Abstract
Hypoxia-inducible factor 1α (HIF-1α) is a transcription factor that regulates cellular responses to hypoxia. It controls the expression of both BCL2/adenovirus E1B 19-kDa protein–interacting protein 3 (BNIP3) and insulin-like growth factor 2 (IGF2). Previous studies have demonstrated that in hypoxia, copper is required for the expression of BNIP3 but not for that of IGF2. Here, using ChIP assays, computational analyses, luciferase reporter assays, and real-time quantitative RT-PCR, we sought to better understand how copper regulates the differential target gene selectivity of HIF-1α. Human umbilical vein endothelial cells (HUVECs) were exposed to CoCl2 or hypoxia conditions to increase HIF-1α accumulation. The binding of HIF-1α to hypoxia-responsive element (HRE) sites in the BNIP3 or IGF2 gene promoter in high- or low-copper conditions was examined. Our analyses revealed three and two potential HRE sites in the BNIP3 and IGF2 promoters, respectively. We identified that HRE (−412/−404) in the BNIP3 promoter and HRE (−354/−347) in the IGF2 promoter are the critical binding sites of HIF-1α. Tetraethelenepentamine (TEPA)-mediated reduction in copper concentration did not affect hypoxia- or CoCl2-induced HIF-1α accumulation. However, the copper reduction did suppress the binding of HIF-1α to the HRE (−412/−404) in BNIP3 but not the binding of HIF-1α to the HRE (−354/−347) in IGF2. In summary, our findings uncovered the mechanistic basis for differential HIF-1α–mediated regulation of BNIP3 and IGF2, indicating that copper regulates target gene selectivity of HIF-1α at least in part by affecting HIF-1α binding to its cognate HRE in the promoters of these two genes.
Keywords: hypoxia-inducible factor (HIF), copper, transcription target gene, chromatin immunoprecipitation (ChIP), gene regulation, BNIP3, hypoxia responsive element, IGF2
Introduction
Hypoxia-inducible factor 1 (HIF-1), 3 a key transcription factor in regulation of cellular metabolism and homeostasis, transactivates the expression of multiple genes involved in anaerobic glycolysis, erythropoiesis, and angiogenesis in response to hypoxic condition that occurs under a diversity of physiological and pathological conditions, such as embryonic development, ischemic disease, pulmonary disease, and cancer (1–3). HIF-1 is composed of HIF-1α and HIF-1β subunits. The cellular stability of HIF-1α determines the ultimate activation of HIF-1. The protein level of HIF-1α is undetectable in most cell types under normoxic condition because of its degradation by the ubiquitin-proteasome pathway (4–6). Under hypoxic conditions, HIF-1α escapes from the degradation pathway, accumulates in the cytosol and is translocated into the nucleus, where it dimerizes with HIF-1β, interacts with cofactors to assemble the HIF-1 transcriptional complex, and binds to the hypoxia-responsive element (HRE) sites of its target genes, leading to transactivation of target genes expression. However, little is known about how the selectivity of the target genes of HIF-1α is regulated.
There are almost 300 genes that are regulated by HIF-1α (7, 8). It appears that not all of the genes controlled by HIF-1α are transactivated at the same time under a set of stress conditions. Previous studies have revealed several mechanisms regulating the target gene selectivity of HIF-1α, such as epigenetic modifications (9–11). Our previous studies found that copper regulates the target gene selectivity of HIF-1α (12). We observed that cobalt-induced accumulation of HIF-1α results in the transactivation of multiple HIF-1α target genes. Among these genes are critical angiogenic factors such as vascular endothelial growth factor (VEGF) (13–15). The treatment of human umbilical vein endothelial cells (HUVECs) with a copper chelator, tetraethylenepentamine (TEPA), does not affect cobalt-induced accumulation of HIF-1α but blocks the expression of VEGF, an effect that is reversible by an addition of copper sulfate (16).
This suppression of HIF-1α–regulated gene expression by copper deficiency is not only limited to VEGF but is also observed in other genes such as BCL2/adenovirus E1B 19-kDa protein–interacting protein 3 (BNIP3) under the same cobalt treatment condition (12). However, the expression of another set of genes, also transactivated by HIF-1α in response to cobalt treatment such as insulin-like growth factor 2 (IGF2), is not affected by copper deprivation (12). This differential regulation of HIF-1α–controlled target gene expression by copper deficiency in vitro was also observed in the monkey model of myocardial ischemic infarction (17). Under a chronic myocardial ischemic condition induced by a permanent coronary artery ligation, copper concentrations were remarkably decreased, but HIF-1α accumulation was significantly increased in the same infarct myocardial tissue (17). The expression of critical angiogenic factors including VEGF controlled by HIF-1α was severely suppressed, but that of other HIF-1α–controlled genes such as angiopoietin-2 (Ang-2) was actually up-regulated (17). These observations thus raise an important question: what is the mechanism by which copper regulates the target gene selectivity of HIF-1α?
Our early studies demonstrated that copper deprivation suppresses the binding of HIF-1α to the HRE of VEGF gene by electrophoretic mobility shift assay (18). Therefore, the present study was undertaken to test the hypothesis that copper regulates the target gene selectivity of HIF-1α by modulating the binding of this transcription factor to HRE sites of the target genes. Because the expression of copper-dependent BNIP3 and copper-independent IGF2 in the HUVECs is well defined (12), the same genes were applied in the present study. The hypoxia (1% O2) and cobalt treatment were used to induce HIF-1α accumulation.
Results
Determination of functional HIF-1α–binding sites in the promoter regions of BNIP3 and IGF2 genes
Three potential HRE sites were found in BNIP3 promoter; they were BNIP3–HRE1 at −412/−404 (5′-CCTGCACGT-3′), BNIP3–HRE2 at −48/−40 (5′-GCCGCACGT-3′), and BNIP3–HRE3 at −43/−35 (5′-ACGTGCCAC-3′). Two potential HRE sites were found in IGF2 promoter; they were IGF2–HRE1 at −648/−641 (5′-TGCGTGGG-3′) and IGF2–HRE2 at −354/−347 (5′-GACGTGAC-3′). BNIP3–HRE2 and BNIP3–HRE3 are overlapped so that these two sites were tested together (BNIP3–HRE2–3) (Fig. 1A).
Figure 1.
Determination of functional HIF-1α–binding sites in the promoters of BNIP3 and IGF2. A, the predicted potential HRE sites of BNIP3 and IGF2 by computational analyses according to the core consensus sequences of HRE sites. B, binding of HIF-1α to HRE site of VEGF detected by ChIP assay. C and D, determination by ChIP assay of the HIF-1α functional binding sites among the predicted possible binding sites of the BNIP3 promoter (C) and the IGF2 promoter (D). E, map of PGL3-promoter vector and sequences inserted into the vectors. The WT and mutated HRE sites along with their flanking sequences in BNIP3 or IGF2 promoter were cloned into the PGL3-promoter vector. The WT and mutated sequences of a same gene were identical (hyphens) except for a 3-bp substitution within the HIF-1α–binding sites (underlined). F and G, luciferase reporter assay analysis of the luciferase activities driven by the functional binding sites or mutated sites of HIF-1α in the BNIP3 (F) and the IGF2 (G). Mu, mutant. HUVECs were treated with CoCl2 for 16 h at a final concentration of 100 μm (Co), with an untreated group as control (Control). At least three independent experiments were carried out. *, significantly different from control group (p < 0.05) detected by unpaired t test.
After treatment with CoCl2, the binding of HIF-1α to the endogenous VEGF HRE site (19, 20) increased markedly compared with the control group as shown in Fig. 1B. CoCl2 caused HIF-1α binding to the endogenous BNIP3–HRE1 and IGF2–HRE2 sites but not to BNIP3–HRE2–3 or IGF2–HRE1 site (Fig. 1, C and D). These HIF-1α–binding sites (BNIP3–HRE1 and IGF2–HRE2) were confirmed by luciferase reporter assay using the vectors of PGL3-promoter and PGL3-basic. The map of luciferase vectors and the sequences that were inserted into the two vectors respectively are shown in Fig. 1E and Fig. S1A. Compared with the response of the control group, CoCl2 induced luciferase activity of plasmids containing sequences with BNIP3–HRE1 or IGF2–HRE2 (WT BNIP3–HRE1 and WT IGF2–HRE2) but not that of plasmids containing mutated sequences (Mu BNIP3–HRE1 and Mu IGF2–HRE2) (Fig. 1, F and G, and Fig. S1, B and C). Thus, BNIP3–HRE1 (5′-CCTGCACGT-3′) and IGF2–HRE2 (5′-GACGTGAC-3′) were identified as the functional HIF-1α–binding sites.
Reduction of nucleus-associated copper concentrations but not HIF-1α accumulation by TEPA
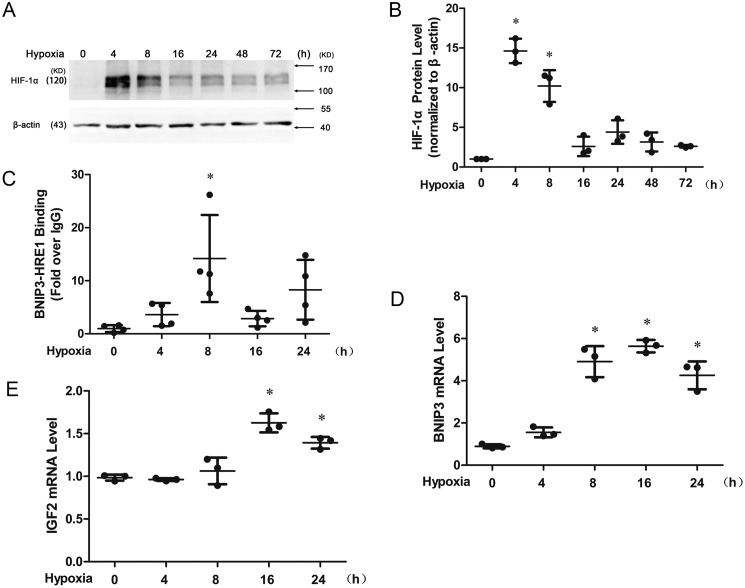
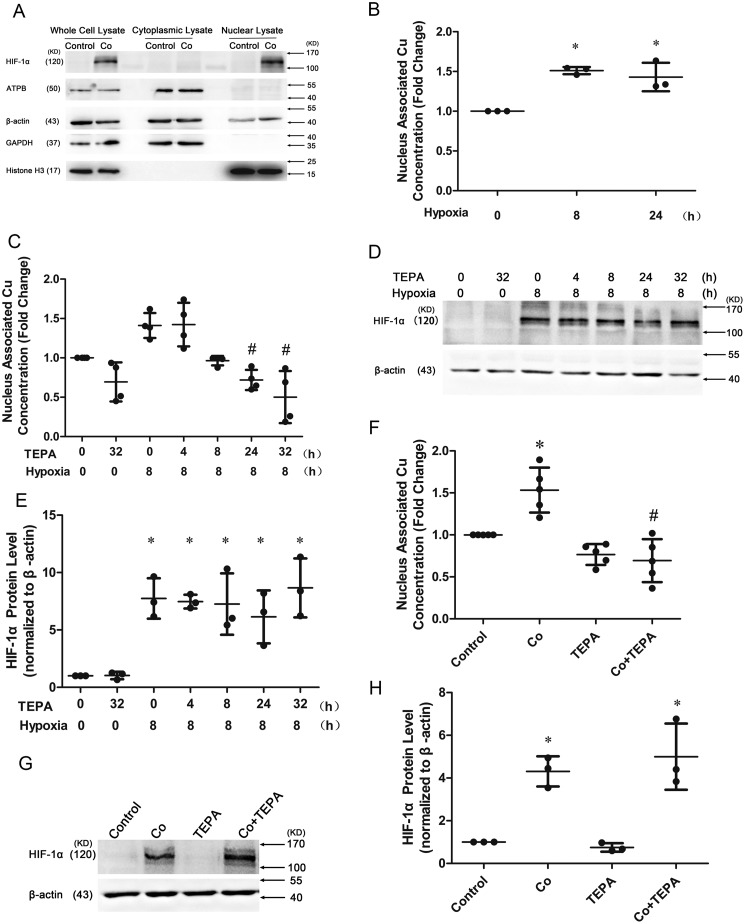
HIF-1α protein accumulation was induced to the highest level at 4 h and decreased after 8 h (Fig. 2, A and B) after hypoxia treatment of HUVECs. The binding of HIF-1α to BNIP3–HRE1 reached to the highest level at 8 h (Fig. 2C), and the expression of BNIP3 and IGF2 mRNA was significantly induced at 16 h (Fig. 2, D and E) after hypoxia. Western blotting analysis showed that the isolated nuclear fraction was free of cytoplasmic contamination because the nuclear lysate was enriched with HIF-1α and histone H3, with no detectable ATPB or GAPDH and with reduced β-actin compared with the whole cell lysate (Fig. 3A). Copper concentrations associated with nuclei were increased when the cells were cultured under hypoxic conditions for 8 or 24 h (Fig. 3B), but it was completely blocked if the cells were pretreated with TEPA 24 h or pretreated with TEPA 24 h with an additional 8-h concomitant treatment under hypoxic conditions (totally 32 h) (Fig. 3C), whereas the accumulation of HIF-1α protein in the nuclei under these conditions was not affected by TEPA treatment (Fig. 3, D and E). In parallel, TEPA also reduced the nucleus-associated copper concentrations but did not affect the HIFs-1α accumulation induced by CoCl2 (Fig. 3, F–H).
Figure 2.
Transcriptional activity of HIF-1α under hypoxic condition. A and B, Western blotting analysis showing the HIF-1α protein accumulation in nuclei (A) and quantified in B. C, binding of HIF-1α to BNIP3–HRE1 detected by ChIP assay. D and E, qRT-PCR determination of the mRNA expression of BNIP3 (D) and IGF2 (E). HUVECs were cultured under hypoxic (1% O2) condition for 4, 8, 16, 24, 48, or 72 h. At least three independent experiments were carried out. *, significantly different from control group (p < 0.05) detected by one-way ANOVA.
Figure 3.
Effects of TEPA on nucleus-associated copper concentrations and HIF-1α protein levels in the nuclei. A, determination of the purity of isolated nuclei by Western blotting analysis of cytoplasmic protein contaminations in the nuclear fraction. Cultured HUVECs were treated with CoCl2 for 16 h at a final concentration of 100 μm (Co), with an untreated group as control (Control). ATPB, mitochondrial marker; GAPDH, cytoplasmic marker; β-actin, whole-cell protein; Histone H3, nuclear marker. B, AAS analysis of the nucleus-associated copper concentrations after cultured cells under hypoxic condition for 8 h or 24 h. C, effects of TEPA on nucleus-associated copper concentrations detected by AAS. HUVECs were treated with 50 μm TEPA for 0, 4, 8, 24, or 32 h, with or without the final 8 h in hypoxia (1% O2). #, significantly different from treatment of hypoxia for 8 h group (line 3) (p < 0.05) detected by Kruskal–Wallis analysis. D, Western blotting detection of HIF-1α protein levels in the nuclei. HUVECs were treated as described above. E, FUSION analysis of Western blotting of HIF-1α protein level. F, AAS detection of the effect of CoCl2 and TEPA on nucleus-associated copper concentrations. HUVECs were treated with 100 μm CoCl2 for 22 h only (Co), treated with 50 μm TEPA for 6 h only (TEPA), or treated with 100 μm CoCl2 for 16 h first, then for additional 6 h with 50 μm TEPA (Co+TEPA), with an untreated group as control (Control). #, significantly different from Co group (line 2) (p < 0.05). G and H, Western blotting detection of HIF-1α protein levels in the nuclei (G) and quantified in H. HUVECs were treated as described above. At least three independent experiments were carried out. *, significantly different from control group (p < 0.05) detected by (B and E) one-way or (F and H) two-way ANOVA.
Suppression of HIF-1α binding to the HRE of BNIP3, but not to that of IGF2, by copper deprivation
In the presence of TEPA, the binding of HIF-1α to the BNIP3–HRE1 site was completely blocked relative to the hypoxia- or cobalt-treated positive controls, as detected by ChIP assay (Fig. 4, A and B) and confirmed by luciferase reporter assay (Fig. 4, C and D, and Fig. S1, D and E) and the detection of BNIP3 mRNA expression by real-time quantitative RT-PCR (qRT-PCR) (Fig. 4E).
Figure 4.
Effects of TEPA on HIF-1α binding to HRE sites of BNIP3 or IGF2. A and B, effect of TEPA on the binding of HIF-1α to the BNIP3–HRE1 detected by ChIP assay under CoCl2 (A) or hypoxia treatment (B). C and D, luciferase reporter assay detection of the effect of TEPA on the transcriptional function mediated by BNIP3–HRE1 under CoCl2 (C) or hypoxia treatment (D). E, effect of TEPA on mRNA expression of BNIP3 in cells treated with CoCl2 detected by qRT-PCR. F and G, effect of TEPA on the binding of HIF-1α to the IGF2–HRE2 detected by ChIP assay under CoCl2 (F) or hypoxia treatment (G). H and I, luciferase reporter assay detection of the effect of TEPA on the transcriptional function mediated by IGF2–HRE2 under CoCl2 (H) or hypoxia treatment (I). J, effect of TEPA on mRNA expression of IGF2 detected by qRT-PCR under CoCl2 treatment. A, C, E, F, H, and J, cells treatments were as described for Fig. 3F. B, D, G and I, HUVECs were cultured under hypoxic condition (1% O2) for 8 h only (Hypoxia), or treated with 50 μm TEPA for 32 h only (TEPA) or cultured cells under hypoxic condition for 8 h, with a pretreatment of 50 μm TEPA, for a total of 32 h (including the final 8 h for hypoxia treatment) (Hypoxia+TEPA); an untreated group served as control (Control). The plasmid used in the luciferase reporter assay (C, D, H, and I) was PGL3-promoter containing WT BNIP3–HRE1 or WT IGF2–HRE2. At least three independent experiments were carried out. *, significantly different from control group (p < 0.05); #, significantly different from Co or hypoxia group (line 2) (p < 0.05) detected by two-way ANOVA.
On the contrary, copper deprivation did not affect the binding of HIF-1α to the IGF2–HRE2 under hypoxia or cobalt treatment (Fig. 4, F and G). As expected, both luciferase reporter assay and the detection of IGF2 mRNA expression confirmed the observation by ChIP assay (Fig. 4, H–J, and Fig. S1, F and G). Thus, copper deprivation decreased the binding of HIF-1α to BNIP3 promoter but not to IGF2 promoter.
Discussion
In response to hypoxia, HIF-1α accumulation leads to transactivation of multiple genes expression. There are almost 300 genes that are regulated by HIF-1α (7, 8). The well tuning of HIF-1α activity under certain conditions is required for cellular responses to different stresses and for maintaining cellular homeostasis. Several studies have focused on the mechanisms that determine the target gene selectivity of HIF-1α. It was found that HIF-1α prefers to bind to genes that are already activated. Under acute hypoxic conditions, more than 80% of genes bound by HIF-1α in HepG2 and U87 cells are those that have already been activated under normal growth condition prior to hypoxia, because the loci in the promoter were transcriptionally activated and available for binding of HIF-1α (21). In addition, HIF-1α cooperates with other transcription factors and selectively binds to the target genes. In HeLa cells, the mutation of the binding sites of transcription factor CCAAT/enhancer-binding protein in glycogen synthase 1 promoter or cAMP-response element-binding protein in lactate dehydrogenase A promoter proximal to the HRE sequence decreased the HIF-1α–induced response to hypoxia, whereas the mutation of the Apetala 1–binding site in carbonic anhydrase IX/CA9 (CA9) promoter increased this response (22). The results obtained from the present study demonstrated that the target gene selectivity of HIF-1α is also regulated by cellular modulators. This copper regulation of target gene selectivity of HIF-1α represents such a mechanism.
In the present study, we observed that copper regulates the target gene selectivity of HIF-1α via at least in part its influence on the binding of HIF-1α to the binding sites of target genes. As previously demonstrated, BNIP3 and IGF2 are two genes controlled by HIF-1α, but their expression is differentially regulated by copper, namely copper-dependent versus copper-independent, respectively (12). Both genes contain effective HRE sequences in their promoter regions, as defined in the present study. Copper deprivation by TEPA did not affect the accumulation and the nuclear entry of HIF-1α but decreased the nucleus-associated copper concentrations. Under such a condition, the binding of HIF-1α to the HRE site of BNIP3 was completely suppressed, along with a complete inhibition of BNIP3 mRNA expression, but the binding of HIF-1α to the HRE site of IGF2 or the expression of IGF2 mRNA was not affected. Therefore, selective modulation of HIF-1α binding to different HRE sites is a determinant factor for copper regulation of the target gene selectivity of HIF-1α.
The effect of copper on the gene selectivity of HIF-1α would play an important role in pathological conditions, such as ischemic heart diseases and cancers. Copper efflux from the chronic ischemic myocardium has been observed both in human and animal models (17, 23, 24). Our recent studies of monkey model of myocardial ischemic infarction have revealed the link between copper deprivation in the ischemic heart and the suppressed expression of HIF-1α–controlled angiogenic factors, including VEGF, tyrosine-protein kinase receptor Tie-2, angiopoietin-1, and fibroblast growth factor-1. The depressed angiogenesis in the chronic ischemic heart is actually accompanied by increased HIF-1α protein level (17) and transactivation of the expression of other genes such as IGF2 (17). This understanding of the relationship between copper and HIF-1α would provide a potential novel approach in clinical setting to selectively activate HIF-1α–controlled angiogenesis as a therapy target for chronic ischemic heart disease.
On the other hand, the effect of copper on the regulation of HIF-1α activity in cancers presents another aspect of the clinical relevance. Copper concentrations in the serum and red blood cells of patients with cancers, such as hepatocellular carcinoma, are higher than those in the healthy people (25, 26). Copper accumulation activates HIF-1α–controlled angiogenesis in the tumor, promoting tumor progression (27–29). A specific targeting to copper-regulated promoter regions of the selective angiogenic genes would provide an alternative approach to anticancer therapy.
It was interesting to notice that under hypoxia or cobalt treatment, nucleus-associated copper concentrations were increased nearly 50% more than that under normoxic conditions. We have observed that copper chaperone for superoxide dismutase-1 is majorly responsible for copper transport from the cytosol to the nucleus under the hypoxic conditions (30). This coordination in the nuclear translocation between copper and HIF-1α would help their functional integration for the target gene selection in response to hypoxia.
In summary, the present study defined that HRE (−412/−404) of BNIP3 and HRE (−354/−347) of IGF2 were functional binding sites for HIF-1α. Copper, by differentially affecting the binding of HIF-1α to the HRE sites, regulates the target gene selectivity of HIF-1α under the hypoxic conditions. This provides a novel insight into the regulation of HIF-1α transactivation of target gene expression. Further studies will examine the molecular components and their interactions that mediate copper regulation of target gene selectivity of HIF-1α.
Experimental procedures
Cell culture and treatment
HUVECs obtained from American Type Culture Collection were cultured in high glucose Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Hyclone) at 37 °C in a 5% CO2 and 95% air incubator. Penicillin and streptomycin, 50 μg/liter each, were added to the medium, and cells were seeded in culture flasks until achieving density of 50% confluence. The cells were treated with CoCl2 (Chron Chemicals), TEPA (Sigma), or cultured cells under hypoxic condition (1% O2, 5% CO2, and 37 °C) with pretreated with TEPA in accordance with the experimental protocol as detailed in the figure legends.
Nucleus isolation
The cells were washed three times using PBS and scraped from the culture flasks with scrapping solution (1% PBS, 0.05% Tween 20). The residual scrapping solution was removed by washing cell pellet with PBS. The cell pellet was washed with sucrose solution (0.3 m sucrose, 10 mm HEPES–NaOH, pH 7.9, 1% Triton X-100, 2 mm MgOAc) to obtain the isolated nuclei and then washed the nucleus pellet with glycerol buffer (25% glycerol, 10 mm HEPES–NaOH, pH 7.9, 0.1 mm EDTA, 5 mm MgOAc). All washing steps were repeated twice except the glycerol buffer, which was performed once, by centrifugation at 2500 × g for 5 min at 4 °C after suspending cell pellet. The final nucleus pellet was stored at −80 °C. All processes above were performed on ice.
Atomic absorption spectrophotometry (AAS) analysis
The cells were collected after being washed with PBS containing EDTA-2Na (10 mm; Sigma) for three times. Then the cell pellet was washed using the scrapping solution, PBS, and sucrose solution as described above. The isolated nucleus pellet was lysed with SDS lysis buffer (1% SDS) containing 1% complete EDTA-free protease inhibitor mixture (Roche DE). The nuclear lysate was digested with nitric acid at 60 °C for 72 h and diluted in 3-fold distilled water, and then detected by AAS (ICE3500; Thermo Fisher). Nucleus-associated copper concentrations were normalized by nuclear protein concentrations.
ChIP assay
The cells were incubated at 37 °C for 15 min in 1% formaldehyde (Sigma), and the reaction was terminated by glycine solution (0.125 m). The cells were scraped from culture flasks after being washed three times with PBS. The nuclei were isolated as described above and lysed in nuclei lysis buffer (50 mm Tris, pH 8.0, 10 mm EDTA, 1% SDS) containing 1% complete EDTA-free protease inhibitor mixture. After being incubated at 4 °C for 10 min and centrifuged at 800 × g for 5 min at 4 °C, the precipitate was suspended in a dilution buffer (20 mm Tris-HCl, pH 8.0, 167 mm NaCl, 1.1 mm EDTA, 0.01% SDS, 1.1% Triton X-100), and DNA in the lysate was interrupted to lengths between 100 and 800 bp by ultrasonic treatment. The endogenous DNA binding to HIF-1α was immunoprecipitated as follows: immunomagnetic beads (Invitrogen; 35 μl) were incubated at 25 °C for 30 min with 5 μg of goat anti-human HIF-1α polyclone antibody (31, 32) (AF1935, KAK0214121; R&D Systems) or 5 μg of nonspecific goat IgG (AB-108-C, ES4115041; R&D Systems), IgG was served as a negative control. Sheared DNA of 7 μg was removed as “input” DNA; 70 μg of DNA was incubated with an antibody bead complex at 25 °C for 40 min. After immunoprecipitation, the nonspecific binding DNA was removed by washing buffer as described in previous study (32), and the HIF-1α-binding DNA was eluted by elution buffer (1% SDS, 0.1 m NaHCO3). The DNA product was incubated with RNase (TaKaRa; 0.2 mg/ml) at 37 °C for 1 h and followed by protease K (Roche, DE) at 65 °C for at least 4 h. Eluted DNA was purified using a TIAN quick mini purification kit (Tiangen). Quantitative real-time PCR was used to detect the purified DNA, and the binding level was analyzed relative to IgG level using the 2−ΔΔCT method in each sample. PROMO (http://alggen.lsi.upc.es/), 4 a program for the prediction of transcription factor binding sites in DNA sequences (33, 34), and data from early studies using ChIP-sequencing with anti-HIF-1α antibody in human cell lines (Fig. S1H) (35, 36) were used to predict the potential binding sites of HIF-1α in the promoter regions of BNIP3 and IGF2 genes (Fig. 1A), and then those sites were verified by ChIP assay.
Primers were designed according to the HRE sites as shown below: forward, 5′-GACCGCGCAGCCCACTCGT-3′, and reverse, 5′-GTGTGGCACGTGCGGCGC-3′ for HRE (−48/−35) site of BNIP3; forward, 5′-GGCCGCTTCCCTGCACGTC-3′ and reverse, 5′-GCCGGGTTCTCCTTTGAAGGG-3′ for HRE (−412/−404) site of BNIP3; forward, 5′-GGATTTTAGGTGCTCCCGGT-3′, and reverse, 5′-GTCCAATCGCCCAATCCAGA-3′ for HRE (−354/−347) site of IGF2; and forward, 5′-TCAGGAAGAGGCAAAGACGTG-3′, and reverse, 5′-ATCCCAACACAGGCTAGTAAAAC-3′ for HRE (−648/−641) site of IGF2. Primers designed according to gene sequences far away from HIF-1α–binding sites in BNIP3 or IGF2 were used as negative controls for the two functional HRE sites: forward, 5′-ATATGGGATTGGTCAAGTCGGC-3′, and reverse, 5′-CAGGAGTCACACAGTCACATCAC-3′ for BNIP3; and forward, 5′-CGAGGCACCCCAAATTACCT-3′, and reverse, 5′-TGAAGACATTGGGGACACGG-3′ for IGF2.
Western blotting
Nucleus pellet was lysed by ice-cold SDS lysis buffer (1% SDS), which contains 1% complete EDTA-free protease inhibitor mixture. Modified Lowry protein assay (Thermo Fisher) was used to measure the protein concentration. An aliquot of 20 μg of protein from each sample was subjected to a 10% SDS-polyacrylamide electrophoresis gel and blotted onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). After blocking the PVDF membrane for an hour with nonfat dry milk (5%) dissolved in TBS/Tween 20 (TBST, 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.1% Tween 20), the PVDF membrane was incubated at 4 °C overnight in block solution containing antibodies as follows: goat anti-human HIF-1α polyclonal antibody (AF1935, KAK0214121; R&D Systems; 1:1000), mouse anti-human β-actin mAb (TA-09, 151110; ZSGB-BIO; 1:1000), mouse anti-human GAPDH mAb (TA-08, 131024; ZSGB-BIO; 1:1000), mouse anti-human histone H3 mAb (051341, 2272711; Millipore; 1:2000), or rabbit anti-human ATPB mAb (ab170947, GR129946-6; Abcam; 1:1000). The membranes were washed in TBST buffer for three times and incubated at 37 °C for 1 h in block solution containing horseradish peroxidase-linked anti-goat IgG (ZB-2306, 113402; ZSGB-BIO; 1:1000), anti-rabbit IgG (ZB-2301, 107015; ZSGB-BIO; 1:1000), or anti-mouse IgG antibody (ZB-2305, 101966; ZSGB-BIO; 1:1000). The antibody validation information was shown in Fig. S2. The protein was visualized using a chemiluminescence HRP substrate (Millipore), and chemiluminescent images were collected by FUSION FX (Vilber Lourmat). The images were then analyzed using FUSION analysis software.
Luciferase reporter assay
Four sequences were cloned into PGL3-promoter (Fig. 1E), which contains a SV-40 promoter or PGL3-basic (Fig. S1A) vector (Promega); they were the WT BNIP3–HRE1 (5′-CCTGCACGT-3′, −412/−404) or the mutated BNIP3–HRE1 (5′-CCTGCTTTT-3′, −412/−404) along with their flanking sequences (−652/−353, 300 bp), and the WT IGF2–HRE2 (5′-GACGTGAC-3′, −354/−347) or the mutated IGF2–HRE2 (5′-GAAAAGAC-3′, −354/−347) along with their flanking sequences (−560/−261, 300 bp) (Fig. 1E). For luciferase assay, the cells were grown in a 6-well tissue culture dish, reaching a density of 60% confluence by the time of transfection. Plasmids containing WT/Mu HRE site of BNIP3 or IGF2 were cotransfected with an internal control pRL-SV40 (Promega) by Lipofectamine 2000 reagent (Invitrogen). 6 h after transfection, the medium was changed to high glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, and the cells were cultured overnight. To test the luciferase activities, the cells were collected and detected by a Dual-Luciferase reporter assay kit (Promega).
qRT-PCR
Total RNA was extracted from HUVECs using a TRIzol reagent (Invitrogen), and 900 ng of RNA was reverse transcribed by an Moloney murine leukemia virus RTase cDNA synthesis kit (TaKaRa). The PCR and data analysis were followed as our previous study (12). Primers were shown below: forward, 5′-TCAGCATGAGGAACACGAGCGT-3′ and reverse, 5′-GAGGTTGTCAGACGCCTTCCAA-3′, for BNIP3; forward, 5′-GACCGCGGCTTCTACTTCAG-3′, and reverse, 5′-AAGAACTTGCCCACGGGGTAT-3′ for IGF2; and forward, 5′-TTCGGAACTGAGGCCATGAT-3′, and reverse, 5′-TTTCGCTCTGGTCCGTCTTG-3′ for RPS18.
Statistical analysis
The data were analyzed by one-way or two-way ANOVA which followed by Tukey's multiple comparison test, unpaired t test, or Kruskal–Wallis analysis where appropriate. At least three independent experiments were carried, and the data from each experiment were expressed as mean values ± S.D. A p value < 0.05 was considered significant.
Author contributions
X. L., W. Z., Z. W., Y. Y., and Y. J. K. data curation; X. L., W. Z., Z. W., and Y. J. K. formal analysis; X. L., Z. W., and Y. Y. methodology; X. L. and W. Z. writing-original draft; X. L., W. Z., and Y. J. K. project administration; X. L., W. Z., and Y. J. K. writing-review and editing; W. Z. and Y. J. K. conceptualization; W. Z. and Y. J. K. funding acquisition; W. Z. and Y. J. K. validation; W. Z. investigation; Y. J. K. resources; Y. J. K. supervision; X. L. carried out the experiments.
Supplementary Material
Acknowledgments
We thank Xiaoming Hou, Zhenghui Luo, and Suzhen Fan for technical support.
This work was supported by National Natural Science Foundation of China Grants 81600214 (to W. Z.) and 81230004 (to Y. J. Kang). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and supporting references.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- HIF-1α
- hypoxia-inducible factor-1α
- BNIP3
- BCL2/adenovirus E1B 19-kDa protein-interacting protein 3
- IGF2
- insulin-like growth factor 2
- HUVEC
- human umbilical vein endothelial cell
- HRE
- hypoxia-responsive element
- TEPA
- tetraethelenepentamine
- VEGF
- vascular endothelial growth factor
- AAS
- atomic absorption spectrophotometry
- qRT-PCR
- real-time quantitative RT-PCR
- PVDF
- polyvinylidene difluoride
- ANOVA
- analysis of variance.
References
- 1. Semenza G. L. (2003) Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 10.1038/nrc1187 [DOI] [PubMed] [Google Scholar]
- 2. Semenza G. L. (2000) HIF-1 and human disease: one highly involved factor. Genes Dev. 14, 1983–1991 [PubMed] [Google Scholar]
- 3. Hirsilä M., Koivunen P., Xu L., Seeley T., Kivirikko K. I., and Myllyharju J. (2005) Effect of desferrioxamine and metals on the hydroxylases in the oxygen sensing pathway. FASEB J. 19, 1308–1310 10.1096/fj.04-3399fje [DOI] [PubMed] [Google Scholar]
- 4. Huang L. E., Gu J., Schau M., and Bunn H. F. (1998) Regulation of hypoxia-inducible factor 1α is mediated by an O-2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 10.1073/pnas.95.14.7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., and Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O-2-regulated prolyl hydroxylation. Science 292, 468–472 10.1126/science.1059796 [DOI] [PubMed] [Google Scholar]
- 6. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., and Kaelin W. G. Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 10.1126/science.1059817 [DOI] [PubMed] [Google Scholar]
- 7. Fedele A. O., Whitelaw M. L., and Peet D. J. (2002) Regulation of gene expression by the hypoxia-inducible factors. Mol. Interv. 2, 229–243 10.1124/mi.2.4.229 [DOI] [PubMed] [Google Scholar]
- 8. Greijer A. E., van der Groep P., Kemming D., Shvarts A., Semenza G. L., Meijer G. A., van de Wiel M. A., Belien J. A., van Diest P. J., and van der Wall E. (2005) Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J. Pathol. 206, 291–304 10.1002/path.1778 [DOI] [PubMed] [Google Scholar]
- 9. Iguchi-Ariga S. M., and Schaffner W. (1989) CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 3, 612–619 10.1101/gad.3.5.612 [DOI] [PubMed] [Google Scholar]
- 10. Perini G., Diolaiti D., Porro A., and Della Valle G. (2005) In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc. Natl. Acad. Sci. U.S.A. 102, 12117–12122 10.1073/pnas.0409097102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bacon A. L., Fox S., Turley H., and Harris A. L. (2007) Selective silencing of the hypoxia-inducible factor 1 target gene BNIP3 by histone deacetylation and methylation in colorectal cancer. Oncogene 26, 132–141 10.1038/sj.onc.1209761 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Z., Qiu L., Lin C., Yang H., Fu H., Li R., and Kang Y. J. (2014) Copper-dependent and -independent hypoxia-inducible factor-1 regulation of gene expression. Metallomics 6, 1889–1893 10.1039/C4MT00052H [DOI] [PubMed] [Google Scholar]
- 13. Gleadle J. M., Ebert B. L., Firth J. D., and Ratcliffe P. J. (1995) Regulation of angiogenic growth factor expression by hypoxia, transition metals, and chelating agents. Am. J. Physiol. 268, C1362–C1368 10.1152/ajpcell.1995.268.6.C1362 [DOI] [PubMed] [Google Scholar]
- 14. Gleadle J. M., Ebert B. L., and Ratcliffe P. J. (1995) Diphenylene iodonium inhibits the induction of erythropoietin and other mammalian genes by hypoxia: implications for the mechanism of oxygen sensing. Eur. J. Biochem. 234, 92–99 10.1111/j.1432-1033.1995.092_c.x [DOI] [PubMed] [Google Scholar]
- 15. Befani C., Mylonis I., Gkotinakou I. M., Georgoulias P., Hu C. J., Simos G., and Liakos P. (2013) Cobalt stimulates HIF-1-dependent but inhibits HIF-2-dependent gene expression in liver cancer cells. Int. J. Biochem. Cell Biol. 45, 2359–2368 10.1016/j.biocel.2013.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qiu L., Ding X., Zhang Z., and Kang Y. J. (2012) Copper is required for cobalt-induced transcriptional activity of hypoxia-inducible factor-1. J. Pharmacol Exp. Ther. 342, 561–567 10.1124/jpet.112.194662 [DOI] [PubMed] [Google Scholar]
- 17. Zhang W., Zhao X., Xiao Y., Chen J., Han P., Zhang J., Fu H., and James Kang Y. (2016) The association of depressed angiogenic factors with reduced capillary density in the Rhesus monkey model of myocardial ischemia. Metallomics 8, 654–662 10.1039/C5MT00332F [DOI] [PubMed] [Google Scholar]
- 18. Feng W., Ye F., Xue W., Zhou Z., and Kang Y. J. (2009) Copper regulation of hypoxia-inducible factor-1 activity. Mol. Pharmacol. 75, 174–182 10.1124/mol.108.051516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu P., and Kodadek T. (2007) Dynamics of the hypoxia-inducible factor-1-vascular endothelial growth factor promoter complex. J. Biol. Chem. 282, 35035–35045 10.1074/jbc.M707557200 [DOI] [PubMed] [Google Scholar]
- 20. Kimura H., Weisz A., Ogura T., Hitomi Y., Kurashima Y., Hashimoto K., D'Acquisto F., Makuuchi M., and Esumi H. (2001) Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J. Biol. Chem. 276, 2292–2298 10.1074/jbc.M008398200 [DOI] [PubMed] [Google Scholar]
- 21. Xia X., and Kung A. L. (2009) Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol. 10, R113 10.1186/gb-2009-10-10-r113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Villar D., Ortiz-Barahona A., Gómez-Maldonado L., Pescador N., Sánchez-Cabo F., Hackl H., Rodriguez B. A., Trajanoski Z., Dopazo A., Huang T. H., Yan P. S., and Del Peso L. (2012) Cooperativity of stress-responsive transcription factors in core hypoxia-inducible factor binding regions. PLoS One 7, e45708 10.1371/journal.pone.0045708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chevion M., Jiang Y., Har-El R., Berenshtein E., Uretzky G., and Kitrossky N. (1993) Copper and iron are mobilized following myocardial ischemia: possible predictive criteria for tissue injury. Proc. Natl. Acad. Sci. U.S.A. 90, 1102–1106 10.1073/pnas.90.3.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He W., and James Kang Y. (2013) Ischemia-induced copper loss and suppression of angiogenesis in the pathogenesis of myocardial infarction. Cardiovasc. Toxicol. 13, 1–8 10.1007/s12012-012-9174-y [DOI] [PubMed] [Google Scholar]
- 25. Balter V., Nogueira da Costa A., Bondanese V. P., Jaouen K., Lamboux A., Sangrajrang S., Vincent N., Fourel F., Télouk P., Gigou M., Lécuyer C., Srivatanakul P., Bréchot C., Albarède F., and Hainaut P. (2015) Natural variations of copper and sulfur stable isotopes in blood of hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. U.S.A. 112, 982–985 10.1073/pnas.1415151112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baharvand M., Manifar S., Akkafan R., Mortazavi H., and Sabour S. (2014) Serum levels of ferritin, copper, and zinc in patients with oral cancer. Biomed. J 37, 331–336 10.4103/2319-4170.132888 [DOI] [PubMed] [Google Scholar]
- 27. Himoto T., Fujita K., Nomura T., Tani J., Miyoshi H., Morishita A., Yoneyama H., Kubota S., Haba R., Suzuki Y., and Masaki T. (2016) Roles of copper in hepatocarcinogenesis via the activation of hypoxia-inducible factor-1α. Biol. Trace Elem. Res. 174, 58–64 10.1007/s12011-016-0702-7 [DOI] [PubMed] [Google Scholar]
- 28. Rigiracciolo D. C., Scarpelli A., Lappano R., Pisano A., Santolla M. F., De Marco P., Cirillo F., Cappello A. R., Dolce V., Belfiore A., Maggiolini M., and De Francesco E. M. (2015) Copper activates HIF-1α/GPER/VEGF signalling in cancer cells. Oncotarget 6, 34158–34177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Finney L., Vogt S., Fukai T., and Glesne D. (2009) Copper and angiogenesis: unravelling a relationship key to cancer progression. Clin. Exp. Pharmacol. Physiol. 36, 88–94 10.1111/j.1440-1681.2008.04969.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L., Ge Y., and Kang Y. J. (2016) Effect of copper on nuclear translocation of copper chaperone for superoxide dismutase-1. Exp. Biol. Med. (Maywood) 241, 1483–1488 10.1177/1535370216645412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robador P. A., San José G., Rodríguez C., Guadall A., Moreno M. U., Beaumont J., Fortuño A., Diez J., Martínez-González J., and Zalba G. (2011) HIF-1-mediated up-regulation of cardiotrophin-1 is involved in the survival response of cardiomyocytes to hypoxia. Cardiovasc. Res. 92, 247–255 10.1093/cvr/cvr202 [DOI] [PubMed] [Google Scholar]
- 32. Lopez-Haber C., Barrio-Real L., Casado-Medrano V., and Kazanietz M. G. (2016) Heregulin/ErbB3 signaling enhances CXCR4-driven Rac1 activation and breast cancer cell motility via hypoxia-inducible factor 1α. Mol. Cell. Biol. 36, 2011–2026 10.1128/MCB.00180-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Messeguer X., Escudero R., Farre D., Nunez O., Martinez J., and Alba M. M. (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 10.1093/bioinformatics/18.2.333 [DOI] [PubMed] [Google Scholar]
- 34. Farré D., Roset R., Huerta M., Adsuara J. E., Roselló L., Albà M. M., and Messeguer X. (2003) Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31, 3651–3653 10.1093/nar/gkg605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang J., Wang C., Chen X., Takada M., Fan C., Zheng X., Wen H., Liu Y., Wang C., Pestell R. G., Aird K. M., Kaelin W. G. Jr., Liu X. S., and Zhang Q. (2015) EglN2 associates with the NRF1-PGC1α complex and controls mitochondrial function in breast cancer. EMBO J. 34, 2953–2970 10.15252/embj.201591437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yao X., Tan J., Lim K. J., Koh J., Ooi W. F., Li Z., Huang D., Xing M., Chan Y. S., Qu J. Z., Tay S. T., Wijaya G., Lam Y. N., Hong J. H., Lee-Lim A. P., et al. (2017) VHL deficiency drives enhancer activation of oncogenes in clear cell renal cell carcinoma. Cancer Discov. 7, 1284–1305 10.1158/2159-8290.CD-17-0375 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.