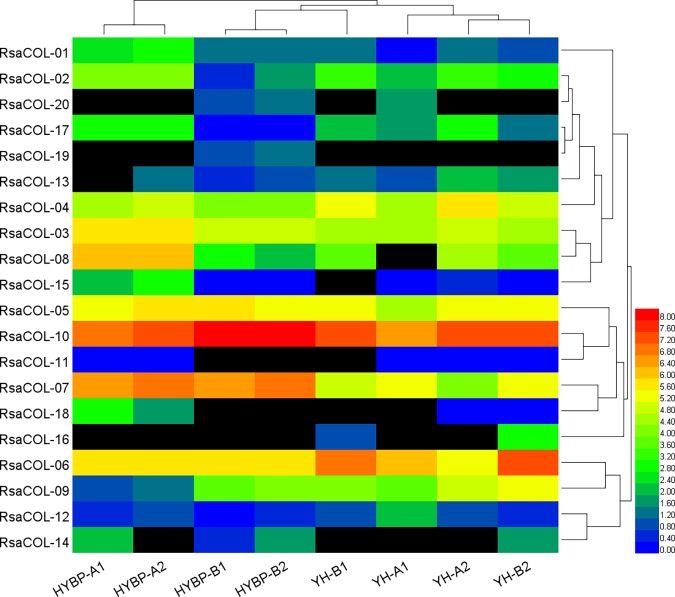
We re-analyzed the expression of each COL gene in radish buds with lengths approximately 1.5 mm from two CMS lines (HYBP-B, YH-B) possessing the CMS-inducing orf138 gene and corresponding to near-isogenic maintainer lines (HYBP-A, YH-A) [43].The clustering of samples showed that the expression of COL genes was not associated with cytoplasmic male sterility (Fig 9, S8 Table). The expression of RsaCOL-01, RsaCOL-02, RsaCOL-08, RsaCOL-15, and RsaCOL-17 significantly declined in CMS line HYBP-B compared with normal line HYBP-A. However, the expression of those genes was insignificantly expressed in CMS line YH-B. The expression of RsaCOL-09 increased in HYBP-B, and the expression of RsaCOL-06 increased in YH-B. These genes were non-significantly expressed in another CMS line, which indicated that the expression levels of the COL genes were not coincident in those two CMS lines. The expression levels of radish COL genes were more closely associated with the cultivars, more than with whether the plant was from a CMS line or not.
Fig 9. The heat map shows the expression profile of the COL genes in radish cytoplasmic male sterility and fertile floral buds.
Expression pattern of the radish COL genes in response to vernalization
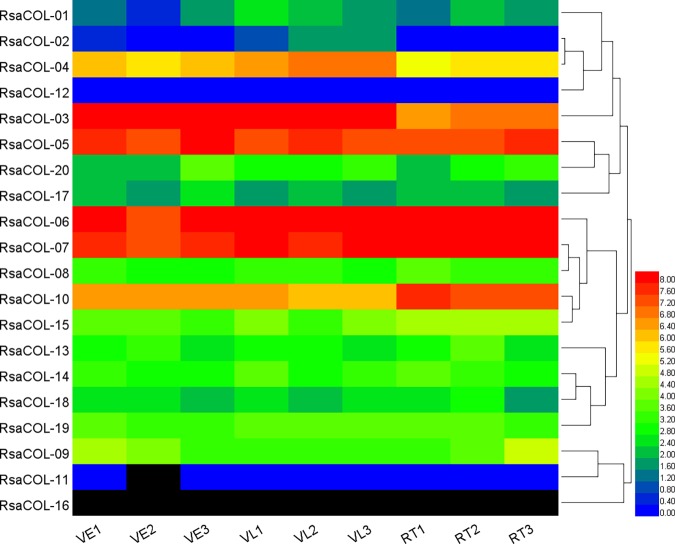
To investigate the potential links between COL genes and radish vernalization, we determined the expression of each COL gene from radish plants that were exposed to low temperatures for 0, 3 and 20 d in a previous report [44]. Substantial numbers of radish COL genes were differentially expressed after vernalization treatment (Fig 10, S9 Table). The expression of RsaCOL-03, RsaCOL-04, RsaCOL-05, RsaCOL-06, RsaCOL-07 and RsaCOL-10 were high in all the samples, of which most were from Group I. The expression levels of RsaCOL-02 and RsaCOL-04 were significantly increased during vernalization treatment, while the expression of RsaCOL-10 was significantly decreased. Using COL5 over-expressing lines, we show that, under short days, the constitutive expression of COL5 affects the flowering time and the expression of the floral integrator genes FT and SOC1 [66]. The expression of the corresponding radish gene RsaCOL-03 was significantly increased at the early and late stages of vernalization. RsaCOL-14 and RsaCOL-15 were orthologs corresponding to Arabidopsis gene COL9, which is regulated by the circadian clock in the photoperiod pathway, by repressing the expression of CO and concomitantly reducing the expression of FT, delaying floral transition [67]. The expression of RsaCOL-15 halved at the early stage of vernalization and then remained steady at the late stage of vernalization, while RsaCOL-14 was not differentially expressed. The RsaCOL-05, an ortholog of COL5 in Arabidopsis, remained steady during all the stages of the vernalization treatment.
Fig 10. The heat map shows the expression profile of the radish COL genes of radish in response to vernalization.
Supporting information
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Nanjing Huasequen Biotechnologies Co, Ltd., China for assistance on bioinformatics analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by the National technology system of commodity vegetable industry-radish (CARS-23-A-14), and New variety breeding project of the Major Science and Technology Projects of Zhejiang (2016C02051-2-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Amasino RM, Michaels SD (2010) The timing of flowering. Plant Physiol 154: 516–520. 10.1104/pp.110.161653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nie S, Xu L, Wang Y, Huang D, Muleke EM, Sun X, et al. (2015) Identification of bolting-related microRNAs and their targets reveals complex miRNA-mediated flowering-time regulatory networks in radish (Raphanus sativus L.). Sci Rep 5: 14034 10.1038/srep14034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037. 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fornara F, de Montaigu A, Coupland G (2010) SnapShot: Control of flowering in Arabidopsis. Cell 141: 550, 550 e551–552. 10.1016/j.cell.2010.04.024 [DOI] [PubMed] [Google Scholar]
- 5.Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J, Lee I (2010) Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot 61: 2247–2254. 10.1093/jxb/erq098 [DOI] [PubMed] [Google Scholar]
- 7.Moon J, Lee H, Kim M, Lee I (2005) Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol 46: 292–299. 10.1093/pcp/pci024 [DOI] [PubMed] [Google Scholar]
- 8.Parcy F (2005) Flowering: a time for integration. Int J Dev Biol 49: 585–593. 10.1387/ijdb.041930fp [DOI] [PubMed] [Google Scholar]
- 9.Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- 10.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, et al. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- 11.Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. 10.1038/35074138 [DOI] [PubMed] [Google Scholar]
- 12.An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626. 10.1242/dev.01231 [DOI] [PubMed] [Google Scholar]
- 13.Robson F, Costa MM, Hepworth SR, Vizir I, Pineiro M, Reeves PH, et al. (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28: 619–631. [DOI] [PubMed] [Google Scholar]
- 14.Borden KL (2000) RING domains: master builders of molecular scaffolds? J Mol Biol 295: 1103–1112. 10.1006/jmbi.1999.3429 [DOI] [PubMed] [Google Scholar]
- 15.Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, et al. (2009) The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420. 10.1105/tpc.109.069088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cockram J, Thiel T, Steuernagel B, Stein N, Taudien S, Bailey PC, et al. (2012) Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS One 7: e45307 10.1371/journal.pone.0045307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu W, Zheng XM, Chen D, Zhang Y, Ma W, Zhang H, et al. (2017) OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci 260: 60–69. 10.1016/j.plantsci.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 18.Jin JP, Zhang H, Kong L, Gao G, Luo JC (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research 42: D1182–D1187. 10.1093/nar/gkt1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131: 1855–1867. 10.1104/pp.102.016188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song X, Duan W, Huang Z, Liu G, Wu P, Liu T, et al. (2015) Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom. Sci Rep 5: 14631 10.1038/srep14631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtis IS (2003) The noble radish: past, present and future. Trends Plant Sci 8: 305–307. 10.1016/S1360-1385(03)00127-4 [DOI] [PubMed] [Google Scholar]
- 22.Mitsui Y, Shimomura M, Komatsu K, Namiki N, Shibata-Hatta M, Imai M, et al. (2015) The radish genome and comprehensive gene expression profile of tuberous root formation and development. Scientific Reports 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong YM, Kim N, Ahn BO, Oh M, Chung WH, Chung H, et al. (2016) Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor Appl Genet 129: 1357–1372. 10.1007/s00122-016-2708-0 [DOI] [PubMed] [Google Scholar]
- 24.Wang SF, Wang XF, He QW, Liu XX, Xu WL, Li LB, et al. (2012) Transcriptome analysis of the roots at early and late seedling stages using Illumina paired-end sequencing and development of EST-SSR markers in radish. Plant Cell Reports 31: 1437–1447. 10.1007/s00299-012-1259-3 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Pan Y, Liu Z, Zhu X, Zhai L, Xu L, et al. (2013) De novo transcriptome sequencing of radish (Raphanus sativus L.) and analysis of major genes involved in glucosinolate metabolism. BMC Genomics 14: 836 10.1186/1471-2164-14-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L, Wang Y, Liu W, Wang J, Zhu X, Zhang K, et al. (2015) De novo sequencing of root transcriptome reveals complex cadmium-responsive regulatory networks in radish (Raphanus sativus L.). Plant Sci 236: 313–323. 10.1016/j.plantsci.2015.04.015 [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Jia H, Yin Y, Wu G, Xia H, Wang X, et al. (2013) Transcriptome analysis of leaf tissue of Raphanus sativus by RNA sequencing. PLoS One 8: e80350 10.1371/journal.pone.0080350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curtis IS, Nam HG, Yun JY, Seo KH (2002) Expression of an antisense GIGANTEA (GI) gene fragment in transgenic radish causes delayed bolting and flowering. Transgenic Res 11: 249–256. [DOI] [PubMed] [Google Scholar]
- 29.Nie S, Li C, Wang Y, Xu L, Muleke EM, Tang M, et al. (2016) Transcriptomic Analysis Identifies Differentially Expressed Genes (DEGs) Associated with Bolting and Flowering in Radish (Raphanus sativus L.). Front Plant Sci 7: 682 10.3389/fpls.2016.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. (2014) Pfam: the protein families database. Nucleic Acids Research 42: D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy SR (2011) Accelerated Profile HMM Searches. Plos Computational Biology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584. 10.1038/nature12211 [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Deng J, Damaris RN, Yang M, Xu L, Yang P (2015) LOTUS-DB: an integrative and interactive database for Nelumbo nucifera study. Database (Oxford) 2015: bav023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31: 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Stoeckert CJ Jr., Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koch MA, Haubold B, Mitchell-Olds T (2000) Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol 17: 1483–1498. 10.1093/oxfordjournals.molbev.a026248 [DOI] [PubMed] [Google Scholar]
- 42.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432. 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mei S, Liu T, Wang Z (2016) Comparative Transcriptome Profile of the Cytoplasmic Male Sterile and Fertile Floral Buds of Radish (Raphanus sativus L.). Int J Mol Sci 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C, Wang S, Xu W, Liu X (2017) Genome-wide transcriptome profiling of radish (Raphanus sativus L.) in response to vernalization. PLoS One 12: e0177594 10.1371/journal.pone.0177594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34: W293–297. 10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitashiba H, Li F, Hirakawa H, Kawanabe T, Zou Z, Hasegawa Y, et al. (2014) Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res 21: 481–490. 10.1093/dnares/dsu014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong YM, Chung WH, Chung H, Kim N, Park BS, Lim KB, et al. (2014) Comparative analysis of the radish genome based on a conserved ortholog set (COS) of Brassica. Theor Appl Genet 127: 1975–1989. 10.1007/s00122-014-2354-3 [DOI] [PubMed] [Google Scholar]
- 48.Karanja BK, Fan L, Xu L, Wang Y, Zhu X, Tang M, et al. (2017) Genome-wide characterization of the WRKY gene family in radish (Raphanus sativus L.) reveals its critical functions under different abiotic stresses. Plant Cell Rep 36: 1757–1773. 10.1007/s00299-017-2190-4 [DOI] [PubMed] [Google Scholar]
- 49.Li C, Wang Y, Xu L, Nie S, Chen Y, Liang D, et al. (2016) Genome-Wide Characterization of the MADS-Box Gene Family in Radish (Raphanus sativus L.) and Assessment of Its Roles in Flowering and Floral Organogenesis. Front Plant Sci 7: 1390 10.3389/fpls.2016.01390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zobell O, Coupland G, Reiss B (2005) The family of CONSTANS-like genes in Physcomitrella patens. Plant Biol (Stuttg) 7: 266–275. [DOI] [PubMed] [Google Scholar]
- 51.Fitch WM (1970) Distinguishing homologous from analogous proteins. Syst Zool 19: 99–113. [PubMed] [Google Scholar]
- 52.Koonin EV (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39: 309–338. 10.1146/annurev.genet.39.073003.114725 [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z, Yu J (2006) Evaluation of six methods for estimating synonymous and nonsynonymous substitution rates. Genomics Proteomics Bioinformatics 4: 173–181. 10.1016/S1672-0229(06)60030-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Xiao J, Wu J, Zhang H, Liu G, Wang X, et al. (2012) ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun 419: 779–781. 10.1016/j.bbrc.2012.02.101 [DOI] [PubMed] [Google Scholar]
- 55.Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S (2010) Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci U S A 107: 18724–18728. 10.1073/pnas.0909766107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438. 10.1038/nature01521 [DOI] [PubMed] [Google Scholar]
- 57.Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, et al. (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18: 1348–1359. 10.1105/tpc.106.041665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Couvreur TL, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K (2010) Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol Biol Evol 27: 55–71. 10.1093/molbev/msp202 [DOI] [PubMed] [Google Scholar]
- 59.Moghe GD, Hufnagel DE, Tang H, Xiao Y, Dworkin I, Town CD, et al. (2014) Consequences of Whole-Genome Triplication as Revealed by Comparative Genomic Analyses of the Wild Radish Raphanus raphanistrum and Three Other Brassicaceae Species. Plant Cell 26: 1925–1937. 10.1105/tpc.114.124297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43: 1035–1039. 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- 61.Tripathi P, Carvallo M, Hamilton EE, Preuss S, Kay SA (2017) Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc Natl Acad Sci U S A 114: 172–177. 10.1073/pnas.1616459114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Preuss SB, Meister R, Xu Q, Urwin CP, Tripodi FA, Screen SE, et al. (2012) Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS One 7: e30717 10.1371/journal.pone.0030717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670. 10.1105/tpc.010332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gene Ontology Consortium Blake JA, Dolan M, Drabkin H, Hill DP, Li N, et al. (2013) Gene Ontology annotations and resources. Nucleic Acids Res 41: D530—D535. 10.1093/nar/gks1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hassidim M, Harir Y, Yakir E, Kron I, Green RM (2009) Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 230: 481–491. 10.1007/s00425-009-0958-7 [DOI] [PubMed] [Google Scholar]
- 67.Cheng XF, Wang ZY (2005) Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J 43: 758–768. 10.1111/j.1365-313X.2005.02491.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.