Abstract
Interleukin (IL)-13 is a signature cytokine of type 2 inflammation important for the pathogenesis of various diseases, including allergic diseases. Signal transducer and activator of transcription (STAT) 6 is a critical transcriptional factor for the IL-13 signals; however, it remains unknown how expression of the IL-13–induced genes is differentiated by the transcriptional machineries. In this study, we identified IL-13–induced transcriptional factors in lung fibroblasts using DNA microarrays in which SOX11 was included. Knockdown of SOX11 down-regulated expression of periostin and CCL26, both of which are known to be downstream molecules of IL-13, whereas enforced expression of SOX11 together with IL-13 stimulation enhanced expression of periostin. Moreover, we found that in DNA microarrays combining IL-13 induction and SOX11 knockdown there exist both SOX11-dependent and -independent molecules in IL-13–inducible molecules. In the former, many inflammation-related and fibrosis-related molecules, including periostin and CCL26, are involved. These results suggest that SOX11 acts as a trans-acting transcriptional factor downstream of STAT6 and that in lung fibroblasts the IL-13 signals are hierarchically controlled by STAT6 and SOX11.
Keywords: allergy, cell signaling, cytokine, fibroblast, fibrosis, interleukin, STAT transcription factor, transcription
Introduction
IL-13 2 is a signature cytokine of type 2 inflammation produced by TH2 cells, follicular helper T cells, group 2 innate lymphoid cells, eosinophils, mast cells, and basophils (1–4). IL-13 plays important roles in the pathogenesis of not only allergic diseases (asthma, allergic rhinitis, and atopic dermatitis) but also other inflammatory diseases, chronic pulmonary obstructive disease, cancers, inflammatory bowel diseases, autoimmune diseases, and pulmonary fibrosis (5–9). Based on this knowledge, antagonists against IL-13 have been developed as potential therapeutic agents against these diseases.
IL-13 is a pleiotropic cytokine acting on various kinds of cells by binding its receptor, which is composed of the IL-13 receptor α1 chain and the IL-4 receptor α chain on the cell surface of B cells, T cells, mast cells, macrophages, epithelial cells, fibroblasts, smooth muscle cells, and endothelial cells, and exerting its pathophysiological roles (10). It is of note that the actions of IL-13 on fibroblasts are important for accelerating inflammation and generating fibrosis. IL-13 induces expression of various chemokines, recruiting inflammatory cells, including CCL26/eotaxin-3 and profibrotic molecules such as tenascin-C and periostin, and activating fibrosis-related molecules such as transforming growth factor-β and several matrix metalloproteins (1, 11).
Signal transducer and activator of transcription (STAT) 6 is a critical transcriptional factor for IL-13 signaling (12, 13). After it is phosphorylated and activated by Janus kinases following the ligation of IL-13 to its receptor, it recognizes the sequence motif TTCN3–4GAA, regulating the transcriptional activities of the IL-13–targeted genes. However, it remains unknown how expression of the IL-13–induced genes is differentiated by the transcriptional machineries.
Periostin is a matricellular protein downstream of the IL-13 signals (14). We found that expression of periostin requires STAT6 in model mice because periostin induction is diminished in STAT6-deficient mice (15) and that periostin is highly expressed in the basement membrane in asthma patients. Crucially, it is positively correlated with poor prognosis of these patients (16, 17). We then showed that periostin acts as a proinflammatory molecule by activating NF-κB, a transcriptional factor important for inflammation, in keratinocytes or fibroblasts in a paracrine or autocrine manner (15, 18). Moreover, because periostin acts as a surrogate marker for IL-13 reflecting type 2 inflammation (14), measurement of serum periostin has been applied to predicting the efficacy of IL-13 antagonists (19, 20). However, the precise mechanism of periostin expression by IL-13 has remained elusive.
In the SOX (SRY-related HMG box) family, SOX11 is a member of the SOX C group as well as SOX4 and SOX12 (21). The members of the SOX C group have DNA-binding HMG domains close to the N terminus and transcriptional activation domains in the C-terminal regions. SOX11 is important for organ development in embryos; SOX11-deficient mice immediately die after birth, probably due to heart defects. These mice manifest craniofacial and skeletal malformations; asplenia; and hypoplasia of the lung, stomach, and pancreas, suggesting widespread roles of SOX11 in tissues or organ development (22). Additionally, SOX11 plays a critical role in neural differentiation in both embryos and adults (23–25). Moreover, it has been reported that SOX11 is expressed in various malignancies, including mantle cell lymphoma, suggesting the oncogenic activity of SOX11 in certain types of malignancies (26). However, there is no report showing that SOX11 exerts immunomodulatory effects.
In this study, we identified IL-13–induced transcriptional factors in lung fibroblasts using DNA microarrays in which SOX11 was included. We then showed that SOX11 acts as a trans-acting transcriptional factor for periostin. Moreover, we found that in DNA microarrays combining IL-13 induction and SOX11 knockdown there exist both SOX11-dependent and -independent molecules in IL-13–inducible molecules. The former involves many inflammation-related and fibrosis-related molecules. These results suggest that SOX11 acts as a trans-acting transcriptional factor downstream of STAT6 and that in lung fibroblasts the IL-13 signals are hierarchically controlled by STAT6 and SOX11.
Results
Involvement of a trans-acting transcriptional mechanism through STAT6 in IL-13–mediated expression in lung fibroblasts
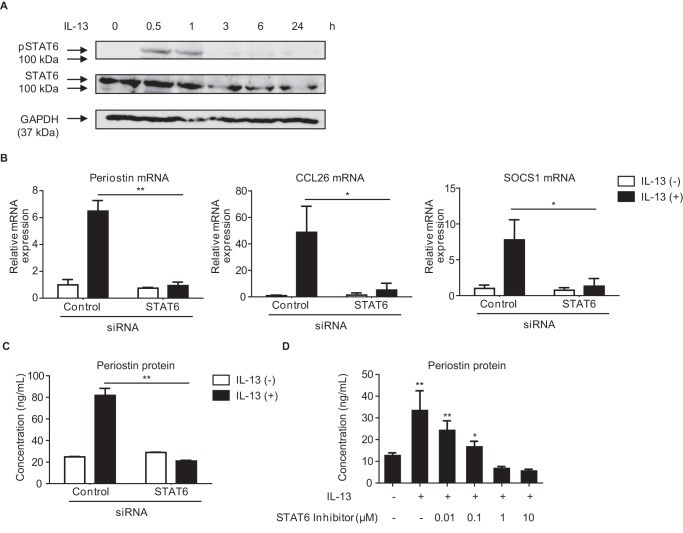
It had already been established that STAT6 plays a critical role in IL-13 signaling (12, 13). We first examined whether STAT6 is required for IL-13–mediated induction of the POSTN, CCL26, and SOCS1 genes, known to encode downstream molecules of IL-13 (15, 27, 28). IL-13 induced transient tyrosine phosphorylation of STAT6 in MRC-5 cells, a human lung fibroblast cell line (Fig. 1A). Knockdown of STAT6 significantly decreased expression of all of POSTN, CCL26, and SOCS1 genes (Fig. 1B). Moreover, secretion of periostin protein in IL-13–stimulated MRC-5 cells was mostly diminished by knockdown of STAT6 or treatment with a STAT6 inhibitor, AS1517499 (Fig. 1, C and D). These results suggest that induction of periostin, CCL26, and SOCS1 by IL-13 in lung fibroblasts requires STAT6.
Figure 1.
Dependence of IL-13–induced genes on STAT6 in lung fibroblasts. A, kinetics of STAT6 phosphorylation (pSTAT6) in MRC-5 cells. After stimulation with 50 ng/ml IL-13 for the indicated times, cells were lysed, and the lysates were immunoblotted with the indicated Abs. B and C, effects of knockdown of STAT6 on expression of periostin, CCL26, and SOCS1. MRC-5 cells were treated with or without 50 ng/ml IL-13 for 24 h in the presence of 20 nm either control siRNA or STAT6 siRNA. Expression of periostin, CCL26, and SOCS1 mRNA (B) and periostin concentrations in the cell supernatants (C) are depicted. D, effects of AS1517499, a STAT6 inhibitor, on IL-13–induced periostin production. MRC-5 cells were precultured in the indicated concentration of AS1517499 for 30 min and then treated with or without 50 ng/ml IL-13 for 24 h. Periostin concentrations in the cell supernatants are depicted. *, p < 0.05; **, p < 0.01. Error bars represent S.D.
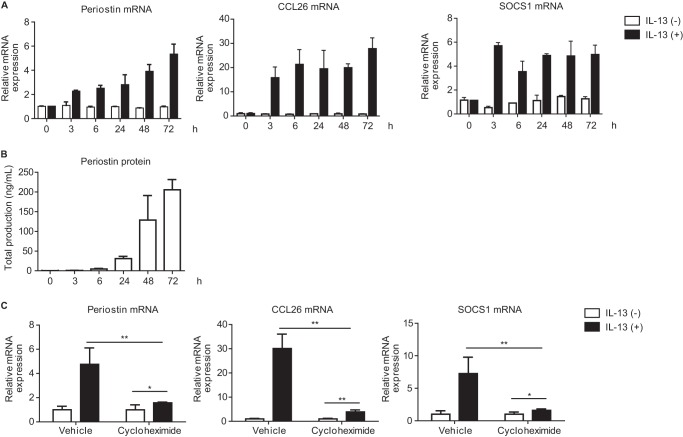
We next examined the kinetics of periostin expression in MRC-5 cells. In contrast to phosphorylation of STAT6, expression of periostin at both the mRNA and protein levels was prolonged and gradually increased after IL-13 stimulation up to 72 h (Fig. 2, A and B). Moreover, expression of SOCS1 and CCL26 at the mRNA level was also prolonged and sustained up to 72 h. This raised the possibility that in lung fibroblasts IL-13–mediated expression requires a trans-acting transcriptional mechanism through STAT6. To address this, we examined the effects of cycloheximide, an inhibitor against de novo protein synthesis, on IL-13–dependent expression of the POSTN, CCL26, and SOCS1 genes. Cycloheximide significantly decreased the expression of all of these IL-13–inducible genes (Fig. 2C), although there was still induction of these genes by IL-13. These results suggest that in lung fibroblasts a trans-acting transcriptional mechanism through STAT6 is involved in the IL-13–mediated expression of periostin, CCL26, and SOCS1.
Figure 2.
Involvement of a trans-acting transcriptional mechanism through STAT6 in IL-13–mediated expression in lung fibroblasts. A and B, kinetics of expression of periostin, CCL26, and SOCS1 mRNA expression (A) and periostin concentrations in the cell supernatants (B). MRC-5 cells were stimulated with 50 ng/ml IL-13 for the indicated times. C, effects of cycloheximide on IL-13–induced expression of periostin, CCL26, and SOCS1. MRC-5 cells were precultured in 10 μg/ml cycloheximide for 30 min and then treated with or without 50 ng/ml IL-13 for 24 h. The values were adjusted by GAPDH expression. The same experiments were performed three times. *, p < 0.05; **, p < 0.01. Error bars represent S.D.
The cis-acting transcriptional mechanism of STAT6 for IL-13–induced periostin expression
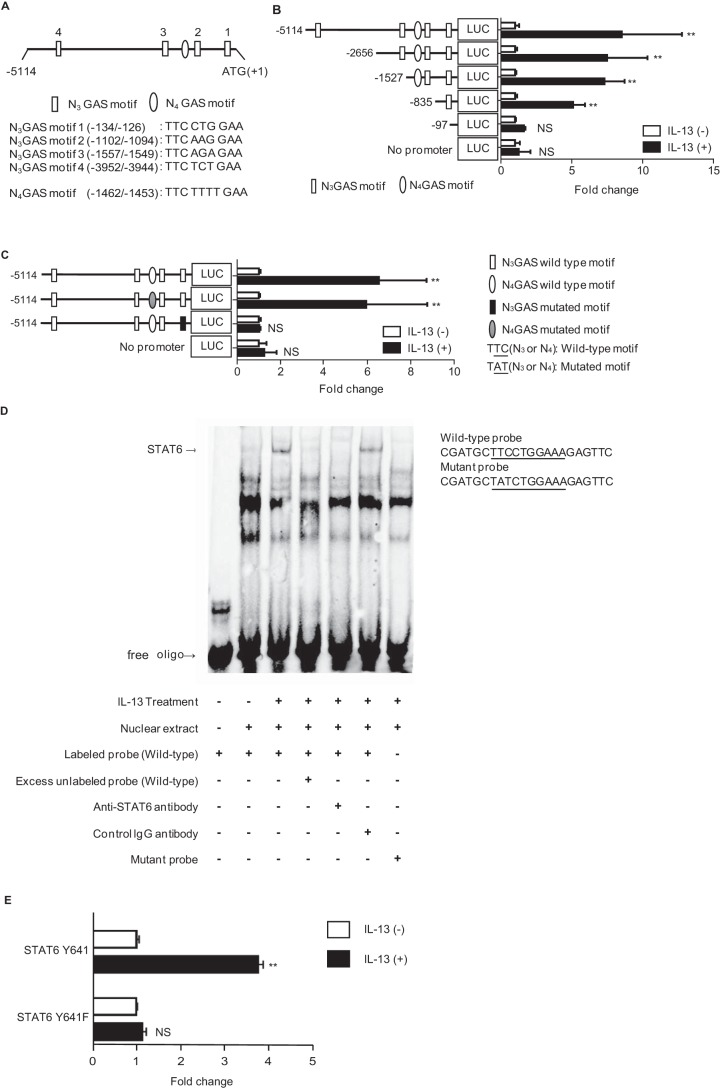
Treatment with cycloheximide significantly, but not completely, diminished IL-13–mediated expression of periostin as shown in Fig. 2C, which suggested that, through STAT6, there might exist a cis-acting transcriptional mechanism in addition to a trans-acting mechanism. To explore this possibility, we prepared a series of plasmids encoding promoter regions in the −5114 to +1 bp site relative to the transcription start site in the POSTN gene. We then applied them to transient transfection and luciferase assay using HEK293T cells. Because HEK293T cells lack STAT6, we cotransfected the plasmid encoding each promoter region together with the plasmid encoding human STAT6. These regions contain one TTCN4GAA sequence (N4 GAS motif), a typical STAT6-binding site, and four TTCN3GAA sequences (N3 GAS motifs 1–4), atypical STAT6-binding sites (Fig. 3A). When we narrowed down the promoter region from −835 to −97 bp, luciferase activity decreased significantly, suggesting that this region contains an IL-13–responsive element (Fig. 3B). Because this same region contains N3 GAS motif 1 (−134/−126), we examined whether this motif was required for IL-13 responsiveness by introducing a mutation. Introduction of a mutation into N3 GAS motif 1 (−134/−126), but not into the N4 GAS motif (−1462/−1453), diminished IL-13 responsiveness (Fig. 3C). Electrophoretic mobility shift assay (EMSA) showed that the probe corresponding to this motif specifically binds to STAT6 (Fig. 3D). Moreover, we confirmed that tyrosyl phosphorylation of STAT6 is required for induction of periostin by IL-13 by using the mutated form of STAT6, Y641F, in which the phosphorylated tyrosyl residue of STAT6 is mutated (Fig. 3E). Taken together, these results demonstrate that STAT6 acts as a cis-regulatory mechanism for transcription of periostin induced by IL-13 via the N3 GAS motif located at −134 to −126 bp.
Figure 3.
The cis-acting transcriptional mechanism of STAT6 for IL-13–induced periostin expression. A, schematic model of the 5′-UTR in the human POSTN gene (−5114/+1). Four N3 GAS motifs (1–4) and one N4 GAS motif in this region are depicted. B and C, effects of truncated or mutated 5′-UTR on luciferase (LUC) activity. HEK293T cells were transiently transfected with the plasmids encoding each construct and STAT6. Six kinds of truncated constructs (B) and two kinds of mutated constructs in N3 GAS motif 1 (−134/−126) or N4 GAS motif (−1462/−1453) (C) together with their activities are depicted. The luciferase activities relative to the vehicle are shown. D, EMSA assay for STAT6 in IL-13–stimulated HEK293T cells. Nuclear extracts from HEK293T cells treated with or without 50 ng/ml IL-13 for 60 min were applied. WT or mutant probes were used. Either excess amounts of unlabeled probe or anti-STAT6 Ab or control IgG Ab was added. The image is representative of three individual experiments. The WT and mutated N3 GAS motif 1 sequences are shown. E, requirement of STAT6 phosphorylation for induction of periostin by IL-13. HEK293T cells were transiently transfected with the plasmid encoding WT or the mutated form of STAT6. Expression of POSTN in the cells treated with 50 ng/ml IL-13 for 24 h is shown. *, p < 0.05; **, p < 0.01; NS, not significant. Error bars represent S.D.
Identifying SOX11 as a candidate for a trans-acting factor for IL-13–induced transcription of periostin in lung fibroblasts
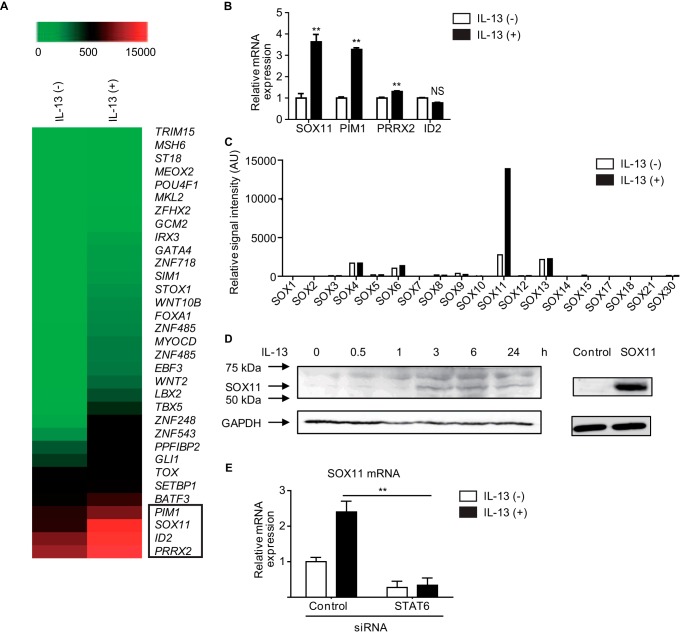
To identify candidates for trans-acting factors for IL-13–induced transcription of periostin in lung fibroblasts, we searched for transcriptional factors among the IL-13–inducible molecules in lung fibroblasts using DNA microarray analyses, finding that expressions of four transcription-related factors—SOX11, PIM1, PRRX2, and ID2—were significantly up-regulated in IL-13–stimulated MRC-5 cells (Fig. 4A). The analyses of quantitative RT-PCR (qRT-PCR) showed that expression of SOX11, PIM1, and PRRX2, but not ID2, was increased by stimulation of IL-13 in MRC-5 cells (Fig. 4B). Among these three molecules, we focused on SOX11 for two reasons. 1) SOX11 was the only SOX family member induced by IL-13 (Fig. 4C), and 2) the Gene Expression Omnibus (GEO) at NCBI shows in a data set that expression of both POSTN and SOX11 are up-regulated in IL-13–treated IMR-90 cells, another human fibroblast cell line (GEO accession number GDS5256; Fig. S1). We confirmed that IL-13 up-regulated expression of SOX11 protein in MRC-5 cells (Fig. 4D). Moreover, knockdown of STAT6 significantly decreased SOX11 expression induced by IL-13 in MRC-5 cells (Fig. 4E). In contrast, IL-13 did not induce expression of SOX11 in mouse primary lung fibroblasts, although it induced expression of periostin and SOCS1 via STAT6, indicating that the expression mechanism of SOX11 is different in humans and mice (data not shown). These results demonstrate that SOX11 is a downstream molecule of the IL-13 signal and indicate that SOX11 may be a candidate for a trans-acting factor for IL-13–induced transcription of periostin in lung fibroblasts.
Figure 4.
Identification of SOX11 as a candidate for a trans-acting factor for IL-13–induced transcription of periostin in lung fibroblasts. A, DNA microarray analysis of the genes in MRC-5 cells treated with or without 50 ng/ml IL-13 for 24 h. Heat maps depict gene expression levels. A total of 44,077 probes were tested, and the results of all 33 transcription-related factors are shown. SOX11, PIM1, PRRX2, and ID2 are boxed. B, expression of SOX11, PIM1, PRRX2, and ID2 genes in MRC-5 cells treated with or without 50 ng/ml IL-13 for 24 h. **, p < 0.01; NS, not significant. Error bars represent S.D. C, gene expression of the SOX family in MRC-5 stimulated with IL-13 for 24 h. D, Western blotting for SOX11 in MRC-5 cells. After MRC-5 cells were stimulated with IL-13 for the indicated times, cells were lysed and immunoblotted with anti-SOX11 Ab (left). Western blotting control for SOX11 in HEK293T cells transfected with control or SOX11 expression plasmid (right) is also shown. The bands of GAPDH are the same as those in Fig. 1A because these were performed at the same time. E, effects of STAT6 knockdown on SOX11 expression in MRC-5 cells. MRC-5 cells were treated with or without 50 ng/ml IL-13 for 24 h in the presence of 20 nm either control siRNA or STAT6 siRNA. AU, arbitrary units.
Regulation of IL-13–induced periostin expression by SOX11 in lung fibroblasts
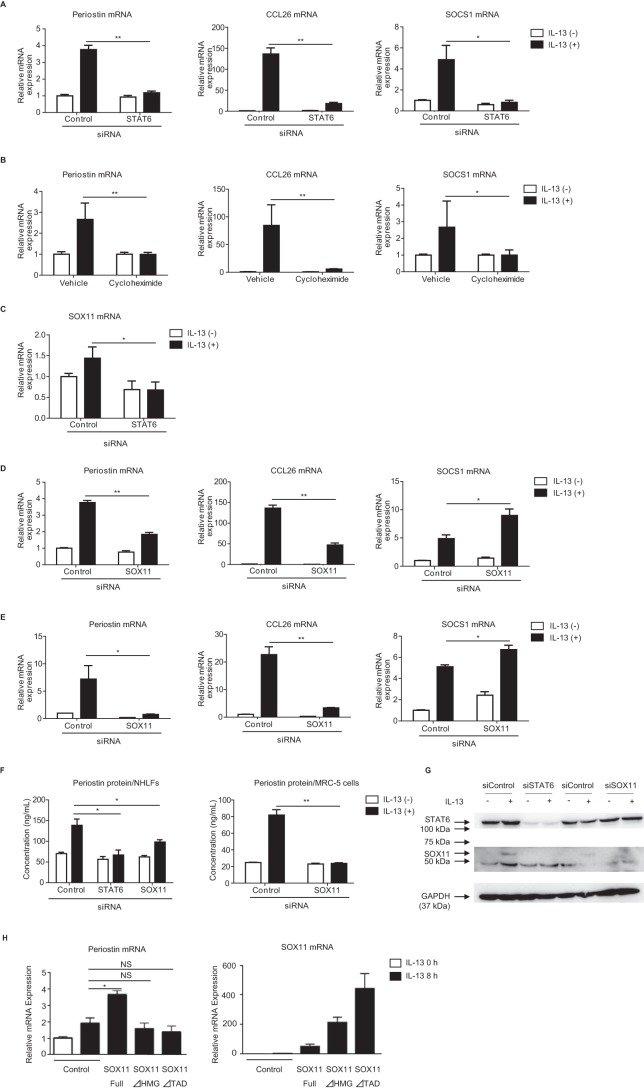
We next examined the regulatory roles of SOX11 in IL-13–induced periostin expression using primary normal human lung fibroblasts (NHLFs). IL-13 induced expression of POSTN, CCL26, and SOCS1, which were significantly down-regulated by either knockdown of STAT6 (Fig. 5A) or treatment with cycloheximide (Fig. 5B), in these cells as well as in MRC-5 cells. Moreover, knockdown of STAT6 also significantly decreased IL-13–induced SOX11 expression in these cells (Fig. 5C). To clarify the role of SOX11 in transcriptional regulation by IL-13, we examined the effects of knockdown of SOX11 on expression of periostin, CCL26, and SOCS1 induced by IL-13. Knockdown of SOX11 significantly diminished expression of periostin and CCL26 at the mRNA level and of periostin at the protein level, whereas expression of SOCS1 did not decrease (Fig. 5, D and F). Knockdown of SOX11 did not change the expression of STAT6, excluding the possibility that down-regulation of periostin and CCL26 was not via down-regulation of STAT6 (Fig. S2). Down-regulated expression of both periostin and CCL26, but not of SOCS1, by knockdown of SOX11 was observed in MRC-5 cells as well (Fig. 5, E and F). Knockdown effects of STAT6 on SOX11 expression was confirmed at the protein level too (Fig. 5G). Knockdown of PIM1 or PRRX2, transcriptional-related factors identified by DNA microarray (Fig. 4A), affected not at all or only slightly the expression of periostin in MRC-5 cells (data not shown). Moreover, overexpression of SOX11 enhanced periostin expression by IL-13 in MRC-5 cells, whereas overexpression of the mutated forms of SOX11 in which the DNA-binding domain or the transactivation domain is deleted did not have such an effect (Fig. 5H). Both forms of SOX11 impaired the induction activity of periostin by IL-13. These results demonstrate that in lung fibroblasts SOX11 acts as a trans-regulatory molecule for expression of periostin and CCL26, but not SOCS1, by IL-13.
Figure 5.
Regulation of IL-13–induced periostin expression by SOX11 in lung fibroblasts. A and B, effects of STAT6 knockdown (A) or cycloheximide (B) on expression of periostin, CCL26, and SOCS1 mRNA in NHLFs. C, effects of STAT6 knockdown on expression of SOX11 in NHLFs. A and C, NHLFs were treated with or without 50 ng/ml IL-13 for 24 h in the presence of 20 nm either control siRNA or STAT6 siRNA. B, NHLFs were precultured in 10 μg/ml cycloheximide for 30 min and treated with or without 50 ng/ml IL-13 for 24 h. D and E, effects of SOX11 knockdown on mRNA levels of periostin, CCL26, and SOCS1 in NHLFs (D) and MRC-5 cells (E). F, effects of SOX11 knockdown on periostin protein in the supernatant of NHLFs and MRC-5 cells. D–F, these cells were treated with or without 50 ng/ml IL-13 for 24 h in the presence of 20 nm either control siRNA, STAT6 siRNA, or SOX11 siRNA. The values were adjusted by GAPDH expression. *, p < 0.05; **, p < 0.01; NS, not significant. Error bars represent S.D. G, effects of STAT6 or SOX11 knockdown on expression of STAT6 or SOX11 protein in MRC-5 cells. After MRC-5 cells were treated with or without 50 ng/ml IL-13 for 6 h in the presence of 20 nm either control siRNA, STAT6 siRNA, or SOX11 siRNA, cells were lysed, and the lysates were immunoblotted with the indicated Abs. H, effects of overexpression of the WT, the DNA-binding domain–deleted, or the transactivation domain–deleted forms of SOX11 on expression of periostin mRNA in MRC-5 cells. After the plasmids were transfected into MRC-5 cells, the cells were treated with or without 5 ng/ml IL-13 for 8 h.
Up-regulation of periostin in lung fibroblasts by enforced expression of SOX11
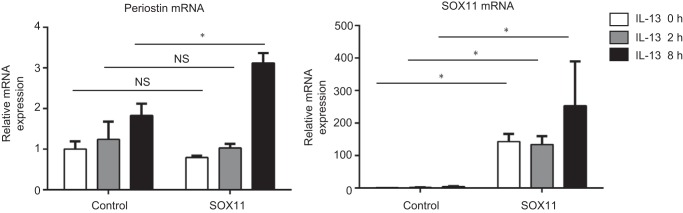
We next examined whether enforced expression of SOX11 reciprocally up-regulates periostin expression in lung fibroblasts. We transfected the plasmid encoding human SOX11 or control plasmid to MRC-5 cells followed by stimulation of IL-13. When we transfected the control plasmid to MRC-5 cells, IL-13 stimulation up-regulated periostin expression as shown previously, which is due to a combination of cis- and trans-regulation by STAT6 and endogenously expressed SOX11 (Fig. 6). When SOX11 was exogenously overexpressed in MRC-5 cells without IL-13 stimulation, expression of periostin was unchanged. However, when SOX11-overexpressing cells were stimulated with IL-13 for 8 h, periostin expression was significantly up-regulated compared with mock-transfected cells. SOX11 was not coimmunoprecipitated with STAT6, excluding the possibility that these two molecules act by forming a heterodimer (data not shown). These results reinforce the role of SOX11 in the trans-regulation mechanism of periostin, indicating that some transcriptional or post-translational modification induced by IL-13 would be required for this mechanism.
Figure 6.
Up-regulation of periostin in lung fibroblasts by enforced expression of SOX11. MRC-5 cells were transiently transfected with the expression plasmid encoding SOX11 and then treated with or without 5 ng/ml IL-13 for 2 and 8 h. Expression of periostin (left) and SOX11 (right) of two experiments is depicted. *, p < 0.05; NS, not significant. Error bars represent S.D.
Differentiating SOX11-dependent and -independent genes in IL-13–inducible genes in lung fibroblasts
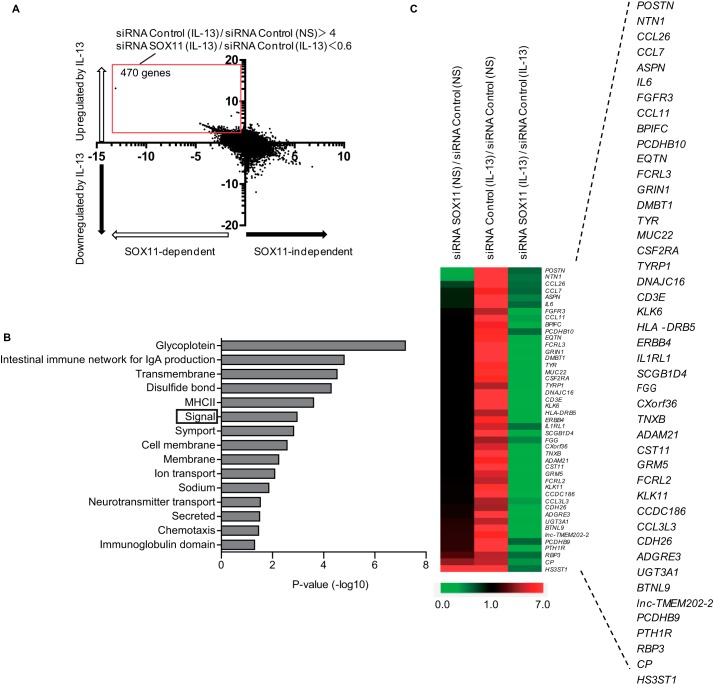
The finding that knockdown of SOX11 down-regulated IL-13–induced expression of periostin and CCL26 but not of SOCS1 suggested that there would be two groups in IL-13–induced genes in lung fibroblasts: SOX11-dependent and -independent genes. To differentiate these two groups, we submitted four groups of genes from NHLFs, with or without stimulation of IL-13 and with or without SOX11 knockdown, to DNA microarray analysis. Based on the up-regulation by stimulation of IL-13 (longitudinal axis) and the down-regulation by SOX11 knockdown (horizontal axis), we divided the genes into four groups (Fig. 7A): 1) IL-13–inducible and SOX11-dependent (upper left panel), 2) IL-13–inducible and SOX11-independent (upper right panel), 3) IL-13–noninducible and SOX11-dependent (lower left panel), and 4) IL-13–noninducible and SOX11-independent (lower right panel). As expected, both POSTN and CCL26 were included in the IL-13–inducible and SOX11-dependent genes, whereas SOCS1 was included in the IL-13–inducible and SOX11-independent genes, validating the classification of the genes by IL-13 induction and SOX11 dependence using DNA microarray analysis.
Figure 7.
Sorting out SOX11-dependent and -independent IL-13–induced genes in lung fibroblasts. A, a dot plot of the genes by IL-13 induction and SOX11 dependence by the DNA microarrays. We incubated NHLFs with or without 50 ng/ml IL-13 for 24 h in the presence of 20 nm either control siRNA or SOX11 siRNA and then subjected them to DNA microarray. The longitudinal axis represents the induction of the genes stimulated by IL-13 (control siRNA/IL-13− versus control siRNA/IL-13+). The horizontal axis represents the down-regulation by SOX11 knockdown (control siRNA/IL-13+ versus SOX11 siRNA/IL-13+). The red square represents the genes with more than 4-fold up-regulation by stimulation of IL-13 and more than 40% down-regulation by SOX11 knockdown. NS, no stimulant. B, GO terms highly correlated with SOX11-dependent and IL-13–induced genes in lung fibroblasts are depicted. Signal is boxed. C, the genes included in signal as a GO term up-regulated by IL-13 and down-regulated by SOX11 knockdown. Heat maps depict the genes in the GO term signal with control siRNA/IL-13− versus SOX11 siRNA/IL-13− (left), control siRNA/IL-13− versus control siRNA/IL-13+ (middle), and control siRNA/IL-13+ versus SOX11 siRNA/IL-13+ (right). MHC, major histocompatibility complex.
We then applied the IL-13–inducible and SOX11-dependent genes, defined by more than 4-fold up-regulation by stimulation of IL-13 and more than 40% down-regulation by SOX11 knockdown (470 genes), to DAVID analysis to enrich biological functions in this group. We found that this procedure picked up “signal” (a GO term) with a high enrichment score in which both POSTN and CCL26 were included (Fig. 7B). This group contained chemokines (CCL3L3, CCL7, CCL11, and CCL26), a proinflammatory cytokine (IL6), cytokine receptors (IL1RL1, CSF2RA, and FGFR3), immune receptors (CD3E, FCRL2, FCRL3, and HLA-DRB5), cadherin molecules (CDH26, PCDHB9, and PCDHB10), and an extracellular matrix protein (TNXB) as well as POSTN. These results suggest that there exist two groups in IL-13–induced genes in lung fibroblasts, SOX11-dependent and -independent genes, and that the POSTN and CCL26 genes are included in the former group.
Discussion
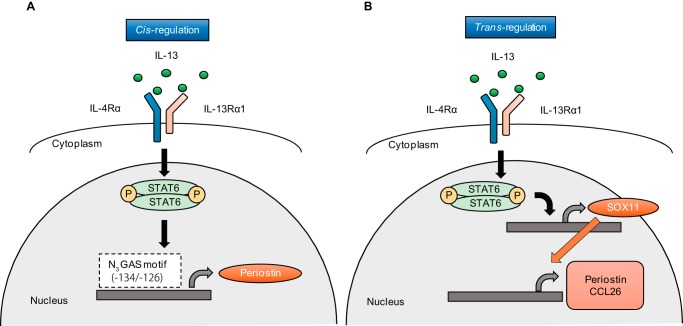
In this study, we showed that in lung fibroblasts there exist both cis- and trans-acting transcriptional mechanisms through STAT6 in IL-13–mediated expression; in the former, STAT6 activated by IL-13 directly binds to the promoter/enhancer region on the targeted genes, whereas in the latter, STAT6 activated by IL-13 induces expression of SOX11, which then induces some IL-13–targeted genes (Fig. 8). This in turn indicates that there exist SOX11-dependent and -independent genes in the IL-13–inducible genes downstream of STAT6. These results have highlighted the importance of SOX11 in the pathogenesis of IL-13–related diseases such as allergic diseases.
Figure 8.
Schematic model of the molecular mechanism of how expression of IL-13–induced periostin is regulated in lung fibroblasts. A, IL-13 induced phosphorylation of STAT6, which binds to N3GAS motif 1 (−134/-126) in the promoter/enhance region of the POSTN gene up-regulating POSTN expression (cis-regulation). B, IL-13 induces SOX11 expression via STAT6, which then induces POSTN and CCL26 expression (trans-regulation). Thus, the cis-regulation and trans-regulation by SOX11 cooperatively act as the transcriptional mechanism of the IL-13 signals.
The SOX family is composed of nine groups (A–H with the B group including B1 and B2) among which the SOX C group contains three members, SOX4, SOX11, and SOX12, having a common genetic structure (21). The members of the SOX family mainly function in tissue or organ development. We found that SOX11 is the only SOX member induced by IL-13 (Fig. 4C) and then that SOX11 plays a role in inducing certain genes downstream of the IL-13/STAT6 signal. To our knowledge, transcriptional regulation downstream of IL-13/STAT6 is the first example of the immune-regulatory effects of the SOX family. This suggests the unique characteristic of SOX11 in the SOX family and, furthermore, that the functions of SOX11 are broader than what are currently known. Thus far, we do not know in detail how SOX11 induces periostin expression. The requirement of IL-13 stimulation in periostin expression by enforced SOX11 expression (Fig. 6) indicates that enforced SOX11 expression alone is not enough for periostin expression but that expression of some cofactor or some post-translational modification induced by IL-13 in lung fibroblasts would be needed. We failed in applying HEK293T cells overexpressing SOX11 to luciferase assay as shown in Fig. 3, which supports this notion (data not shown). Further examination aiming at this point is needed.
The transcriptional activities of STAT6 are exerted by coactivation with other transcriptional factors such as CBP/p300, p100/staphylococcal nuclease and tudor domain–containing 1, and nuclear receptor coactivator-3 (12, 13). Moreover, it is known that STAT6 cooperates with other transcriptional factors such as CCAAT/enhancer–binding protein β (29), NF-κB (30), Ets-1 (31), and peroxisome proliferator–activated receptor γ (32). However, the trans-activation mechanism of IL-13–inducible transcription through STAT6 has been poorly understood. It is known that in TH2 cells STAT6 induces GATA-binding protein 3, which in turn induces type 2 cytokines IL-4, IL-5, and IL-13 (33). However, to our knowledge, no trans-activating transcriptional system downstream of STAT6 via a transcriptional factor other than GATA-binding protein 3, in cells other than TH2 cells, or targeting genes other than type 2 cytokines is known. The present finding has added a novel transcriptional mechanism to the signal transduction pathway of IL-13.
It has been established that IL-13 is a pleiotropic cytokine exerting various actions in many kinds of cells, including B cells, T cells, mast cells, macrophages, epithelial cells, fibroblasts, smooth muscle cells, and endothelial cells (10). In the present study, we showed that SOX11 acts as a trans-acting transcriptional factor downstream of the IL-13/STAT6 signals in lung fibroblasts. Moreover, we propose that the IL-13/STAT6/SOX11 pathway in lung fibroblasts is important for both inflammation and fibrosis by inducing inflammation- or fibrosis-related molecules, including periostin and CCL26. However, it remains undetermined whether the IL-13/STAT6/SOX11 pathway is adopted in other cells. It is known that SOX11 is expressed in various cells, including neural cells, kidney cells, and osteoblasts (21, 34), and that epithelial-mesenchymal transition up-regulates SOX11 expression, suggesting that SOX11 expression in fibroblasts would be relatively abundant (35). As far as we could determine from GEO Profiles searches, SOX11 was not listed as an IL-13–inducible gene in bronchial or esophageal epithelial cells. Overall, the IL-13/STAT6/SOX11 pathway may not be common in different cell lineages but more cell lineage–specific, at least in fibroblasts.
We identified the IL-13–inducible genes as either SOX11-dependent or -independent (Fig. 7A). The upper left and upper right panels corresponding to the IL-13–inducible/SOX11-dependent and IL-13–inducible/SOX11-independent genes contained ∼7700 and ∼6700 dots, respectively, suggesting that roughly more than half of the IL-13–inducible genes are SOX11-dependent. To establish the role of SOX11 downstream of the IL-13/STAT6 signals in lung fibroblasts, we await the comparison of the phenotypes of mice deficient in STAT6 or SOX11 specifically in fibroblasts; it has been shown that global deficiency of SOX11 is lethal in mice (22). However, in the present study, we show that, based on microarray analysis, the IL-13/STAT6/SOX11 pathway is important for induction of genes related to inflammation and/or fibrosis, including chemokines, proinflammatory cytokines, cytokine receptors, immune receptors, cadherin molecules, and extracellular matrix proteins (Fig. 7, B and C). Consistent with this assumption, we and others have reported that deficiency in either IL-13 or periostin, a downstream molecule of the IL-13 signal, shows comparable protection of pulmonary fibrosis by bleomycin (36, 37). Thus, the discovery of involvement of SOX11 in the IL-13/STAT6 signals would clarify the pathophysiological roles of the IL-13/STAT6 signals. Alternatively, various kinds of agents targeting IL-13 or its receptor to inhibit STAT6 activation could target asthma and pulmonary fibrosis (Clinical Trial NCT01872689) (20, 38). Our present finding may give us a clue to finding another option to develop an inhibitor for IL-13/STAT6 signals.
Experimental procedures
Cell culture
HEK293T cells and MRC-5 cells (normal embryonic lung fibroblast cells; Riken BioResource Center) were maintained in Dulbecco's modified Eagle's medium (Millipore-Sigma) supplied with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Meiji Seika Pharma). NHLFs were purchased from Lonza and maintained as the vendor recommends. These cells were stimulated with 50 ng/ml IL-13 (PeproTech), and then the supernatants were applied to ELISA. For knockdown experiments, MRC-5 cells and NHLFs were cultured with small interfering RNA (siRNA) for 24 h followed by stimulation with 50 ng/ml IL-13 for 24 h. Then RNA extracts and the supernatants were applied to qRT-PCR or ELISA, respectively. For some experiments, MRC-5 cells and NHLFs were pretreated with the indicated concentration of a STAT6-selective inhibitor, AS1517499 (AXON Medchem), or 10 μg/ml cycloheximide for 30 min and then treated with 50 ng/ml IL-13 for 24 h. For overexpression experiments, MRC-5 cells were transfected with plasmid for 24 h followed by stimulation with 5 ng/ml IL-13 for 8 h. Then RNA extracts were applied to qRT-PCR.
Knockdown of mRNA by siRNA
siRNA oligonucleotides were purchased from Dharmacon/GE Healthcare. Cells were transfected with ON-TARGETplus siRNA for STAT6, SOX11, PIM1, PRRX2, or control at the indicated concentrations and for the indicated times in the presence of RNAiMAX reagent (Thermo Fisher Scientific). The nucleotide sequences are depicted in Table S1. Silencing of target genes was confirmed by qRT-PCR.
Western blotting
Western blotting was performed as described previously (18). The antibodies (Abs) used in this study were against STAT6 (Santa Cruz Biotechnology), phosphorylated STAT6 (Tyr-641; Cell Signaling Technology), SOX11 (Merck Millipore), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Merck Millipore). In SOX11 overexpression experiments, pME18S encoding human SOX11 cDNA was transfected into HEK293T cells, which were harvested for Western blotting 24 h after transfection.
ELISA for periostin production
ELISA for periostin was performed using two kinds of anti-periostin Abs, SS18A and SS17B (Shino-Test) as described previously (39).
qRT-PCR
qRT-PCR was performed as described previously (18). Primers for qRT-PCR are described in Table S2.
Expression vectors and reporter gene constructs
We subcloned the proximal 5′ region of the POSTN gene (−5114/+1 bp relative to the transcription start site) into the XhoI and NcoI sites of the firefly luciferase reporter vector pGL4.10 (Promega). This construct was used as a template to generate a series of deletion plasmid constructs by inverse PCR. The mutation in the STAT6-binding site of the promoter region was introduced by PCR-based mutagenesis (40). We generated two mutation constructs in the N4 GAS motif (−1462/−1453, TTCTTTTGAA to TATTTTTGAA), and in the N3 GAS motif (−134/−126, TTCCTGGAA to TATCTGGAA). The plasmid pME18S containing human STAT6 cDNA was prepared as described previously (41).
STAT6 Y641F and the mutated form of SOX11 deleting the DNA-binding domain (ΔHMG) were generated using inverse PCR with primers designed to alter tyrosine to phenylalanine (TAT to TTT) or to delete HMG box domain, respectively. The mutated form of SOX11 deleting the transactivation domain (ΔTAD) was constructed by PCR with primers to delete the C-terminal 34 amino acids. Primers for introducing mutant constructs are as follows: 5′-TTTGTCCCAGCTACCATCAAGATG-3′ for STAT6 Y641F forward, 5′-ACCCCTGCCATCCTTACCCATCT-3′ for STAT6 Y641F reverse, 5′-AAGCCCAAAATGGACCCCTCG-3′ for SOX11 ΔHMG forward, 5′-CGACGCCGTCTTGCACCAGTCTG-3′ for SOX11 ΔHMG reverse, 5′-ACTCGAGATCATGGTGCAGCAGGCGGA-3′for SOX11 ΔTAD forward, and 5′-AATACTAGTTCACAGGCTGCCCTCGCTGAACG-3′ for SOX11 ΔTAD reverse.
Transient transfection and luciferase assay
Transfection was performed using HEK293T cells with FuGENE 6 reagent (Promega). Both the plasmids encoding each POSTN promoter and human STAT6 cDNA were cotransfected into the HEK293T cells. The pRL-TK plasmid (Promega) was also cotransfected as an internal control to normalize transfection by Renilla luciferase activity.
Overexpression of SOX11 protein
MRC-5 cells were transfected with 2.5 μg of SOX11 expression plasmid or control in the presence of 100 μl of nucleofector solution specifically formulated for transfection of fibroblasts (Lonza) using the Amaxa Nucleofection system (Lonza). Immediately after pulsing, the cells were transferred to 12-well plates and cultured for 24 h followed by stimulation with 5 ng/ml IL-13 for 8 h. Then RNA extracts were applied to qRT-PCR.
EMSA
Nuclear extracts were prepared from HEK293T cells transiently transfected with pME18S encoding human STAT6 cDNA. The primer sequences in this assay were as follows: biotin-5′-CGATGCTTCCTGGAAAGAGTTC-3′ for the wildtype (WT) POSTN promoter and biotin-5′-CGATGCTATCTGGAAAGAGTTC-3′ for the mutant probe. For competitive inhibition, a 100-fold molar excess of nonbiotinylated WT probe was added to the reaction mixture 15 min before adding the WT probe. For the supershift assay, nuclear extracts were preincubated with anti-STAT6 Ab or control IgG for 15 min. The samples were separated on a 5% nondenaturing polyacrylamide gel containing 0.4× Tris-buffered saline at 4 °C. Samples were transferred to nylon membranes (Hybond-N, GE Healthcare) for 50 min at 380 mA with stirring. Membranes were cross-linked at 120 mJ/cm2 for 1 min with a UV Stratalinker 2400 cross-linker (Agilent Technologies). Immunodetection was performed with a Chemiluminescent Nucleic Acid Detection Module kit (Thermo Fisher Scientific).
DNA microarray analysis
MRC-5 cells were stimulated with or without 50 ng/ml IL-13 for 24 h. NHLFs were cultured with siRNA for 24 h followed by stimulation with 50 ng/ml IL-13 for 24 h. One replicate array for each experimental condition was performed. Total RNA with an RNA integrity number greater than 9.0 was applied to an Agilent Expression Array (SurePrint G3 Human GE8x60K v2 Microarray for MRC-5 and v3 Microarray for NHLFs, Takara Bio). The signal intensity values themselves or their relative ratios were presented on a heat map and subjected to MultiExperiment Viewer (MeV) v4.9 software (Dana–Farber Cancer Institute). The identified genes are registered in the NCBI Gene Expression Omnibus (GEO) (for MRC-5 cells, GEO accession number GSE104936; for NHLFs, GEO accession number GSE104937). For gene ontology analysis, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) tool (National Cancer Institute) was used. This database includes the Gene Ontology Database (http://geneontology.org/). 3
Statistical analysis
Data are presented as mean ± S.D. Statistical analyses were performed using Prism 5.0 software (GraphPad Software). The significance of differences was assessed using an unpaired or paired Student's t test. Values of p < 0.05 were considered statistically significant.
Author contributions
Y. M. data curation; Y. M., Y. N., T. Y., K. K., and S. F. formal analysis; Y. M. and K. I. writing-original draft; S. N., K. A., M. F., and K. I. conceptualization; S. N., Y. N., T. Y., K. K., S. F., and M. F. writing-review and editing; H. T. and H. N. resources; K. I. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. Dovie R. Wylie for critical review of the manuscript. We also thank Miho Miyake and Maki Watanabe for technical assistance.
This work was supported by Japan Society for the Promotion of Science KAKENHI Grants JP15K15372 and JP16H05343 (to K. I.) and by AstraZeneca (to K. I.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Tables S1 and S2.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE104936, GSE104937, and GSE104938.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- IL
- interleukin
- STAT
- Signal transducer and activator of transcription
- SOX
- SRY-related HMG box
- HMG
- high mobility group
- HEK
- human embryonic kidney
- GAS
- interferon γ–activated site
- EMSA
- electrophoretic mobility shift assay
- qRT-PCR
- quantitative RT-PCR
- NHLF
- normal human lung fibroblast
- GO
- gene ontology
- CBP
- cAMP-response element–binding protein (CREB)-binding protein
- Ab
- antibody
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TAD
- transactivation domain.
References
- 1. Izuhara K., Arima K., Kanaji S., Ohta S., and Kanaji T. (2006) IL-13: a promising therapeutic target for bronchial asthma. Curr. Med. Chem. 13, 2291–2298 10.2174/092986706777935140 [DOI] [PubMed] [Google Scholar]
- 2. Kubo M. (2017) T follicular helper and TH2 cells in allergic responses. Allergol. Int. 66, 377–381 10.1016/j.alit.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 3. Kabata H., Moro K., Koyasu S., and Asano K. (2015) Group 2 innate lymphoid cells and asthma. Allergol. Int. 64, 227–234 10.1016/j.alit.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 4. Miyake K., and Karasuyama H. (2017) Emerging roles of basophils in allergic inflammation. Allergol. Int. 66, 382–391 10.1016/j.alit.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 5. Walsh G. M. (2017) Biologics targeting IL-5, IL-4 or IL-13 for the treatment of asthma—an update. Expert Rev. Clin. Immunol. 13, 143–149 10.1080/1744666X.2016.1216316 [DOI] [PubMed] [Google Scholar]
- 6. May R. D., and Fung M. (2015) Strategies targeting the IL-4/IL-13 axes in disease. Cytokine 75, 89–116 10.1016/j.cyto.2015.05.018 [DOI] [PubMed] [Google Scholar]
- 7. Matsunaga M. C., and Yamauchi P. S. (2016) IL-4 and IL-13 inhibition in atopic dermatitis. J. Drugs Dermatol. 15, 925–929 [PubMed] [Google Scholar]
- 8. Huang X. L., Wang Y. J., Yan J. W., Wan Y. N., Chen B., Li B. Z., Yang G. J., and Wang J. (2015) Role of anti-inflammatory cytokines IL-4 and IL-13 in systemic sclerosis. Inflamm. Res. 64, 151–159 10.1007/s00011-015-0806-0 [DOI] [PubMed] [Google Scholar]
- 9. Passalacqua G., Mincarini M., Colombo D., Troisi G., Ferrari M., Bagnasco D., Balbi F., Riccio A., and Canonica G. W. (2017) IL-13 and idiopathic pulmonary fibrosis: possible links and new therapeutic strategies. Pulm. Pharmacol. Ther. 45, 95–100 10.1016/j.pupt.2017.05.007 [DOI] [PubMed] [Google Scholar]
- 10. Izuhara K., Ohta S., Shiraishi H., and Suzuki S. (2011) Interleukin 4, interleukin 13, and interleukin 9, in Inflammation and Allergy Drug Design (Izuhara K., Holgate S. T., and Wills-Karp M., eds) pp. 175–185, Wiley-Blackwell, London [Google Scholar]
- 11. Wills-Karp M. (2004) Interleukin-13 in asthma pathogenesis. Immunol. Rev. 202, 175–190 10.1111/j.0105-2896.2004.00215.x [DOI] [PubMed] [Google Scholar]
- 12. Hebenstreit D., Wirnsberger G., Horejs-Hoeck J., and Duschl A. (2006) Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev. 17, 173–188 10.1016/j.cytogfr.2006.01.004 [DOI] [PubMed] [Google Scholar]
- 13. Goenka S., and Kaplan M. H. (2011) Transcriptional regulation by STAT6. Immunol. Res. 50, 87–96 10.1007/s12026-011-8205-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Izuhara K., Arima K., Ohta S., Suzuki S., Inamitsu M., and Yamamoto K. (2014) Periostin in allergic inflammation. Allergol. Int. 63, 143–151 10.2332/allergolint.13-RAI-0663 [DOI] [PubMed] [Google Scholar]
- 15. Masuoka M., Shiraishi H., Ohta S., Suzuki S., Arima K., Aoki S., Toda S., Inagaki N., Kurihara Y., Hayashida S., Takeuchi S., Koike K., Ono J., Noshiro H., Furue M., et al. (2012) Periostin promotes chronic allergic inflammation in response to Th2 cytokines. J. Clin. Investig. 122, 2590–2600 10.1172/JCI58978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takayama G., Arima K., Kanaji T., Toda S., Tanaka H., Shoji S.,McKenzie A. N., Nagai H., Hotokebuchi T., and Izuhara K. (2006) Periostin: a novel component of subepithelial fibrosis of bronchial asthma downstream of IL-4 and IL-13 signals. J Allergy Clin. Immunol. 118, 98–104 10.1016/j.jaci.2006.02.046 [DOI] [PubMed] [Google Scholar]
- 17. Kanemitsu Y., Ito I., Niimi A., Izuhara K., Ohta S., Ono J., Iwata T., Matsumoto H., and Mishima M. (2014) Osteopontin and periostin are associated with a 20-year decline of pulmonary function in patients with asthma. Am. J. Respir. Crit. Care Med. 190, 472–474 10.1164/rccm.201403-0562LE [DOI] [PubMed] [Google Scholar]
- 18. Taniguchi K., Arima K., Masuoka M., Ohta S., Shiraishi H., Ontsuka K., Suzuki S., Inamitsu M., Yamamoto K. I., Simmons O., Toda S., Conway S. J., Hamasaki Y., and Izuhara K. (2014) Periostin controls keratinocyte proliferation and differentiation by interacting with the paracrine IL-1α/IL-6 loop. J. Invest. Dermatol. 134, 1295–1304 10.1038/jid.2013.500 [DOI] [PubMed] [Google Scholar]
- 19. Corren J., Lemanske R. F., Hanania N. A., Korenblat P. E., Parsey M. V., Arron J. R., Harris J. M., Scheerens H., Wu L. C., Su Z., Mosesova S., Eisner M. D., Bohen S. P., and Matthews J. G. (2011) Lebrikizumab treatment in adults with asthma. N. Engl. J. Med. 365, 1088–1098 10.1056/NEJMoa1106469 [DOI] [PubMed] [Google Scholar]
- 20. Brightling C. E., Chanez P., Leigh R., O'Byrne P. M., Korn S., She D., May R. D., Streicher K., Ranade K., and Piper E. (2015) Efficacy and safety of tralokinumab in patients with severe uncontrolled asthma: a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir. Med. 3, 692–701 10.1016/S2213-2600(15)00197-6 [DOI] [PubMed] [Google Scholar]
- 21. Kamachi Y., and Kondoh H. (2013) Sox proteins: regulators of cell fate specification and differentiation. Development 140, 4129–4144 10.1242/dev.091793 [DOI] [PubMed] [Google Scholar]
- 22. Sock E., Rettig S. D., Enderich J., Bösl M. R., Tamm E. R., and Wegner M. (2004) Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell. Biol. 24, 6635–6644 10.1128/MCB.24.15.6635-6644.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin L., Lee V. M., Wang Y., Lin J. S., Sock E., Wegner M., and Lei L. (2011) Sox11 regulates survival and axonal growth of embryonic sensory neurons. Dev. Dyn. 240, 52–64 10.1002/dvdy.22489 [DOI] [PubMed] [Google Scholar]
- 24. Wang Y., Lin L., Lai H., Parada L. F., and Lei L. (2013) Transcription factor Sox11 is essential for both embryonic and adult neurogenesis. Dev. Dyn. 242, 638–653 10.1002/dvdy.23962 [DOI] [PubMed] [Google Scholar]
- 25. Haslinger A., Schwarz T. J., Covic M., and Lie D. C. (2009) Expression of Sox11 in adult neurogenic niches suggests a stage-specific role in adult neurogenesis. Eur. J. Neurosci. 29, 2103–2114 10.1111/j.1460-9568.2009.06768.x [DOI] [PubMed] [Google Scholar]
- 26. Beà S., and Amador V. (2017) Role of SOX11 and genetic events cooperating with cyclin D1 in mantle cell lymphoma. Curr. Oncol. Rep. 19, 43 10.1007/s11912-017-0598-1 [DOI] [PubMed] [Google Scholar]
- 27. Hoeck J., and Woisetschläger M. (2001) Activation of eotaxin-3/CCLl26 gene expression in human dermal fibroblasts is mediated by STAT6. J. Immunol. 167, 3216–3222 10.4049/jimmunol.167.6.3216 [DOI] [PubMed] [Google Scholar]
- 28. Egwuagu C. E., Yu C. R., Zhang M., Mahdi R. M., Kim S. J., and Gery I. (2002) Suppressors of cytokine signaling proteins are differentially expressed in Th1 and Th2 cells: implications for Th cell lineage commitment and maintenance. J. Immunol. 168, 3181–3187 10.4049/jimmunol.168.7.3181 [DOI] [PubMed] [Google Scholar]
- 29. Delphin S., and Stavnezer J. (1995) Characterization of an interleukin 4 (IL-4) responsive region in the immunoglobulin heavy chain germline ϵ promoter: regulation by NF-IL-4, a C/EBP family member and NF-κ B/p50. J. Exp. Med. 181, 181–192 10.1084/jem.181.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Messner B., Stütz A. M., Albrecht B., Peiritsch S., and Woisetschläger M. (1997) Cooperation of binding sites for STAT6 and NFκB/rel in the IL-4-induced up-regulation of the human IgE germline promoter. J. Immunol. 159, 3330–3337 [PubMed] [Google Scholar]
- 31. Travagli J., Letourneur M., Bertoglio J., and Pierre J. (2004) STAT6 and Ets-1 form a stable complex that modulates Socs-1 expression by interleukin-4 in keratinocytes. J. Biol. Chem. 279, 35183–35192 10.1074/jbc.M403223200 [DOI] [PubMed] [Google Scholar]
- 32. Szanto A., Balint B. L., Nagy Z. S., Barta E., Dezso B., Pap A., Szeles L., Poliska S., Oros M., Evans R. M., Barak Y., Schwabe J., and Nagy L. (2010) STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity 33, 699–712 10.1016/j.immuni.2010.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maier E., Duschl A., and Horejs-Hoeck J. (2012) STAT6-dependent and -independent mechanisms in Th2 polarization. Eur. J. Immunol. 42, 2827–2833 10.1002/eji.201242433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lefebvre V., and Bhattaram P. (2016) SOXC genes and the control of skeletogenesis. Curr. Osteoporos. Rep. 14, 32–38 10.1007/s11914-016-0296-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Venkov C., Plieth D., Ni T., Karmaker A., Bian A., George A. L. Jr., and Neilson E. G. (2011) Transcriptional networks in epithelial-mesenchymal transition. PLoS One 6, e25354 10.1371/journal.pone.0025354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uchida M., Shiraishi H., Ohta S., Arima K., Taniguchi K., Suzuki S., Okamoto M., Ahlfeld S. K., Ohshima K., Kato S., Toda S., Sagara H., Aizawa H., Hoshino T., Conway S. J., et al. (2012) Periostin, a matricellular protein, plays a role in the induction of chemokines in pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 46, 677–686 10.1165/rcmb.2011-0115OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu T., Jin H., Ullenbruch M., Hu B., Hashimoto N., Moore B., McKenzie A., Lukacs N. W., and Phan S. H. (2004) Regulation of found in inflammatory zone 1 expression in bleomycin-induced lung fibrosis: role of IL-4/IL-13 and mediation via STAT-6. J. Immunol. 173, 3425–3431 10.4049/jimmunol.173.5.3425 [DOI] [PubMed] [Google Scholar]
- 38. Wenzel S., Castro M., Corren J., Maspero J., Wang L., Zhang B., Pirozzi G., Sutherland E. R., Evans R. R., Joish V. N., Eckert L., Graham N. M., Stahl N., Yancopoulos G. D., Louis-Tisserand M., et al. (2016) Dupilumab efficacy and safety in adults with uncontrolled persistent asthma despite use of medium-to-high-dose inhaled corticosteroids plus a long-acting β2 agonist: a randomised double-blind placebo-controlled pivotal phase 2b dose-ranging trial. Lancet 388, 31–44 10.1016/S0140-6736(16)30307-5 [DOI] [PubMed] [Google Scholar]
- 39. Kanemitsu Y., Matsumoto H., Izuhara K., Tohda Y., Kita H., Horiguchi T., Kuwabara K., Tomii K., Otsuka K., Fujimura M., Ohkura N., Tomita K., Yokoyama A., Ohnishi H., Nakano Y., et al. (2013) Increased periostin associates with greater airflow limitation in patients receiving inhaled corticosteroids. J. Allergy Clin. Immunol. 132, 305–312.e3 10.1016/j.jaci.2013.04.050 [DOI] [PubMed] [Google Scholar]
- 40. Hebenstreit D., Luft P., Schmiedlechner A., Regl G., Frischauf A. M., Aberger F., Duschl A., and Horejs-Hoeck J. (2003) IL-4 and IL-13 induce SOCS-1 gene expression in A549 cells by three functional STAT6-binding motifs located upstream of the transcription initiation site. J. Immunol. 171, 5901–5907 10.4049/jimmunol.171.11.5901 [DOI] [PubMed] [Google Scholar]
- 41. Arima K., Sato K., Tanaka G., Kanaji S., Terada T., Honjo E., Kuroki R., Matsuo Y., and Izuhara K. (2005) Characterization of the interaction between interleukin-13 and interleukin-13 receptors. J. Biol. Chem. 280, 24915–24922 10.1074/jbc.M502571200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.