Abstract
CCAAT/enhancer binding protein β (C/EBPβ) is required for murine mammary ductal morphogenesis and alveologenesis. Progesterone is critical for proliferation and alveologenesis in adult mammary glands, and there is a similar requirement for progesterone receptor isoform B (PRB) in alveologenesis. We examined C/EBPβ regulation of PR expression. All three C/EBPβ isoforms, including typically inhibitory LIP, transactivated the PR promoter. LIP, particularly, strongly synergized with c-Jun to drive PR transcription. Endogenous C/EBPβ and c-Jun stimulated a PR promoter-reporter and these two factors showed promoter occupancy on the endogenous PR gene. Additionally, LIP overexpression elevated endogenous PR protein expression. In pregnancy, both PRB and the relative abundance of LIP among C/EBPβ isoforms increase. Consistent with a role in PRB expression, in vivo C/EBPβ and PR isoform A expression showed mutually exclusive localization in mammary epithelium, while C/EBPβ and PRB largely co-localized. We suggest a critical role for C/EBPβ, particularly LIP, in PRB expression.
1. Introduction
CCAAT/enhancer binding protein β (C/EBPβ) is a critical transcription factor in the regulation of murine mammary gland proliferation and development. Experiments with C/EBPβ-deficient mice demonstrate a requirement for C/EBPβ in ductal morphogenesis and alveologenesis (Robinson et al., 1998; Seagroves et al., 1998). C/EBPβ occurs in three isoforms in mammary and other tissues: C/EBPβ p38 (LAP1) and C/EBPβ p35 (LAP2), both potent transcriptional activators, and C/EBPβ p20 (LIP), a truncated form generally reported to inhibit C/EBP-dependent transcription (reviewed in Zahnow, 2002). During the course of pregnancy in the mouse, C/EBPβ protein expression increases, with LIP expression being particularly elevated, in contrast to not being detectable in the virgin mammary gland (Seagroves et al., 1998).
Progesterone signaling through the progesterone receptor (PR) is also a critical factor for proliferation, ductal morphogenesis and alveologenesis in the adult mammary gland (reviewed in Fendrick et al., 1998; Shyamala et al., 1998; Aupperlee et al., 2005; Aupperlee et al., 2007). Furthermore, experiments with mice deficient in PR isoform B (PRB) show a requirement of PRB for alveologenesis (Lydon et al., 1995; Mulac-Jericevic et al., 2003).
The block to alveologenesis in both C/EBPβ- and PRB-deficient mice suggests that these transcription factors might act in the same pathway or may regulate overlapping sets of downstream target genes. An overall decrease in PR observed in sexually mature wildtype mice fails to occur in C/EBPβ-deficient mice, while no alterations in C/EBPβ expression are observed in PR-deficient mice (Seagroves et al., 2000). This is consistent with C/EBPβ acting upstream of PR. PR isoform A (PRA) is the predominant isoform of PR expressed in the mammary glands of virgin adults, with its expression dramatically decreasing at pregnancy and PRB being expressed with alveolar development during pregnancy (Aupperlee et al., 2005). This raises the possibility that C/EBPβ is required for the differential upregulation and localization of PRB expression that is observed during pregnancy.
In this report, we examined whether C/EBPβ participates in the transcriptional regulation of PR expression in the murine mammary gland. Transient co-transfection of a PR promoter-reporter with expression vectors that individually express C/EBPβ isoforms into a murine mammary tumor cell line revealed that all C/EBPβ isoforms, surprisingly including LIP, can transactivate the PR promoter. Importantly, we found that LIP, in particular, robustly synergizes with c-Jun to drive PR transcription. Consistent with significant roles for C/EBPβ and c-Jun in PR expression, knockdown experiments showed that endogenous levels of C/EBPβ and c-Jun expression are sufficient to stimulate the PR promoter-reporter. Additionally, overexpression of LIP elevates PR protein expression from the intact endogenous gene encoding PR. Furthermore, in vivo immunofluorescence studies showed that the localization of C/EBPβ and PRA expression are mutually exclusive in the mammary epithelium, while PRB is expressed in cells that express C/EBPβ. Collectively, our data suggest a critical role for C/EBPβ, particularly LIP, in PRB expression.
2. Materials and methods
2.1. Mice
Female BALB/c mice were purchased from Harlan (Indianapolis, IN, USA). All animal experimentation was conducted in accord with accepted standards of humane animal care under guidelines approved by the All University Committee on Animal Use and Care at Michigan State University
2.2. Cells and cell culture
MC7-L1, MC4-L2, and MC4-L3 are mammary cell lines of epithelial origin, derived from murine mammary ductal carcinomas (Lanari et al., 2001; a gift from Dr. Claudia Lanari, Universidad de Buenos Aires, Buenos Aires, Argentina). These cell lines express both ER and PR. MC7-L1 and MC4-L2 are hormone responsive in vitro and MC4-L3 is hormone responsive in vivo. Cells were maintained in DMEM-F12 (1:1) medium supplemented to 5% FCS, 100 units/ml penicillin and 100mg/ml streptomycin. Experiments were carried out with charcoal-stripped FCS in the absence of antibiotics. Cells were cultured at 37°C with 5% CO2.
2.3. Expression vectors and promoter-reporters
For transient transfections, murine C/EBPβ isoforms were individually expressed from pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). pcDNA-LIP has been described (Dearth et al., 2001). pcDNA-LAP2 (plasmid 12557; Addgene, Cambridge, MA, USA) has been described (Basu et al., 2011). pcDNA-LAP1 was derived from a plasmid containing the complete coding sequence of murine C/EBPβ inserted between the EcoRI and HindIII sites of pcDNA3.1(−) (a gift from Dr. Peter Johnson, NCI-Frederick, Frederick, MD, USA). The ATG translational start sites for LAP2 and LIP were mutated to GCG and a consensus Kozak sequence introduced upstream of the LAP1 translational start, mutating GCGTTCATG to GCCACCATG (mutated bases underlined) by site-directed mutagenesis. pCMV-c-jun has been described (McCabe et al., 1996). Murine PRA and PRB were individually expressed from vectors derived from the complete murine PR cDNA (Schott et al., 1991). For PRA, a HincII/EcoRV fragment of the PR cDNA, lacking the PRB ATG translational start site, was ligated into the EcoRV site of pcDNA3.1(−). For PRB, a NheI/NotI fragment of the PR cDNA was ligated into the cognate sites of pcDNA3.1(+). The PRA ATG translational start site was mutated to GCG.
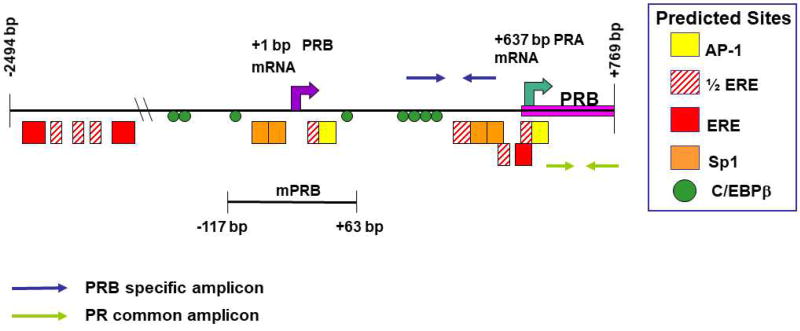
The tandem PR promoter-reporter (Fig. 1) consists of the region −2494 to +769 base pairs (bp) in relation to the predicted PRB transcriptional start site inserted between the Asp718 and NcoI sites of pGL3-Basic (pGL3B) (Promega, Madison, WI, USA). This region contains the putative transcriptional start sites for both PRB and PRA. The murine PR promoter region was isolated by PCR amplification of C57/Bl6 genomic mouse DNA using the following primers: − 2502 to −2471 bp, 5’-ACATGGTACCAGCGTGTCACCTGGCACAGA-3’ (containing an underlined Asp718 site); +771 to +753 bp, 5’-CTGTCCATGGACACGTCCGAGTGCTGGCT-3’ (containing an underlined NcoI site). TA cloning placed the PCR fragment into pCR2.1 (TA Cloning Kit; Invitrogen, Life Technologies, Grand Island, NY, USA). The promoter fragment was then excised with Asp718 and NcoI, and inserted into pGL3B. The minimal PR promoter-reporter consists of the region −117 to +63 base pairs (bp) in relation to the predicted PRB transcriptional start site (Fig. 1) and was constructed in two steps. First, PCR amplification using 5’-AGCACCTGCAACTTCACCTCTG-3’ (−444 to −423 bp) and 5’-TAGCAGAATGTCAGAA TCCTC-3’ (+43 to +63 bp) produced a promoter fragment that was placed into pCR2.1 by TA cloning, after which the promoter fragment was excised with Sac I and Xho I, and inserted into pGL3B. This construct was then digested with Spe I and religated to produce the −117/+63 PR promoter-reporter. 2×PRE-TK-luc (plasmid 11350; Addgene) (Giangrande et al., 2000) contains two copies of a consensus progesterone response element (PRE) upstream of the human thymidine kinase promoter. pRL-SV40 expresses Renilla luciferase from the SV40 early enhancer-promoter (Promega).
Fig. 1.
Map of the murine tandem PR promoter region (−2494 to +769 bp) used in these studies. Predicted AP-1, Sp-1, and estrogen-responsive element (ERE) half-sites are analogous on the basis of sequence comparisons to transcription factor binding sites in the human PR gene that have been experimentally confirmed (Petz at al., 2002; Schultz et al., 2003; Petz et al., 2004a; Petz et al., 2004b; Schultz et al., 2005). Also using sequence comparisons, the location of the 5 putative EREs mapped for the rat PR gene (Kraus et al., 1994) are indicated. The putative start sites for transcription of PRB and PRA mRNAs are analogous to those experimentally defined in human studies (Kastner et al., 1990). Examination of the DNA sequence in the region from −2494 to +769 bp of the murine PRB gene with MatInspector (Quandt et al., 1995; Cartharius et al., 2005) predicted potential C/EBPβ binding sites.
2.4. Transient transfections
Transient transfections were conducted in either 12-well cell culture plates or 6 cm culture plates containing 1 ml/well or 4 ml/plate, respectively, of DMEM-F12 supplemented to 5% charcoal-stripped FCS. 12-well cell culture plates were seeded with 5×104 cells/well and 6 cm culture plates with 5×105 cells 24 h prior to transfection. FuGENE 6 Transfection Reagent (Promega) and plasmid DNAs were mixed at a 2:1 ratio (volume:weight) in 50 or 100 µl of serum-free medium for 12-well and 6 cm culture plates, respectively. This mixture was incubated at room temperature for 30 min before addition to cell cultures. The DNA, totaling 500 ng or 5 µg per individual 12-well and 6 cm culture plate transfection, respectively, comprised 100 ng or 1 µg of the −2494/+769 PR promoter-reporter, 1 ng or 10 ng of pRL-SV40, and varying amounts of control “empty” pcDNA3.1, C/EBPβ expression vector and/or c-Jun expression vectors, as indicated in figure legends. All quantities of expression vector are expressed as ng per 5×104 cells. Cells were assayed 24 h after transfection. In experiments involving the −117/+63 PR promoter-reporter, luciferase values derived from pGL3B lacking any inserted sequences were subtracted from those of the −117/+63 PR promoter-reporter to eliminate background levels of luciferase expression driven by C/EBPβ and c-Jun responsive sequences in the parent vector. For experiments utilizing 2×PRE-TK-luc, a slightly modified transfection protocol was used. 400 ng of the 2×PRE-TK-luc promoter-reporter was transiently co-transfected with expression vector DNA totaling 200 ng, comprising 100 ng pcDNA-LIP and/or 100 ng pCMV-c-jun, or 200 ng pcDNA3.1. For experiments examining PRA and PRB activity, 2×PRETK-luc was co-transfected with expression vector DNA totaling 500 ng, comprising ratios of PRA and PRB vectors as described in Fig. 4B. These transfections were performed in a 12-well plate format and assayed 36 h after transfection. In some cases, progestin R5020 (PerkinElmer, Waltham, MA, USA) was added to transfected cells 3 h after transfection at a final concentration of 20 nM. Transfected cells were harvested, lysed, and analyzed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase values were normalized to the Renilla luciferase values as a control for transfection efficiency between individual samples. All transfections were carried out in duplicate and repeated at least three times.
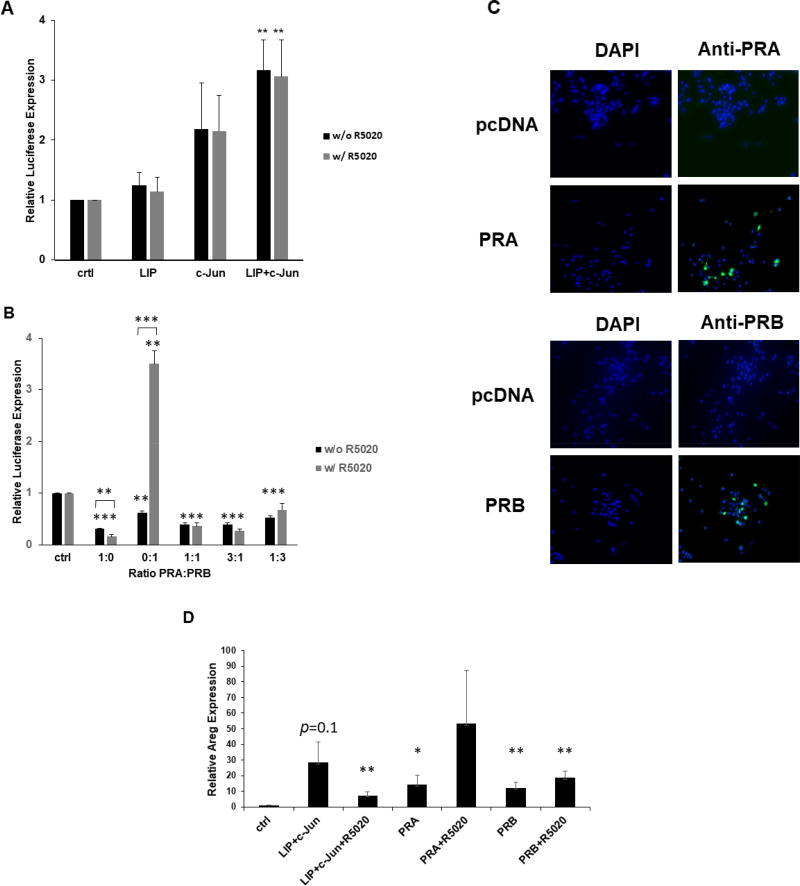
Fig. 4.
Co-expression of LIP and c-Jun increases 2×PRE promoter activity independently of R5020. (A) Transient co-transfection of C/EBPβ and c-Jun expression vectors were carried out in duplicate and repeated at least three times in MC7-L1 cells with the 2×PRE promoter-reporter. Values were normalized and statistics performed as described in Fig. 2. **, p < 0.05. (B) Transient co-transfection of PRA and PRB expression vectors, either singly or in various ratios as indicated, were carried out in duplicate or repeated at least three times in MC7-L1 cells with the 2×PRE promoter-reporter. Values were normalized and statistics performed as described in Fig. 2. **, p < 0.05; ***, p < 0.01. (C) Immunofluorescent detection of PRA and PRB in respectively transfected MC7-L1 cells. Transfection with empty pcDNA3.1 (pcDNA) vector served as a control. Left panels display DAPI (blue) staining. Right panels display PRA and PRB immunofluorescence (green) superimposed on the DAPI staining. (D) Transient co-transfection of 1 µg C/EBPβ and 1 µg c-Jun expression vectors, and 2 µg PRA and PRB expression vectors per 5×105 cells were carried out in duplicate and repeated three times in MC7-L1 cells. Transfected cultures were treated with and without R5020. Amphiregulin (Areg) and GAPDH RNA expression were quantitated by qRT-PCR. Amphiregulin values were normalized to GAPDH levels, and then normalized to a relative value of 1.0 for “ctrl” cells receiving only “empty” pcDNA3.1. *, p<0.1; **, p<0.05 for comparisons to “ctrl”.
In transient transfections assayed by the isolation of RNA or nuclear protein extracts, 5×105 cells were transfected in 6 cm culture plates under conditions similar to those described above. The quantities of expression plasmids are indicated in figure legends. Cells were harvested for RNA isolation or nuclear extracts as described below.
In transient transfections assayed by immunofluorescence, cells were grown to 30% confluence in 6-well cell culture plates on 22 mm round cover slips (Fisher Scientific, Pittsburgh, PA, USA) in 2 ml DMEM-F12 supplemented to 5% charcoal-stripped FCS. Cells were either transfected using the FuGENE 6 Transfection Reagent (Promega), as described above, or using the K2® Transfection System (Biontex, Munich, Germany). For the K2® Transfection System, cells were treated with 12 µl K2® Multiplier for 2 h before DNA transfection. K2® Multiplier was dripped slowly onto the medium with gentle mixing. For each transfection, 1 µg of plasmid DNA encoding LIP was mixed with 4µl of K2® Transfection Reagent in 100 µl DMEM-F12 medium and incubated at room temperature for 20 min before addition to the cells. After 24 h incubation, the cells on the cover slips were fixed, and then stained for immunofluorescent analysis.
2.5. Plasmid Immunoprecipitation (PIP)
The −117/+63 PR promoter-reporter was digested with ScaI to separate promoter sequences from the pGL3B backbone. Transient transfections were performed similarly to promoter-reporter assays, except the DNA transfection mix comprised 2 µg of Sca I-digested promoter-reporter, 1.5 µg of each expression plasmid, an amount of “empty” pcDNA3.1 to total 5 µg of total DNA, and 10 µl FuGENE 6 Transfection Reagent in 100 µl of serum-free DMEM/F12 medium. PIP was performed 24 h after transfection.
Trypsinized cells were suspended in DMEM/F-12 medium supplemented to 5% FCS. The cells were then crosslinked at 1% formaldehyde for 10 min at room temperature. Crosslinking was stopped by the addition of glycine to a final concentration of 0.125 M. The cells were washed twice with ice cold PBS, resuspended in cell-lysis buffer (5 mM Pipes [pH 8.0], 85 mM KC1, 0.5% NP40 with protease inhibitors [1mM dithiothreitol, 0.5 mM PMSF, 2.5 ug/ml leupeptin, 5 ug/ml antipain, 5 ug/ml aprotinin and 1 uM pepstatin A]), and then incubated on ice for 10 min. Nuclei were pelleted by centrifugation at 5000 rpm for 5 min at 4°C. The nuclei were resuspended in nuclear lysis buffer (50 mM Tris-Cl [pH 8.1], 10 mM EDTA, 1% SDS with protease inhibitors). The cells were then sonicated 10 times at 30% amplitude, 0.7 s on and 1.3 s off, over 30-s intervals on ice.
Each 100 µl protein-DNA complex was pre-cleared at 4°C overnight in 900 µl PIP dilution buffer (0.01% SDS, 1.1% Triton X100, 1.2 mM EDTA, 16.7 mM Tris-Cl [pH8.1], 167 mM NaCl with protease inhibitors) and 80 µl of a 50% (v/v) Protein G-Agarose (Thermo Scientifc Pierce, Waltham, MA) slurry containing 20 µg salmon sperm DNA and 1 mg/ml BSA, and stored in 4°C for later use. Equal amounts of pre-cleared protein-DNA complex in PIP dilution buffer were incubated with 2 µg either rabbit anti-C/EBPβ (C-19; Santa Cruz Biotechnology, Dallas, TX), rabbit anti-c-Jun (H-79; Santa Cruz Biotechnology), or normal rabbit IgG (Santa Cruz Biotechnology) at 4°C overnight. Immunoprecipitated products were collected after 2 h incubation with 50 µl of pre-coated Protein G-Agarose beads. Protein G Agarose beads were pre-coated with 20 µg salmon sperm DNA and l mg/ml BSA at 4°C overnight, and stored at 4°C for later use.
Immunoprecipitated material was washed sequentially in TSE buffer (20 mM Tris [pH 8.1], 0.1% SDS, 2mM EDTA, 1% Triton X-100), TSE buffer plus 150 mM NaCl, and TSE buffer plus 500 mM NaCl, and finally in buffer III (10 mM Tris [pH 8.0], 1 mM EDTA, 0.25 M LiCl, 1% NP-40, 1% deoxycholate). Each wash step was performed twice for 15 min at room temperature with a rotating plate. Beads were then washed twice in TE buffer (20 mM Tris [pH 8.0], 2 mM EDTA), and protein-DNA complexes were eluted in 500 µl of 0.1 M NaHC03-l% SDS for 30 min at 65°C. Crosslinking of the protein-DNA complexes was reversed, and RNA and protein removed by overnight incubation with 2 µl of 10 mg/ml RNAse A at 65°C, followed by incubation with 2 µl of 20 mg/ml Proteinase K at 42°C for 2 h. DNA was then purified by phenol-chloroform extraction and ethanol precipitation, and subsequently dissolved in 50 µl of TE buffer.
DNA from the input and immunoprecipitated samples were subjected to quantitative polymerase chain reaction (qPCR) with specific primers in triplicate. The primers for the −117/+63 PR promoter were 5’-CTAGCAAAATAGGCTGTCCC-3’ and 5’-CTTTATGTTTTTG GCGTCTTCCA-3’. The primers for the pGL3B backbone region were 5’-GCGACACGGAAATGTTGAATAC-3’ and 5’-CTACGTGAACCATCACCCTAATC-3’. qPCR was performed with the ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Inc., Foster City, CA) using the following program: step 1, 95°C for 10 min; step 2, 40 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s; step 3, 72°C for 5 min. qPCR was also performed with additional primers for the pGL3B backbone region as a control. The amplified qPCR products from immunoprecipitated DNA are presented as fold-enrichment compared to normal rabbit IgG immunoprecipitation products.
2.6. Chromatin immunoprecipitation (ChIP)
ChIP was performed using the EZ-Magna ChIP™ A/G Chromatin Immunoprecipitation Kit (Millipore Sigma) according to manufacturer’s protocol. MC7-L1 cells were harvested at 80% confluence, crosslinked, lysed, and then sonicated to produce 500–1000 bp chromatin fragments. Chromatin complexes were precipitated with 5µg of rabbit anti-C/EBP beta (C-19; Santa Cruz Biotechnology), anti-c-Jun (ab31419; Abcam, Cambridge, MA, USA), or normal rabbit IgG (Abcam) in 50 µl of the sonicated samples. DNA from the input and immunoprecipitated samples were subjected to qPCR with specific primers in triplicate. The primers were: −2220/−2008 bp: 5'-CACTGCTTCACACAGGTGCT-3” and 5'-TCCCAGAATCCTCTTGGCTA -3’; −1621/−1448 bp: 5' -TGCAAAGAAAGGAGGAGGAA- 3’ and 5'-TGGGAAGCAAAGAAATGAGG-3’; −1082/−867 bp: 5'-GGCATTTCCCAAACGACTAA-3’ and 5'-AATGGCATT TTCTCCCCTCT-3’; −645/−485 bp: 5'-GGCAGCCAGTAAGGTTGTTC and 5'-GCAGCAAGT GCTCTGTGTGT-3’; −202/−23 bp: 5'- GGGCTGGCATGCTTCTTAAT and 5'- CGTTGGCGCTCTAGGAAG-3’. qPCR was performed with the ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Inc.) using the following program: step 1, 95°C for 10 min; step 2, 40 cycles of 95°C for 15 sec, 50°C for 30 sec, and 65°C for 45 sec.
2.7. RNA isolation and quantitative RT-PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. For RNA isolation in the experiments examining PR promoter-reporter, the Agilent Total RNA Isolation Mini Kit (Agilent Technologies, Santa Clara, CA, USA) was used to eliminate residual transfected plasmid DNA. RNA was reverse-transcribed into cDNA using the RT2 First Strand Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s protocol. cDNA reactions lacking reverse transcriptase were performed to confirm elimination of DNA. cDNAs (20 µL) were diluted to 60 µl with deionized H2O. For qRT-PCR, each reaction (15 µl) included 7.5 µl of RT2 SYBR Green qPCR Mastermix (Qiagen), 1 µl of diluted first-stand cDNA synthesis reaction, 1 µl of primer, and 5.5 µl of deionized H2O. qRT-PCR was performed with the ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Inc.) using the following program: step 1: 95°C, 10 min; step 2: 40 cycles of 95°C for 15 sec, 56°C for 30 sec, and 72°C for 45 sec; step 3: dissociation curve at 95°C for 1 min, 65°C for 2 min (optics off), and 65–95°C at 2°C per min (optics on). Primers 5’-AAAGGATCCGCAGGTTCTC-3’ and 5’-GTTCCATCTTCCAGCGGATA-3’ were used for amplification of total PR transcripts. Primers 5’-CCCAGTTCTCAGACCAGACC-3’ and 5’-GTGGGATCTCCACCTCCTG-3’ were used for amplification of PRB transcripts. The positions of these primers are indicated in Fig. 1. Detection of amphiregulin and GAPDH utilized commercial primers (Real Time Primers, Elkins Park, PA, USA). qRT-PCR was performed with the ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Inc.) using the following program: step 1: 95°C, 10 min; step 2: 40 cycles of 95°C for 15 sec and 60°C for 1 min.
2.8. Small interfering RNA (siRNA)
C/EBPβ siRNA (sc-29862), c-Jun siRNA (sc-29224), and control siRNA (sc-37007) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Transient transfections of siRNAs were performed in 12-well cell culture plates containing 1 ml/well of DMEM-F12 supplemented to 5% charcoal-stripped FCS. Each well was seeded with 5×104 MC7-L1 cells 24 h prior to transfection. 200 ng plasmid DNA of the −2494/+769 PR promoter-reporter, 1 ng of pRL-SV40, and either 10 pmol C/EBPβ siRNA, 25 pmol c-Jun siRNA, both siRNAs, or 25 pmol control siRNA were diluted in 60 µl serum-free medium (solution A). 2 or 3 µl of Lipofectamine 2000 (Life Technologies), respectively for C/EBPβ or c-Jun and control siRNAs, was diluted in 60 µl serum-free medium, and incubated for 5 min (solution B). Then, solutions A and B were combined and incubated for another 20 min before being added into culture wells. Transfections were performed in duplicate, and repeated at least three times. Transfected cells were analyzed for luciferase activity 36 h after transfection as described for transient transfections.
2.9. Western analysis
Nuclear extracts were prepared as follows. Cells were washed in phosphate-buffered saline (PBS) and lysed in 15 mM KCl, 10 mM HEPES (pH 7.6), 2 mM MgCl2, 0.1 mM EDTA, 1mM DTT, 0.1% (volume:volume) Nonidet P-40, 0.5mM PMSF, 2.5 µg/ml leupeptin, 5 µg/ml antipain, and 5 µg/ml aprotinin for 10 min on ice. Nuclei were pelleted by centrifugation at 14,000×g for 20 sec at room temperature. Proteins were extracted from nuclei by 20 min incubation at 4°C with vigorous vortexing in buffer C (420mM NaCl, 20mM HEPES (pH 7.9), 0.2mM EDTA, 25% [volume:volume] glycerol, 1 mM DTT, 0.5 mM PMSF, 2.5 µg/ml leupeptin, 5 µg/ml antipain, and 5 µg/ml aprotinin). Nuclear debris was pelleted by centrifugation at 13,000 rpm for 30 min at 4°C, and the supernatant extract was collected and stored at −80°C. The extracts (10 µg) were adjusted to 1× Laemmli sample buffer (Laemmli, 1970) and processed by SDS-12% PAGE. The gel was transferred to a Protran membrane (Schleicher and Schuell Bio-Science, Inc., Keene, NH, USA), and antigen-antibody complexes were visualized with the Enhanced Chemiluminescence Kit (Amersham Biosciences, Piscataway, NJ, USA). The primary antibodies utilized were rabbit anti-C/EBPβ specific to the carboxyl terminus (C-19; 1:1000; Santa Cruz Biotechnology) and rabbit anti-β-tubulin (H-235; 1:1000; Santa Cruz Biotechnology). Anti-rabbit IgG horseradish peroxidase (HRP)-conjugate was used as a secondary antibody (1:3000; Promega).
To detect endogenous PRA and PRB, 200ug of nuclear extract from MC7-L1 cells were subjected to immunoprecipitation with 20ug of anti-PR (Alpha PR6, ChIP Grade, ab2765, Abcam) or normal mouse IgG. This was diluted to 600 µl final volume with TE (10 mM Tris [pH 8.0] and 1 mM EDTA) plus 1% Triton X100. NaCl was then added to a final concentration at 150mM. This mixture was incubated at 4°C overnight. Immunoprecipitates were collected on fast flow protein G agarose (Millipore Sigma, Burlington, MA, USA), and then washed 3 times with TE plus 1% Triton X100 and 150mM NaCl, eluted with 2× Laemmli sample buffer (Laemmli, 1970) at 95°C for 5 min, and electrophoresed through 4–20% gradient PAGE. The gel was transferred to a Protran membrane (Schleicher and Schuell Bio-Science, Inc), and antigen-antibody complexes were visualized using the LI-COR® Odyssey Western Blotting Kits (LI-COR Biotechnology, Lincoln, NE, USA). The anti-PR detected both PRA and PRB (Alpha PR6; 1:50; Abcam). Rabbit anti-mouse IgG conjugated to Alexa 800 (1:7,500; Molecular Probes, Invitrogen) was used as a secondary antibody with a 1 h incubation.
2.10. Immunofluorescence
Dual immunofluorescence detection of PRA and C/EBPβ or PRB and C/EBPβ was performed using rabbit anti-PRA (1:100; A0098, Dako, Carpinteria, CA, USA) or rabbit anti-PRB (1:800; B15) (Kariagina et al., 2007) with mouse anti-C/EBPβ (1:50; sc7962, Santa Cruz Biotechnology) primary antibodies followed by appropriate secondary antibodies conjugated to Alexa 488 or Alexa 546 (Molecular Probes Eugene, OR, USA). Nuclei were counterstained with 4’,6-diamidino-2-phenylindole, dilactate (DAPI) (1:10,000 weight:volume in H2O).
The anti-PRB (B15) was raised against the N-terminal region of the mouse PRB protein. The specificity of B15 was confirmed by its failure to co-localize with anti-PRA staining in virgin mouse mammary gland, while at the same time co-localizing with staining of pregnant mouse mammary gland by commercially available anti-PRB (Kariagina et al., 2007). B15 also detects a protein of similar size to that detected by commercially available anti-PRB in western blots (Kariagina et al., 2007). The specificity of the anti-PRA was confirmed by its ability to detect PR in the virgin mouse mammary gland, while being unable to detect PRB in pregnant mouse mammary gland. The specificity of the anti-PRA is explained by availability of the recognized epitope being dependent on protein folding and masked in native PRB; this anti-PRA does not recognize PRA under denaturing conditions (Mote et al., 2001).
For tissue sections, mice were killed and their mammary glands were fixed and paraffin-embedded for immunohistochemistry (Aupperlee et al, 2005). MC7-L1 cells were grown on a round microscope cover glass (Fisher Scientific, Pittsburgh, PA, USA) with a starting density of 2×105 cells in each well of 6-well cell culture plates. After 20 h, transfections were performed as described above, using 0.5 µg of pcDNA3.1 or pcDNA-LIP. Transfected cells were incubated for 24 h prior to staining. Cell-coated cover glasses were fixed in 3.7% formalin for 30 min, rinsed twice in PBS for 5min, permeabilized in 0.5% (weight:volume) Triton X-100 for 15 min, rinsed again in PBS, and then blocked with PBS containing 2% BSA for 15 min. After blocking, cells were treated with primary antibodies overnight. The next day, slides were washed with PBS, incubated with secondary antibody for 30 min. After another rinse in PBS, slides were stained with DAPI for 5 min. Slides were mounted with fluorescence mounting medium and dried overnight. Stained tissue sections and cells were visualized, and images captured using a Nikon inverted epifluorescence microscope (Mager Scientific, Dexter, MI, USA) with MetaMorph software (Molecular Devices Corporation, Downington, PA, USA).
3. Results
3.1. C/EBPβ and c-Jun synergize in stimulating PR promoter activity
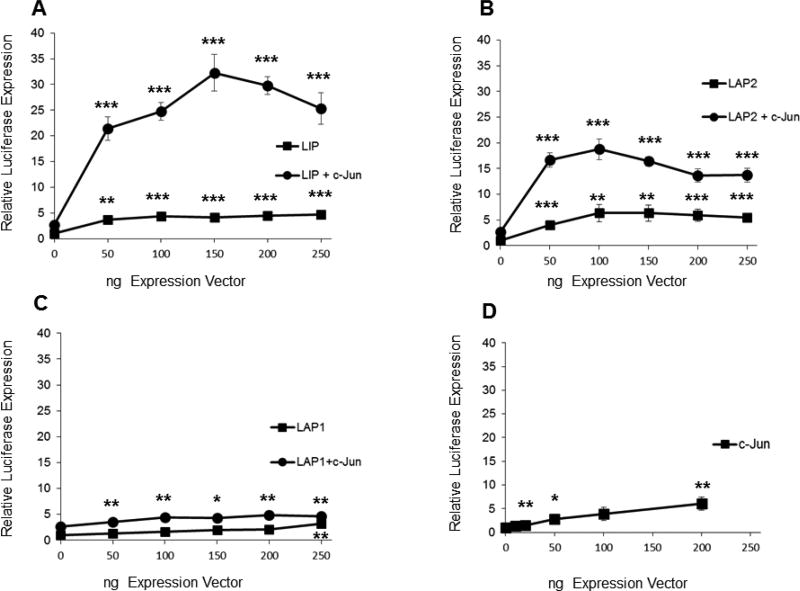
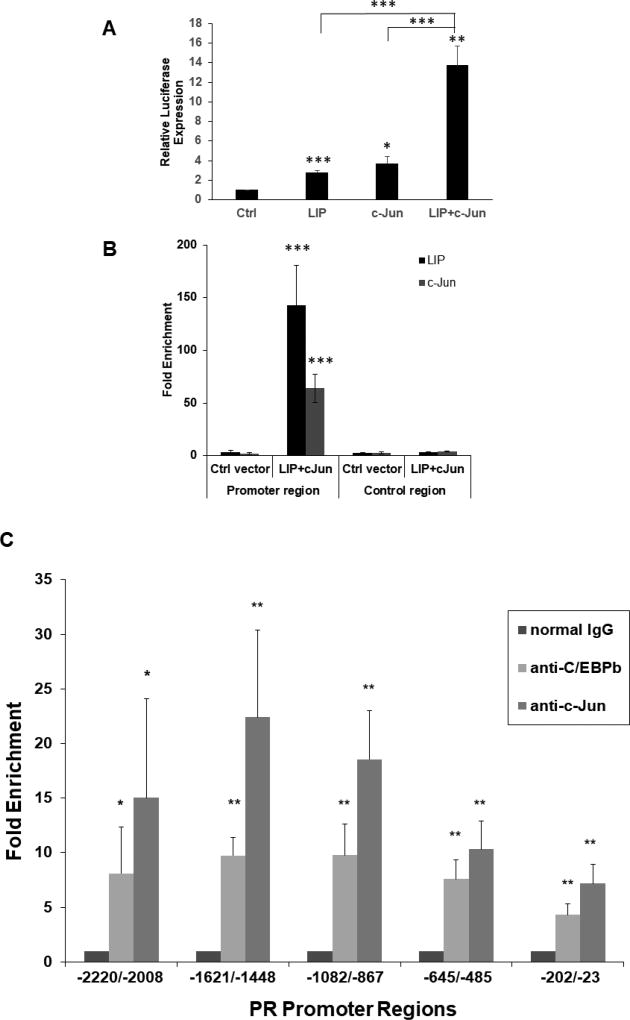
Examination of the DNA sequence in the region from −2502 to +753 bp of the murine PRB gene with MatInspector (Quandt et al., 1995; Cartharius et al., 2005) revealed a number of predicted C/EBPβ binding sites (Fig. 1). In order to investigate the ability of C/EBPβ isoforms to stimulate transcription of the murine PR gene, we performed transient co-transfections of vectors that singly expressed LAP1, LAP2, and LIP into murine mammary carcinoma cell line MC7-L1 (Lanari et al., 2001) with the −2494/+769 PR promoter-reporter. MC7-L1 is of mammary epithelial origin and is known to express PR. Thus. It offers a cell culture system that may provide insight into the mechanisms that regulate PR expression in normal mammary epithelial cells. The −2494/+769 PR promoter-reporter retains sequences allowing transcription from both the PRB and PRA transcriptional start sites. We chose MC7-L1 because its endogenous expression of PR assured its ability to support a response from the −2494/+769 PR promoter-reporter. All three C/EBPβ isoforms displayed modest activity (maximally 3 to 6-fold) in stimulating the PR promoter (Fig. 2A, B, and C).
Fig. 2.
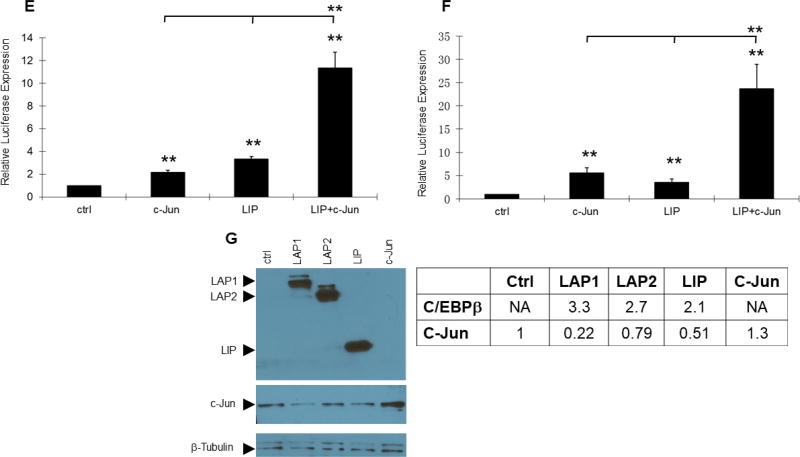
Transient co-expression of LIP and c-Jun synergize to induce expression from the PR promoter-reporter. Transient co-transfection of C/EBPβ and c-Jun expression vectors were carried out in duplicate and repeated at least three times in murine mammary tumor cell lines of epithelial origin with the −2494/+769 PR promoter-reporter. (A, B, C, D) MC7-L1 cells. The varied ng quantities of the expression vector per 5×104 cells are indicated below the x-axis. For panels A, B, and C, C/EBPβ expression vectors were co-transfected with 100 ng of c-Jun expression vector in some transfections. Luminometer values were normalized for expression from a co-transfected SV40 early enhancer/promoter-Renilla luciferase reporter. These values were then normalized to a relative value of 1.0 for cells receiving only “empty” pcDNA3.1 and neither C/EBPβ nor c-Jun expression vector. The data presented are the means of at least three experiments with SEM. p was calculated by Student’s t-Test in comparison to the control transfection for transfections involving a single transfected expression vector, and in comparison to transfections involving a C/EBPβ vector alone for co-transfections involving both C/EBPβ and c-Jun expression vectors. *, p < 0.1; **, p < 0.05; ***, p < 0.01. (E) MC4-L2 cells. 100 ng LIP and 100 ng c-Jun expression vectors per 5×104 cells. Values were normalized and statistics performed as described above. (F) MC4-L3 cells. 50 ng LIP and 50 ng c-Jun expression vectors per 5×104 cells. Values were normalized and statistics performed as described above. (G) A western blot confirms similar levels of C/EBPβ expression among the various isoforms in transfection of MC7-L1 cells. c-Jun is overexpressed in cells transfected for c-Jun expression. β-tubulin was detected as a loading control.
Previous investigators demonstrated a role for AP-1 (i.e., c-Jun/c-Fos) in stimulating human PR promoter activity in conjunction with estrogen receptor and Sp1 binding (Petz et al., 2002; Schultz et al., 2005). Additionally, transgenic mice expressing inducible TAM67, a dominant negative truncated form of c-Jun that lacks a transactivation domain, showed reduced mammary ductal development at puberty, as well as in hormonally stimulated adults (Shen et al., 2006). Since the −2494/+769 PR promoter-reporter contains two putative AP-1 binding sites, we decided to examine how AP-1 overexpression might interact with C/EBPβ activation of the promoter-reporter. To that end, LAP1, LAP2, and LIP were transiently co-expressed with c-Jun. Co-expression of LIP with c-Jun showed a striking synergy in activation of the PR promoter (Fig. 2A). Peak stimulation of the PR promoter averaged 32-fold under conditions where LIP and c-Jun singly provided 4-fold and 2.7-fold stimulation, respectively. This synergy was more modestly evident with LAP2 (Fig. 2B), while LAP1 (Fig. 2C) only showed additive levels of stimulation with co-expression of the two transcription factors. Expression of c-Jun by itself (Fig. 2D) showed only modest stimulation of the PR promoter over a range similar to that of the individual C/EBPβ isoforms. Differences in activity were not due to differential levels of expression, as all of the C/EBPβ isoforms showed similar levels of expression in parallel transfections, and c-Jun did not induce C/EBPβ expression (Fig. 2G). Interestingly, all isoforms of C/EBPβ suppress c-Jun expression to some degree. Transient transfections were also performed with expression vectors for JunD and c-Fos without any indication of synergy with or augmentation of LIP transactivation (data not shown). A similar synergy between LIP and c-Jun on the PR promoter was also observed in MC4-L2 and MC4-L3 (Fig. 2E and F), two other mammary carcinoma cell lines that show endogenous PR expression (Lanari et al., 2001). MC7-L1 expresses estrogen receptor and its proliferation is stimulated by 17β-estradiol in culture (Lanari et al. 2001). Given the literature describing AP-1 stimulation of human PR promoter activity in conjunction with estrogen receptor (Petz et al., 2002; Schultz et al., 2005), we tested the response of the −2494/+769 PR promoter-reporter to 17β-estradiol with and without co-expression of LIP and/or c-Jun. No effects were observed (data not shown).
3.2. C/EBPβ and c-Jun bind directly to the proximal promoter region of the PR promoter-reporter
We performed plasmid immunoprecipitation (PIP) on a PR promoter-reporter transfected into MC7-L1 cells to confirm that its synergistic transactivation by LIP and c-Jun was associated with their direct binding to the promoter-reporter. In our examination of the DNA sequence of the murine PR gene (Fig. 1), we found multiple putative C/EBPβ binding sites and at least one putative C-Jun binding site in the proximal region of the promoter between nucleotides −117 and +63 in relation to the predicted PRB transcriptional start site. We constructed a minimal −117/+63 PR promoter-reporter containing that region and showed that it reiterated the synergism of LIP and c-Jun observed with the longer −2494/+769 PR promoter-reporter (Fig. 3A). We next examined the physical binding of LIP and c-Jun to the −117/+63 promoter region by a transient transfection PIP assay. MC7-L1 cells were transfected with the −117/+63 PR promoter-reporter and expression vectors for LIP and c-Jun or an “empty” control expression vector. We detected LIP and c-Jun on the −117/+63 PR promoter with 143-fold and 64-fold enrichment, respectively, compared to precipitation with normal IgG (Fig. 3B). These levels of occupancy were much higher than the occupancy observed with transfection of control expression vector or in a region of the −117/+63 PR promoter-reporter plasmid distal to the promoter, showing the specificity of the occupancy and consistent with LIP and c-Jun acting directly on the PR promoter-reporter.
Fig. 3.
Plasmid immunoprecipitation (PIP) and chromatin immunoprecipitation (ChIP), respectively, show binding of C/EBPβ and c-Jun to the promoter regions of the −117/+63 PR promoter-reporter and the endogenous PR gene, consistent with their transactivation of the PR promoter. (A) Transient co-transfections of LIP and c-Jun expression vectors were carried out in MC7-L1 with the −117/+63 PR promoter-reporter. 50 ng LIP and 50 ng c-Jun expression vectors were transfected per 5×104 cells. Luminometer values were normalized for expression from a co-transfected SV40 early enhancer/promoter-Renilla luciferase reporter. These values were then normalized to a relative value of 1.0 for cells receiving only “empty” pcDNA3.1 and neither C/EBPβ nor c-Jun expression vector. The data presented are the means of at least three experiments with SEM. p was calculated by Student’s t-Test in comparison to the control transfection. *, p < 0.1; **, p < 0.05; ***, p < 0.01. (B) Occupancy of C/EBPβ-LIP and c-Jun on wildtype −117/+63 PR promoter in PIP assays. MC7-L1 cells were transfected with the −117/+63 PR promoter-reporter that had been linearized by ScaI digestion and expression vectors for both C/EBPβ-LIP and c-Jun, or control expression vector (as indicated). Plasmid-protein complexes were precipitated with either C/EBPβ antibody or c-Jun antibody, or normal IgG. The DNA from precipitated plasmid was quantitated by qPCR, either with a primer pair for the promoter region (left panel) or with another primer pair for the control region (right panel). The value of qPCR-amplified product is calculated as the percentage of input, and then the fold enrichment is presented as the ratio of amplified product with specific antibody to that with nonspecific IgG antibody. Results are the mean of three independent experiments with SEM. p was calculated for each PIP with specific antibody by Student’s t-Test in comparison to the control vector transfection, and in comparison to the control region. ***, p<0.01. (C) Occupancy of the PR gene promoter region by C/EBPβ and c-Jun was determined by ChIP. Chromatin-protein complexes were precipitated with either C/EBPβ antibody or c-Jun antibody, or normal IgG. The DNA from precipitated chromatin was quantitated by qPCR with primer pairs for regions upstream of the predicted PRB transcriptional start site: −2220 to −2008 bp, −1621 to −1448 bp, −1082 to −867 bp, −645/−485 bp, and −202 to −23 bp. The value of qPCR-amplified product is calculated as the percentage of input, and then the fold enrichment is presented as the ratio of amplified product with specific antibody to that with nonspecific normal IgG antibody. Results are the mean of three independent experiments with SEM. p was calculated by Student’s t-Test in comparison to normal IgG. *, p<0.1; **, p<0.05.
3.3. C/EBPβ and c-Jun bind directly to the promoter region of the endogenous PR gene in MC7-L1 cells
Having observed the direct binding of LIP and c-Jun to the promoter region of the PR promoter-reporter, we examined whether endogenous levels of C/EBPβ and c-Jun bound to the endogenous promoter region of the PR gene in MC7-L1 cells in a chromatin immunoprecipitation (ChIP) assay. We examined promoter occupancy by C/EBPβ and c-Jun at 5 sites in a region extending approximately 2100 bp upstream of the putative PRB transcriptional start site, a region encompassed within the PR −2494/+769 promoter-reporter and including the −117/+63 PR promoter (Figure 3C). We detected C/EBPβ and c-Jun across all 5 sites with enrichments ranging from about 4 to 8-fold and 7 to 22-fold, respectively, in comparison to the enrichment produced with normal IgG.
3.4. Overexpression of LIP and c-Jun increase endogenous PR expression and activity
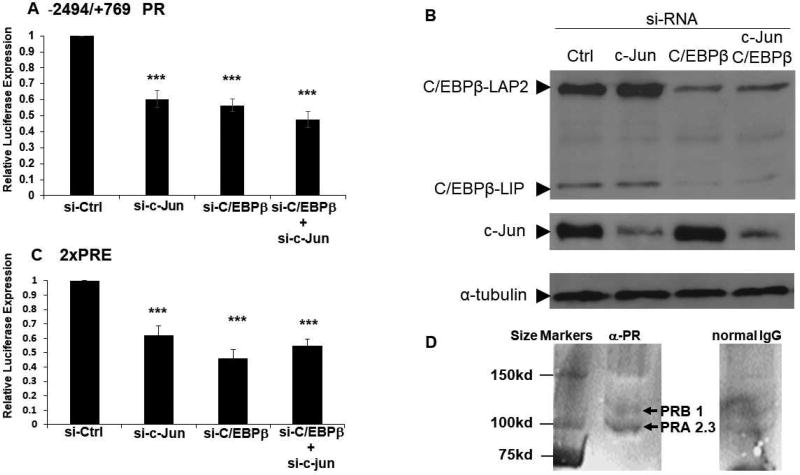
Having observed the robust synergy between LIP and c-Jun in activating the PR promoter in a transient transfection system and confirmed their promoter occupancy in both PR promoter-reporter and the endogenous PR promoter region, we tested whether the intact endogenous PR gene would respond to these transcription factors. To that end, LIP and c-Jun overexpression was introduced transiently into MC7-L1 cells in co-transfections with the 2×PRE-TK-luc promoter-reporter. The activity of the 2×PRE promoter is dependent upon PR expression. While LIP or c-Jun overexpression by itself did not significantly increase 2×PRE activity, co-expression of LIP and c-Jun increased 2×PRE promoter activity 3.2-fold (Fig. 4A). We also tested whether R5020, a PR agonist (Haslam et al., 2008; Santos et al., 2009), could augment promoter activity, but it did not.
The failure of R5020 to stimulate the 2×PRE promoter may be due to the induction of PRA along with PRB, as even relatively low amounts of PRA are capable of repressing PRB activity (Mohamed et al., 1994; Pieber et al., 2001). In order to test this in MC7-L1 cells, we performed transient co-transfection of the 2×PRE promoter with PRA and PRB expression vectors, either singly or in various ratios. R5020 was only able to stimulate 2×PRE activity with co-transfection of the PRB expression vector by itself (Fig. 4B). However, even a 3:1 abundance of the PRB vector over the PRA vector did not support R5020 stimulation of 2×PRE. Furthermore, only PRB in the absence of PRA stimulated the 2×PRE promoter; transfections that included PRA expression vector suppressed basal activity of the promoter (Fig. 4B). Note that MC7-L1 cells in the absence of transfection express higher levels of PRA than PRB (Fig. 6D) and, thus, R5020 does not stimulate endogenous PR to drive the 2×PRE promoter. Low transfection efficiency precluded detection of transfected PRA and PRB expression by western analysis, so PR expression was examined by immunofluorescence. Immunofluorescent detection of PRA and PRB in respectively transfected MC7-L1 cells demonstrated the expression of the PR isoforms above endogenous levels (Fig. 4C). We sought to confirm that transfection of LIP and c-Jun expression into MC7-L1 could induce expression of an endogenous PR target gene, amphiregulin (Aupperlee et al., 2013). Co-transfection of LIP and c-Jun, PRA, and PRB with and without R5020 treatment all induced amphiregulin expression, although the induction with LIP and c-Jun without R5020 only approached significance (Fig. 4D). R5020 did not exert statistically significant effects.
Fig. 6.
siRNAs directed against endogenous C/EBPβ or c-Jun expression similarly suppress expression from the −2494/+769 PR and 2×PRE promoter-reporters. (A) Transient transfections of siRNAs directed against C/EBPβ (si-C/EBPβ) and c-Jun (si-c-Jun), as well as control siRNA (si-Ctrl), were carried out in MC7-L1 cells with the −2494/+769 PR promoter-reporter. Luminometer values were normalized for expression from a co-transfected SV40 early enhancer/promoter-Renilla luciferase reporter. These values were then normalized to a relative value of 1.0 for cells receiving control siRNA. The data presented are the means of at least three experiments with SEM. p was calculated by Student’s t-Test in comparison to the control transfection. ***, p < 0.01. (B) A western blot confirms decreased C/EBPβ and c-Jun expression with transfection of their cognate siRNAs. β-tubulin was detected as a loading control. (C) Transient transfections of siRNAs directed against C/EBPβ (si-C/EBPβ) and c-Jun (si-c-Jun), as well as control siRNA (si-Ctrl), were carried out in MC7-L1 cells with the 2×PRE promoter-reporter. Data were processed as in (A). (D) A western blot confirms expression of PRA and PRB in MC7-L1 cells. Whole cell lysate of MC7-L1 cells was immunoprecipitated with either anti-PR (αPR) or normal IgG, and then PRA and PRB detected by western blot. The relative densitometry of PRA and PRB is presented.
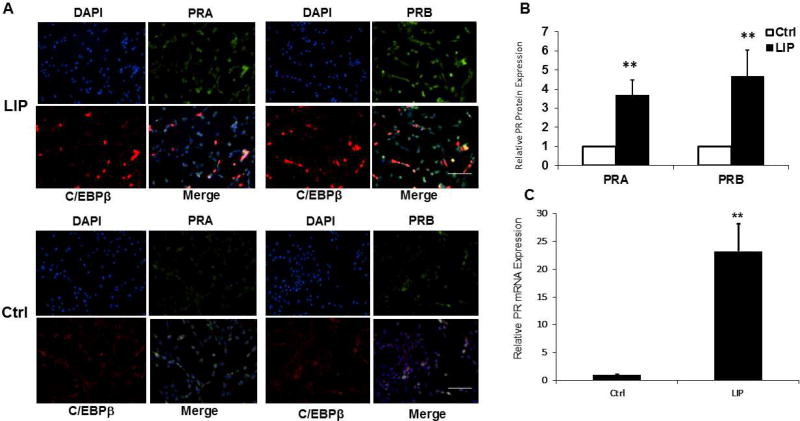
As a more direct assessment of the effects of LIP and c-Jun overexpression on endogenous PR gene expression, we introduced LIP and c-Jun expression transiently into MC7-L1 cells, and then PRA and PRB expression were examined by immunofluorescence. Consistent with the results reported in Fig. 4, both PRA and PRB expression were increased about 4-fold in the LIP-transfected populations (Fig. 5A and B). c-Jun neither increased PR expression nor enhanced LIP stimulation of PR expression (data not shown). We explored the possibility that overexpression of c-Jun interferes with its phosphorylation, but this was not observed (data not shown). The lack of an effect from c-Jun overexpression in this assay may reflect a surfeit of c-Jun expression in the context of the diploid endogenous gene as opposed to more limited availability for the multiple copies of the transfected −2494/+769 PR promoter-reporter. Consistent with the notion that LIP transcriptionally activates the endogenous PR gene, PR RNA levels were increased 23-fold by LIP overexpression (Fig. 5C).
Fig. 5.
LIP expression induces endogenous PRA and PRB expression. Transient transfections of the LIP expression vector were carried out in MC7-L1 cells. (A) Immunofluorescent detection of PRA (green) and C/EBPβ (red) in cells transfected for LIP expression (LIP) and in cells transfected with empty pcDNA3.1 vector (Ctrl). Nuclei were counterstained with DAPI (blue). 20× magnification; scale bar, 50 µm. (B) The immunofluorescence of PRA and PRB were measured in cells showing LIP overexpression after transfection with LIP expression vector (LIP) in comparison to the level of immunofluorescence in cells transfected with empty vector (Ctrl). Control transfections showed no change in PR expression compared to untransfected cells (data not shown). Values from LIP overexpressing cells were normalized to a relative value of 1.0 for cells transfected with empty vector. The data presented are the means of at least three experiments with SEM. Approximately 100 LIP overexpressing cells were scored in each experiment. p was calculated by Student’s t-Test in comparison to the control transfection. **, p<0.05. (C) The quantities of PR mRNA were measured by qRT-PCR in cells transfected for LIP expression (LIP) in comparison to cells transfected with empty vector (Ctrl). Values were normalized to a relative value of 1.0 for cells receiving the empty vector control. The data presented are the means of nine experiments with SEM. p was calculated by Student’s t-Test in comparison to the control transfection. **, p<0.05.
3.5. Endogenous levels of C/EBPβ and c-Jun contribute to PR expression
In order to examine whether the levels of C/EBPβ and c-Jun normally present in MC7-L1 cells can support PR expression, “gene knockdown” experiments were performed with siRNAs directed against the mRNAs of these transcription factors. SiRNAs directed against C/EBPβ and c-Jun mRNAs were transiently transfected, either singly or together, with the −2494/+769 PR promoter-reporter. The activity of this promoter was reduced by about 40% by either siRNA alone, while co-transfection with both siRNAs reduced promoter activity by about 50%, not significantly different from either siRNA alone (Fig. 6A). Similar reductions in basal activity were found for control promoter-reporters specifically dependent on C/EBP or c-Jun family transcription factors (data not shown). At the same time, protein levels of C/EBPβ isoforms and c-Jun were dramatically reduced by their cognate siRNAs, but were unaffected by control and non-specific siRNAs (Fig. 6B). Furthermore, this demonstrates endogenous expression of C/EBPβ and c-Jun in MC7-L1 cells at levels that are sufficient to support PR transcription. Additionally, the fact that co-transfection with siRNAs for both C/EBPβ and c-Jun suppressed expression from the PR promoter little more than each siRNA alone is consistent with the notion that the activity of these transcription factors on the PR promoter is largely derived from their synergy. The fact that protein levels of C/EBPβ and c-Jun were only affected by their cognate siRNAs demonstrates the specificity of siRNA activity. Western analysis of MC7-L1 also verifies endogenous expression of PRA and PRB in the absence of transfected expression of C/EBPβ and c-Jun (Fig. 6D). PRA appears in greater abundance than PRB, densitometry finding a ratio or 2.3 to 1.
In order to extend our examination of the effects of gene knockdown to the intact endogenous PR gene, we repeated the transfection of siRNAs directed against C/EBPβ and c-Jun mRNAs with the 2×PRE-TK-luc promoter-reporter. Similarly to the case with the −2494/+769 PR promoter, activity of the 2×PRE promoter was reduced by about 40 to 50% whether transfected singly or with both siRNAs (Fig. 6C). This suggests a similar dependence of the intact endogenous PR promoter for endogenous levels of C/EBPβ and c-Jun expression as for the −2494/+769 PR promoter-reporter.
3.6. C/EBPβ is associated with PRB expression in vivo
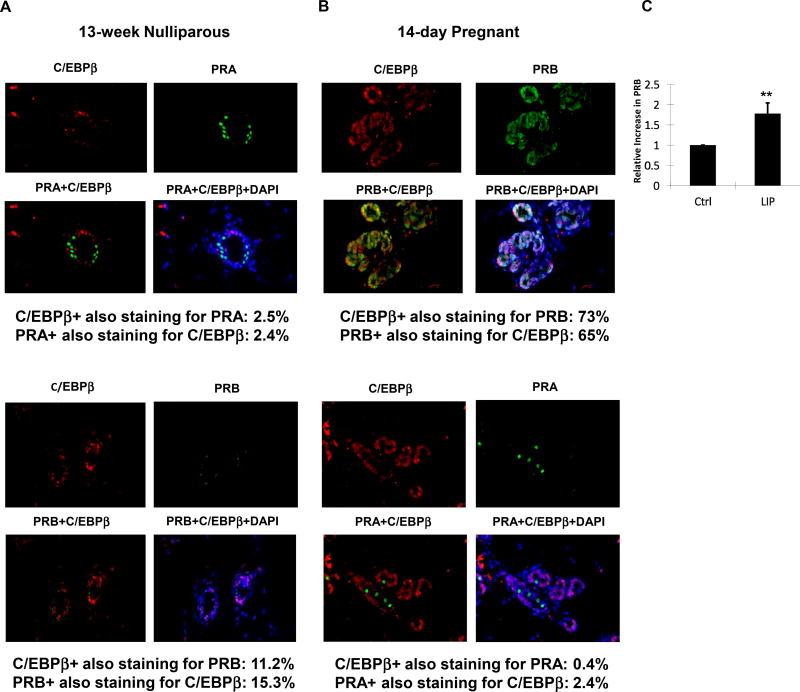
C/EBPβ expression, particularly LIP, increases during the course of pregnancy in the mouse (Seagroves et al., 1998). The block to alveologenesis in both C/EBPβ- (Robinson et al., 1998; Seagroves et al., 1998) and PRB-deficient mice (Lydon et al, 1995; Mulac-Jericevic et al., 2003), coupled with PRB being expressed with alveolar development during pregnancy (Aupperlee et al., 2005), led us to hypothesize that LIP may be associated with PRB expression during pregnancy. Immunofluorescent staining of PRA, PRB and C/EBPβ was performed on the mammary glands of nulliparous compared to 14-day pregnant mice. Staining revealed that the localization of PRA, the predominant PR isoform in the nulliparous mammary gland, was mutually exclusive of C/EBPβ (Fig. 7A). In contrast, PRB, the predominant PR isoform in the pregnant mammary gland, largely co-localized with C/EBPβ (Fig. 7B), consistent with a role for C/EBPβ in PRB expression. Consistent with the specificity of the anti-PRA and anti-PRB antibodies that we used in this analysis, anti-PRA detected an exceedingly small number of cells in pregnant mammary gland (Fig. 7B), while anti-PRB detected similarly exceeding small number of cells in nulliparous mammary gland (Fig. 7A).
Fig. 7.
PRA expression is mutually exclusive of C/EBPβ, while PRB and C/EBPβ largely co-localize. (A, B) PRA (green) or PRB (green) and C/EBPβ (red) were detected by immunofluorescence in the mammary glands of 13-week old nulliparous mice and 14-day pregnant mice. Nuclei were counterstained with DAPI (blue). Representative tissue sections are shown with the percentages of PR and C/EBPβ co-localization. For 13-week old nulliparous mice (n=2), PRA-C/EBPβ staining: 11 visual fields, 646 cells; PRB-C/EBPβ staining: 9 visual fields, 304 cells. For pregnant mice (n=2), PRB-C/EBPβ staining: 4 visual fields, 482 cells; PRA-C/EBPβ staining: 7 visual fields; 507 cells. Difference between nulliparous PRA-C/EBPβ co-localization and pregnant PRB-C/EBPβ co-localization: p<0.001. 40× magnification; scale bar, 25 mm. (C) The quantities of PR RNA transcribed from the −2494/+769 PR promoter-reporter were measured by qRT-PCR in cells transfected for LIP expression (LIP) in comparison to cells transfected with empty pcDNA3.1 vector (Ctrl). A ratio of PRB to total PR RNA was calculated and these values were normalized to a relative value of 1.0 for cells receiving the empty vector control. The data presented are the means of seven experiments with SEM. p was calculated by Student’s t-Test in comparison to the control transfection. **, p < 0.05.
Overexpression of LIP increased both PRA and PRB expression in MC7-L1 cells (Fig. 5A and B), but the relative expression of PRA and PRB proteins is uncertain as these proteins were detected with different antibodies. While LIP overexpression clearly increased PR expression, protein expression levels were not high enough to allow reliable western blot analysis assessing relative expression of PRA and PRB based on their size (data not shown). In order to assess differential expression of PRA and PRB, we performed a PCR assay utilizing primers specific to either a region common to both PRA and PRB transcripts or to an upstream region specific to PRB transcripts. Transcripts from the endogenous gene were not abundant enough to allow that determination (data not shown), so we instead performed assays on RNAs isolated from cells transfected with the −2494/+769 PR promoter-reporter. Consistent with a role for LIP in PRB expression, LIP overexpression increased the ratio of PRB transcripts to total PR transcripts by 1.8-fold (Fig. 7C).
4. Discussion
The data presented in this paper support an important role for C/EBPβ in the transcriptional regulation of PR in mice. The LIP isoform of C/EBPβ was particularly active in synergy with c-Jun on a murine PR promoter-reporter, and was capable of increasing PR protein expression in a murine mammary carcinoma cell line. Overexpressed LIP and c-Jun also clearly bound directly to the promoter region of the −117/+63 PR promoter-reporter and endogenous levels of C/EBPβ and c-Jun bound directly to the intact endogenous PR promoter region. While tumor cell lines may not reiterate the behavior of normal tissue, the cell line primarily utilized in this study is mammary-derived and expresses PR, C/EBPβ, and c-Jun, making it a suitable cell culture model. Importantly, PRB expression in the mammary glands of pregnant mice co-localized with C/EBPβ expression, while PRA and C/EBPβ expression in nulliparous mice were mutually exclusive in their localization. While our expression experiments in cultured cells neither showed a very strong one-to-one relationship between LIP overexpression and PR expression at the level of individual cells, nor as exclusive a relationship between C/EBPβ and PRB expression as seen in vivo, those experiments suggest that C/EBPβ LIP favors longer transcripts that can express PRB. This is also consistent with the LIP and c-Jun stimulation of the 2×PRE promoter-reporter; only a PRB expression vector without co-transfection of a PRA expression vector elicited stimulation, suggesting that LIP plus c-Jun elicits largely PRB expression. Further, the ability of LIP plus c-Jun to induce the PR target gene amphiregulin suggests that functional PR expression is induced by these transcription factors. Consistent with our immunofluorescent detection of both PRA and PRB, the lack of response to R5020 suggests that some PRA expression is induced by LIP and c-Jun. Further, modest increases in LIP expression that would not be obvious by staining in individual cells may elicit significant changes in PR expression, as LIP is the most potent of C/EBPβ isoforms in driving expression from the PR promoter. Discrepancies between high LIP expression and high PR expression may also be due to differential expression between individual cells of other factors that regulate PR expression. LIP expression is particularly increased in the mammary glands of pregnant mice (Seagroves et al., 1998) at the same time when PRB becomes the predominant PR isoform in the murine mammary gland (Aupperlee et al., 2005). Both LIP (Seagroves et al., 1998) and PRB (Aupperlee et al., 2005) are also downregulated at lactation and involution. Viewed in this context, our results suggest a specific role for LIP in the expression of PRB. Observations in knockout mice are consistent with a role for C/EBPβ in regulating PRB expression. PRB-deficient mice are defective in alveologenesis (Lydon et al., 1995; Mulac-Jericevic et al., 2003), as are C/EBPβ-deficient mice (Robinson et al., 1998; Seagroves et al., 1998). Further, in contrast to alterations in PR expression that are observed in C/EBPβ-deficient mice, no alterations in C/EBPβ expression are observed in PR-deficient mice (Seagroves et al., 2000). This is consistent with our conclusion that C/EBPβ acts upstream of PR.
It is interesting that 17β-estradiol did not affect LIP and c-Jun stimulation of the −2494/+769 PR promoter-reporter. Either this promoter-reporter excludes a critical estrogen-responsive element(s), or the elements included in this promoter-reporter are not active outside the context of chromatin, or, unlike the case in human cells (Petz et al., 2002; Schultz et al., 2005), estrogen does not play a direct role in murine PR regulation.
Our results also showed that overexpression of C/EBPβ isoforms reduced c-Jun expression; LIP overexpression reduced c-Jun by half in MC7-L1 cells. This reduction of c-Jun is relatively modest against the level of synergy observed in our transient transfection system, where LIP and c-Jun independently exerted 4-fold and 2.7-fold stimulation, while synergistically providing 32-fold stimulation. Extensive in vivo studies will likely be required to fully resolve the potential effects of regulatory crosstalk between these two transcription factors.
At the same time that knockout studies support the notion of an involvement of C/EBPβ in PRB regulation, there are, at least superficially, some inconsistencies. C/EBPβ-deficient mice show elevated numbers of PR-positive cells rather than a decrease in PR expression that might be expected (Seagroves et al., 2000). However, that study did not distinguish between PRA and PRB, and, as those observations were made in nulliparous mice, they most likely represent PRA expression. The number of PR-positive cells did not decrease in sexually mature C/EBPβ-deficient mice and is not decreased by E plus P treatment that mimics pregnancy, as is the case in wildtype mice (Seagroves et al., 2000). Again, the interpretation of this lies with specific isoform expression that was not assessed. In wildtype animals, PRA decreases with maturity and is further diminished with pregnancy, and PRB is not evident until day 14 of pregnancy (Aupperlee et al., 2005). The failure to decrease PR expression in C/EBPβ-deficient mice may reflect a block in the transition to PRB expression that occurs with pregnancy. It would be informative to analyze the developmental expression of PRA and PRB in C/EBPβ-deficient mice.
Another question arises from studies showing that, in contrast to complete C/EBPβ deficiency, mice that fail to express LIP do not show defects in mammary gland development (Wethmar et al., 2010). Mice lacking LAP2 expression also have normal mammary gland development (Uematsu et al., 2007). These findings are entirely consistent with our findings that although LIP is the most robust isoform for transactivation of the PR promoter-reporter, LAP2 also has significant activity. As our results shows little activity for LAP1 on the PR promoter, a LAP1 knock-in on a C/EBPβ-deficient background would not be expected to rescue mammary gland development.
It is interesting that LIP, generally described as an inhibitor of LAP-mediated transcription (Descombes and Schibler, 1991), is the C/EBPβ isoform that most robustly activates the PR promoter. There is precedent for activation of other genes by LIP, and, as in this report, LIP activity is associated with synergy with other transcription factors. LIP can transactivate the IL-6 promoter reporter in manner dependent upon an intact NF-κB site (Hu et al., 2000; Spooner et al., 2007). IL-8 is similarly activated through a synergy between C/EBPβ and NF-κB, and this is, at least in part, mediated through cooperative DNA binding (Stein and Baldwin, 1993). LIP and PRB cooperate to transactivate the decidual prolactin and MMTV promoters (Christian et al., 2002). LIP and Runx2 cooperate to transactivate the promoters for alkaline phosphatase and osteocalcin (Hata et al., 2005). This latter activity is based upon the enhanced ability of ATF4-C/EBPβ (i.e., LIP) heterodimers to bind DNA and interact with Runx2 (Tominaga et al., 2008). Knockdown of C/EBPβ, c-Jun, or both, all inhibited the PR promoterreporter to a similar extent, implying a mutual dependence of the LIP and c-Jun for their synergistic activity. The basis for this synergy remains to be elucidated.
Progesterone acting through PRB is reported to upregulate C/EBPβ expression in a human breast cancer cell line (Richer et al., 2002) and progesterone upregulates C/EBPβ expression in mouse mammary glands (Aupperlee et al., 2009). The progestin medroxyprogesterone acetate is also reported to upregulate c-Jun in a human breast cancer cell line (Alkhalaf et al., 1992). If progesterone acts to upregulate both C/EBPβ and c-Jun expression in mouse mammary glands, our finding of C/EBPβ and c-Jun synergy in regulating PR presents a robust mechanism for a positive feedback loop in PRB expression in the pregnant mammary gland. Progesterone, indeed, decreases PRA and increases PRB levels in the normal adult murine mammary gland (Aupperlee and Haslam, 2007). Such a positive feedback loop may be critical in the action of P to promote mammary gland PRB expression during murine pregnancy. While, unlike humans (Mote et al., 2002), expression of PRA and PRB are largely temporally and spatially dissociated in the mouse (Aupperlee et al., 2005), the conservation of transcription factor binding sites between the mouse and human suggests the potential relevance of the findings herein to understanding the regulation of PR expression in humans and in breast cancer. Alterations in the ratio of PRA to PRB is associated with breast cancer progression and this alteration is often observed to increase the ratio of PRA to PRB (Graham et al., 1995). The regulatory mechanisms reported here suggest C/EBPβ and c-Jun as candidates for further study to understand the dysregulation of PR expression that can occur in breast cancer.
C/EBPβ LIP and c-Jun synergize to activate the murine progesterone receptor promoter.
C/EBPβ LIP and c-Jun exhibit promoter occupancy on the progesterone receptor gene.
C/EBPβ and progesterone receptor B co-localize to the pregnant mammary epithelium.
Acknowledgments
This work was made possible by the Breast Cancer and the Environment Research Program (BCERP) award number 1UO1ESO19434 and the Breast Cancer and the Environment Research Centers (BCERC) award number 1U01ES12800 from the National Institute of Environmental Health Sciences (NIEHS) and the National Cancer Institute (NCI), NIH, DHHS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS or NCI, the National Institutes of Health. We also gratefully acknowledge support of the Avon Foundation, Ladies Auxiliary to the Veterans of Foreign Wars, and the Helen L. Kay Charitable Trust for this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alkhalaf M, Murphy LC. Regulation of c-jun and jun-B by progestins in T-47D human breast cancer cells. Mol Endocrinol. 1992;6:1625–1633. doi: 10.1210/mend.6.10.1448115. [DOI] [PubMed] [Google Scholar]
- 2.Aupperlee MD, Smith KT, Kariagina A, Haslam SZ. Progesterone receptor isoforms A and B: temporal and spatial differences in expression during murine mammary gland development. Endocrinology. 2005;146:3577–3588. doi: 10.1210/en.2005-0346. [DOI] [PubMed] [Google Scholar]
- 3.Aupperlee MD, Haslam SZ. Differential hormonal regulation and function of progesterone receptor isoforms in normal adult mouse mammary gland. Endocrinology. 2007;148:2290–2300. doi: 10.1210/en.2006-1721. [DOI] [PubMed] [Google Scholar]
- 4.Aupperlee MD, Drolet AA, Durairaj S, Wang W, Schwartz RC, Haslam SZ. Strain-specific differences in the mechanisms of progesterone regulation of murine mammary gland development. Endocrinology. 2009;150:1485–1494. doi: 10.1210/en.2008-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aupperlee MD, Leipprandt JR, Bennett JM, Schwartz RC, Haslam SZ. Amphiregulin mediates progesterone-induced mammary ductal development during puberty. Breast Cancer Res. 2013;15:R44. doi: 10.1186/bcr3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basu SK, Malik R, Huggins CJ, Lee S, Sebastian T, Sakchaisri K, Quiñones OA, Alvord WG, Johnson PF. 3'UTR elements inhibit Ras-induced C/EBPβ post-translational activation and senescence in tumour cells. EMBO J. 2011;30:3714–3728. doi: 10.1038/emboj.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 8.Christian M, Pohnke Y, Kempf R, Gellersen B, Brosens JJ. Functional association of PR and CCAAT/enhancer-binding protein beta isoforms: promoter-dependent cooperation between PR-B and liver-enriched inhibitory protein, or liver-enriched activatory protein and PR-A in human endometrial stromal cells. Mol Endocrinol. 2002;16:141–154. doi: 10.1210/mend.16.1.0763. [DOI] [PubMed] [Google Scholar]
- 9.Dearth LR, Hutt J, Sattler A, Gigliotti A, DeWille J. Expression and function of CCAAT/enhancer binding proteinβ (C/EBPβ) LAP and LIP isoforms in mouse mammary gland, tumors and cultured mammary epithelial cells. J Cell Biochem. 2001;82:357–370. doi: 10.1002/jcb.1167. [DOI] [PubMed] [Google Scholar]
- 10.Descombes P, Schibler U. A liver-enriched transcriptional activator protein LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 11.Fendrick JL, Raafat AM, Haslam SZ. Mammary gland growth and development from the postnatal period to postmenopause: ovarian steroid receptor ontogeny and regulation in the mouse. J Mammary Gland Biol Neoplasia. 1998;3:7–22. doi: 10.1023/a:1018766000275. [DOI] [PubMed] [Google Scholar]
- 12.Giangrande PH, Kimbrel EA, Edwards DP, McDonnell DP. The opposing transcriptional activities of the two isoforms of the human progesterone receptor are due to differential cofactor binding. Mol Cell Biol. 2000;20:3102–3115. doi: 10.1128/mcb.20.9.3102-3115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham JD, Yeates C, Balleine RL, Harvey SS, Milliken JS, Bilous AM, Clarke CL. Characterization of progesterone receptor A and B expression in human breast cancer. Cancer Res. 1995;55:5063–5068. [PubMed] [Google Scholar]
- 14.Haslam SZ, Drolet A, Smith K, Tan M, Aupperlee M. Progestin-regulated luminal cell and myoepithelial cell-specific responses in mammary organoid culture. Endocrinology. 2008;149:2098–2107. doi: 10.1210/en.2007-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hata K, Nishimura R, Ueda M, Ikeda F, Matsubara T, Ichida F, Hisada K, Nokubi T, Yamaguchi A, Yoneda T. A CCAAT/enhancer binding protein β isoform, liver-enriched inhibitory protein, regulates commitment of osteoblasts and adipocytes. Mol Cell Biol. 2005;25:1971–1979. doi: 10.1128/MCB.25.5.1971-1979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu HM, Tian Q, Baer M, Spooner CJ, Williams SC, Johnson PF, Schwartz RC. The C/EBP bZIP domain can mediate lipopolysaccharide induction of the proinflammatory cytokines interleukin-6 and monocyte chemoattractant protein-1. J Biol Chem. 2000;275:16373–16381. doi: 10.1074/jbc.M910269199. [DOI] [PubMed] [Google Scholar]
- 17.Kariagina A, Aupperlee MD, Haslam SZ. Progesterone receptor isoforms and proliferation in the rat mammary gland during development. Endocrinology. 2007;148:2723–2736. doi: 10.1210/en.2006-1493. [DOI] [PubMed] [Google Scholar]
- 18.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus WL, Montano MM, Katzenellenbogen BS. Identification of multiple, widely spaced estrogen-responsive regions in the rat progesterone receptor gene. Mol Endocrinol. 1994;8:952–969. doi: 10.1210/mend.8.8.7997237. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lanari C, Lüthy I, Lamb CA, Fabris V, Pagano E, Helguero LA, Sanjuan N, Merani S, Molinolo AA. Five novel hormone-responsive cell lines derived from murine mammary ductal carcinomas: in vivo and in vitro effects of estrogens and progestins. Cancer Res. 2001;61:293–302. [PubMed] [Google Scholar]
- 22.Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Jr, Shyamala G, Conneely OM, O'Malley BW. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–2278. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- 23.McCabe LR, Banerjee C, Kundu R, Harrison RJ, Dobner PR, Stein JL, Lian JB, Stein GS. Developmental expression and activities of specific fos and jun proteins are functionally related to osteoblast maturation: role of Fra-2 and Jun D during differentiation. Endocrinology. 1996;137:4398–4408. doi: 10.1210/endo.137.10.8828501. [DOI] [PubMed] [Google Scholar]
- 24.Mohamed MK, Tung L, Takimoto GS, Horwitz KB. The leucine zippers of c-fos and c-jun for progesterone receptor dimerization: A-dominance in the A/B heterodimer. J Steroid Biochem Mol Biol. 1994;51:241–250. doi: 10.1016/0960-0760(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 25.Mote PA, Johnston JF, Manninen T, Tuohimaa P, Clarke CL. Detection of progesterone receptor forms A and B by immunohistochemical analysis. J Clin Pathol. 2001;54:624–630. doi: 10.1136/jcp.54.8.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mote PA, Bartow S, Tran N, Clarke CL. Loss of co-ordinate expression of progesterone receptors A and B is an early event in breast carcinogenesis. Breast Cancer Res Treat. 2002;72:163–172. doi: 10.1023/a:1014820500738. [DOI] [PubMed] [Google Scholar]
- 27.Mulac-Jericevic B, Lydon JP, DeMayo FJ, Conneely OM. Defective mammary gland morphogenesis in mice lacking the progesterone receptor B isoform. Proc Natl Acad Sci U S A. 2003;100:9744–9749. doi: 10.1073/pnas.1732707100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petz LN, Ziegler YS, Loven MA, Nardulli AM. Estrogen receptor alpha and activating protein-1 mediate estrogen responsiveness of the progesterone receptor gene in MCF-7 breast cancer cells. Endocrinology. 2002;143:4583–4591. doi: 10.1210/en.2002-220369. [DOI] [PubMed] [Google Scholar]
- 29.Petz LN, Ziegler YS, Schultz JR, Kim H, Kemper JK, Nardulli AM. Differential regulation of the human progesterone receptor gene through an estrogen response element half site and Sp1 sites. J Steroid Biochem Mol Biol. 2004a;88:113–122. doi: 10.1016/j.jsbmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Petz LN, Ziegler YS, Schultz JR, Nardulli AM. Fos and Jun inhibit estrogen-induced transcription of the human progesterone receptor gene through an activator protein-1 site. Mol Endocrinol. 2004b;18:521–532. doi: 10.1210/me.2003-0105. [DOI] [PubMed] [Google Scholar]
- 31.Pieber D, Allport VC, Bennett PR. Progesterone receptor isoform A inhibits isoform B-mediated transactivation in human amnion. Eur J Pharmacol. 2001;427:7–11. doi: 10.1016/s0014-2999(01)01189-x. [DOI] [PubMed] [Google Scholar]
- 32.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richer JK, Jacobsen BM, Manning NG, Abel MG, Wolf DM, Horwitz KB. Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J Biol Chem. 2002;277:5209–5218. doi: 10.1074/jbc.M110090200. [DOI] [PubMed] [Google Scholar]
- 34.Robinson GW, Johnson PF, Hennighausen L, Sterneck E. The C/EBPβ transcription factor regulates epithelial cell proliferation and differentiation in the mammary gland. Genes Dev. 1998;12:1907–1916. doi: 10.1101/gad.12.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos SJ, Aupperlee MD, Xie J, Durairaj S, Miksicek R, Conrad SE, Leipprandt JR, Tan YS, Schwartz RC, Haslam SZ. Progesterone receptor A-regulated gene expression in mammary organoid cultures. J Steroid Biochem Mol Biol. 2009;115:161–172. doi: 10.1016/j.jsbmb.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schott DR, Shyamala G, Schnneider W, Parry G. Molecular cloning, sequence analyses, and expression of complementary DNA encoding murine progesterone receptor. Biochemistry. 1991;30:7014–7011. doi: 10.1021/bi00242a029. [DOI] [PubMed] [Google Scholar]
- 37.Schultz JR, Petz LN, Nardulli AM. Estrogen receptor alpha and Sp1 regulate progesterone receptor gene expression. Mol Cell Endocrinol. 2003;201:165–175. doi: 10.1016/s0303-7207(02)00415-x. [DOI] [PubMed] [Google Scholar]
- 38.Schultz JR, Petz LN, Nardulli AM. Cell- and ligand-specific regulation of promoters containing activator protein-1 and Sp1 sites by estrogen receptors α and β. J Biol Chem. 2005;280:347–354. doi: 10.1074/jbc.M407879200. [DOI] [PubMed] [Google Scholar]
- 39.Seagroves TN, Krnacik S, Raught B, Gay J, Burgess-Beusse B, Darlington GJ, Rosen JM. C/EBPβ, but not C/EBPα, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev. 1998;12:1917–1928. doi: 10.1101/gad.12.12.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seagroves TN, Lydon JP, Hovey RC, Vonderhaar BK, Rosen JM. C/EBPβ (CCAAT/enhancer binding protein) controls cell fate determination during mammary gland development. Mol Endocrinol. 2000;14:359–368. doi: 10.1210/mend.14.3.0434. [DOI] [PubMed] [Google Scholar]
- 41.Shen Q, Zhang Y, Uray IP, Hill JL, Kim HT, Lu C, Young MR, Gunther EJ, Hilsenbeck SG, Chodosh LA, Colburn NH, Brown PH. The AP-1 transcription factor regulates postnatal mammary gland development. Dev Biol. 2006;295:589–603. doi: 10.1016/j.ydbio.2006.03.042. [DOI] [PubMed] [Google Scholar]
- 42.Shyamala G, Yang X, Silberstein G, Barcellos-Hoff MH, Dale E. Transgenic mice carrying an imbalance in the native ratio of A to B forms of progesterone receptor exhibit developmental abnormalities in mammary glands. Proc Natl Acad Sci U S A. 1998;95:696–701. doi: 10.1073/pnas.95.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spooner CJ, Guo X, Johnson PF, Schwartz RC. Differential roles of C/EBP beta regulatory domains in specifying MCP-1 and IL-6 transcription. Mol Immunol. 2007;44:1384–1392. doi: 10.1016/j.molimm.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Stein B, Baldwin AS., Jr Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-κB. Mol Cell Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tominaga H, Maeda S, Hayashi M, Takeda S, Akira S, Komiya S, Nakamura T, Akiyama H, Imamura T. CCAAT/enhancer-binding protein β promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol Biol Cell. 2008;19:5373–5386. doi: 10.1091/mbc.E08-03-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uematsu S, Kaisho T, Tanaka T, Matsumoto M, Yamakami M, Omori H, Yamamoto M, Yoshimori T, Akira S. The C/EBPβ isoform 34-kDa LAP is responsible for NF-IL-6-mediated gene induction in activated macrophages, but is not essential for intracellular bacteria killing. J Immunol. 2007;179:5378–5386. doi: 10.4049/jimmunol.179.8.5378. [DOI] [PubMed] [Google Scholar]
- 47.Wethmar K, Bégay V, Smink JJ, Zaragoza K, Wiesenthal V, Dörken B, Calkhoven CF, Leutz A. C/EBPβΔuORF mice--a genetic model for uORF-mediated translational control in mammals. Genes Dev. 2010;24:15–20. doi: 10.1101/gad.557910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zahnow CA. CCAAT/enhancer binding proteins in normal mammary development and breast cancer. Breast Cancer Res. 2002;4:113–121. doi: 10.1186/bcr428. [DOI] [PMC free article] [PubMed] [Google Scholar]