Abstract
Optogenetics is possibly the most revolutionary advance in neuroscience research techniques within the last decade. Here, we describe lab modules, presented at a workshop for undergraduate neuroscience educators, using optogenetic control of neurons in the fruit fly Drosophila melanogaster. Drosophila is a genetically accessible model system that combines behavioral and neurophysiological complexity, ease of use, and high research relevance. One lab module utilized two transgenic Drosophila strains, each activating specific circuits underlying startle behavior and backwards locomotion, respectively. The red-shifted channelrhodopsin, CsChrimson, was expressed in neurons sharing a common transcriptional profile, with the expression pattern further refined by the use of a Split GAL4 intersectional activation system. Another set of strains was used to investigate synaptic transmission at the larval neuromuscular junction. These expressed Channelrhodopsin 2 (ChR2) in glutamatergic neurons, including the motor neurons. The first strain expressed ChR2 in a wild type background, while the second contained the SNAP-25ts mutant allele, which confers heightened evoked potential amplitude and greatly increased spontaneous vesicle release frequency at the larval neuromuscular junction. These modules introduced educators and students to the use of optogenetic stimulation to control behavior and evoked release at a model synapse, and establish a basis for students to explore neurophysiology using this technique, through recapitulating classic experiments and conducting independent research.
Keywords: optogenetics, Drosophila, neuromuscular junction, behavior, SNAP-25, electrophysiology
The recent optogenetics revolution has changed the landscape of neuroscience research (Boyden et al., 2005; Fenno et al., 2011; Sjulson et al., 2016). Remote control of neuronal activity using light has given researchers an unprecedented level of access to specific, transcriptionally-defined neural circuits, even within intact and freely behaving animals (Arenkiel et al., 2007; Pulver et al., 2009; Nieh et al., 2013; Smith and Graybiel, 2013). From the first widely applicable system, reported in 2005 (Boyden et al., 2005), the optogenetic toolbox has greatly expanded to include excitatory, inhibitory, and biochemically active molecules. In addition to their usefulness in research, optogenetics-based clinical applications are being actively explored. These include potential treatments for macular degeneration and retinal disease (Scholl et al., 2016), deafness (Moser, 2015), peripheral pain (Liu et al., 2016), and as a substitute for deep brain stimulation for diseases such as Parkinson’s and Tourette’s (Kalanithi and Henderson, 2012).
Most optogenetic methods consist of co-opting a photosensitive ion channel or pump from a bacterium or algae and expressing them in genetically accessible organism under the control of a particular promoter or enhancer. With the appropriate enhancer, selected neurons express the photosensitive ion channel which can be activated by a light stimulus such as a laser, or a high-power LED, the latter of which is well within the capabilities of a budget-conscious research laboratory or an undergraduate teaching lab (Pulver et al., 2011; Titlow et al., 2015; Rose, 2018, this issue; Pokala and Glater, 2018).
Here, we use Drosophila melanogaster as a model organism to demonstrate optogenetic control of neural circuits and use this technique as a foundation for investigation of neurophysiology and behavior. In Drosophila, transgenes can be expressed with exquisite control by utilizing the binary system, GAL4/UAS, which utilizes the tissue-specific expression of the yeast transcriptional activator (GAL4) to drive expression of genes that are under control of the GAL4 upstream activation sequence (UAS). GAL4 and UAS lines can be mixed and matched to express virtually any gene in a multitude of patterns (Pulver and Berni, 2012). In this case, channelrhodopsins are expressed in adult neuronal circuits to elicit behavioral responses and to trigger action potentials in motor neurons of the larva. Further refinement of the GAL4 system is also possible by expressing the two halves of the protein with different promoters, and only the intersecting cells would contain the functional GAL4, further restricting the neurons being activated (Luan et al., 2006; Pfeiffer et al., 2010; Dionne et al., 2018).
At the 2017 Faculty for Undergraduate Neuroscience Workshop at Dominican University, participants used optogenetic activation of neural circuits in transgenic and mutant Drosophila lines to observe behavioral and neurophysiological effects of activating specific neurons, and to conduct simple experiments demonstrating basic principles of neurophysiology and behavior. Two transgenic lines expressed CsChrimson, an algal opsin with an activation peak in the orange-red range at about 600 nm (Klapoetke et al., 2014), in restricted groups of neurons within the CNS (Jenett et al., 2012). Of the two lines exhibiting light-triggered behavior, the first, Moonwalker, drives expression in a specific cluster of descending neurons that integrate input from the visual system and drive a backward walking behavior (Sen et al., 2017). The second has an expression pattern that includes bilateral neurons, which elicit startle and escape response when activated. These were used for behavioral observation. A third line expressed Channelrhodopsin 2 (ChR2), the first widely implemented optogenetic construct with an activation peak in the blue range at 460 nm, in glutamatergic neurons (Hornstein et al., 2009). This expression pattern includes motor neurons innervating the body wall muscles of the Drosophila third instar larva, a classic neuromuscular junction research prep often used as a genetically accessible synapse (Zhang and Stewart, 2010). The line expressing ChR2 in glutamatergic neurons causes tetanic muscle contraction when activated and can be used to record evoked potentials and miniature endplate potentials in the body wall muscles innervated by motor neurons.
The workshop had two broad teaching aims. The first was familiarization of the participants with Drosophila as a model organism for investigating and teaching neuroscience. Participants began by directly handling Drosophila, in both larval and adult forms. Those who had no experience with this model system were guided in handling food vials containing fly stocks, transferring the animals into clear vials, anesthetizing them on ice, and observing them under the dissection scope. Hands-on familiarization with the system is often key to overcoming the barrier between conceptually understanding the value of Drosophila modules, and actually trying the experiments for the first time in their own teaching labs. Participants then observed free roaming behavior of adult Drosophila, followed by their response to specific circuit activation. Then, they observed third instar larval locomotion, and the response of larvae to motor neuron activation. Finally, they practiced dissecting larvae to expose the body wall muscles and associated nerves and observed evoked junctional potentials (EJPs) and miniature endplate potentials (minis) in muscle cells through intracellular recording, while stimulating presynaptic motor neurons with brief pulses of blue light. Participants could in this way merge theoretical understanding of genetic manipulation with actual data that they personally obtained in the workshop.
The second aim was to familiarize participants with the principles of optogenetics, and the application of these principles in a lab setting. By experimenting with the techniques required to prepare animals or neuromuscular junction preps, to set up intracellular recording, and to deliver light stimuli so as to elicit tightly controlled responses, participants could put their theoretical understating into an immediate, applied context. They experienced the physical setup, the range of light amplitudes, pulse durations, and stimulation frequencies that would result in the desired physiological effects. Participants could explore the dynamics of specific elicited behaviors, and experiment with short-term synaptic plasticity in a genetically accessible model synapse. As with handling flies, this served to overcome another barrier to implementation of the demonstrated exercises.
MATERIALS AND METHODS
Fly Genetics
All optogenetic behavior lines were raised on standard cornmeal-molasses fly food with the addition 0.5mM all-trans retinal at 25 degrees Celsius in foil-covered vials. The adult behavior lines used were the “Moonwalker”-20XUAS-IVS-CSChrimson.mVenus attP18;; VT050660-GAL4 attP2 and the “Jumping”-20XUAS-IVS-CSChrimson.mVenus attP18;; R42E06-GAL4 attP2. The optogenetic line used in the electrophysiology recording from the larval NMJ is w; OK371-GAL4, UAS-ChR2-H134-Cherry, MHC-GFP;+, in a wild type background and with the mutant allele SNAP-25ts on chromosome III.
Adult Behavior
Adult flies, 3–5 days old were anesthetized on ice and loaded into glass test tubes with cotton caps and covered with aluminum foil. Behavioral activity was observed by eye or under a dissecting microscope during optogenetic stimulation.
Larval Electrophysiology
Wandering third instar larvae (large white, mobile larva found on the walls of the vial) were selected with forceps, placed in a sylgard petri dish, and rinsed once in HL3 saline lacking Ca2+ consisting of (in mM): 70 NaCl, 20 MgCl2, 5 KCl, 10 NaHCO3, 5 trehalose, 115 sucrose (Stewart et. al., 1994). Larvae were prepared as described in Hornstein et al., 2009 and Pulver et al., 2011. Briefly, larvae were oriented trachea side up, pinned at the tail and mouth hooks with Minuten pins (Fine Science Tools). HL3 saline lacking Ca2+ was added, a superficial cut was made with micro-scissors from tail to mouth hooks, and the internal organs were carefully removed without damaging the body wall muscles or the CNS and its associated axons. Four more pins were used to splay out the larva at each corner so it resembled a fillet (Hornstein et. al., 2009). After rinsing the “fillet” with HL3 saline lacking Ca2+ (to prevent muscle contractions), the prep bathing solution was replaced with HL3 saline containing 3 mM CaCl2. Sharp glass electrodes (5–30 MΩ) filled with 3M KCl were used for intracellular recordings. A ground electrode was placed in the dish and the intracellular electrode was advanced slowly into one of the body wall muscles. Altering the fiber lights so they illuminate from the side often helps to visualize the musculature. Commonly, muscle 6 or 7 were used as shown in Fig. 2A. A resting potential below −40 mV is usually sufficient to see minis in wild type and SNAP-25ts larvae. Signals were amplified with a model AM-Systems 1600. The resulting signals were digitized at 20 KHz with a Powerlab 8/30 data acquisition system (ADInstruments, Colorado Springs, CO, USA) and recorded and analyzed in LabChart 8 (ADInstruments). Once a satisfactory resting potential was achieved, the dissection scope illumination was extinguished, and minis could be recorded (Fig. 3). To elicit evoked release, a high intensity blue LED was positioned closed to the preparation with the light aimed directly at the larva. A single stimulus of 2–5 ms, at maximum intensity, was usually sufficient to observe evoked release. The length of the pulse could be increased to 50 ms if no initial response was observed.
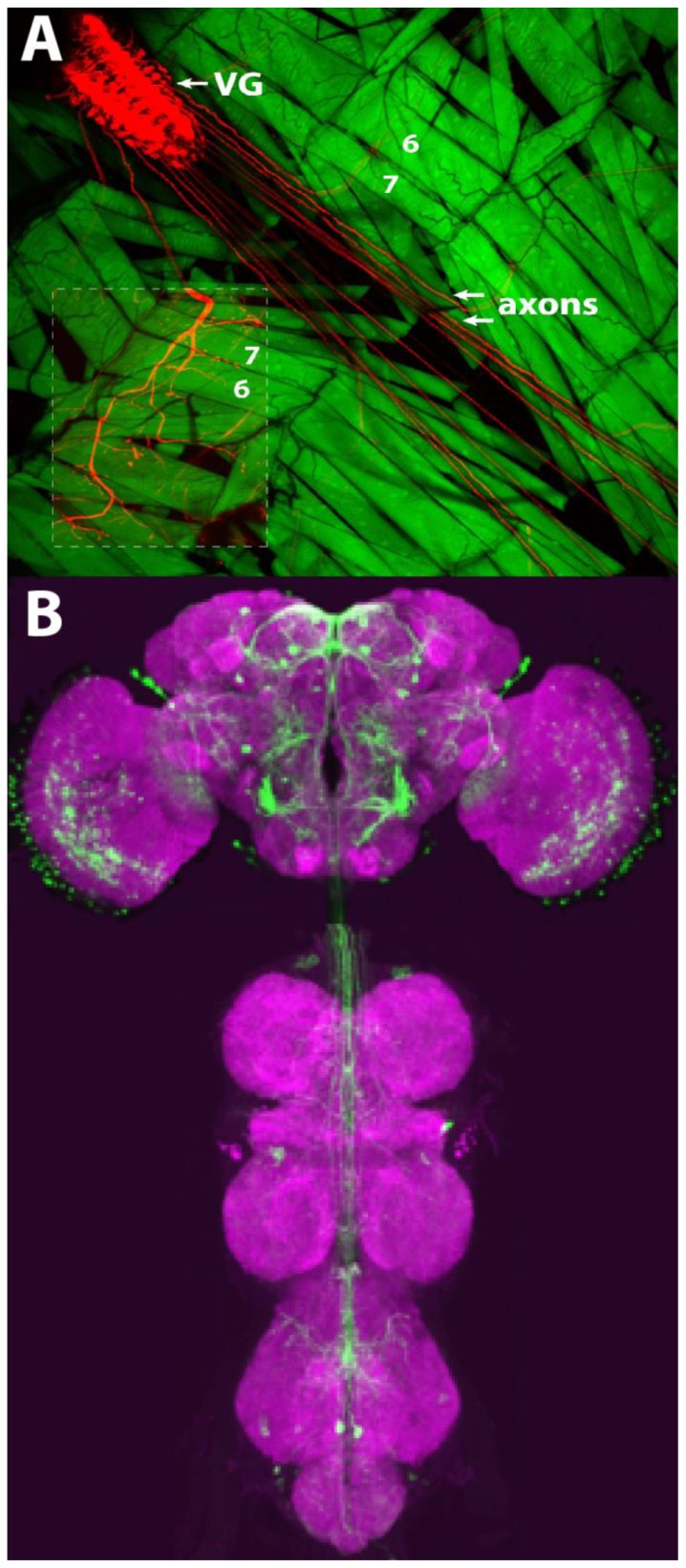
Figure 2.
Drosophila adult and larval CNS. (A) Larval OK371-GAL4 driving expression of mCherry-tagged channelrhodopsin2 in motor neurons of the ventral ganglion (VG) and GFP expression in muscle. Note expression of channelrhodopsin2 in motor neuron axons. Location of muscles 6 and 7 are indicated. The inset has increased brightness of mCherry to show the innervation of the neurons at the muscles. (B) Confocal image of Moonwalker-GAL4 driving expression of GFP in adult brain (green) and the presynaptic marker BRP (magenta).
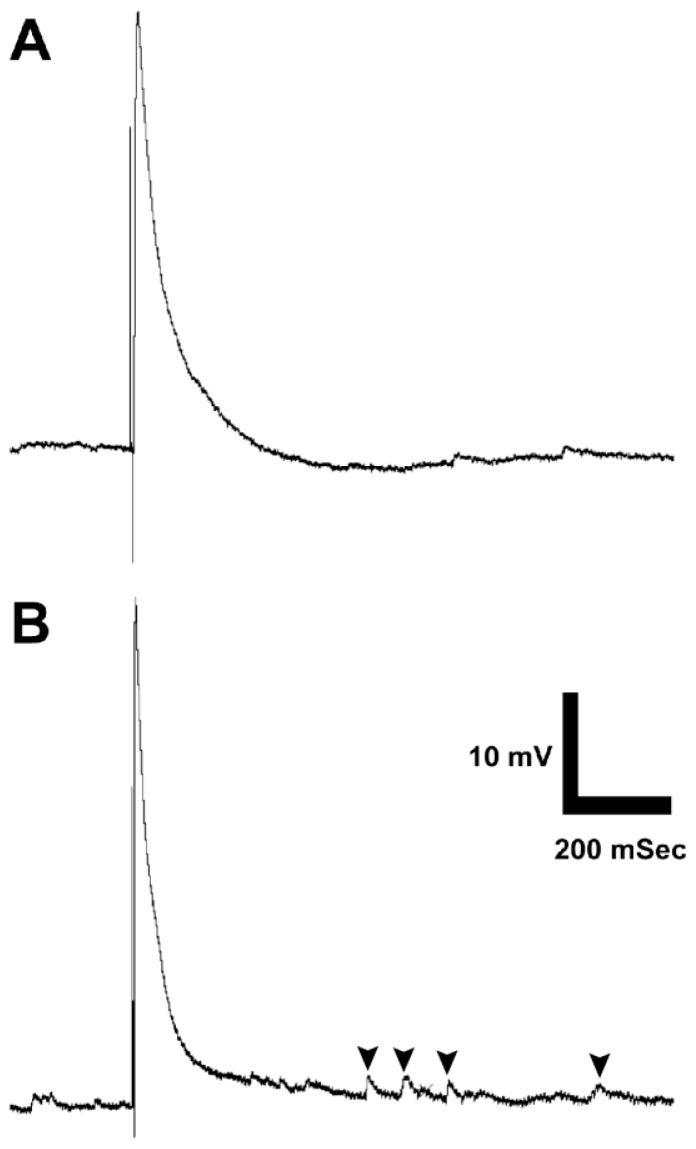
Figure 3.
Sample student recordings from the Drosophila third install larva neuromuscular junction. Evoked potentials are elicited by brief pulses of blue light from a high power LED in transgenic strains expressing Channelrhodopsin 2 in glutamatergic neurons. (A) Wild type evoked potential and miniature endplate potentials. (B) SNAP-25ts, showing characteristically elevated frequency of miniature endplate potentials indicated by arrowheads.
Optogenetic Stimulation
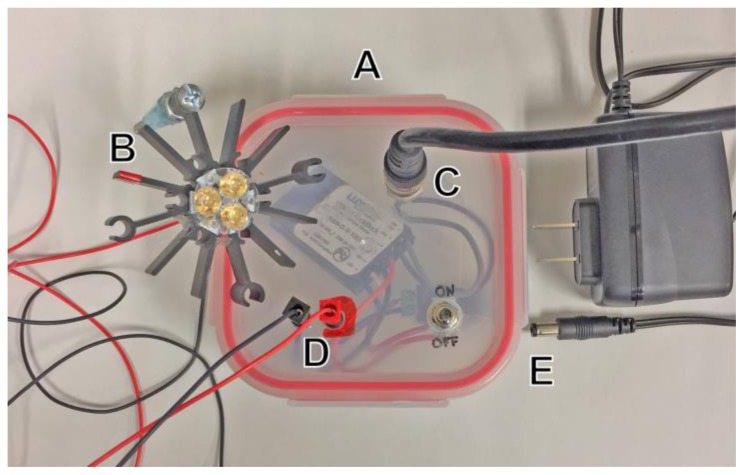
Construction of a simple, voltage controllable LED current driver (Fig. 1) was based on the BuckBLock DC LED driver by LEDdynamics (LEDsupply.com, part # 0A009-D-V-1000). The blue and red LEDs used were Luxeon Rebel 3up LED packages on star bases (LEDsupply, parts # 07007-PB000-D and 07007-PD000-F, respectively). LEDs were mounted with thermal adhesive on a star heat sink (Digikey.com, part # 345-1105-ND). Light intensity was controlled by the 0–10 Volt analog output of a PowerLab 26T (ADInstruments, Sydney, Australia).
Figure 1.
LED and power supply assembly. (A) A voltage controlled current source is mounted in an enclosure that holds input, output, power, and switch components. (B) Blue (460 nm) or Red (625 nm) LED is mounted on a heatsink. (C) Port for voltage control. (D) Power output ports for LED. (E) Switching power supply and input. LED light source apparatus modified and upgraded from Pulver et al., 2011.
Free crawling larvae were stimulated with 500 ms pulses of 460 nm blue light using a computer controlled LED (Fig. 1). Dissected larval fillets were stimulated with this wavelength, but with brief pulses lasting 2–50 ms. Optogenetic stimulation for behavioral responses was induced with brief exposure to light activation with a red (620–625nm) CREE XP-E LED Flashlight available from Amazon or from a computer controlled red LED light source (Fig. 1).
Imaging
The adult CNS was imaged with the methods detailed in Jennet et al., 2012. OK371-GAL4, UAS-ChR2-H134-Cherry, MHC-GFP;+, larvae was dissected as described for electrophysiology in HL3 saline lacking Ca2+. The native mCherry and GFP were imaged with a Zeiss LSM 880 with a 10X water immersion objective. The mCherry signal in the inset of the image was intensified using ImageJ.
Workshop Participant Evaluations
Feedback on our fly workshop sessions was gathered by asking participants to answer the following questions that the workshop organizers provided, on a Likert Scale of 1–5, with 1-strongly agree to 5-strongly disagree: (1) this workshop met my expectations, 2) this workshop increased my understanding of the topic material, 3) this workshop was well organized with clear objectives, 4) the exercise presented at this workshop is a good vehicle for teaching undergraduates principles of neuroscience, 5) I am likely to incorporate this material as a classroom activity, and 6) I am likely to use the material in this workshop as a resource for my class lectures and my own neuroscience background.
RESULTS
Behavioral responses to optogenetic stimulation
The first exercise examined the effects of red LED light on the adult flies as they freely moved in an empty glass test tubes and the effects of blue light on crawling larvae. Illumination was accomplished through a high intensity blue or red LED powered by a simple current source and controlled via software, using the device depicted in Figure 1. Adult flies could also be stimulated by an inexpensive red LED flashlight. The larvae were of the same strain used later for intracellular recording at the NMJ, which expressed ChR2 under control of the OK371 promoter, driving expression in glutamatergic neurons (Fig. 2A). Illumination of optogenetically activatable larvae resulted in a characteristic “seizure” behavior. Larvae stopped crawling and contracted into a tight oval for the duration of the light stimulus. Sometimes, if the larvae were crawling in a particular direction, the brief seizure acted as an aversive shock, causing them to quickly change direction. For adult behavior, two different genotypes were tested: the Moonwalker-GAL4 and the Jumping-GAL4, each expressing CsChrimson, the red-sensitive channelrhodopsin, in different sets of neurons. Red-shifted channelrhodopsin was used in the adult experiment since red light is easily transmitted through the adult cuticle than blue. After removing the foil and observing fly behavior under low light conditions, a red light beam from an inexpensive LED flashlight or a high intensity LED was used to briefly illuminate the vial (see Supplementary Video for a depiction of Jumping-GAL4 stimulation). Workshop participants observed, either with the naked eye or under a dissecting scope, changes in behavior when the light was on. Vials were tested several times to determine the reproducibility and the nuances of the behavior. The red light triggers the “moonwalker” flies to walk backwards while the “jumping” flies jump excitedly (as if exhibiting a startle response) in the tube as long as the light shone. The expression pattern of Moonwalker-GAL4 is shown in Figure 2B. Here, the Moonwalker Gal4 driven GFP is expressed in a set of central neurons in sensory and motor neuropils. These neurons are sufficient to elicit the backward walking escape response.
Electrophysiological responses to optogenetic stimulation
The second exercise examined neurotransmitter release at the third instar larval neuromuscular junction (NMJ). Instead of electrically stimulating specific motor neuron axons to trigger neurotransmitter release, blue light is used to generate action potentials in all the motor neurons and an intracellular electrode inserted in the muscle records the synaptic events. This is a much easier way to stimulate Drosophila motor neurons than using conventional direct electrical stimulation (Pulver et al., 2011). Either wild type larva or SNAP-25ts mutant larva expressing the channelrhodopsin2 can be used. The SNAP-25ts mutant has the advantage of having a very high frequency of spontaneous vesicle release (minis) allowing students to observe these small synaptic events easily. Wandering third instar larva from vials expressing mCherry tagged channelrhodopsin2 in motor neurons (Fig. 2A) were investigated.
A sample fillet can be viewed under a fluorescent dissecting scope to acquaint students with the anatomy of the ventral ganglion and body wall muscles with the fluorescent proteins mCherry and GFP, respectively (Fig. 2A). A typical evoked response from wild type animals is observed in Fig. 3A. The amplitude of the evoked potential varies with resting potential: the best recordings were obtained with resting potentials of −60 mV or more negative, but reasonable data could be obtained even with suboptimal resting potentials of as low as −30. Minis were generally not visible at resting potentials less negative than −40 mV. Fig. 3B shows a typical response in SNAP-25ts larvae, which displays a characteristically elevated mini frequency, and occasionally demonstrates increased evoked potential amplitude. Students may vary the frequency of the stimulus to observe either synaptic potentiation or depression.
Workshop Session Feedback
Feedback on our fly workshop sessions by 42 of the 75 participants who attended and signed our mailing list indicated an overall very positive response to our activities. Participants rated our workshop sessions with the following questions on a scale of 1 (strongly agree) to 5 (strongly disagree); mean responses +− SD after each question: 1) this workshop met my expectations (1.44 +− 0.709); 2) This workshop increased my understanding of the topic material (1.48 +− 0.724); 3) This workshop was well organized with clear objectives (1.56 +− 0.709; 4) The exercise presented at this workshop is a good vehicle for teaching undergraduates principles of neuroscience (1.67 +− 0.972); 5) I am likely to incorporate this material as a classroom activity (2.04 +− 1.075), and 6) I am likely to use the material in this workshop as a resource for my class lectures and my own neuroscience background (1.83 +− 1.028). Mean participant responses expressed satisfaction with the workshop material as useful and interesting neuroscience teaching resources. Less positive scores for using the material in class or as a teaching resource may be due to inexperience with fly biology and the lack of electrophysiological equipment in some neuroscience programs.
DISCUSSION
Neuroscience research is characterized by constant change, adaptation, and adoption of new techniques. The described laboratories at the 2017 Faculty for Undergraduate Neuroscience Workshop highlighted the use of optogenetics, arguably the most influential and transformative technology to have been developed in recent years. This method uses molecular genetic techniques to make a wide variety of neural circuits controllable remotely by pulses of light. Though powerful, the technique is easily implemented in the teaching environment as well as the research lab. The prep used was Drosophila, the classical genetic model organism that combines ease of use, cost effectiveness, and unparalleled versatility in demonstrating behavior and neuronal physiology.
Optogenetic control of specific central circuits in the Drosophila brain can drive a variety of behaviors. Our lab exercises focused on locomotor behaviors, because these feature the most robust responses and are most easily observed by students and researchers new to the model system. The startle response is elicited by a small group of neurons within the brain, and is readily apparent, even without the use of a dissection scope. The collection of transgenic lines that drive expression in specific circuits and elicit specific behaviors is increasing, and these can often be readily incorporated into teaching labs. Well-characterized examples include a strain that expresses ChR2 in gustatory neurons and drives a proboscis extension reflex upon activation (Inagaki et al., 2014), and a strain that expresses CsChrimson in a central pattern that includes the giant fiber neurons, driving an escape response similar to the one presented here (Klapoetke et al., 2014, Titlow et al., 2015). Many lines driving expression in sparse groups of neurons remain to be characterized (Jenett et al., 2012). Undergraduates in a lab setting can observe and record videos of the response using their now-ubiquitous cell phones. Behavioral responses can be categorized by the use of ethogram analysis, where discrete, stereotyped actions are defined, quantified, and linked into series of more complex behavioral response repertoires (Chen et al., 2002). Students can use their observations, whether live or reviewed on video, and develop their own ethogram definitions. These can then be compared to literature reports (Branson et al., 2009), and be used as a basis for self-directed research projects comparing different transgenic and mutant lines in an optogenetically activatable background (McKellar and Wyttenbach, 2017).
Synaptic plasticity is a primary component of development, learning, and memory (Harris and Littleton, 2015). A key goal of neuroscience education is to give students the training and the platform to discover and experiment with these fundamental processes. The Drosophila third instar neuromuscular junction demonstrates the principles of synaptic transmission in an easily accessible and controllable prep. By varying pulse number and frequency, the participants could demonstrate short-term synaptic dynamics. At typical physiological levels of extracellular calcium (about 3 mM), the neuromuscular junction at the large central muscles in Drosophila larvae tend to show mild synaptic depression (Kidokoro et al., 2004). Reducing extracellular calcium or recording from some smaller muscles within the larval segment, yields smaller amplitude EJPs, but allows observation of short-term synaptic facilitation, as each evoked potential amplitude is larger than the previous one. Participants began to conduct their own explorations, testing the precise dynamics of the synapse they were recording. Experiments included testing the frequency dependence of synaptic depression or facilitation, the maximal firing rate of the synapse, the recovery time required to return to baseline synaptic strength, and longer-term adaptation or potentiation dynamics. In addition, these synapses often show detectable minis. Thus, participants could estimate the mini frequency and potentially perform simple quantal analysis to determine the average quantal content of an EJP.
Recent studies have begun to address questions of synaptic transmission and synaptic plasticity using optogenetics (Watanabe et al., 2013). However, these processes remain relatively little studied in the Drosophila larval NMJ model synapse. This offers an opportunity for students to design protocols to explore various aspects of synaptic transmission with this novel way of remotely controlling neuronal activity (Pulver et al., 2011). The workshop participants had the opportunity to try optogenetic activation of motor neurons in an electrophysiologically interesting mutant, SNAP-25ts, a temperature sensitive paralytic mutant allele of the SNAP-25 gene (Rao et al., 2001). In the larval stage, this mutation results in a greatly elevated mini frequency, and often a slightly elevated EJP amplitude. Other mutants that affect synaptic transmission and synaptic dynamics are open to exploration by students using the application of optogenetic techniques. For example, ion channel mutants such as Shaker (Salkoff et al., 1992), or vesicle recycling mutants such as Shibire (van der Bliek and Meyerowitz, 1991), are prime candidates for application in the teaching lab. These experiments are all the more impactful because all of the molecular components listed have direct homologues in humans, with fundamentally conserved function. Taken together, these exercises gave the participants a way to demonstrate and explore the molecular and physiological principles of synaptic transmission though optogenetic stimulation.
Tying together genetics, molecular biology, neurophysiolology and behavior was a key aim of our workshop presentation. Organismal behavior can be directly observed, and neuronal activity can be directly recorded, but the genes and proteins underlying these processes are, for most undergraduates, mostly hypothetical and inaccessible. Optogenetics applied to a highly genetically manipulable animal allows students to make direct connections between molecules and behavior. These connections become tangible as students perform these experiments themselves, using the same techniques that are at the cutting edge of current research. This link between theory and practice not only promotes learning and understanding, but also serves as a platform for discovery in the teaching lab that can be used to conduct original investigations. Finally, the lab modules presented serve to familiarize participants with the tools of optogenetic stimulation, and the Drosophila prep, in both the adult and larval form, in order to overcome the novelty barrier that can delay adoption of new approaches to teaching and research.
Footnotes
This work was supported by the College of Arts and Sciences, University of Cincinnati and the Departmental of Neurobiology and Behavior, Cornell University. Support for Karen Hibbard comes from HHMI, Janelia Research Farms Campus. We thank Rajyashree Sen for the Moonwalker image, Gudrun Ihrke for assistance with imagining the larval CNS, and Annette Stowasser for technical advice on constructing the LED current source circuit. The lab exercises presented here are part of the CrawFly Faculty Workshops supported by ADInstruments, Inc. as part of the Classroom of Excellence program at Cornell, Emory, the University of Cincinnati and other CrawFly hosting institutions.
REFERENCES
- Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, Ehlers MD, Feng G. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Branson K, Robie AA, Bender J, Perona P, Dickinson MH. High-throughput ethomics in large groups of Drosophila. Nat Methods. 2009;6:451–457. doi: 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lee AY, Bowens NM, Huber R, Kravitz EA. Fighting fruit flies: a model system for the study of aggression. Proc Natl Acad Sci U S A. 2002;99:5664–5668. doi: 10.1073/pnas.082102599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne H, Hibbard KL, Cavallaro A, Kao J-C, Rubin GM. Genetic reagents for making split-GAL4 lines in Drosophila. Genetics. 2018;209:31–35. doi: 10.1534/genetics.118.300682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KP, Littleton JT. Transmission, development, and plasticity of synapses. Genetics. 2015;201:345–375. doi: 10.1534/genetics.115.176529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein NJ, Pulver SR, Griffith LC. Channelrhodopsin2 mediated stimulation of synaptic potentials at Drosophila neuromuscular junctions. J Vis Exp. 2009;16(25) doi: 10.3791/1133. pii:1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki HK, Jung Y, Hoopfer ED, Wong AM, Mishra N, Lin JY, Tsien RY, Anderson DJ. Optogenetic control of Drosophila using a red-shifted channelrhodopsin channelrhodopsin reveals experience-dependent influences on courtship. Nat Methods. 2014;11:325–332. doi: 10.1038/nmeth.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo T-TB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalanithi PS, Henderson JM. Optogenetic neuromodulation. Int Rev Neurobiol. 2012;107:185–205. doi: 10.1016/B978-0-12-404706-8.00010-3. [DOI] [PubMed] [Google Scholar]
- Kidokoro Y, Kuromi H, Delgado R, Maureira C, Oliva C, Labarca P. Synaptic vesicle pools and plasticity of synaptic transmission at the Drosophila synapse. Brain Res Brain Res Rev. 2004;47:18–32. doi: 10.1016/j.brainresrev.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li C, Xing Y, Wang Y, Tao F. Role of neuromodulation and optogenetic manipulation in pain Treatment. Curr Neuropharmacol. 2016;14:654–661. doi: 10.2174/1570159X14666160303110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKellar CE, Wyttenbach RA. A protocol demonstrating 60 different Drosophila behaviors in one assay. J Undergrad Neurosci Educ. 2017;15:A110–A116. [PMC free article] [PubMed] [Google Scholar]
- Moser T. Optogenetic stimulation of the auditory pathway for research and future prosthetics. Curr Opin Neurobiol. 2015;34:29–36. doi: 10.1016/j.conb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Nieh EH, Kim S-Y, Namburi P, Tye KM. Optogenetic dissection of neural circuits underlying emotional valence and motivated behaviors. Brain Res. 2013;1511:73–92. doi: 10.1016/j.brainres.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokala K, Glater EE. Using optogenetics to understand neuronal mechanisms underlying behavior in C. elegans. J Undergrad Neurosci Educ. 2018;16:A152–A158. [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Berni J. The fundamentals of flying: simple and inexpensive strategies for employing Drosophila genetics in neuroscience teaching laboratories. J Undergrad Neurosci Educ. 2012;11:A139–A148. [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Pashkovski SL, Hornstein NJ, Garrity PA, Griffith LC. Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J Neurophysiol. 2009;101:3075–3088. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Hornstein NJ, Land BL, Johnson BR. Optogenetics in the teaching laboratory: Using channelrhodopsin-2 to study the neural basis of behavior and synaptic physiology in Drosophila. Adv Physiol Educ. 2011;35:82–91. doi: 10.1152/advan.00125.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Stewart BA, Rivlin PK, Vilinsky I, Watson BO, Lang C, Boulianne G, Salpeter MM, Deitcher DL. Two distinct effects on neurotransmission in a temperature-senitive SNAP-25 mutant. EMBO J. 2001;20:6761–6771. doi: 10.1093/emboj/20.23.6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK. Demonstrating connections between neuron signaling and behavior using C. elegans learning assays and optogenetics in a laboratory class. J Undergrad Neurosci Educ. 2018;16:A223–A231. [PMC free article] [PubMed] [Google Scholar]
- Salkoff L, Baker K, Butler A, Covarrubias M, Pak MD, Wei A. An essential “set” of K+ channels conserved in flies, mice and humans. Trends Neurosci. 1992;15:161–166. doi: 10.1016/0166-2236(92)90165-5. [DOI] [PubMed] [Google Scholar]
- Scholl HPN, Strauss RW, Singh MS, Dalkara D, Roska B, Picaud S, Sahel JA. Emerging therapies for inherited retinal degeneration. Sci Transl Med. 2016;8:368rv6. doi: 10.1126/scitranslmed.aaf2838. [DOI] [PubMed] [Google Scholar]
- Sen R, Wu M, Branson K, Robie A, Rubin GM, Dickson BJ. Moonwalker descending neurons mediate visually evoked retreat in Drosophila. Curr Biol. 2017;27:766–771. doi: 10.1016/j.cub.2017.02.008. [DOI] [PubMed] [Google Scholar]
- Sjulson L, Cassataro D, DasGupta S, Miesenböck G. Cell-specific targeting of genetically encoded tools for neuroscience. Annu Rev Genet. 2016;50:571–594. doi: 10.1146/annurev-genet-120215-035011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KS, Graybiel AM. Using optogenetics to study habits. Brain Res. 2013;1511:102–114. doi: 10.1016/j.brainres.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- Titlow JS, Johnson BR, Pulver SR. Light activated escape circuits: a behavior and neurophysiology lab module using Drosophila optogenetics. J Undergrad Neurosci Educ. 2015;13:A166–A173. [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Rost BR, Camacho-Pérez M, Davis MW, Söhl-Kielczynski B, Rosemund C, Jorgensen EM. Ultrafast endocytosis at mouse hippocampal synapses. Nature. 2013;504(7479):242–247. doi: 10.1038/nature12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Stewart B. Synaptic electrophysiology of the Drosophila neuromuscular junction. In: Zhang B, Freeman MR, Waddell S, editors. Drosophila neurobiology: a laboratory manua. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2010. pp. 171–187. [DOI] [PubMed] [Google Scholar]