Abstract
Bone morphogenetic proteins (BMPs) induce mesenchymal–epithelial transition (MET) and enhance the generation of induced pluripotent stem cells (iPSCs). However, BMPs are also signaling molecules critical for arresting reprogramming in the pre-iPSC state. In this study, using mouse embryonic fibroblasts, we found that the time- and concentration-dependent effects of BMPs on reprogramming are mediated by Msh homeobox 2 (MSX2), a homeobox-containing transcription factor. BMPs up-regulated Msx2 by activating SMAD1/5, and MSX2 then directly bound to the promoters and up-regulated the expression of the cadherin 1 (Cdh1, also known as E-cadherin), GATA-binding protein 3 (Gata3), and Nanog genes. Cdh1 contributed to BMP4- and MSX2-induced MET and subsequently promoted reprogramming. On the other hand, GATA3 promoted reprogramming, possibly by up-regulating Spalt-like transcription factor 4 (SALL4) expression. As key transcriptional factors in maintaining pluripotency, up-regulation of SALL4 and NANOG enhanced reprogramming. Moreover, the ability of MSX2 to up-regulate Cdh1, Gata3, Nanog, and Sall4 was further potentiated in the presence of Krüppel-like factor 4 (KLF4). However, MSX2 did not mediate the effects of BMP4 signaling on activation of the microRNAs miR-205 and miR-200 or the inhibitory effects that arrested reprogramming in the pre-iPSC state. In conclusion, MSX2 partially mediates the effects of BMP4 signaling during reprogramming, improving our understanding of the role of BMP signaling in MET and of the connection between cell lineage specifiers such as MSX2 and GATA3 and pluripotency.
Keywords: reprogramming, bone morphogenetic protein (BMP), cadherin-1 (CDH1), epithelial cadherin (E-cadherin), GATA transcription factor, Krüppel-like factor 4 (KLF4), homeobox transcription factor, MET, Msx2, Sall4, stem cell
Introduction
The mechanisms underlying the generation of induced pluripotent stem cells (iPSCs) 3 have been studied for a long time. At first, only pluripotency-associated factors were considered as potential inducers or mediators of reprogramming (1–3). Later studies suggested that lineage specifiers are able to substitute for particular Yamanaka factors in promoting iPSC generation (4, 5). Thus, the balance of different lineage-specifying forces, such as for the mesoendodermal lineage and ectodermal lineage, may maintain pluripotency (5, 6).
MSX2, a homeobox-containing transcription factor (7, 8), controls the mesendoderm lineage commitment of human PSCs by suppressing Sox2 expression and inducing Nodal expression (9). In addition, Msx2 plays a crucial role in craniofacial, limb, and ectodermal organogenesis, and knockout of Msx2 results in developmental defects (10, 11). Thus, it is possible that Msx2 also has beneficial roles during reprogramming as a lineage specifier. In addition, as both a transcriptional repressor and activator (9, 14, 15), Msx2 is a downstream target of bone morphogenetic protein (BMP) signaling and possibly mediates the effects of BMP signaling during reprogramming (16, 17).
BMPs play important roles in early embryonic patterning (18, 19) and modulate lineage commitment toward trophoblast or mesoderm in embryonic stem (ES) cells (16, 20, 21). However, BMPs have complex and sometimes controversial functions during reprogramming, from mouse embryonic fibroblasts (MEFs) to iPSCs (22). BMP-dependent induction of miR-205 and miR-200a/b/c leads to mesenchymal–epithelial transition (MET), which has been suggested to be a critical step during the initial phase of reprogramming (23). In addition to MET, BMP-dependent signaling also enhances the expression of Nanog and Sall4, which are required for the establishment of pluripotency in MEFs (24, 25).
Colonies with ESC-like morphology but without activation of the endogenous locus encoding Oct4 were identified as pre-iPSCs during reprogramming (26, 27). BMPs have been identified as critical signaling molecules in arresting reprogramming in the pre-iPSC state (28). Thus, not all BMP-dependent signaling is beneficial for reprogramming. In addition, the BMP4–MSX2 axis promotes mesodermal commitment by inducing epithelial–mesenchymal transition (EMT), which is not consistent with BMP's functions during reprogramming. The ability of Msx2 to induce both MET and EMT has been observed in different experimental systems (29, 30), possibly because of the different partners with which MSX2 associates. Therefore, in this study, by using reprogramming from MEFs to iPSCs as a major experimental model, we elucidated the mechanisms underlying modulation of the BMP4–MSX2 axis during reprogramming and how Msx2 mediates the beneficial or inhibitory roles of BMPs.
Results
Msx2 mediates the beneficial effects of BMP signaling
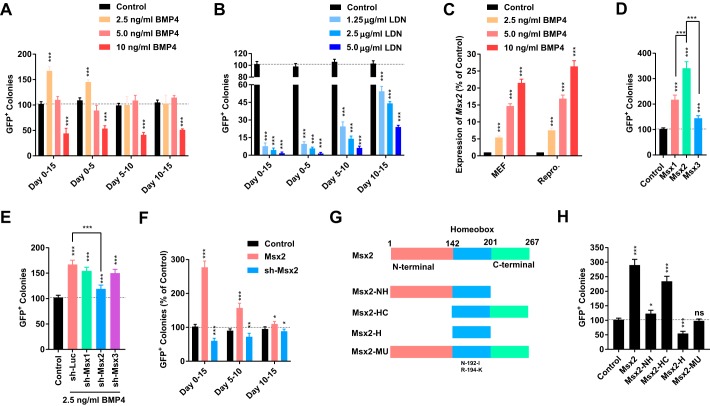
Different concentrations of BMP4 and LDN-193189, an inhibitor of the BMP signaling pathway, were used at different periods during reprogramming from MEFs to iPSCs. A low concentration of BMP4 (2.5 ng/ml) promoted reprogramming at the early stage (days 0–5) but had little effect at the middle (days 5–10) or late (days 10–15) stages (Fig. 1A). A high concentration of BMP4 (10 ng/ml) inhibited reprogramming constantly (Fig. 1A). However, LDN-193189 impaired reprogramming at all tested concentrations and mainly at the early stage during reprogramming (Fig. 1B), which suggested that a low to medium level of activation of BMP signaling was required for reprogramming. In summary, the beneficial roles of BMP signaling could be observed only at relatively low concentrations and at an early stage during reprogramming.
Figure 1.
Msx2 mediated the beneficial functions of BMP4. A–C, different concentrations of BMP4 and LDN-193189 were used for different periods during reprogramming. The ability of BMP4 to up-regulate Msx2 were determined by qPCR on day 4 in both in MEFs and during reprogramming. D and E, Msx1/2/3 were overexpressed during reprogramming without BMP4 or suppressed during reprogramming with 2.5 ng/ml BMP4. GFP+ colonies were counted on day 15. F, the expression of Msx2 was modulated for different periods during reprogramming. GFP+ colonies were counted on day 15. G and H, mutations of MSX2 with deletion of different domains were constructed. Their functions during reprogramming were determined. All experiments were repeated at least five times. One-way ANOVA with Dunnett's post hoc test was used. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
We then checked the expression of various factors in the BMP pathway (Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway 04350) based on a previously reported microarray dataset (GSE21757) (22). Of the 36 genes analyzed, only four genes were up-regulated to more than 2-fold or down-regulated to below 50% of basal levels (Fig. S1A). This phenomenon supports the hypothesis that low to medium levels of activation of the BMP pathway are required for reprogramming.
We then determined the ability of BMP4 to up-regulate downstream effectors such as Msx1/2/3 (16, 17). As indicated in Fig. 1C and Fig. S1, B and C, BMP4 up-regulated Msx1/2/3 in concentration-dependent manner and up-regulated Msx2 more than Msx1 and Msx3. Retroviruses encoding Msx genes and shRNAs against them were delivered into MEFs. Msx2 had a higher ability to promote reprogramming than Msx1 and Msx3 (Fig. 1D). In addition, only shRNA against Msx2, but not against Msx1 or Msx3, significantly attenuated the ability of 2.5 ng/ml BMP4 to promote reprogramming (Fig. 1E). The GFP+ colonies generated with Msx2 overexpression had typical characteristics of iPSCs, including suppressed expression of exogenous Yamanaka factors, high expression of endogenous pluripotent factors, normal karyotypes, low DNA methylation of Nanog and Oct4 promoters, and the ability to generate chimera mice (Fig. S2).
When retroviruses encoding Msx2 and the corresponding shRNA were delivered into MEFs at different time points during reprogramming, larger modulations on reprogramming were observed when infections were performed at the beginning of reprogramming (Fig. 1F). Therefore, Msx2 also regulates reprogramming at the early stage, which is consistent with the functions of BMP4.
Retroviruses encoding different truncations of MSX2 were prepared and tested (Fig. 1G). Both the C terminus and homeobox domain, a DNA binding domain, were required for MSX2 to promote reprogramming (Fig. 1H) (15). In addition, mutating the DNA binding site of MSX2 (N192A and T194K) totally destroyed its ability to promote reprogramming, and the homeobox domain functioned as a dominant-negative factor when used alone. Therefore, Msx2 mediates the beneficial roles of BMP signaling during reprogramming by functioning as a transcriptional factor.
BMP4 up-regulates Msx2 via the conventional Smad1/5 cascade
Phosphorylation of SMAD1/5/8 are conventional downstream cascades of the BMP pathway. It was reasonable to suggest a direct pathway from BMP4 to Msx2 via phosphorylated SMAD1/5/8. To confirm this, shRNAs against Smad1 and Smad5 were used. On one hand, either shRNA against Smad1 or against Smad5 inhibited the ability of 2.5 ng/ml BMP4 to promote reprogramming, possibly because of the ability of these shRNAs to block BMP4-induced up-regulation of Msx2 both in MEFs and during reprogramming (Fig. 2, A and B).
Figure 2.
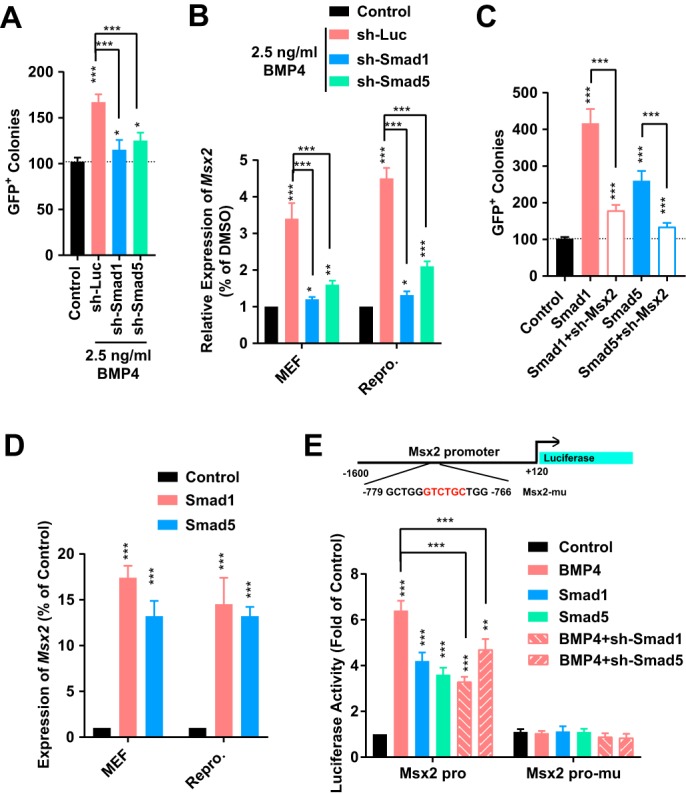
Msx2 was downstream of the BMP4–SMAD1/5 axis. A and B, shRNA against luciferase, Smad1, or Smad5 was used in MEFs or during reprogramming in the presence of 2. 5 ng/ml BMP4. Msx2 expression and reprogramming efficiency were determined. C and D, a lentivirus encoding Smad1, or Smad5 was used in MEFs or during reprogramming with or without shRNA against Msx2. Msx2 expression and reprogramming efficiency were determined. E, a promoter of Msx2 (−1.6 to +0.12 kb relative to the TSS) was used to drive the expression of luciferase, which was determined when 2.5 ng/ml BMP4 was used or the expression of Smad1/5 was modulated. All experiments were repeated at least five times. One-way ANOVA with Dunnett's post hoc test was used. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
On the other hand, overexpression of Smad1 or Smad5 promoted the generation of iPSCs, which could be attenuated by suppressing Msx2 (Fig. 2C). This phenomenon was consistent with the ability of Smad1 and Smad5 to increase the expression of Msx2 both in MEFs and during reprogramming (Fig. 2D). A luciferase assay with the Msx2 promoter further supported targeting of SMAD1/5 to Msx2. Overexpression of Smad1 or Smad5 increased the activity of the WT Msx promoter but not that of the mutated promoter (deletion in −773 to −768 relative to the transcriptional start site (TSS)) (Fig. 2E). Therefore, Msx2 is a direct downstream target of BMP4 and SMAD1/5 not only in MEFs but also during reprogramming.
Msx2 slightly promoted MET by targeting Cdh1
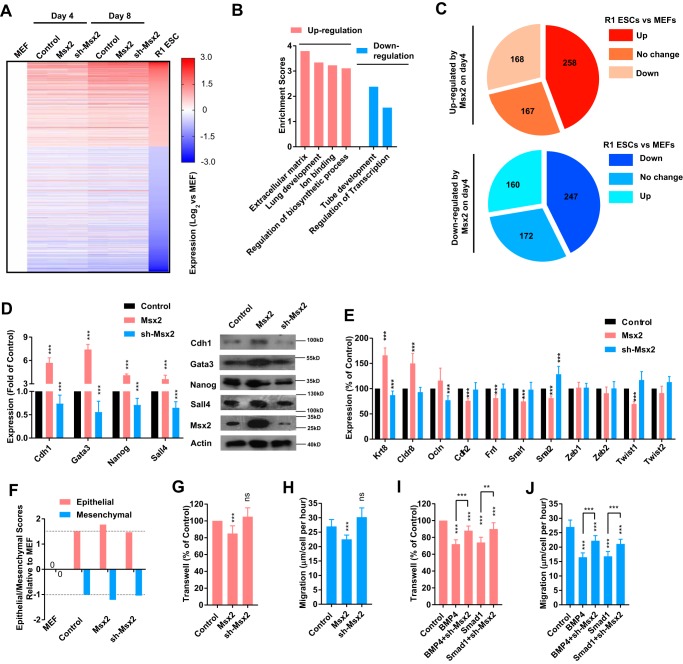
Retroviruses encoding Msx2 and the corresponding shRNAs were used during reprogramming, and gene expression profiles were analyzed on day 4 and day 8 (GSE114120) (Table S1). MEFs and R1 ES cells were used as controls.
We focused on genes whose expression in R1 ES cells was more than 2-fold or less than 50% of that in MEFs (Fig. 3A). Introducing Msx2 and the corresponding shRNA did not change the overall progress of reprogramming but did induce some expression changes (Fig. 3A).
Figure 3.
Msx2 up-regulated genes with the ability to promote reprogramming. A–C, a lentivirus encoding Msx2 or shRNA against Msx2 was delivered to MEFs. The expression profiles of cells were assayed on day 4 and day 8 during reprogramming with RNA-seq. MEFs and R1 ES cells were used as controls. Genes having significantly higher (over 2-fold) or lower (below 50%) expression in R1 ES cells than in MEFs were analyzed (A). Genes that were significantly regulated by Msx2 on day 4 were subjected to GO analysis (B) and categorized into three groups based on their expression in R1 ES cells and MEFs (C). D and E, a lentivirus encoding Msx2 or shRNA against Msx2 was delivered to MEFs. The expression of the indicated genes was determined with qPCR on day 4 of reprogramming. F, epithelial and mesenchymal scores relative to MEFs were calculated basing current RNA-seq. G–J, the ability of cells to migrate was determined with a transwell assay or with a live-cell tracing system that measured the migration distances of cells. Cells were modulated from different aspects of BMP4–Smad1–Msx2. All experiments except RNA-seq were repeated at least five times. One-way ANOVA with Dunnett's post hoc test was used. **, p < 0.01; ***, p < 0.001; ns, not significant.
Genes that were significantly regulated by Msx2 (expression in the Msx2 group was more than 2-fold or less than 50% of that in the control group) were subjected to gene ontology (GO) analysis. Genes up-regulated by Msx2 on day 4 were enriched for GO terms linked to the extracellular matrix, lung development, ion binding, and regulation of the biosynthetic process, whereas down-regulated genes were enriched for tube development and regulation of transcription (Fig. 3B). Of the 593 up-regulated genes, 425 genes (71.7%) had higher or equal expression in R1 ES cells compared with MEFs. Of the 579 down-regulated genes, 419 genes (72.4%) had lower or equal expression in R1 ES cells compared with MEFs (Fig. 3C). Thus, the changes in gene expression profiles were consistent with the ability of Msx2 to promote reprogramming. Among the genes regulated by Msx2, we identified multiple genes, like Cdh1, Gata3, Nanog, and Sall4 (Fig. 3D), that are beneficial for reprogramming (3, 23, 25, 31).
Cdh1, also known as E-cadherin, is an important marker of epithelial cells and plays critical roles in MET during early reprogramming (23, 32). In addition, BMP signaling promotes reprogramming by facilitating MET at least partially (22). Thus, it was reasonable for us to suggest that Msx2 mediates the ability of BMP signaling to facilitate MET.
Three epithelial markers and eight mesenchymal markers were monitored after Msx2 overexpression with qPCR. Seven markers were significantly regulated by Msx2, and all detected modulations on these markers suggested promoted MET (Fig. 3E). Another method that calculates the changes of epithelial and mesenchymal characteristics of cells using the expression of selected markers was also used (33). Based on current RNA-seq results, an increase in the epithelial score and decrease in the mesenchymal score were observed after Msx2 overexpression (Fig. 3F).
The migration ability of cells was determined with a transwell assay and live imaging system. Overexpression of Msx2 decreased the migration ability of cells, and shRNA against Msx2 attenuated the ability of BMP4 to modulate cell migration (Fig. 3, G–J). Therefore, Msx2 mediates the ability of BMP signaling to facilitate partial MET.
Because shRNA against Msx2 did not fully attenuate the abilities of BMP4 to promote reprogramming and to modulate cell migration (Figs. 1E and 3, I and J), BMP signaling might facilitate MET via additional pathways, such as the miR-205 and miR-200 families, as suggested in a previous report (22). As indicated in Fig. S3, 2.5 ng/ml BMP4, but not Msx2, increased the expression of miR-205 and miR-200a/b/c. In addition, shRNA against Msx2 did not affect the ability of BMP signaling to affect miRNAs. Therefore, Msx2 partially mediates the ability of BMP signaling to facilitate MET, possibility by up-regulating Cdh1.
Msx2 mediates the effect of BMP4 on Cdh1, Gata3, Nanog, and Sall4
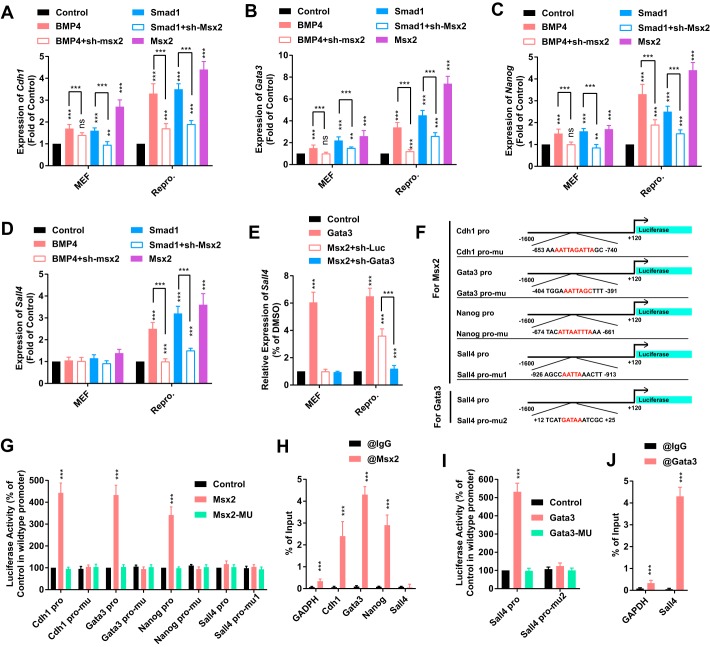
Overexpression of Msx2 up-regulates Cdh1, Gata3, Nanog, and Sall4 and subsequently promotes reprogramming. However, whether BMP signaling promotes reprogramming similarly requires further investigation.
BMP4 at a 2.5 ng/ml concentration and a retrovirus encoding Smad1 up-regulated the expression of Cdh1, Gata3, and Nanog both in MEFs and during reprogramming (Fig. 4, A–C). shRNA against Msx2 attenuated this up-regulation, which confirmed that both BMP signaling and Msx2 promoted reprogramming by activating Cdh1, Gata3, and Nanog.
Figure 4.
Msx2 promoted reprogramming by up-regulating several pluripotency-related genes. A–D, 2.5 ng/ml BMP4 and a lentivirus encoding Msx2, sh-Msx2, or sh-Smad1 were used in MEFs or during reprogramming. The expression of Cdh1, Gata3, Nanog, and Sall4 was determined with qPCR on day 4. E, a lentivirus encoding Msx2, Gata3, or sh-Gata3 was used in MEFs or during reprogramming. The expression of Sall4 was determined with qPCR on day 4. F, promoters (−1.6 to +0.12 kb relative to the TSS) of Cdh1, Gata3, Nanog, and Sall4 were constructed. The binding sites of MSX2 or GATA3 with the best prediction were indicated and deleted. G and H, the responses of WT and mutated promoters of Cdh1, Gata3, Nanog, and Sall4 to WT and mutated MSX2 were determined (G). The binding between MSX2 and regions on the promoters indicated in F was assayed with ChIP–qPCR (H). I and J, the responses of WT and mutated promoters of Sall4 to WT and mutated GATA3 were determined (I). The binding between GATA3 and regions on the promoters indicated in F was assayed with ChIP–qPCR (J). All experiments were repeated at least five times. Two-tailed Student's t test (H–J) and one-way ANOVA with Dunnett's post hoc test were used. **, p < 0.01; ***, p < 0.001; ns, not significant.
However, significant up-regulation of Sall4 was only observed during reprogramming, but not in MEFs (Fig. 4D), possibly because Gata3, but not Msx2, was the direct upstream regulator of Sall4 (31). Actually, overexpression of Gata3 up-regulated Sall4 both in MEFs and during reprogramming (Fig. 4E). shRNA against Gata3 attenuated the ability of Msx2 to up-regulate Sall4 during reprogramming (Fig. 4E). Thus, Msx2 up-regulates Sall4 by up-regulating Gata3.
We then used the Cdh1, Gata3, Nanog, and Sall4 promoters (−1. 6 to +0.12 kb relative to the TSS) to drive luciferase expression. The potential binding sites of MSX2 were predicted and deleted on these promoters based on the information provided by JASPAR 2018 (Fig. 4F) (12). A luciferase assay confirmed direct targeting of MSX2 to Cdh1, Gata3, and Nanog promoters but not Sall4 promoter (Fig. 4G). By using primers surrounding the predicted binding sites (Table S2), ChIP–qPCR analysis further confirmed the targeting (Fig. 4H).
The targeting of GATA3 to the Sall4 promoter was confirmed in a similar manner. Overexpression of Gata3 activated the WT Sall4 promoter. Mutating either the binding domain of GATA3 or the binding motif on the Sall4 promoter diminished the increase in luciferase expression (Fig. 4I). ChIP–qPCR experiments further confirmed the binding of GATA3 to the Sall4 promoter (Fig. 4J). Therefore, both BMP4 and Msx2 up-regulate Cdh1, Gata3, and Nanog and up-regulates Sall4 via Gata3.
In addition, overexpression of Msx1 during reprogramming up-regulated Cdh1 and Nanog (Fig. S4). Overexpression of Msx3 during reprogramming only up-regulated Cdh1 (Fig. S4). This up-regulation was less than that induced by Msx2 (Fig. 3D). Therefore, although Msx1/2/3 are expressed in overlapping domains and function similarly under certain circumstances, Msx2 is a more important mediator for BMP signaling during reprogramming.
Klf4 potentiates the ability of Msx2 to activate downstream factors
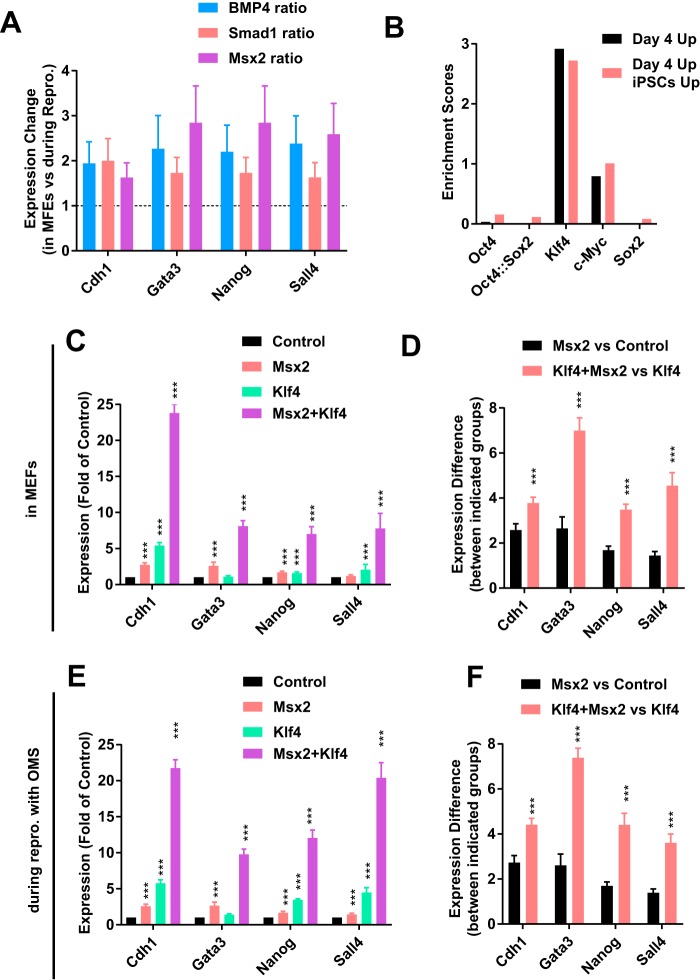
When the abilities of Msx2 to up-regulate Cdh1, Gata3, Nanog, and Sall4 were compared, it was found that Msx2 induced larger up-regulation during reprogramming than in MEFs (Fig. 5A). Thus, it was reasonable to suggest cooperation between MSX2 and an additional factor or factors in activating downstream factors.
Figure 5.
Klf4 potentiated the functions of Msx2. A, the ability of 2. 5 ng/ml BMP4, Smad1, and Msx2 to up-regulate Cdh1, Gata3, Nanog, and Sall4 during reprogramming was normalized to that in MEFs based on the results summarized in Fig. 4, A–D. B, enrichment of the binding motifs of OCT4, KLF4, c-MYC, and SOX2 was determined with Pscan software. C and D, Klf4, Msx2, or both Klf4 and Msx2 were overexpressed in MEFs. The expression of Cdh1, Gata3, Nanog, and Sall4 was determined on day 4 (C). The ability of Msx2 to up-regulate these genes was calculated by performing the comparisons indicated in D. E and F, Klf4, Msx2, or both Klf4 and Msx2 were overexpressed during reprogramming with only OMS. The expression of Cdh1, Gata3, Nanog, and Sall4 was determined on day 4 (C). The ability of Msx2 to up-regulate these genes was calculated by performing the comparisons indicated in D. All experiments were repeated at least five times. One-way ANOVA with Dunnett's post hoc test was used. ***, p < 0.001.
Two groups of genes, genes that were up-regulated by Msx2 on day 4 during reprogramming (day 4 up) and genes that were up-regulated by Msx2 and also had higher expression in R1 ES cells than in MEFs (day 4 up and ES cells up), were then analyzed with Pscan (13). Of the four Yamanaka factors, only the KLF4 binding motif was enriched in the promoters of both groups of genes (Fig. 5B). Thus, Klf4 could help Msx2 to activate downstream factors.
We further tested whether KLF4 and MSX2 cooperate to up-regulate genes like Cdh1, Gata3, Nanog, and Sall4. Msx2 had a higher ability to up-regulate Cdh1, Gata3, Nanog, and Sall4 in MEFs in the presence of Klf4 (Fig. 5, C and D). In addition, Msx2 induced larger up-regulation of Cdh1, Gata3, Nanog, and Sall4 during reprogramming with Oct4, Klf4, c-Myc, and Sox2 (OKMS) than during reprogramming with Oct4, c-Myc, and Sox2 (OMS) (Fig. 5, E and F). Thus, Klf4 potentiated the ability of Msx2 to activate downstream factors. Similar observations were not made when potential cooperation between Msx2 and other three Yamanaka factors was tested (Fig. S5).
Members of the GATA family can induce somatic cell reprogramming independent of Oct4 (31). Whether Msx2 can replace Oct4 or other Yamanaka factors to reprogram somatic cells to iPSCs was determined. Msx2 could replace Oct4, Klf4, or Sox2 to reprogram somatic cells to iPSCs (Fig. S6). More iPSCs were generated when Oct4, Klf4, or Sox2 was replaced with Gata3, possibly because overexpression of Msx2 only up-regulated Gata3 by about 6- to 8-fold (Fig. 4B). However, Msx2 can replace Oct4 to induce reprograming.
Msx2 does not mediate the inhibitory roles of BMP during reprogramming
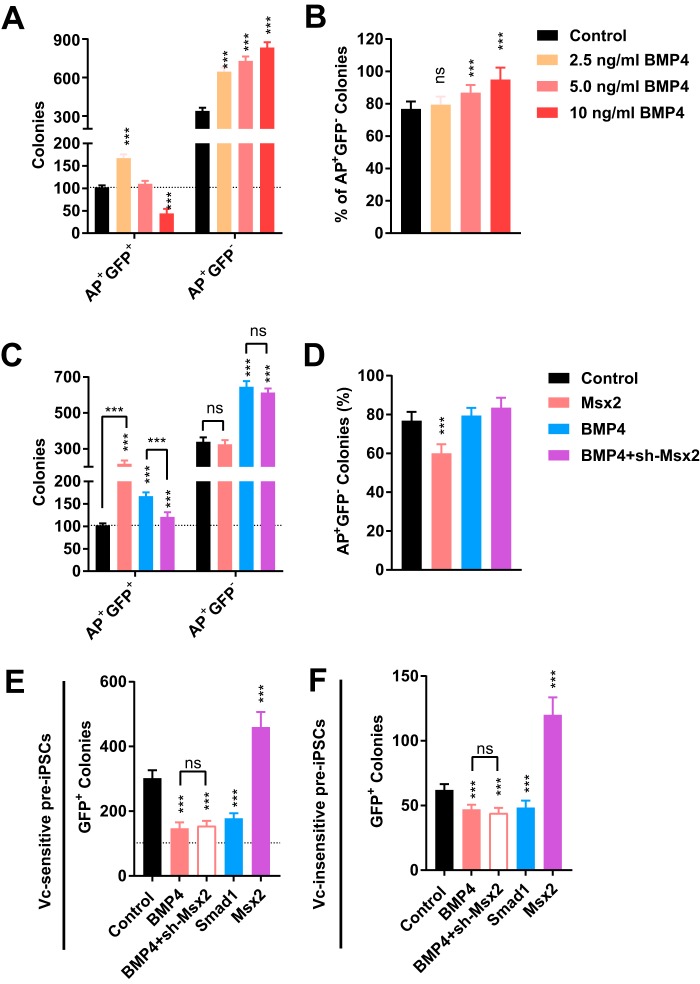
BMPs have been identified as critical molecules to arrest reprogramming in the pre-iPSC state (28). This function of the BMP pathway explains the fact that 2.5 ng/ml BMP4 not only increased the number of AP+GFP+ colonies but also increased that of AP+GFP− colonies (pre-iPSCs colonies) (Fig. 6, A and B). In addition, high concentrations of BMP4 decreased the number of AP+GFP+ colonies but increased that of AP+GFP− colonies (Fig. 6, A and B). However, overexpression of Msx2 did not increase the number of AP+GFP− colonies, and shRNA against Msx2 did not attenuate the ability of BMP4 to increase the number of AP+GFP− colonies (Fig. 6, C and D).
Figure 6.
Msx2 did not mediate the functions of BMP4 to generate more pre-iPSCs. A and B, different concentrations of BMP4 were used during reprogramming. AP+GFP+ and AP+GFP− colonies were counted on day 15 (A). The percentages of AP+GFP− colonies (pre-iPSCs) were calculated (B). C and D, the expression of Msx2 was modulated during reprogramming with or without 2.5 ng/ml BMP4. AP+GFP+ and AP+GFP− colonies were counted on day 15 (C). The percentages of AP+GFP− colonies (pre-iPSCs) were calculated (D). E and F, vitamin C–sensitive (E) and -insensitive (F) pre-iPSCs colonies were converted to iPSCs with 50 μg/ml vitamin C. The ability of BMP4, Smad1, and Msx2 to affect this conversion was determined. All experiments were repeated at least five times. One-way ANOVA with Dunnett's post hoc test was used. ***, p < 0.001; ns, not significant.
Actually, overexpression of Msx2 facilitated the conversion from pre-iPSCs to iPSCs. Vitamin C-sensitive and -insensitive pre-iPSCs colonies established in a previous report were used (28). Vitamin C at 50 μg/ml concentration was used to induce the conversion from pre-iPSCs to iPSCs. BMP4 attenuated and Msx2 facilitated this process (Fig. 6, E and F). Therefore, Msx2 does not mediate the ability of BMP signaling to arrest cells in the pre-iPSC state.
Discussion
In summary, Msx2 mediated the ability of BMP signaling (at low concentration and at an early stage) to up-regulate Cdh1, Gatas-Sall4, and Nanog and to promote reprogramming from MEFs to iPSCs (Fig. 7). The inability of Msx2 to up-regulate miR-205 and miR-200a/b/c and to arrest cells in the pre-iPSC state suggested that Msx2 did not mediate the inhibitory effects of BMP signaling during reprograming (Figs. S3 and S6). Although we did not test all functions of BMP signaling during reprogramming, it was still reasonable to conclude that Msx2 mediated the beneficial roles of BMP signaling during reprogramming.
Figure 7.
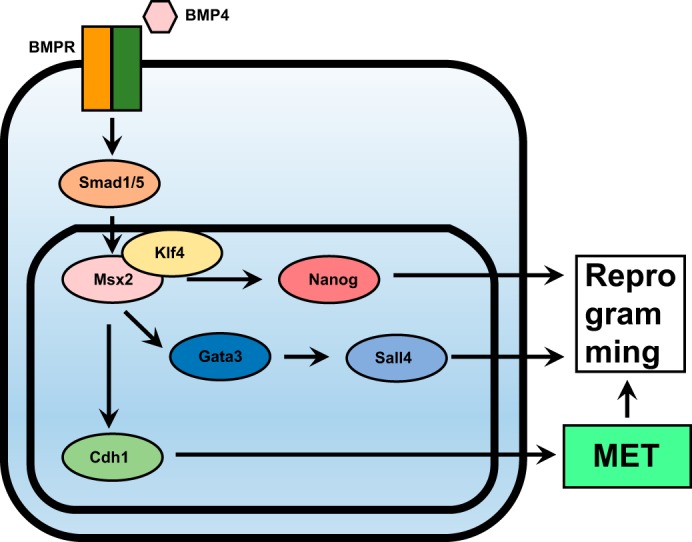
Msx2 mediated the functions of BMP4 to up-regulate several pluripotency-related genes. BMP4 induces up-regulation of Msx2 via SMAD1/5. MSX2 cooperates with KLF4 to up-regulate Cdh1, Gata3-Sall4, and Nanog. Cdh1 promotes reprogramming by facilitating MET, whereas Sall4 and Nanog promote reprogramming by functioning as transcriptional factors that are critical in maintaining pluripotency.
Msx2 is just one member of the Msh family, and the Msh family has two additional members, Msx1 and Msx3. MSX1/2/3 have similar binding motifs and similar functions (7, 12). Thus, we speculated that these three proteins have a similar function during reprogramming. However, although BMP4 did up-regulate Msx1 and Msx3 in a concentration-dependent manner, and Msx1 and Msx3 did promote reprogramming, their connection with BMP4 and reprogramming was not as tight as that of Msx2. For example, shRNAs against Msx1 and Msx3 did not affect the function of BMP4. Thus, we suggested that Msx2 mediated the majority of the beneficial roles of BMP signaling. However, we did not exclude the possibility of Msx1 and Msx3 mediating the function of BMP signaling, such as up-regulating Cdh1, Gata3, Nanog, and Sall4. In addition, Msx1 and Msx3 may mediate the other functions of BMP signaling, such as up-regulating miR-205 and miR-200a/b/c and arresting cells in the pre-iPSC state. Therefore, further investigation is required to answer these questions.
In addition to core pluripotency regulators such as Oct4, Sox2, and Nanog, lineage specifiers such as GATA family members have also been suggested to substitute for particular Yamanaka factors and to promote iPSCs generation (4, 5). Thus, the balance of different lineage-specifying forces, such as those for the mesoendodermal and ectodermal lineages, may maintain pluripotency (5, 6). Both Msx2 and BMP signaling have been reported to modulate lineage commitment toward mesoderm in ES cells (9, 16, 20, 21). Therefore, it was expected to observe beneficial roles of Msx2 and BMP4 in promoting reprogramming.
As an upstream regulator, BMP4 has more functions than Msx2, which partially explains why Msx2 only mediates part of the beneficial roles of BMP signaling. Another explanation, based on the “seesaw” model (5), is that BMP signaling is a strong committer for the mesoderm lineage, which results in imbalance when used at high concentrations. As a downstream effect of BMP signaling, Msx2 has a lower ability to induce mesoderm lineage commitment and imbalance between pluripotency factors and/or counteracting lineage specifiers, which leads to the observation that Msx2 only mediates the beneficial roles of BMP signaling during reprogramming.
Other homeobox transcription factors that also function as lineage specifiers may also affect reprogramming. Their function in cell fate conversion may provide a new understanding of pluripotency and homeobox transcription factors and will enrich the foundation of regenerative medicine.
Experimental procedures
Materials
The chemicals, antibodies, and primers used in this study are listed in Table S2.
Animal studies
Experiments with mice were performed to obtain MEFs and to determine the pluripotency of the generated iPSCs. Our study followed the Guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Committee on the Ethics of Animal Experiments of the Guangzhou Institutes of Biomedicine and Health. All efforts were made to minimize animal discomfort.
Generation of mouse iPSCs
MEFs were derived from 13.5-day-old mouse embryos carrying the Oct4-GFP transgenic allele. MEFs, human embryonic kidney 293T cells, and Plat-E cells were maintained in FBS medium. FBS medium was supplemented with Dulbecco's modified Eagle's medium (Gibco), 10% FBS (Gibco), 1% nonessential amino acids (Gibco), and 1% l-glutamine.
MEFs within two passages were seeded into a 6-well or 12-well plate (2.5 × 104 or 1.5 × 104 cells/well). MEFs were infected with a retrovirus (produced with Plat-E cells and pMXs-based retroviral vectors as reported previously). MES medium was used during reprogramming and was supplemented with Dulbecco's modified Eagle's medium, 15% FBS, 1% Glutamax, 1% nonessential amino acids, 1% sodium pyruvate, 0.1 mm 2-mercaptoentanol, leukemia inhibitory factor, and 50 μg/ml vitamin C (Sigma).
After adding Polybrene to 4 μg/ml, the viral supernatant was used for infection. iPSC colonies were counted or picked according to their Oct4-GFP expression and ES cell–like morphology.
Cell line characterization
Characterization of iPSCs was performed as reported previously (32). Primary antibodies against NANOG and REX1 were used together with the appropriate Alexa-conjugated secondary antibodies. Immunofluorescence results were obtained using a Zeiss LSM 800 confocal microscope.
For karyotype analysis, demecolcine (50 μg/ml, Sigma) was added to the cells for 1 h. The cells were trypsinized, pelleted, resuspended in 0.075 m KCl, and incubated for 20 min at 37 °C. The cells were then fixed with acetic acid:methanol (1:3) for 10 min at 37 °C. The cells were collected by centrifugation, resuspended in fixative solution, dropped on a cold slide, and incubated at 75 °C for 3 h. The slides were treated with trypsin and colorant, and metaphase chromosomes were analyzed using an Olympus BX51 microscope.
RNA-seq
RNA was extracted from cells using TRIzol reagent (Invitrogen). Illumina mRNA-seq libraries were prepared for each RNA sample using the TruSeq RNA Sample Preparation Kit v2; the mRNA-seq libraries were then sequenced on an Illumina NextSeq 500 instrument with the NextSeq 500 Mid Output Kit v2.
qPCR
Total RNA was extracted from cells using TRIzol (Invitrogen), and 5 μg of RNA was used to synthesize complementary DNA with ReverTra Ace® (Toyobo) and oligo(dT) (Takara) according to the manufacturer's instructions. The transcript levels of genes were determined using SYBR Premix Ex TaqII (Tli RNaseH Plus) (Takara) and a CFX-96 real-time system (Bio-Rad). The primers used are listed in Table S2.
ChIP–qPCR
Cells were cross-linked with 1% formaldehyde for 10 min at room temperature. After quenching formaldehyde with 125 mm glycine for 5 min, cells were washed twice with cold PBS. Cells were then lysed in ChIP buffer A (50 mm HEPES–KOH (pH 7.3), 140 mm NaCl, 1 mm EDTA (pH 8.0), 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and protease inhibitor mixture) for 10 min at 4 °C. Samples were centrifuged at 1,400 × g for 5 min at 4 °C. Pellets were resuspended in ChIP buffer B (1% SDS, 50 mm Tris-HCl (pH 8.0), 10 mm EDTA, and protease inhibitor mixture) for 10 min at 4 °C and sheared by sonication.
Sheared chromatin was centrifuged (12,000 × g for 1 min at 4 °C) to discard the pellets, and the supernatant was diluted with ChIP buffer (0.01% SDS, 1% Triton X-100,2 mm EDTA, 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, and protease inhibitor mixture) to make the final SDS concentration lower than 0.1%. Antibodies were coupled to Dynabeads with protein A or G for more than 3 h at 4 °C in PBS supplemented with 0.01% Tween 20, and beads were washed with PBS supplemented with 0.01% Tween 20.
Antibodies were incubated with sheared chromatin overnight at 4 °C. After immunoprecipitation, beads were washed with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), and 150 mm NaCl), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), and 500 mm NaCl), LiCl wash buffer (0.25 m LiCl, 1% NP-40, 1% deoxycholate, 1 mm EDTA, and 10 mm Tris-HCl (pH 8.1)), and TE buffer (10 mm Tris-HCl and 1 mm EDTA (pH 8.0)). DNA was extracted and used for analysis. The sequences of all primers used in this study are given in Table S2.
Data deposition
The current RNA-seq were performed on days 4 and 8 during reprogramming with modulations on the expression of Msx2. The results were deposited in GEO under accession number GSE114120. A previously reported microarray dataset (GSE21757) was also used (22).
Statistical analysis
All experiments were repeated at least five times (n ≥ 5), with the exception of RNA-seq and chimera experiments. The data were analyzed and compared using a two-tailed t test or one-way ANOVA with Dunnett's post hoc test in GraphPad Prism 7.0. Error bars and n represent the standard deviation (standard error if indicated) and the number of independent experiments, respectively. Asterisks represent significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001) from the indicated control groups.
For control samples, when BMP4 was dissolved in DMSO and was used to treat cells, we used the same amount of DMSO to treat control samples; when Msx2 or other genes were overexpressed, a retrovirus encoding FLAG was introduced to control samples; when shRNA against Msx2 or other genes was overexpressed, a retrovirus encoding shRNA against luciferase was introduced to control samples; and control samples were both treated with DMSO and infected with a retrovirus encoding FLAG and shRNA against luciferase when complex treatment was studied.
Author contributions
L. Lin, L. Liang, X. Y., H. S., and Y. L. formal analysis; L. Lin, L. Liang, X. Y., H. S., Y. L., and H. Z. investigation; X. Y. writing-original draft; D. P. and H. Z. funding acquisition; D. P. and H. Z. project administration; H. Z. supervision; H. Z. writing-review and editing.
Supplementary Material
This work was supported by the National Natural Science Foundation of China (U1601228, 31671475, 31422032, and 31421004); the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16000000); the Key Research Program of Frontier Sciences, Chinese Academy of Sciences (QYZDB-SSW-SMC031); the Guangdong Natural Science Foundation (2014A030308002); and the Science and Technology Planning Project of Guangdong Province (2017B030314056). This work was also supported by the Guangzhou branch of the Supercomputing Center of the Chinese Academy of Sciences. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6 and Tables S1 and S2.
The results of the current RNA-seq experiments were deposited in the NCBI Gene Expression Omnibus database under accession number GSE114120.
- iPSC
- induced pluripotent stem cell
- AP
- alkaline phosphatase
- BMP
- bone morphogenetic protein
- ES cell
- embryonic stem cell
- MEF
- mouse embryonic fibroblast
- miR
- microRNA
- MET
- mesenchymal–epithelial transition
- EMT
- epithelial–mesenchymal transition
- shRNA
- short hairpin RNA
- TSS
- transcription start site
- GO
- gene ontology
- qPCR
- quantitative PCR
- RNA-seq
- RNA sequencing
- FBS
- fetal bovine serum
- ANOVA
- analysis of variance.
References
- 1. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 2. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 3. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., and Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- 4. Montserrat N., Nivet E., Sancho-Martinez I., Hishida T., Kumar S., Miquel L., Cortina C., Hishida Y., Xia Y., Esteban C. R., and Izpisua Belmonte J. C. (2013) Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell 13, 341–350 10.1016/j.stem.2013.06.019 [DOI] [PubMed] [Google Scholar]
- 5. Shu J., Wu C., Wu Y., Li Z., Shao S., Zhao W., Tang X., Yang H., Shen L., Zuo X., Yang W., Shi Y., Chi X., Zhang H., Gao G., et al. (2013) Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell 153, 963–975 10.1016/j.cell.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loh K. M., and Lim B. (2011) A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell 8, 363–369 10.1016/j.stem.2011.03.013 [DOI] [PubMed] [Google Scholar]
- 7. Davidson D. (1995) The function and evolution of Msx genes: pointers and paradoxes. Trends Genet. 11, 405–411 10.1016/S0168-9525(00)89124-6 [DOI] [PubMed] [Google Scholar]
- 8. Bell J. R., Noveen A., Liu Y. H., Ma L., Dobias S., Kundu R., Luo W., Xia Y., Lusis A. J., Snead M. L., and Maxson R. (1993) Genomic structure, chromosomal location, and evolution of the mouse Hox 8 gene. Genomics 16, 123–131 10.1006/geno.1993.1149 [DOI] [PubMed] [Google Scholar]
- 9. Wu Q., Zhang L., Su P., Lei X., Liu X., Wang H., Lu L., Bai Y., Xiong T., Li D., Zhu Z., Duan E., Jiang E., Feng S., Han M., et al. (2015) MSX2 mediates entry of human pluripotent stem cells into mesendoderm by simultaneously suppressing SOX2 and activating NODAL signaling. Cell Res. 25, 1314–1332 10.1038/cr.2015.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. MacKenzie A., Ferguson M. W., and Sharpe P. T. (1992) Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development 115, 403–420 [DOI] [PubMed] [Google Scholar]
- 11. Satokata I., Ma L., Ohshima H., Bei M., Woo I., Nishizawa K., Maeda T., Takano Y., Uchiyama M., Heaney S., Peters H., Tang Z., Maxson R., and Maas R. (2000) Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 24, 391–395 10.1038/74231 [DOI] [PubMed] [Google Scholar]
- 12. Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J. A., van der Lee R., Bessy A., Chèneby J., Kulkarni S. R., Tan G., Baranasic D., Arenillas D. J., Sandelin A., Vandepoele K., Lenhard B., et al. (2018) JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 10.1093/nar/gkx1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zambelli F., Pesole G., and Pavesi G. (2009) Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37, W247–W252 10.1093/nar/gkp464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y., Liu J., Hudson M., Kim S., and Hatch N. E. (2010) FGF2 promotes Msx2 stimulated PC-1 expression via Frs2/MAPK signaling. J. Cell. Biochem. 111, 1346–1358 10.1002/jcb.22861 [DOI] [PubMed] [Google Scholar]
- 15. Newberry E. P., Latifi T., Battaile J. T., and Towler D. A. (1997) Structure-function analysis of Msx2-mediated transcriptional suppression. Biochemistry 36, 10451–10462 10.1021/bi971008x [DOI] [PubMed] [Google Scholar]
- 16. Xu R. H., Chen X., Li D. S., Li R., Addicks G. C., Glennon C., Zwaka T. P., and Thomson J. A. (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264 10.1038/nbt761 [DOI] [PubMed] [Google Scholar]
- 17. Zhou J., Su P., Li D., Tsang S., Duan E., and Wang F. (2010) High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor β superfamily receptors. Stem Cells 28, 1741–1750 10.1002/stem.504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kishigami S., and Mishina Y. (2005) BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev. 16, 265–278 10.1016/j.cytogfr.2005.04.002 [DOI] [PubMed] [Google Scholar]
- 19. Tam P. P., and Loebel D. A. (2007) Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 8, 368–381 10.1038/nrg2084 [DOI] [PubMed] [Google Scholar]
- 20. Richter A., Valdimarsdottir L., Hrafnkelsdottir H. E., Runarsson J. F., Omarsdottir A. R., Ward-van Oostwaard D., Mummery C., and Valdimarsdottir G. (2014) BMP4 promotes EMT and mesodermal commitment in human embryonic stem cells via SLUG and MSX2. Stem Cells 32, 636–648 10.1002/stem.1592 [DOI] [PubMed] [Google Scholar]
- 21. Zhang P., Li J., Tan Z., Wang C., Liu T., Chen L., Yong J., Jiang W., Sun X., Du L., Ding M., and Deng H. (2008) Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood 111, 1933–1941 10.1182/blood-2007-02-074120 [DOI] [PubMed] [Google Scholar]
- 22. Samavarchi-Tehrani P., Golipour A., David L., Sung H. K., Beyer T. A., Datti A., Woltjen K., Nagy A., and Wrana J. L. (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 10.1016/j.stem.2010.04.015 [DOI] [PubMed] [Google Scholar]
- 23. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., et al. (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 10.1016/j.stem.2010.04.014 [DOI] [PubMed] [Google Scholar]
- 24. Brambrink T., Foreman R., Welstead G. G., Lengner C. J., Wernig M., Suh H., and Jaenisch R. (2008) Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2, 151–159 10.1016/j.stem.2008.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsubooka N., Ichisaka T., Okita K., Takahashi K., Nakagawa M., and Yamanaka S. (2009) Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells 14, 683–694 10.1111/j.1365-2443.2009.01301.x [DOI] [PubMed] [Google Scholar]
- 26. Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., Horvath S., Zhou Q., and Plath K. (2009) Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377 10.1016/j.cell.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., and Meissner A. (2008) Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 10.1038/nature07056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Chen J., Wu Y., Guo L., Zhu J., Zhao X., Peng T., Zhang Y., et al. (2013) H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45, 34–42 [DOI] [PubMed] [Google Scholar]
- 29. Cheng S. L., Shao J. S., Behrmann A., Krchma K., and Towler D. A. (2013) Dkk1 and MSX2-Wnt7b signaling reciprocally regulate the endothelial-mesenchymal transition in aortic endothelial cells. Arterioscler. Thromb. Vasc. Biol. 33, 1679–1689 10.1161/ATVBAHA.113.300647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. di Bari M. G., Ginsburg E., Plant J., Strizzi L., Salomon D. S., and Vonderhaar B. K. (2009) Msx2 induces epithelial-mesenchymal transition in mouse mammary epithelial cells through upregulation of Cripto-1. J. Cell. Physiol. 219, 659–666 10.1002/jcp.21712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shu J., Zhang K., Zhang M., Yao A., Shao S., Du F., Yang C., Chen W., Wu C., Yang W., Sun Y., and Deng H. (2015) GATA family members as inducers for cellular reprogramming to pluripotency. Cell Res. 25, 169–180 10.1038/cr.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu X., Sun H., Qi J., Wang L., He S., Liu J., Feng C., Chen C., Li W., Guo Y., Qin D., Pan G., Chen J., Pei D., and Zheng H. (2013) Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat. Cell Biol. 15, 829–838 10.1038/ncb2765 [DOI] [PubMed] [Google Scholar]
- 33. Liang L., Sun H., Zhang W., Zhang M., Yang X., Kuang R., and Zheng H. (2016) Meta-analysis of EMT datasets reveals different types of EMT. PLoS ONE 11, e0156839 10.1371/journal.pone.0156839 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.