Abstract
αB-Crystallin (αBc) is a small heat shock protein that protects cells against abnormal protein aggregation and disease-related degeneration. αBc is also a major structural protein that forms polydisperse multimers that maintain the liquid-like property of the eye lens. However, the relationship and regulation of the two functions have yet to be explored. Here, by combining NMR spectroscopy and multiple biophysical approaches, we found that αBc uses a conserved β4/β8 surface of the central α-crystallin domain to bind α-synuclein and Tau proteins and prevent them from aggregating into pathological amyloids. We noted that this amyloid-binding surface can also bind the C-terminal IPI motif of αBc, which mediates αBc multimerization and weakens its chaperone activity. We further show that disruption of the IPI binding impairs αBc self-multimerization but enhances its chaperone activity. Our work discloses the structural mechanism underlying the regulation of αBc chaperone activity and self-multimerization and sheds light on the different functions of αBc in antagonizing neurodegeneration and maintaining eye lens liquidity.
Keywords: crystallin, amyloid, α-synuclein, molecular chaperone, protein misfolding, protein structure, protein aggregation, Tau protein (Tau), αB-crystallin, protein quality control
Introduction
Molecular chaperones are key players in the protein quality-control system that governs protein homeostasis in cells (1–4). Under proteostasis stress, small heat shock proteins (sHsps) 3 are considered to be the first cellular defenders that prevent abnormal protein aggregation in an ATP-independent manner (5–7). As a ubiquitous and abundant mammalian sHsp, αB-crystallin (αBc) prevents different pathological amyloid aggregations that are closely associated with various human diseases, including Alzheimer's disease (AD) (8, 9), Parkinson's disease (PD) (10–13), and multiple sclerosis (14). αBc was found to be dramatically up-regulated and to colocalize with α-synuclein (αSyn) in Lewy bodies and Tau in neurofibrillary tangles from the brains of PD and AD patients (8, 10, 15), respectively. Mounting evidence shows that αBc can inhibit the pathological aggregation of various amyloid proteins (e.g. αSyn, Tau, and Aβ) (9, 11, 13, 16). It has been reported that αBc utilizes its central α-crystallin domain (CαBc) to capture Aβ40 (17), although it remains unclear how αBc recognizes different pathological amyloid clients under disease conditions.
In addition to its function as a chaperone, αBc is also an important structural protein in the vertebrate eye lens (18, 19). Life-long transparency and refraction of eye lens require extra high concentrations of soluble crystallins (up to 450 mg/ml) that pack with a short-range order while resisting crystallization and phase separation (20–22). During aging or under pathological conditions, crystallins may misfold and aggregate, which is causative to cataract, a common cause of blindness (23–25).
αBc consists of 175 amino acids, which are divided into three regions (see Fig. 1A). CαBc is flanked by a hydrophobic N-terminal region (NR) and a flexible C terminus (CT) containing a conserved IPI motif (26, 27). CαBc, a hallmark of the sHsp family, features an Ig-like topology and induces the formation of αBc dimers as the building units of higher-order multimers (28–31). αBc forms polydisperse and heterogeneous multimers (10–40 subunits) with rapid subunit exchange, which suggests a highly dynamic nature of αBc (32–35). In addition to CαBc-mediated dimerization, NR–NR and CT–CαBc interactions also contribute to the formation of higher-order αBc multimers (31, 34, 36). Interestingly, it was reported that dissociation of αBc multimers can stimulate the chaperone activity of αBc against amyloid aggregation (37, 38). Therefore, it appears that the two functions of αBc (a structural multimer versus an amyloid chaperone) are negatively correlated, and the mechanism and regulation underlying the switch of the two functions have yet to be investigated.
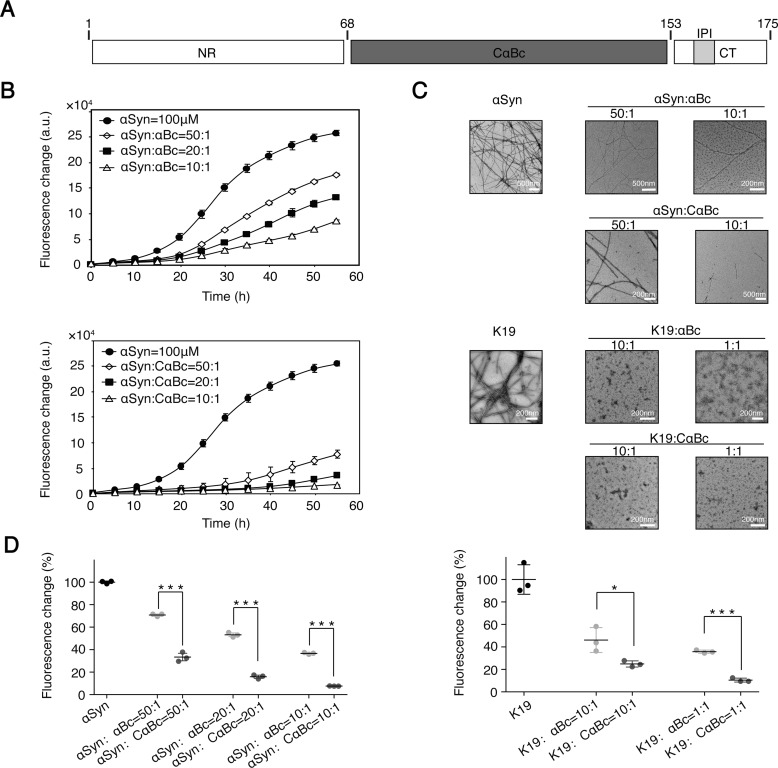
Figure 1.
αBc and CαBc inhibit aggregation of αSyn and K19. A, domain architecture of αBc. The CαBc is flanked by a flexible NR and a flexible CT containing a conserved IPI motif. B, ThT kinetics of αSyn aggregation inhibited by αBc (top) and CαBc (bottom), respectively. C, negative-stain EM images of αSyn (top) and K19 (bottom) fibrils with and without αBc/CαBc at different concentrations. D, comparison of chaperone activity of αBc and CαBc for preventing aggregation of αSyn (left) and K19 (right). The ThT value was taken at the 55-h time point from the ThT kinetics curves. Error bars correspond to mean ± S.E. with n = 3. * indicates p < 0.05, and *** indicates p < 0.005. a.u., absorbance units.
In this study, we found that αBc interacts with αSyn and Tau and prevents their amyloid aggregation by the conserved β4/β8 surface of CαBc. Interestingly, it is known that the C-terminal IPI motif of αBc also binds with the β4/β8 surface to mediate αBc multimerization. Thus, we further revealed that the interaction of IPI with αBc diminishes the chaperone activity of αBc; however, disruption of the interaction between IPI and αBc, which impairs αBc multimerization, in turn enhances its binding with amyloid clients and inhibits amyloid aggregation. Our work demonstrates that β4/β8 strands of αBc provide an interacting surface for the binding of different proteins/motifs that regulates αBc's activities between chaperoning amyloid clients and constructing eye lens via self-multimerization.
Results
CαBc is more potent than full-length αBc in preventing amyloid fibril formation
We first characterized the chaperone activity of αBc in preventing the aggregation of different amyloid clients, including αSyn of Parkinson's disease and K19 (the repeat region of 3R-Tau) of Alzheimer's disease. αBc exhibits potent chaperone activity in inhibiting fibril formation of both αSyn and K19 in a dose-dependent manner as monitored by a thioflavin T (ThT) fluorescence kinetic assay and negative-stain electron microscopy (EM) (Fig. 1, B–D). Consistent with previous reports (33, 34), we observed that αBc assembled into higher-order multimers in solution as measured by multiangle laser light scattering (Fig. S1A, left). Negative-stain EM further showed that αBc multimers are highly heterogeneous and feature spherical architectures with a diameter ranging from 15 to 30 nm (Fig. S1B). In sharp contrast to αBc, CαBc mainly populates as a dimer in solution with a molecular mass of ∼24.9 kDa (Fig. S1A, right). The ion mobility mass spectrum further showed an ensemble of CαBc monomer and dimer (Fig. S1C), indicating the dynamic nature of CαBc dimer. Intriguingly, compared with full-length αBc, CαBc exhibited a significantly enhanced chaperone activity in preventing both αSyn and K19 aggregation (Fig. 1, B–D). These results suggest that CαBc serves as a key region of αBc in preventing aggregation of different amyloid clients, but the chaperone activity is somehow weakened once CαBc is in the context of full-length αBc.
Structural characterization of the interaction between αBc and amyloid clients
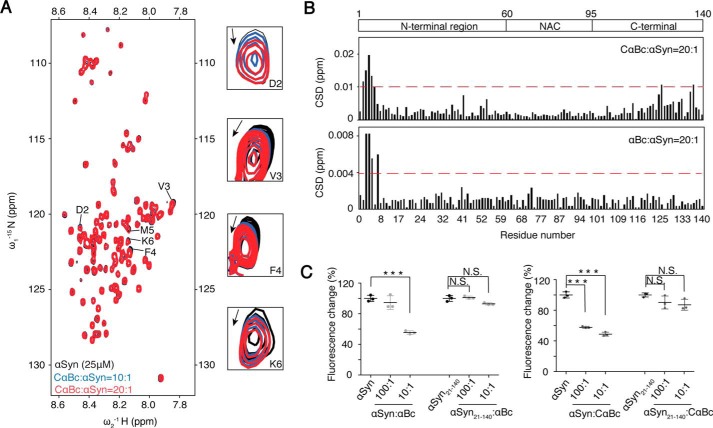
To understand the molecular mechanism underlying the chaperone activity of αBc, we conducted nuclear magnetic resonance (NMR) spectroscopy to investigate the interaction between CαBc/αBc and αSyn. By titration of CαBc into 15N-labeled acetylated αSyn, we found that the N terminus of αSyn, especially residues Asp2, Val3, Phe4, Met5, and Lys6, exhibited subtle chemical shift perturbations (Figs. 2, A and B, and S2A). Titration of full-length αBc to αSyn induced chemical shift changes of the same N-terminal region of αSyn but with smaller perturbations (Figs. 2B and S2B), which is consistent with the stronger inhibitory effect of CαBc on αSyn aggregation than that of full-length αBc (Fig. 1D). These NMR results indicate a weak binding of CαBc and αBc to the N terminus of αSyn. Indeed, as we deleted the N-terminal 20 residues of αSyn (αSyn(21–140)), the inhibitory effects of both CαBc and αBc on αSyn(21–140) aggregation was completely abolished (Figs. 2C and (S2C). Notice that the N terminus of αSyn is involved in membrane binding (39). Thus, in addition to inhibiting αSyn aggregation, αBc may also regulate binding of αSyn to membranes.
Figure 2.
N terminus of αSyn binds to both CαBc and αBc. A, an overlay of the 2D 1H-15N HSQC spectra of 25 μm αSyn in the absence (black) and presence of CαBc at molar ratios (αSyn:CαBc) of 1:10 (blue) and 1:20 (red), respectively. Resonances with relatively large chemical shift perturbations are highlighted on the right. B, CSDs of 25 μm αSyn titrated by CαBc (top) and αBc (bottom), respectively. The CSD values were calculated using the empirical equation CSD = [ΔHN2 + 0.0289(ΔN)2]1/2 where ΔHN and ΔN represent the chemical shift differences of 1H and 15N, respectively. The domain organization of αSyn is shown on the top of the graph. NAC stands for non amyloid-β component. C, the inhibitory effects of αBc and CαBc on the aggregation of αSyn and αSyn(21–140), respectively. The ThT value was taken at the 60-h time point from the ThT kinetics curves. Error bars correspond to mean ± S.E. with n = 3. *** indicates p < 0.005, and N.S. indicates not significant.
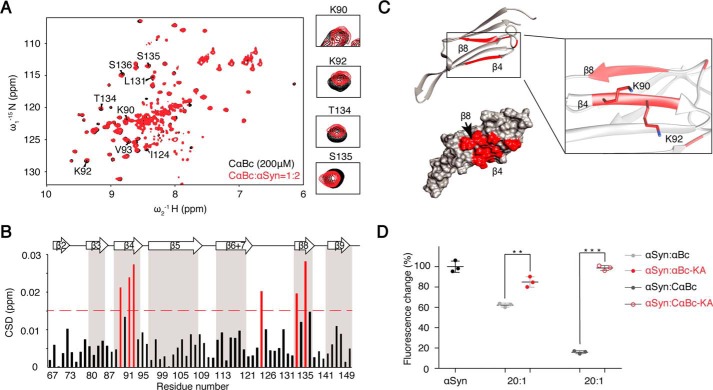
To identify the interacting surface of αBc, we inversely titrated [15N]CαBc with αSyn. The result showed significant chemical shift perturbations of residues including Lys90, Lys92, Val93, Ile124, Thr134, Ser135, Ser136, and Leu137 (Figs. 3, A and B, and S3A). Most perturbed residues cluster on the β4/β8 strands of CαBc (Fig. 3C), implying that the interface of CαBc interacts with αSyn. The apparent Kd value for CαBc–αSyn complex was 275 ± 105 μm as determined by NMR titrations (Fig. S3C), confirming a weak binding between CαBc and its client αSyn. Intriguingly, the binding affinity was enhanced when the temperature was increased (Fig. S3D), indicating that environmental factors (e.g. temperature, pH, and salt) may be involved in regulating the interaction between CαBc and its client. To validate the NMR result, we mutated Lys90 and Lys92 in β4 to alanine (the double mutation is named “KA”). The KA mutation in both CαBc and αBc severely disrupted the chaperone activity of inhibiting αSyn aggregation (Fig. 3D). A previous study showed that the β4/β8 strands of αBc are also involved in Aβ40 binding (17). Thus, we asked whether αBc utilizes a common surface for the binding of different amyloid clients. To address this question, we titrated Tau K19 to [15N]CαBc. The result showed that residues involved in K19 binding are also located within the β4/β8 strands of CαBc, including Lys92, Val93, Leu94, Thr134, Ser136, and Leu137 (Fig. S3E). Taken together, these results demonstrate that αBc utilizes a common surface consisting of the β4/β8 strands to bind different amyloid clients, including αSyn, Aβ, and Tau.
Figure 3.
Identification of the binding surface of CαBc and αBc to αSyn. A, an overlay of the 2D 1H-15N HSQC spectra of 200 μm CαBc in the absence (black) and presence of 400 μm αSyn (red). Residues with significant resonances changing are labeled. Resonances of the four key interacting residues, Lys90, Lys92, Thr134, and Ser135, are highlighted on the right. B, CSD profile of CαBc upon addition of αSyn. Deviations higher than 0.015 ppm are highlighted in red. Secondary structure assignment of CαBc is on the top of the graph. C, residues with large CSD (>0.015 ppm) upon αSyn titration are highlighted in red on the structure of CαBc (Protein Data Bank (PDB) code 2klr) with ribbon (top) and surface representation (bottom). Two key interacting residues, Lys90 and Lys92, are shown in a zoomed-in view on the right. D, inhibitory effects of αBc and its variants on the amyloid aggregation of αSyn (100 μm). The ThT value was taken at the 58-h time point from the ThT kinetics curves. Error bars correspond to mean ± S.E. with n = 3. *** indicates p < 0.005, and ** indicates p < 0.01.
Intriguingly, the β4/β8 surface has been previously identified to interact with the C-terminal IPI motif of αBc (residues 156–164) to mediate αBc self-multimerization (36, 40, 41). Therefore, the β4/β8 surface is essential for both αBc multimerization and chaperone activity, and αBc multimers may represent a self-inhibitory conformation that hinders αBc from binding to amyloid clients. However, in CαBc, which does not contain the IPI motif, the β4/β8 surface is fully exposed to interact with amyloid clients, explaining its enhanced chaperone activity.
The competitive binding of αSyn and the IPI motif to CαBc
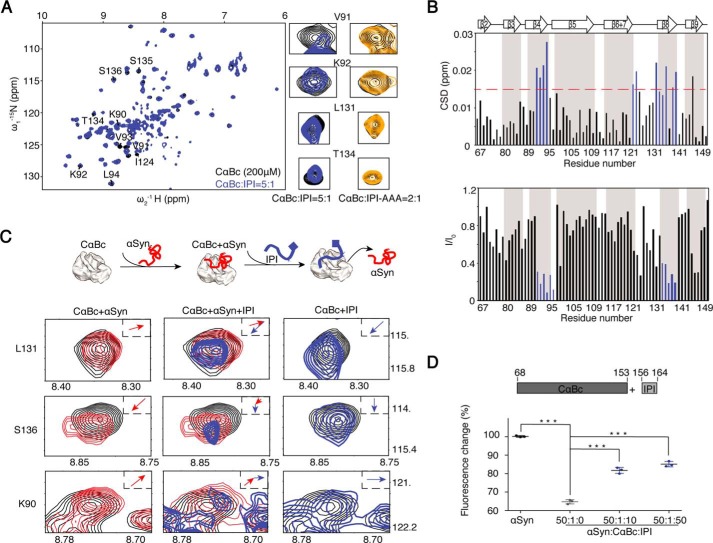
We next investigated the competition between αSyn and the IPI motif in binding the β4/β8 surface of CαBc and its influence in modulating chaperone activity. First, we titrated synthetic peptide 156ERTIPITRE164 (named “IPI” peptide) to [15N]CαBc. The 2D 1H-15N HSQC spectra showed significant chemical shift perturbations and intensity changes of residues, including Lys90, Val91, Lys92, Val93, Leu94, Ile124, Thr134, Ser135, and Ser136 (Fig. 4, A and B), which is consistent with a previous report (40), indicating that the IPI peptide binds to the β4/β8 strands of CαBc in solution. Intriguingly, 40 μm IPI peptide induced significant HSQC spectral changes of CαBc (Fig. 4, A and B); such changes were only achieved by αSyn at 400 μm (Fig. 3, A and B). The result indicates that CαBc binds to the IPI peptide much tighter than to αSyn, which is consistent with previous studies by mass spectrometry showing that the Kd value for the IPI peptide binding to CαBc was 70 μm (36). We further mutated the central residues 159IPI161 of the IPI peptide to AAA (named “AAA” mutation) and observed that the changes on NMR spectra of CαBc were diminished, which indicates the vital role of the 159IPI161 segment in the binding of the IPI peptide to CαBc (Figs. 4A and S4, A and B).
Figure 4.
Competitive binding of the CαBc β4/β8 surface by IPI peptide and αSyn. A, an overlay of the 2D 1H-15N HSQC spectra of 200 μm CαBc alone (black) and after incubation with 40 μm IPI peptide (blue). Resonances of four residues, Val91, Lys92, Leu131, and Thr134, that underwent significant changes are displayed in a zoomed-in view. The resonances of the same residues of CαBc (200 μm) in the presence of 100 μm IPI-AAA peptide (dark yellow) are shown on the right. B, residue-specific CSD (top) and intensity changes (I/I0; bottom) of CαBc in the presence of the IPI peptide. Residues with CSD >0.015 ppm and I/I0 <0.4 are highlighted in blue, respectively. C, resonance changes of Leu131, Ser136, and Lys90 of CαBc (black) in the presence of αSyn alone (left column; red), αSyn (middle column; red) followed by titration of the IPI peptide (middle column; blue), and the IPI peptide alone (right column; blue), respectively. The inset shows the direction of chemical shift changes upon titration. A cartoon of the sequential titrations of αSyn and the IPI peptide to CαBc is shown on top. D, addition of the IPI peptide weakens the chaperone activity of CαBc for inhibiting αSyn aggregation. The ThT value was taken at the 80-h time point from the ThT kinetics curves. Error bars correspond to mean ± S.E. with n = 3. *** indicates p < 0.005.
Notably, although similar residues of the β4/β8 surface are involved in the binding to the IPI peptide and αSyn, their binding patterns are significantly different as probed by NMR spectroscopy. αSyn binding induced a global intensity decrease of the entire CαBc (Fig. S3B). In contrast, binding of the IPI peptide resulted in a significant intensity drop (I/I0 < 0.4) of the interacting residues of the β4/β8 surface (Fig. 4B) in addition to a global decrease, implying that the interaction between IPI and CαBc is in the fast to intermediate exchange on the NMR time scale. Moreover, three residues, namely Lys90, Leu131, and Ser136, exhibit distinct chemical shift perturbation patterns for the two partners (Fig. 4C, left and right).
These differences enabled us to directly monitor the competition between αSyn and the IPI peptide for binding CαBc at the residue level. We premixed [15N]CαBc (200 μm) and αSyn (400 μm) in solution, and then by the addition of IPI peptide we observed sequential chemical shift changes of residues Lys90, Leu131, and Ser136 as shown in Fig. 4C (middle), which indicates the replacement of αSyn by the IPI peptide from the binding of CαBc β4/β8. Only 40 μm IPI peptide, 10% of αSyn, was required to replace αSyn for CαBc binding, further validating that the binding affinity of the IPI peptide to CαBc is much higher than that of αSyn. Consistently, the IPI peptide significantly weakened the chaperone activity of CαBc against αSyn aggregation in a dose-dependent manner (Fig. 4D). These data suggest that αSyn and the free IPI peptide competitively bind to the same β4/β8 surface of CαBc, which indicates that this competition may regulate the two different functions of CαBc.
IPI motif regulates αBc self-multimerization and client binding
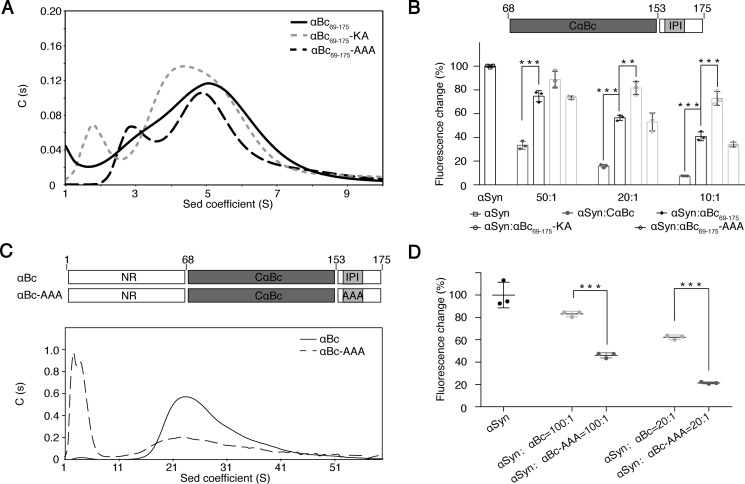
To investigate the regulation of the dual functions of αBc as structural multimers and an amyloid chaperone, we first constructed αBc(69–175), which contains CαBc followed by the C terminus with the IPI motif. Similar to full-length αBc, αBc(69–175) formed higher-order multimers as characterized by analytical size exclusion chromatography (Fig. S5). However, the multimerization of αBc(69–175) was severely impaired by both the AAA and KA mutations that disrupt the interaction between the IPI motif and the β4/β8 surface as monitored by analytical ultracentrifugation (Fig. 5A). These results demonstrate the importance of the IPI–β4/β8 surface interaction in mediating αBc(69–175) multimerization.
Figure 5.
Influence of the IPI-β4/β8 interaction in αBc multimerization and chaperone activity. A, sedimentation velocity analysis of αBc(69–175), αBc(69–175)-KA, and αBc(69–175)-AAA at 20 °C at a concentration of 5 mg/ml. B, comparison of the chaperone activity of CαBc, αBc(69–175), αBc(69–175)-KA, and αBc(69–175)-AAA for preventing αSyn aggregation. The ThT value was taken at the 58-h time point from the ThT kinetics curves. Error bars correspond to mean ± S.E. with n = 3. *** indicates p < 0.005, and ** indicates p < 0.01. C, sedimentation (Sed) velocity analysis of αBc (0.7 mg/ml) and αBc-AAA (0.7 mg/ml) at 20 °C. D, comparison of the chaperone activities of αBc and αBc-AAA for inhibiting αSyn aggregation. Error bars correspond to mean ± S.E. with n = 3. *** indicates p < 0.005.
Notably, αBc(69–175) multimers exhibited decreased chaperone activity against αSyn aggregation compared with CαBc (Fig. 5B). However, the AAA mutation, which prevents the IPI motif from binding to β4/β8, restored the chaperone activity (Fig. 5B). In contrast, the KA mutation on the β4/β8 surface that disrupts the interaction of αBc with both αSyn and the IPI motif abolished the chaperone activity as well as αBc self-assembly (Fig. 5, A and B). Furthermore, similar to that of αBc(69–175), the AAA mutation of full-length αBc significantly disrupted the self-multimerization of αBc but increased the chaperone activity against αSyn aggregation (Fig. 5, C and D). CD spectral analysis confirmed that both KA and AAA mutations retain native structures similar to that of WT αBc (Fig. S6). Taken together, these results demonstrate that as the C-terminal IPI motif binds to the β4/β8 surface, αBc undergoes higher-order self-multimerization that may serve as structural protein ensembles in maintaining eye lens. As the IPI motif releases the β4/β8 surface, αBc may depolymerize, and its function may switch to chaperoning amyloid clients.
Discussion
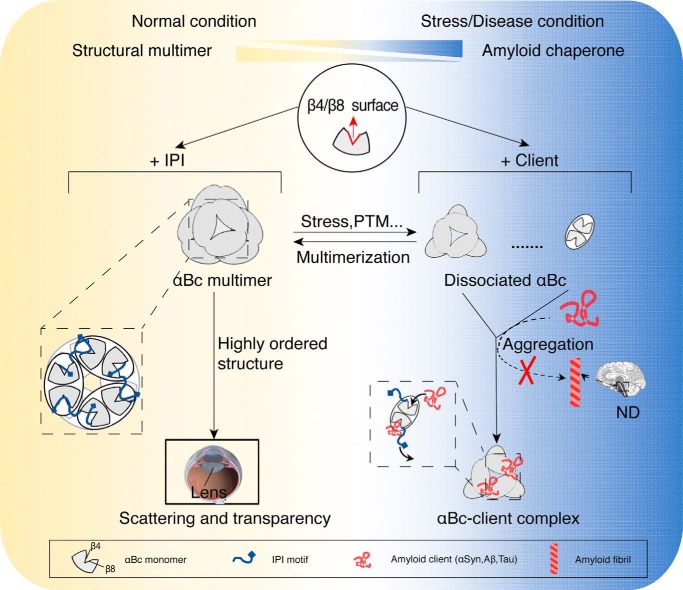
To maintain proteostasis, the activities of different chaperones, especially the stress-activated chaperones, are under elaborate control by distinct regulatory mechanisms in response to different stimuli and numerous clients (3). For instance, chaperone activities can be controlled by large conformational changes trigged by pH (HdeA/HdeB) or cysteine oxidation (Hsp33) or entire quaternary structural rearrangement (Bri2 BRICHOS) (42–45). Previous studies have shown that αBc multimerization is negatively correlated with its chaperone activity (37, 38, 46). However, the structural basis underlying this switch and the regulation of the two functions remain unclear. In this study, we found that αBc utilizes the same conserved β4/β8 surface for both self-assembly and chaperoning different amyloid clients, which enables a competitive regulation between the two functions. Based on our finding in this study and previous results (17, 19), we propose a working model of how αBc functions under distinct biological conditions (Fig. 6). Under normal conditions, αBc mainly forms large, polydisperse multimers to maintain the liquid-like property of lens at extra high local concentrations and to retain its autoinhibited state with minimal chaperone activity in brain and other tissues. However, under stress or disease conditions (e.g. AD and PD) where the amyloid clients (e.g. Tau and αSyn) accumulate, αBc may dissociate from higher-order multimers to release accessible β4/β8 surface with enhanced chaperone activity for capturing amyloid clients and preventing amyloid aggregation in brain.
Figure 6.
Schematic diagram of the regulation of αBc for chaperone activity and multimerization. Under normal conditions, αBc forms polydisperse multimers (left) with limited chaperone activity in which the β4/β8 surface is occupied by the neighboring IPI peptide. In lens, multimerization enables αBc to act as a structural protein that packs into higher-order structures to maintain the scattering and transparency of lens. Under stress or disease conditions, αBc disassembles to small multimers (e.g. dimers and hexamers) in response to different stimuli, e.g. stress or phosphorylation (PTM), and exhibits much enhanced chaperone activity. The activated αBc (right) may capture different pathological amyloid clients (e.g. αSyn, Aβ, and Tau) with a more exposed β4/β8 surface and prevent them from forming irreversible amyloid aggregations, which are closely associated with a variety of neurodegenerative diseases (ND). The regulation of αBc between these two functions is accomplished by the competitive binding of the IPI motif and amyloid clients to the key β4/β8 surface of αBc.
However, the regulation of αBc disassembly is not fully understood. Previously, the NR–NR interaction was found to contribute to αBc multimerization (31), whereas phosphorylation of residues from the NR can depolymerize αBc multimers and increase its chaperone activity (38, 47). We also found that, without NR, αBc(69–175) forms multimers of smaller average size compared with that of full-length αBc (∼5S compared with ∼20S), confirming the importance of NR in αBc multimer formation. Thus, it is important to study how different interactions (e.g. NR–NR, IPI–β4/β8, amyloid client–β4/β8) interplay for controlling (dis)assembly of αBc and its chaperone activity under different conditions and external stimuli (e.g. stress, aging, and diseases).
In addition to forming homomultimers, αBc also forms heteromultimers with different sHsps in vivo (e.g. with αA-crystallin in lens and with Hsp27 outside lens) to fulfill different functions (48, 49). Sequence alignment revealed that the β4/β8 interface and the IPI motif, but not the NR, are highly conserved in αBc, αA-crystallin, and Hsp27 (Fig. S7), suggesting that the hetero-β4/β8–IPI interaction may also play an important role in regulation of the formation of heteromultimers and their chaperone activities under different conditions. As Hsp27 was also found to prevent aggregation of different amyloid proteins (50, 51), it will be of great interest to explore the potential competition between the hetero-β4/β8–IPI interaction and amyloid client binding by αBc and Hsp27 heteromultimers and its role in maintaining protein homeostasis under stress and disease conditions.
Experimental procedures
Plasmid construction
Genes encoding αBc and CαBc were amplified and inserted into pET-28a vector with an N-terminal His6 tag following a tobacco etch virus protease cleavage site. The gene encoding CαBc(69–175) was cloned into pET-32a vector with an N-terminal thioredoxin tag and His6 tag following a PreScission protease recognition site. Mutations KA (K90A/K92A) and AAA (I159A/P160A/I161A) were constructed by site-directed mutagenesis using Q5® site-directed mutagenesis kit (New England Biolabs). All resulting constructs were verified by DNA sequencing (GENEWIZ, Inc., Suzhou, China).
Protein purification
All proteins were expressed in Escherichia coli BL21(DE3) cells. αBc and its variants all contained a His6 tag and were purified on a 5-ml HisTrapTM FF column (GE Healthcare) with buffer containing 50 mm Tris-HCl, 100 mm NaCl, and a gradient of 0–300 mm imidazole, pH 8.0. The N-terminal His6 tag of αBc was removed by tobacco etch virus protease in a cleavage buffer containing 100 mm Tris-HCl and 100 mm NaCl, pH 8.0, and the cleaved proteins were further purified by a Superdex 75 26/60 column (GE Healthcare) equilibrated with buffer containing 50 mm PBS and 50 mm NaCl, pH 7.0. PreScission protease in a cleavage buffer containing 50 mm Tris-HCl and 100 mm NaCl, pH 8.0, was used to remove the N-terminal thioredoxin tag of CαBc(69–175) and its variants. Expression and purification of amyloid proteins αSyn and K19 were the same as described previously (52, 53). 15N-Labeled proteins for solution NMR studies were grown in M9 minimal medium with [15N]NH4Cl (1 g/liter) and/or [13C]glucose as the sole nitrogen and carbon source. Purification was the same as that for the unlabeled proteins.
ThT fluorescence assay
ThT fluorescence of αSyn/K19 fibril formation was monitored by a Varioskan Flash spectral scanning multimode reader (Thermo Fisher Scientific) with excitation at 440 nm and emission at 485 nm. Purified αSyn/K19 monomer was filtered through 0.2-μm membranes (Millipore) and then was mixed with or without αBc and its variants at the indicated concentration in aggregation buffer (50 mm PBS, 50 mm NaCl, and 0.05% NaN3, pH 7.0). A final concentration of 50 μm ThT was added to each sample. Fibril growth was initiated by 0.5% freshly prepared fibril seeds (the seeds were prepared by sonicating fibrils for 15 s) and monitored over 300 runs (5 min for each run) at 37 °C with a shaking speed of 600 rpm. Three to five repeats were performed for each experiment for statistical analysis.
Transmission electron microscopy
Images were collected on Tecnai G2 Spirit transmission electron microscope operated at an accelerating voltage of 120 kV. Samples (8 μl) were deposited on carbon-coated grids for 45 s. The grids were then washed twice with double distilled H2O (8 μl) and incubated with 8 μl of uranyl acetate (2%, v/v) for staining. Images were recorded using a 4000 × 4000 charge-coupled device camera (BM-Eagle, FEI Tecnai). For visualization of αBc oligomers, 50 μm αBc was prepared in phosphate buffer (50 mm PBS and 50 mm NaCl, pH 7.0).
Size exclusion chromatography and multiangle laser light scattering
αBc and its variants were analyzed using an in-line Agilent 1260 HPLC coupled with a Superdex 75 10/300 GL column (GE Healthcare) and a miniDAWN TREOS instrument (Wyatt Technology). Three angles (45°, 90°, and 135°) were used for monitoring light scattering at 690 nm. 100 μl of αBc (1 mg/ml) and CαBc (5 mg/ml) in phosphate buffer were loaded to the column with a flow rate of 0.4 ml/min at room temperature.
Ion mobility mass spectrometry
CαBc was buffer-exchanged into 10 mm ammonium acetate using a desalting column and analyzed by positive ion nano-electrospray ionization with a flow rate of 3 nl/min. An Agilent 6560 ion mobility quadrupole TOF mass spectrometer (Agilent Technologies) equipped with a drift tube before the quadrupole and the TOF analyzers (54) was used for ion mobility MS analyses. The instrumental parameters were as follows: gas temperature, 60 °C; drying gas, 5 liters/min; nebulizer, 15 p.s.i.; capillary voltage, 3500 V; TOF mass range, 300–3200 Da; high pressure funnel RF, 200 V; trap funnel RF, 200 V; drift tube entrance voltage, 1300 V; drift tube exit voltage, 250 V; rear funnel RF, 150 V; ion mobility spectrometry cell pressure, 4.00 torr.
NMR spectroscopy
All NMR samples were prepared in a buffer containing 50 mm sodium phosphate and 50 mm NaCl, pH 7.0, with 10% D2O. All NMR spectra were acquired on a Bruker Avance 900- or 600-MHz spectrometer equipped with cryogenically cooled probes at 25 °C. Backbone assignments of CαBc, αSyn, and K19 were accomplished based on the collected 3D HNCACB and CBCACONH spectra and assignments from previous studies (55–57). 3D experiments were performed using ∼1 mm 15N/13C-labeled NMR samples, respectively. For titration experiments, each 2D 1H-15N HSQC spectrum was collected with 16 scans per transient and complex points of 2048 × 160. Each NMR sample was freshly prepared from high-concentration protein stocks with a total volume of 500 μl. 25 μm 15N-labeled acetylated αSyn was used to conduct the titration experiments with CαBc concentrations of 250 and 500 μm and αBc concentrations of 500 μm. 200 μm 15N-labeled CαBc was mixed in the absence or presence of αSyn (200 and 400 μm), the IPI peptide (40 and 100 μm), and the IPI-AAA peptide (40 and 100 μm), respectively. Chemical shift deviations (CSDs; Δδ) were calculated using the following equation,
| (Eq. 1) |
where Δδ1H and Δδ15N are the chemical shift differences of amide proton and amide nitrogen between free and bound states of the protein, respectively. The Kd for αSyn binding to CαBc was determined by NMRViewJ at 25 and 35 °C, respectively. Six peaks corresponding to residues Lys90, Val91, Lys92, Leu131, Thr134, and Ser135 from titrations were fit to a quadratic binding curve using a base 10 quadratic fit and 250 simulations, and then an average Kd for all peaks fitted was calculated. For the competition experiments between αSyn and IPI peptide binding to CαBc, 200 μm CαBc was first incubated with 400 μm αSyn, and then 40 μm IPI peptide was added. All NMR spectra were processed using NMRPipe (58) and analyzed by SPARKY (59) and NMRView (60).
Analytical ultracentrifugation
The sizes of αBc and its variants were determined by analytical ultracentrifugation using sedimentation velocity analysis. All samples were prepared in a buffer containing 50 mm sodium phosphate and 50 mm NaCl, pH 7.0. The concentration of αBc and αBc-AAA used in this study was 0.7 mg/ml. The concentration of αBc(69–175), αBc(69–175)-KA, and αBc(69–175)-AAA was 5 mg/ml. Sedimentation velocity experiments were performed at 50,000 rpm using a Beckman Coulter XL-I ultracentrifuge (Beckman Instruments) with an An60Ti eight-hole rotor at 25 °C. The absorbance data were collected at 280 nm in continuous mode for at least 12 h. Data were analyzed with the program SEDFIT (61) with a continuous size-distribution (c(s)) model.
Circular dichroism
The secondary structure of αBc and variants was measured by a Chirascan CD spectrometer (Applied Photophysics, UK). The samples (20 μm) were prepared in a buffer containing 50 mm PBS and 50 mm NaCl, pH 7.0. Spectra were recorded at 200–260 nm with a step size of 1 nm and a cell path length of 1 mm. Each sample was scanned three times. All data were analyzed by Pro-Data Viewer. Secondary structural content of each protein was determined by analysis of the CD spectrum using CDNN and BeStSel (62), respectively.
Author contributions
Z. L., S. Z., and C. L. conceptualization; Z. L. data curation; Z. L. software; Z. L. formal analysis; Z. L. and C. L. validation; Z. L. and S. Z. methodology; Z. L. and S. Z. writing-original draft; Z. L., D. L., S. Z., and C. L. writing-review and editing; S. Z. and C. L. supervision; S. Z. and C. L. investigation; S. Z. and C. L. project administration; C. L. resources; C. L. funding acquisition; C. L. visualization; C. W. data collection; Y. L. data collection; C. Z. protein purification; T. L. data analysis.
Supplementary Material
Acknowledgments
We thank Dr. Zhijun Liu, Dr. Songzi Jiang, and other staff members of the National Center for Protein Science Shanghai for assistance in NMR data collection.
This work was supported by Major State Basic Research Development Program Grant 2016YFA0501902, National Natural Science Foundation (NSF) of China Grant 31470748, the Chinese Academy of Sciences, and The “1000 Talents Plan” of China. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7.
- sHsp
- small heat shock protein
- αBc
- αB-crystallin
- CαBc
- central α-crystallin domain
- NR
- N-terminal region
- CT
- C terminus
- αBc(69–175)
- CαBc and the following C terminus of αBc
- IPI peptide
- 156ERTIPITRE164
- αSyn
- α-synuclein
- αSyn(21–140)
- N-terminal 20-residue-deletion mutant of αSyn
- K19
- the repeat region of 3R-Tau
- AD
- Alzheimer's disease
- PD
- Parkinson's disease
- ThT
- thioflavin T
- Aβ
- amyloid β
- HSQC
- heteronuclear single quantum coherence
- RF
- radio frequency
- CSD
- chemical shift deviation.
References
- 1. Gidalevitz T., Prahlad V., and Morimoto R. I. (2011) The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harb. Perspect. Biol. 3, a009704 10.1101/cshperspect.a009704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartl F. U., Bracher A., and Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- 3. Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., and Hartl F. U. (2013) Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- 4. Lindberg I., Shorter J., Wiseman R. L., Chiti F., Dickey C. A., and McLean P. J. (2015) Chaperones in neurodegeneration. J. Neurosci. 35, 13853–13859 10.1523/JNEUROSCI.2600-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor R. P., and Benjamin I. J. (2005) Small heat shock proteins: a new classification scheme in mammals. J. Mol. Cell. Cardiol. 38, 433–444 10.1016/j.yjmcc.2004.12.014 [DOI] [PubMed] [Google Scholar]
- 6. Tyedmers J., Mogk A., and Bukau B. (2010) Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 11, 777–788 10.1038/nrm2993 [DOI] [PubMed] [Google Scholar]
- 7. Treweek T. M., Meehan S., Ecroyd H., and Carver J. A. (2015) Small heat-shock proteins: important players in regulating cellular proteostasis. Cell. Mol. Life Sci. 72, 429–451 10.1007/s00018-014-1754-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shinohara H., Inaguma Y., Goto S., Inagaki T., and Kato K. (1993) αB crystallin and HSP28 are enhanced in the cerebral cortex of patients with Alzheimer's disease. J. Neurol. Sci. 119, 203–208 10.1016/0022-510X(93)90135-L [DOI] [PubMed] [Google Scholar]
- 9. Björkdahl C., Sjögren M. J., Zhou X., Concha H., Avila J., Winblad B., and Pei J. J. (2008) Small heat shock proteins Hsp27 or αB-crystallin and the protein components of neurofibrillary tangles: tau and neurofilaments. J. Neurosci. Res. 86, 1343–1352 10.1002/jnr.21589 [DOI] [PubMed] [Google Scholar]
- 10. Renkawek K., Stege G. J., and Bosman G. J. (1999) Dementia, gliosis and expression of the small heat shock proteins hsp27 and αB-crystallin in Parkinson's disease. Neuroreport 10, 2273–2276 10.1097/00001756-199908020-00009 [DOI] [PubMed] [Google Scholar]
- 11. Rekas A., Adda C. G., Andrew Aquilina J., Barnham K. J., Sunde M., Galatis D., Williamson N. A., Masters C. L., Anders R. F., Robinson C. V., Cappai R., and Carver J. A. (2004) Interaction of the molecular chaperone αB-crystallin with α-synuclein: effects on amyloid fibril formation and chaperone activity. J. Mol. Biol. 340, 1167–1183 10.1016/j.jmb.2004.05.054 [DOI] [PubMed] [Google Scholar]
- 12. Cox D., Selig E., Griffin M. D., Carver J. A., and Ecroyd H. (2016) Small heat-shock proteins prevent α-synuclein aggregation via transient interactions and their efficacy is affected by the rate of aggregation. J. Biol. Chem. 291, 22618–22629 10.1074/jbc.M116.739250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cox D., and Ecroyd H. (2017) The small heat shock proteins αB-crystallin (HSPB5) and Hsp27 (HSPB1) inhibit the intracellular aggregation of α-synuclein. Cell Stress Chaperones 22, 589–600 10.1007/s12192-017-0785-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lowe J., McDermott H., Pike I., Spendlove I., Landon M., and Mayer R. J. (1992) αB crystallin expression in non-lenticular tissues and selective presence in ubiquitinated inclusion bodies in human disease. J. Pathol. 166, 61–68 10.1002/path.1711660110 [DOI] [PubMed] [Google Scholar]
- 15. López-Gonzalez I., Carmona M., Arregui L., Kovacs G. G., and Ferrer I. (2014) αB-Crystallin and HSP27 in glial cells in tauopathies. Neuropathology 34, 517–526 10.1111/neup.12134 [DOI] [PubMed] [Google Scholar]
- 16. Raman B., Ban T., Sakai M., Pasta S. Y., Ramakrishna T., Naiki H., Goto Y., and Rao Ch. M. (2005) αB-Crystallin, a small heat-shock protein, prevents the amyloid fibril growth of an amyloid β-peptide and β2-microglobulin. Biochem. J. 392, 573–581 10.1042/BJ20050339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mainz A., Peschek J., Stavropoulou M., Back K. C., Bardiaux B., Asami S., Prade E., Peters C., Weinkauf S., Buchner J., and Reif B. (2015) The chaperone αB-crystallin uses different interfaces to capture an amorphous and an amyloid client. Nat. Struct. Mol. Biol. 22, 898–905 10.1038/nsmb.3108 [DOI] [PubMed] [Google Scholar]
- 18. Fagerholm P. P., Philipson B. T., and Lindström B. (1981) Normal human lens—the distribution of protein. Exp. Eye Res. 33, 615–620 10.1016/S0014-4835(81)80101-7 [DOI] [PubMed] [Google Scholar]
- 19. Andley U. P. (2007) Crystallins in the eye: function and pathology. Prog. Retin. Eye Res. 26, 78–98 10.1016/j.preteyeres.2006.10.003 [DOI] [PubMed] [Google Scholar]
- 20. Tardieu A. (1988) Eye lens proteins and transparency: from light transmission theory to solution X-ray structural analysis. Annu. Rev. Biophys. Biophys. Chem. 17, 47–70 10.1146/annurev.bb.17.060188.000403 [DOI] [PubMed] [Google Scholar]
- 21. Tardieu A. (1998) α-Crystallin quaternary structure and interactive properties control eye lens transparency. Int. J. Biol. Macromol. 22, 211–217 10.1016/S0141-8130(98)00018-X [DOI] [PubMed] [Google Scholar]
- 22. Delaye M., and Tardieu A. (1983) Short-range order of crystallin proteins accounts for eye lens transparency. Nature 302, 415–417 10.1038/302415a0 [DOI] [PubMed] [Google Scholar]
- 23. Zhao L., Chen X. J., Zhu J., Xi Y. B., Yang X., Hu L. D., Ouyang H., Patel S. H., Jin X., Lin D., Wu F., Flagg K., Cai H., Li G., Cao G., et al. (2015) Lanosterol reverses protein aggregation in cataracts. Nature 523, 607–611 10.1038/nature14650 [DOI] [PubMed] [Google Scholar]
- 24. Makley L. N., McMenimen K. A., DeVree B. T., Goldman J. W., McGlasson B. N., Rajagopal P., Dunyak B. M., McQuade T. J., Thompson A. D., Sunahara R., Klevit R. E., Andley U. P., and Gestwicki J. E. (2015) Pharmacological chaperone for α-crystallin partially restores transparency in cataract models. Science 350, 674–677 10.1126/science.aac9145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clark A. R., Lubsen N. H., and Slingsby C. (2012) sHSP in the eye lens: crystallin mutations, cataract and proteostasis. Int. J. Biochem. Cell Biol. 44, 1687–1697 10.1016/j.biocel.2012.02.015 [DOI] [PubMed] [Google Scholar]
- 26. Liu Z., Zhang S., Li D., and Liu C. (2017) A structural view of αB-crystallin assembly and amyloid aggregation. Protein Pept. Lett. 24, 315–321 10.2174/0929866524666170206122616 [DOI] [PubMed] [Google Scholar]
- 27. Basha E., O'Neill H., and Vierling E. (2012) Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem. Sci. 37, 106–117 10.1016/j.tibs.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bagnéris C., Bateman O. A., Naylor C. E., Cronin N., Boelens W. C., Keep N. H., and Slingsby C. (2009) Crystal structures of α-crystallin domain dimers of αB-crystallin and Hsp20. J. Mol. Biol. 392, 1242–1252 10.1016/j.jmb.2009.07.069 [DOI] [PubMed] [Google Scholar]
- 29. Laganowsky A., Benesch J. L., Landau M., Ding L., Sawaya M. R., Cascio D., Huang Q., Robinson C. V., Horwitz J., and Eisenberg D. (2010) Crystal structures of truncated αA and αB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 19, 1031–1043 10.1002/pro.380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hochberg G. K., Ecroyd H., Liu C., Cox D., Cascio D., Sawaya M. R., Collier M. P., Stroud J., Carver J. A., Baldwin A. J., Robinson C. V., Eisenberg D. S., Benesch J. L., and Laganowsky A. (2014) The structured core domain of αB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc. Natl. Acad. Sci. U.S.A. 111, E1562–E1570 10.1073/pnas.1322673111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jehle S., Vollmar B. S., Bardiaux B., Dove K. K., Rajagopal P., Gonen T., Oschkinat H., and Klevit R. E. (2011) N-terminal domain of αB-crystallin provides a conformational switch for multimerization and structural heterogeneity. Proc. Natl. Acad. Sci. U.S.A. 108, 6409–6414 10.1073/pnas.1014656108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aquilina J. A., Benesch J. L., Bateman O. A., Slingsby C., and Robinson C. V. (2003) Polydispersity of a mammalian chaperone: mass spectrometry reveals the population of oligomers in αB-crystallin. Proc. Natl. Acad. Sci. U.S.A. 100, 10611–10616 10.1073/pnas.1932958100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baldwin A. J., Lioe H., Hilton G. R., Baker L. A., Rubinstein J. L., Kay L. E., and Benesch J. L. (2011) The polydispersity of αB-crystallin is rationalized by an interconverting polyhedral architecture. Structure 19, 1855–1863 10.1016/j.str.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 34. Braun N., Zacharias M., Peschek J., Kastenmüller A., Zou J., Hanzlik M., Haslbeck M., Rappsilber J., Buchner J., and Weinkauf S. (2011) Multiple molecular architectures of the eye lens chaperone αB-crystallin elucidated by a triple hybrid approach. Proc. Natl. Acad. Sci. U.S.A. 108, 20491–20496 10.1073/pnas.1111014108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jehle S., Rajagopal P., Bardiaux B., Markovic S., Kühne R., Stout J. R., Higman V. A., Klevit R. E., van Rossum B. J., and Oschkinat H. (2010) Solid-state NMR and SAXS studies provide a structural basis for the activation of αB-crystallin oligomers. Nat. Struct. Mol. Biol. 17, 1037–1042 10.1038/nsmb.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hilton G. R., Hochberg G. K., Laganowsky A., McGinnigle S. I., Baldwin A. J., and Benesch J. L. (2013) C-terminal interactions mediate the quaternary dynamics of αB-crystallin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110405 10.1098/rstb.2011.0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peschek J., Braun N., Rohrberg J., Back K. C., Kriehuber T., Kastenmüller A., Weinkauf S., and Buchner J. (2013) Regulated structural transitions unleash the chaperone activity of αB-crystallin. Proc. Natl. Acad. Sci. U.S.A. 110, E3780–E3789 10.1073/pnas.1308898110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ecroyd H., Meehan S., Horwitz J., Aquilina J. A., Benesch J. L., Robinson C. V., Macphee C. E., and Carver J. A. (2007) Mimicking phosphorylation of αB-crystallin affects its chaperone activity. Biochem. J. 401, 129–141 10.1042/BJ20060981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fusco G., De Simone A., Gopinath T., Vostrikov V., Vendruscolo M., Dobson C. M., and Veglia G. (2014) Direct observation of the three regions in α-synuclein that determine its membrane-bound behaviour. Nat. Commun. 5, 3827 10.1038/ncomms4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Delbecq S. P., Jehle S., and Klevit R. (2012) Binding determinants of the small heat shock protein, αB-crystallin: recognition of the ‘IxI’ motif. EMBO J. 31, 4587–4594 10.1038/emboj.2012.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pasta S. Y., Raman B., Ramakrishna T., and Rao Ch. M. (2004) The IXI/V motif in the C-terminal extension of α-crystallins: alternative interactions and oligomeric assemblies. Mol. Vis. 10, 655–662 [PubMed] [Google Scholar]
- 42. Winter J., Linke K., Jatzek A., and Jakob U. (2005) Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol. Cell 17, 381–392 10.1016/j.molcel.2004.12.027 [DOI] [PubMed] [Google Scholar]
- 43. Ding J., Yang C., Niu X., Hu Y., and Jin C. (2015) HdeB chaperone activity is coupled to its intrinsic dynamic properties. Sci. Rep. 5, 16856 10.1038/srep16856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rajagopal P., Tse E., Borst A. J., Delbecq S. P., Shi L., Southworth D. R., and Klevit R. E. (2015) A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis. Elife 4, e07304 10.7554/eLife.07304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen G., Abelein A., Nilsson H. E., Leppert A., Andrade-Talavera Y., Tambaro S., Hemmingsson L., Roshan F., Landreh M., Biverstål H., Koeck P. J. B., Presto J., Hebert H., Fisahn A., and Johansson J. (2017) Bri2 BRICHOS client specificity and chaperone activity are governed by assembly state. Nat. Commun. 8, 2081 10.1038/s41467-017-02056-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benesch J. L., Ayoub M., Robinson C. V., and Aquilina J. A. (2008) Small heat shock protein activity is regulated by variable oligomeric substructure. J. Biol. Chem. 283, 28513–28517 10.1074/jbc.M804729200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ahmad M. F., Raman B., Ramakrishna T., and Rao Ch. M. (2008) Effect of phosphorylation on αB-crystallin: differences in stability, subunit exchange and chaperone activity of homo and mixed oligomers of αB-crystallin and its phosphorylation-mimicking mutant. J. Mol. Biol. 375, 1040–1051 10.1016/j.jmb.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 48. Zantema A., Verlaan-De Vries M., Maasdam D., Bol S., and van der Eb A. (1992) Heat shock protein 27 and αB-crystallin can form a complex, which dissociates by heat shock. J. Biol. Chem. 267, 12936–12941 [PubMed] [Google Scholar]
- 49. Mymrikov E. V., Seit-Nebi A. S., and Gusev N. B. (2012) Heterooligomeric complexes of human small heat shock proteins. Cell Stress Chaperones 17, 157–169 10.1007/s12192-011-0296-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cox D., Whiten D. R., Brown J. W. P., Horrocks M. H., San Gil R., Dobson C. M., Klenerman D., van Oijen A. M., and Ecroyd H. (2018) The small heat shock protein Hsp27 binds α-synuclein fibrils, preventing elongation and cytotoxicity. J. Biol. Chem. 293, 4486–4497 10.1074/jbc.M117.813865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Abisambra J. F., Blair L. J., Hill S. E., Jones J. R., Kraft C., Rogers J., Koren J. 3rd, Jinwal U. K., Lawson L., Johnson A. G., Wilcock D., O'Leary J. C., Jansen-West K., Muschol M., Golde T. E., et al. (2010) Phosphorylation dynamics regulate Hsp27-mediated rescue of neuronal plasticity deficits in tau transgenic mice. J. Neurosci. 30, 15374–15382 10.1523/JNEUROSCI.3155-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Daebel V., Chinnathambi S., Biernat J., Schwalbe M., Habenstein B., Loquet A., Akoury E., Tepper K., Müller H., Baldus M., Griesinger C., Zweckstetter M., Mandelkow E., Vijayan V., and Lange A. (2012) β-Sheet core of tau paired helical filaments revealed by solid-state NMR. J. Am. Chem. Soc. 134, 13982–13989 10.1021/ja305470p [DOI] [PubMed] [Google Scholar]
- 53. Singh P. K., Kotia V., Ghosh D., Mohite G. M., Kumar A., and Maji S. K. (2013) Curcumin modulates α-synuclein aggregation and toxicity. ACS Chem. Neurosci. 4, 393–407 10.1021/cn3001203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soper M. T., DeToma A. S., Hyung S. J., Lim M. H., and Ruotolo B. T. (2013) Amyloid-β-neuropeptide interactions assessed by ion mobility-mass spectrometry. Phys. Chem. Chem. Phys. 15, 8952–8961 10.1039/c3cp50721a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eliezer D., Barré P., Kobaslija M., Chan D., Li X., and Heend L. (2005) Residual structure in the repeat domain of tau: echoes of microtubule binding and paired helical filament formation. Biochemistry 44, 1026–1036 10.1021/bi048953n [DOI] [PubMed] [Google Scholar]
- 56. Bermel W., Bertini I., Felli I. C., Lee Y. M., Luchinat C., and Pierattelli R. (2006) Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J. Am. Chem. Soc. 128, 3918–3919 10.1021/ja0582206 [DOI] [PubMed] [Google Scholar]
- 57. Jehle S., van Rossum B., Stout J. R., Noguchi S. M., Falber K., Rehbein K., Oschkinat H., Klevit R. E., and Rajagopal P. (2009) αB-crystallin: a hybrid solid-state/solution-state NMR investigation reveals structural aspects of the heterogeneous oligomer. J. Mol. Biol. 385, 1481–1497 10.1016/j.jmb.2008.10.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 59. Lee W., Tonelli M., and Markley J. L. (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 10.1093/bioinformatics/btu830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 10.1385/1-59259-809-9:313 [DOI] [PubMed] [Google Scholar]
- 61. Gabrielson J. P., Randolph T. W., Kendrick B. S., and Stoner M. R. (2007) Sedimentation velocity analytical ultracentrifugation and SEDFIT/c(s): limits of quantitation for a monoclonal antibody system. Anal. Biochem. 361, 24–30 10.1016/j.ab.2006.11.012 [DOI] [PubMed] [Google Scholar]
- 62. Micsonai A., Wien F., Kernya L., Lee Y. H., Goto Y., Réfrégiers M., and Kardos J. (2015) Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 112, E3095–E3103 10.1073/pnas.1500851112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.