Abstract
Staphylococcus aureus is part of the human nasal and skin microbiomes along with other bacterial commensals and opportunistic pathogens. Nutrients are scarce in these habitats, demanding effective nutrient acquisition and competition strategies. How S. aureus copes with phosphate limitation is still unknown. Wall teichoic acid (WTA), a polyol-phosphate polymer, could serve as a phosphate source, but whether S. aureus can utilize it during phosphate starvation remains unknown. S. aureus secretes a glycerophosphodiesterase, GlpQ, that cleaves a broad variety of glycerol-3-phosphate (GroP) headgroups of deacylated phospholipids, providing this bacterium with GroP as a carbon and phosphate source. Here we demonstrate that GlpQ can also use glycerophosphoglycerol derived from GroP WTA from coagulase-negative Staphylococcus lugdunensis, Staphylococcus capitis, and Staphylococcus epidermidis, which share the nasal and skin habitats with S. aureus. Therefore, S. aureus GlpQ is the first reported WTA-hydrolyzing enzyme, or teichoicase, from Staphylococcus. Activity assays revealed that unmodified WTA is the preferred GlpQ substrate, and the results from MS analysis suggested that GlpQ uses an exolytic cleavage mechanism. Importantly, GlpQ did not hydrolyze the ribitol-5-phosphate WTA polymers of S. aureus, underscoring its role in interspecies competition rather than in S. aureus cell wall homeostasis or WTA recycling. glpQ expression was strongly up-regulated under phosphate limitation, and GlpQ allowed S. aureus to grow in the presence of GroP WTA as the sole phosphate source. Our study reveals a novel and unprecedented strategy of S. aureus for acquiring phosphate from bacterial competitors under the phosphate-limiting conditions in the nasal or skin environments.
Keywords: phosphodiesterases, teichoic acid, Staphylococcus aureus (S. aureus), glycerophospholipid, glycerol, GlpQ, glycerophosphodiesterase, pathogen, ribitol-5-phosphate, teichoicase
Introduction
Staphylococcus aureus colonizes the nares of approximately 30% of the human population, which represents a major risk factor for invasive infections (1). In addition, S. aureus is a transient or permanent member of the skin microbiome in healthy persons or atopic dermatitis patients (2). Competition with other bacteria, in particular with coagulase-negative staphylococci (CoNS) 4 of the nasal or skin microbiome, is thought to enable or prevent S. aureus carriage, depending on the success of individual microbiome members, but the mechanism of S. aureus interaction with nasal commensals remain superficially understood (3, 4). The nose and skin are known to be very nutrient-poor environments, suggesting that bacteria compete for scant nutrients (5–9). Along this line, we have recently shown that S. aureus utilizes glycerophosphodiesters released from host cell lipids using the secreted enzyme GlpQ, which is not produced by most CoNS (10). Our findings have revealed that GlpQ from S. aureus is able to cleave a broad variety of glycerol-3-phosphate (GroP) headgroups from deacylated phospholipids, such as glycerophosphocholine, present in different body fluids, into GroP and the corresponding headgroup and plays an important role under nutrient limitation (10). Additionally, GlpQ is a major S. aureus antigen in infection, indicating that it may have an important role in invasive infections (11, 12).
The bacterial cell wall polymer wall teichoic acid (WTA) could be a rich store for alditols and phosphate under conditions of nutrient limitation, but it is not known whether staphylococci can recycle WTA from their own or from competitors' cell walls. WTAs are attached to the peptidoglycan layer via a highly conserved linkage unit followed by a poly-alditol-phosphate repeat unit, composed of 40 to 60 monomers, building up the actual WTA chain (13). In most Staphylococcus species, these monomers are either ribitol-5-phosphate (RboP), as for S. aureus, or GroP, as in most CoNS. The monomers are linked via phosphodiester bonds and are often substituted with a variety of (amino) sugars and d-alanine (14). Although WTA biosynthesis is well-characterized in S. aureus and Bacillus subtilis, little is known about WTA degradation (13).
Recently it has been shown that WTA can be recycled in B. subtilis when phosphate becomes limiting, a process that requires two secreted phosphodiesterases, the exolytic GlpQ and the endolytic PhoD enzymes (15). It has been suggested that the B. subtilis GlpQ acts on WTA fragments released by the PhoD phosphodiesterase and that the two enzymes have a cooperative role in the recycling of phosphate from the GroP WTA. Although S. aureus does not secrete a PhoD homolog, we recently showed that its GlpQ enzyme, which is 50% identical to the B. subtilis GlpQ, has only glycerophosphodiesterase activity but no activity against S. aureus' own RboP WTA or own lipoteichoic acid (10). The GlpQ enzymes belong to the family of glycerophosphodiester phosphodiesterases (EC 3.1.4.46) (16).
We explored here the substrate spectrum of GlpQ toward phosphodiester bonds of purified GroP WTA derived from Staphylococcus lugdunensis, Staphylococcus capitis, and Staphylococcus epidermidis. We demonstrate that GlpQ can be classified as a teichoicase, a WTA-hydrolyzing enzyme. Teichoicase activity was first described in a soil isolate of Acinetobacter spp. (17). Such activities have also been reported before in Bacillus (18) and in some bacteriophages (19–21), but the enzymes have hardly been isolated and never before described in staphylococci. Thus, S. aureus GlpQ and the recently described homologous enzyme from B. subtilis are the first identified bacterial teichoicases important for phosphate supply. GlpQ was induced by phosphate limitation and enabled S. aureus to thrive in medium with limited phosphate content by mobilizing GroP from WTA polymers of CoNS.
Results
GlpQ cleaves GroP-type but not RboP-type WTA
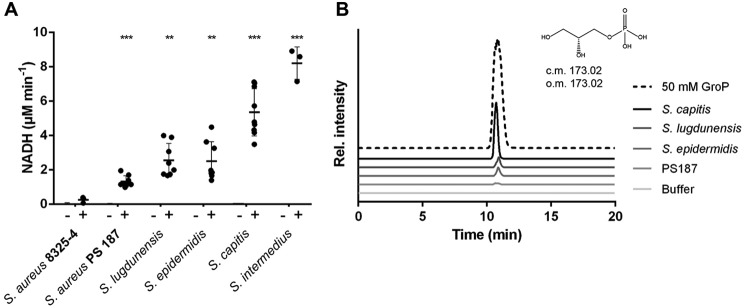
We showed previously that S. aureus GlpQ has glycerophosphodiesterase activity and cleaves phospholipid headgroups to produce GroP, which can be utilized by S. aureus. Although the homologous enzyme of B. subtilis can also cleave its own GroP WTA polymer, such an activity was not observed for the S. aureus RboP WTA. To analyze whether the S. aureus GlpQ may be able to hydrolyze the GroP WTA polymers of co-colonizing CoNS, GroP WTA was isolated from S. lugdunensis, S. epidermidis, S. capitis, Staphylococcus intermedius, and the unusual S. aureus PS187 strain producing GroP WTA (22). GroP WTA was then incubated with purified recombinant GlpQ (expressed in Escherichia coli without signal peptide and fused to the C terminus of a maltose-binding protein (MBP) (10). The release of GroP was monitored via a coupled NADH detection assay, and the identity of reaction products was confirmed by MS analysis (Fig. 1). GlpQ was indeed active toward WTA from all tested GroP WTA types, confirming that it has a broad substrate range for glycerophosphodiesters, including GroP WTA. S. intermedius and S. capitis WTAs were the best substrates, whereas WTA from S. aureus PS187, S. lugdunensis, and S. epidermidis were less efficiently degraded. In contrast, RboP WTA from S. aureus 8325-4 was not hydrolyzed (Fig. 1), which is in agreement with our previous data (10) and demonstrates that GlpQ cannot degrade ribitolphosphodiesters.
Figure 1.
S. aureus GlpQ cleaves glycerol-type WTA derived from S. aureus PS187, S. lugdunensis, S. epidermidis, S. capitis, and S. intermedius. A, glycerophosphodiesterase activity was measured at pH 9, with 2.5 mm isolated WTA from different staphylococcal species in the presence of 5 mm CaCl2 and 5 μg of MBP-GlpQ (black bars, +) or MBP alone as a negative control (gray bars, −). Error bars represent standard deviations of at least three independent assays done in triplicate. Significant differences between S. aureus 8325-4– and GroP WTA–type strains were calculated with Student's t test. **, p < 0.01; ***, p < 0.001. B, extracted ion chromatograms of WTAs before (Buffer) and after treatment with MBP-GlpQ; the GroP standard was analyzed by LC/MS. Diagrams show calculated (c.m.) and observed masses (o.m.) of GroP. The chromatograms of all WTA samples without incubation with MBP-GlpQ (Buffer) overlap.
MS analysis showed that monomeric GroP is the only product of the reaction, as no GroP dimers, trimers, or other polymers were found (Tables S1 and S2). Moreover, no GroP with d-alanine or sugar substitutions was detected, suggesting that GlpQ preferentially hydrolyzes unsubstituted WTA repeating units. Quantification of the released GroP by MS supported the results from the NADH activity assay and confirmed that S. capitis WTA is more readily degraded by GlpQ than S. lugdunensis or S. epidermidis WTA (Table 1).
Table 1.
MS quantification of GroP released from WTA or from cell walls, extracted from different Staphylococcus species after incubation with GlpQ
| GlpQ substrate | S. lugdunensis N920243 | S. epidermidis RP62A | S. capitis ATCC 27840 | S. aureus PS187 | S. aureus PS187 GN1 |
|---|---|---|---|---|---|
| WTA (mm GroP) | 0.80 | 0.64 | 6.82 | 0.60 | 19.55 |
| Cell wall (nmol GroP/ mg cell wall) | 87.52 | 99.80 | 6573.07 | 60.82 | 12631.02 |
GlpQ releases GroP from cell wall–bound WTA
To investigate whether WTA that is still attached to the bacterial cell wall can be directly degraded by GlpQ, cell wall samples were extracted from GroP-WTA–producing bacteria without exposure to harsh acidic conditions to keep WTA attached to peptidoglycan. The insoluble cell walls were incubated with recombinant GlpQ linked to MBP, and the release of soluble organic phosphate to the supernatant was determined and compared with the negative control using MBP protein alone or buffer (Fig. 2). MBP-GlpQ, but not MBP, released phosphate from the cell walls of S. lugdunensis, S. epidermidis, and S. capitis, demonstrating that GlpQ is not only active against released but also against cell wall–attached GroP WTA polymers. The fact that S. capitis WTA did not lead to a pronounced release of phosphate, as observed in the NADH-coupled activity assay where GroP release is measured, might be related to a much lower amount of phosphate in the cell wall of S. capitis, which has been described previously (23).
Figure 2.
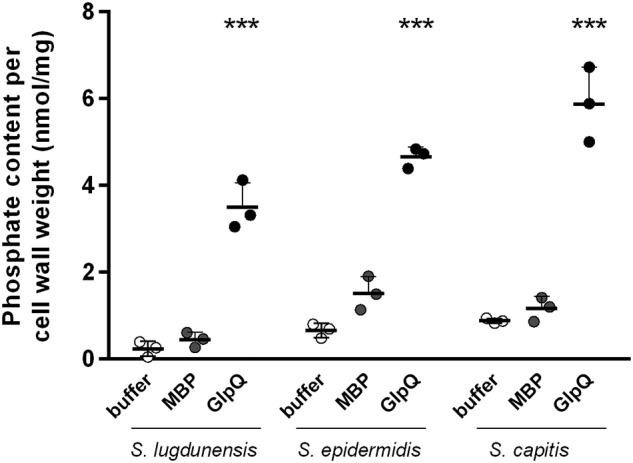
GlpQ is active on cell wall–attached WTA. Shown is phosphate released to the supernatant after incubation of cell wall extracts of S. lugdunensis, S. epidermidis, and S. capitis with recombinant MBP-GlpQ, negative control MBP, or only buffer. Error bars represent standard deviations of at least three independent assays done in triplicate. Significant differences between MBP control and GlpQ treated samples were calculated with Student's t test. ***, p < 0.001.
Cell wall–derived soluble products were also analyzed by MS (Table 1) and, similar to what was observed for the degradation of released WTA, only monomeric GroP but no dimers, polymers, or modified GroP units could be identified. Quantification of the amount of GroP revealed a higher release of GroP from cell walls of S. capitis compared with S. lugdunensis and S. epidermidis, which is in accordance to what was observed for released soluble WTA molecules (Table 1).
GlpQ prefers nonglycosylated WTA as substrate
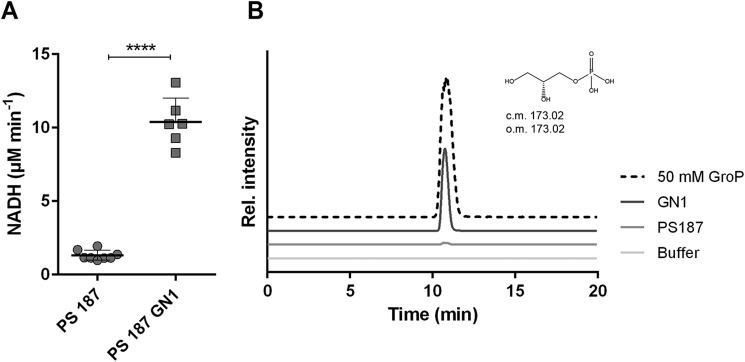
Because only unsubstituted GroP residues were detected after incubation of GlpQ with GroP WTA, we hypothesized that the different GlpQ activities observed for the tested WTA samples may be correlated to the degree of substitution of the WTA backbones with sugar residues. WTAs are usually modified with a variety of hexoses, such as GlcNAc, GalNAc, or glucose, in a species- and strain-specific manner (14, 23, 24). Therefore, differences in glycosylation could explain the different activities of GlpQ for the different CoNS WTA types (Table 2). To verify whether sugar modification indeed affects GlpQ activity, we compared the degradation of GroP WTA from the model strain S. aureus PS187, which is substituted with GalNAc, and from the isogenic mutant GN1 lacking glycosylation (22). Remarkably, the GlpQ activity for purified WTA isolated from strain GN1 showed a drastic increase in the NADH-based (Fig. 3A) and MS-based enzyme assays compared with WTA from the WT (Fig. 3B and Table 1). Similarly, the cell wall from strain GN1 could be more effectively degraded by GlpQ and released more GroP than cell walls of the parental strain PS187 (Table 1). Hence, our data indicate that WTA glycosylation has a strong inhibitory influence on GlpQ activity in degrading GroP-WTA.
Table 2.
Reported WTA architecture of the investigated strains
| Species | Strain | Polyol backbone | Reported sugar modifications and anomeric configurationa | References | GlpQ activityb |
|---|---|---|---|---|---|
| S. lugdunensis | N920243 | GroP | NA | – | ++ |
| S. epidermidis | RP62A | GroP | α-Glc, α-GlcNAc, d-Ala and α-Glc6Ala | 36 | ++ |
| S. intermedius | CCM5739 | GroP | GlcNAc | 24 | +++ |
| S. capitis | ATCC 27840 | GroP | α-GlcNAc | 24 | +++ |
| S. aureus | PS187 | GroP | α-GalNAc | 29 | + |
| S. aureus | PS187 GN1 | GroP | NA | 22 | ++++ |
| S. aureus | 8325-4 | RboP | GlcNAc | 37 | − |
| S. aureus | RN4220 | RboP | α, ß-GlcNAc | 25, 38 | − |
| S. aureus | RN4220 ΔtarMΔtarS | RboP | NA | 25, 29 | − |
a Anomeric configurations are shown only for available data; NA, data not available.
b GlpQ activity refers to the initial kinetic experiment and the formation of NADH in μm min−1. −, <0.8; +, >1; ++, >5; +++, >7.5; ++++, >10 μm min−1.
Figure 3.
Influence of WTA sugar modifications on GlpQ activity. A, 2.5 mm WTA was incubated with 5 μg of recombinant MBP-GlpQ in the presence of 5 mm CaCl2. Means and standard deviations from at least three independent experiments done in triplicates are shown. Significant differences between PS187 WT and PS187 GN1 WTA were calculated with Student's t test. ****, p < 0.0001. B, extracted ion chromatograms of WTA from S. aureus PS187 and nonglycosylated mutant GN1 before (Buffer, light gray) and after treatment with MBP-GlpQ, analyzed by LC/MS. The diagrams show calculated (c.m.) and observed masses (o.m.) of GroP. Data were plotted relative to the intensity of the GroP peak of S. aureus GN1 to allow comparison.
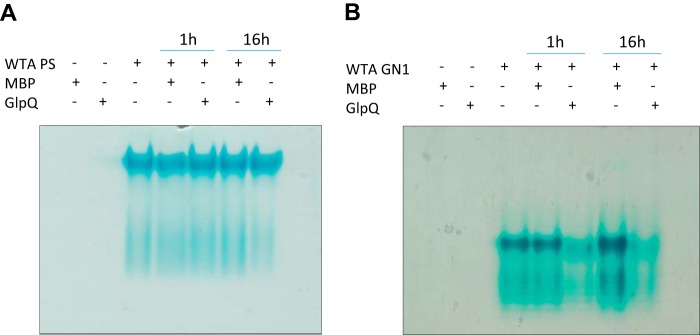
To further analyze to which extent WTA is degraded by GlpQ, we analyzed the end products of WTA-GlpQ reactions by PAGE. Degradation was visible only for the nonglycosylated GN1 strain, where the amount of WTA was clearly reduced (Fig. 4), corroborating the previous findings that GlpQ activity is higher for nonglycosylated WTA, whereas the presence of GalNAc modifications in PS187 might inhibit cleavage of the full chain, allowing only the release of probably unmodified GroP from the distal ends.
Figure 4.
Activity of GlpQ visualized by WTA-PAGE. A and B, 1 mm WTA from parental S. aureus PS187 WT (PS, A) or nonglycosylated GN1 mutant strain (B) was incubated for 1 h or 16 h at room temperature with purified GlpQ or with MBP as a negative control in the presence of 5 mm CaCl2.
To ascertain that lack of GlpQ activity toward RboP-type WTA was not due to the glycosylation of WTA, we used S. aureus strains lacking the enzymes TarM and TarS, responsible for GlcNAc modification in this strain (25, 26). Importantly, unglycosylated RboP WTA from the S. aureus RN4220ΔtarMΔ tarS double mutant strain did not serve as a substrate for GlpQ (Fig. S1), confirming that RboP WTA is not a substrate for GlpQ.
S. aureus profits from WTA as phosphate source under phosphate limitation
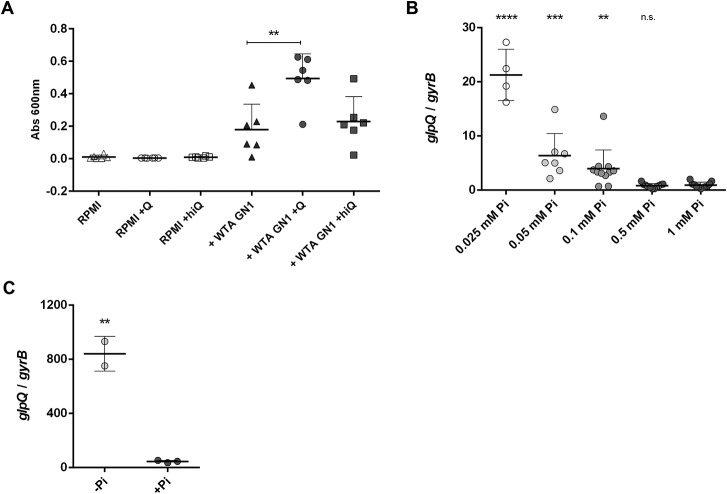
Although B. subtilis GlpQ is known to recycle WTA phosphate from the producer's own cell wall, we hypothesized that S. aureus GlpQ might also help to cope with phosphate limitation by using the GroP WTA of competing CoNS as a substrate instead of its own RboP WTA. We tested this hypothesis by analyzing the growth of a glpQ mutant strain (S. aureus USA300 ΔglpQ) in phosphate-deficient synthetic medium (RPMI-Pi). We observed no growth either with or without GlpQ, as expected. However, when nonglycosylated GroP-WTA from strain PS187 GN1 was added, it restored growth in the presence of GlpQ (Fig. 5A). Heat-inactivated GlpQ was not able to improve growth under such conditions (Fig. 5A), confirming that GlpQ activity is responsible for the observed growth improvement when GroP WTA is the only source of phosphate. The slight growth observed in the absence of GlpQ could be justified by the presence of other teichoicases. However, we did not detect any additional putative enzymes with our activity assays.
Figure 5.
A, nonglycosylated purified GroP WTA (from strain GN1) restores growth of S. aureus USA300 ΔglpQ under phosphate-limiting conditions. Purified, active GlpQ (+Q), but not heat-inactivated GlpQ (hiQ), improves growth when WTA is the only phosphate source in RPMI-Pi medium. Means represent at least three independent assays done in triplicate. Growth after 6 h is shown. B and C, GlpQ expression is induced under phosphate-limiting conditions. Relative amounts of the glpQ transcript were obtained from RNA extracted from cultures grown for 20 h in RPMI-Pi supplemented with different phosphate concentrations (B) or after a 30-min shock without phosphate (C). Values are given as means and standard deviations (n ≥ 3). Statistically significant differences, calculated using an unpaired two-tailed Student's t test, are indicated as follows: n.s., not significant, p > 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
GlpQ expression is induced under phosphate-limiting conditions
The observation that growth under phosphate-limiting conditions could be rescued by WTA and that GlpQ permitted the utilization of WTA suggests that GlpQ could be a component of the phosphate limitation stress response of S. aureus. How S. aureus copes with phosphate limitation has hardly been studied in the past. To determine whether glpQ expression was induced by phosphate-limiting conditions, we quantified the amount of glpQ transcript by RT-PCR from S. aureus USA300 cultures grown in RPMI-Pi supplemented with different phosphate concentrations. glpQ expression levels were substantially higher at low (0.025 mm) phosphate than for high (1 mm) Pi, indicating that GlpQ is a component of the Pi stress response of S. aureus (Fig. 5B). In a different approach, a 30-min incubation in Pi-free medium was enough to strongly induce the expression of glpQ (Fig. 5C).
Discussion
In this study, we characterized the capacity of the secreted GlpQ enzyme from S. aureus to release GroP from GroP polymers in addition to glycerophospholipid headgroups. We show that GlpQ hydrolyzes GroP WTA from a variety of staphylococcal strains that co-inhabit the human nares with S. aureus, including the commensal S. capitis and the opportunistic pathogens S. lugdunensis, S. epidermidis, and S. intermedius (Table 2). Although all tested WTA molecules from different CoNS species were found to be substrates of GlpQ, the amounts of GroP monomers released from WTA varied between the sources of WTA. Enzymatic activity assays revealed unmodified WTA as the most potent substrate for the enzyme, suggesting that differences in the degree and pattern of WTA glycosylation between the various CoNS species may lead to different susceptibility to GlpQ degradation.
MS analysis shed light on the cleavage mechanism of GlpQ by identifying GroP as the only reaction product, whereas oligomers of GroP or substituted monomers could not be detected as products of GlpQ activity on WTA. Consequently, the enzyme most likely follows an exolytic cleavage mechanism that may be inhibited by modified GroP monomers, similar to the B. subtilis GlpQ homolog (15). Importantly, GlpQ could also act directly on crude cell wall extracts, which mimic natural substrates, demonstrating the ability of GlpQ to directly cleave WTA attached to the peptidoglycan layer. This finding emphasizes the classification of GlpQ as a GroP teichoicase.
B. subtilis uses the closely related GlpQ enzyme during Pi starvation to recycle its phosphate-containing GroP WTA, which is at the same time replaced by the phosphate-free polymer teichuronic acid (27). Staphylococci lack teichuronic acid biosynthetic genes and cannot switch to production of a phosphate-free polymer as a phosphate starvation response. Instead, S. aureus appears to use GlpQ to “steal” phosphate from GroP WTA of CoNS colonizing the same habitats. This idea is supported by our observation that GroP WTA could restore growth of S. aureus when phosphate was limiting and that glpQ expression levels were induced by phosphate-limiting conditions. Indeed, the concentrations of phosphate vary strongly in different parts of the human body, being as low as 0.057 ± 0.03 mm in sweat (28), 0.38 ± 0.03 mm in human blood (6), 28 ± 36 mm in saliva (7), or between 0.75 and 7 mm in nasal secretions (8, 9). In certain environments S. aureus may depend on GlpQ to ensure sufficient availability of phosphate for robust growth. We suggest here that S. aureus secretes GlpQ to thrive in phosphate-poor habitats encountered on the host, where it can profit from the GroP WTA co-habitating bacteria.
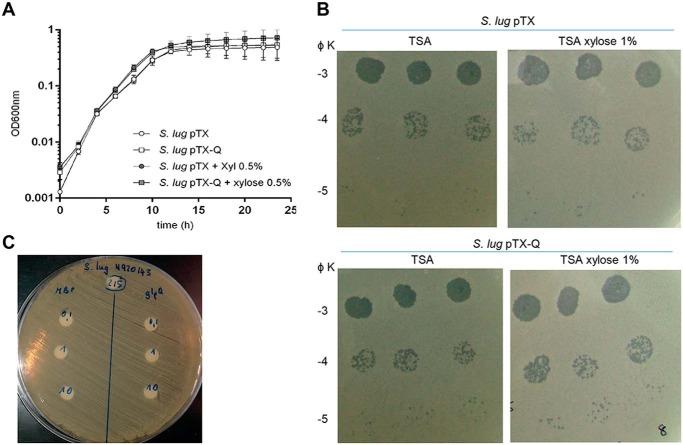
The fact that GlpQ seems to be unable to completely degrade sugar-modified WTA suggests that GlpQ may only trim CoNS WTA without causing complete WTA loss or impairment of cell wall integrity. This idea is supported by the fact that CoNS growth or phage absorption was never found to be affected by treatment of CoNS cells with S. aureus GlpQ or by the heterologous overexpression of GlpQ in CoNS (Fig. 6). Along this line, S. aureus PS187 from the rare clonal complex CC395 with GroP WTA also secretes active GlpQ without notable inhibitory effects on growth or phage susceptibility (Fig. S2).
Figure 6.
A and B, heterologous expression of S. aureus GlpQ in S. lugdunensis does not affect S. lugdunensis (S. lug) growth (A) or susceptibility to phage K (B). S. lugdunensis harboring a xylose-inducible glpQ expression plasmid (pTX-Q) or empty plasmid (pTX), was grown in MHB (A) or on TSA (B) with or without xylose. Serial dilutions of phage K (10−3 to 10−5) were spotted in triplicate on S. lugdunensis lawns (B). C, treatment of S. lugdunensis with recombinant GlpQ is not growth-inhibitory. S. lugdunensis grown on TSA was treated with 0.1, 1, or 10 μg of recombinant purified MBP (negative control) or GlpQ.
It remains unclear why S. aureus evolved a specific RboP-type WTA that distinguishes it from most CoNS. Although our study suggests that it might be a mechanism of immunity against its own GlpQ, RboP WTA could also be an evolutionary strategy to govern attachment of specific transducing phages with an advantage for horizontal gene transfer (29). Additionally, it could shape the interaction with epithelial binding partners such as the EGF-domain containing scavenger receptor SR-F1 (SREC-I) (30). The existence of teichoicases such as GlpQ has long been suggested but difficult to prove. Here we provide evidence for the existence of such an enzyme in S. aureus specific for GroP WTA, similar to the one found in B. subtilis but with a somewhat different function. Our findings that S. aureus GlpQ cannot cleave its own RboP WTA and that most CoNS do not secrete active GlpQ-like enzymes (10) suggests that staphylococci may have different, currently unknown WTA recycling and Pi mobilization strategies. Nevertheless, even bacteria targeted by the S. aureus GlpQ may profit from the release of phosphate from GroP WTA.
The phosphate starvation response in S. aureus has not been analyzed in detail. Studies are limited to the knowledge that phosphate is essential that and its limitation does not lead to a starvation survival state as in glucose limitation (31). Also, so far, there are no studies evaluating the transcriptome or proteome of S. aureus from phosphate-starved cultures. Such studies could unravel the existence of a teichoicase involved in the utilization of its own RboP and/or of other mechanisms of phosphate mobilization. S. aureus might be constantly exposed to phosphate-limiting conditions in specific colonization sites, such as skin-related habitats. Furthermore, the available form of phosphate is pH-dependent. In human body fluids, phosphate is present mostly as HPO42− (32). However, the preferred phosphate form transported by bacteria, the orthophosphate anion PO43−, is only present in trace amounts. As a result, phosphate limitation is likely to occur frequently in natural S. aureus habitats where S. aureus may compete with other commensals for phosphate. Recently, an S. aureus transporter for acquisition of phosphate has been described (33). In contrast to many other staphylococcal species, S. aureus has three dedicated transporters that enable the provision of phosphate under a multitude of distinct conditions, such as pH changes or phosphate limitation, thereby maintaining phosphate homeostasis. Moreover, loss of the transporter NptA in conjunction with a second transporter reduces the ability of S. aureus to cause infection. This study together with our findings demonstrates for the first time how phosphate homeostasis is of particular importance for S. aureus to thrive in the competitive host-associated microbiomes. Phosphate-sensing systems have been described to be involved in virulence in other bacteria and in cross-talk with other nutrition-regulatory pathways (34). Our findings shed light on the phosphate starvation response of S. aureus and expand the knowledge of a barely analyzed although crucial system that might be of importance as a therapeutic target. Moreover, our study reveals a sophisticated strategy used by S. aureus to increase its fitness under challenging conditions at the expense of competing microbiome members.
Experimental procedures
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S3. E. coli was grown in Luria broth (LB) (1% tryptone, 0.5% yeast extract, and 0.5% NaCl) supplemented with 100 μg ml−1 ampicillin and with 0.2% glucose for overexpression of MBP-GlpQ. S. aureus was grown in Müller–Hinton broth (MHB, Carl Roth; 0.2% beef extract, 1.75% acid hydrolysate of casein, and 0.15% starch) or in basic medium (BM) (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, and 0.1% glucose) at 37 °C with shaking at 160 rpm. For phosphate-limiting growth conditions, RPMI medium without phosphate (RPMI-Pi) (c-c-pro, Oberdorla, Germany) was used. For growth curves, bacteria from overnight cultures grown in MHB were centrifuged, washed four times with RPMI-Pi, reinoculated in fresh medium to an initial A600 nm of 0.05 in microtiter plates, and grown in an Epoch2 BIOTEK reader with orbital shaking (180 rpm) at 37 °C with measurement of optical densities every 30 min.
Purification of MBP-GlpQ
Heterologous expression of MBP-GlpQ was accomplished as described recently (10). Briefly, E. coli MT56 transformed with pMAL-glpQ was grown in LB with 0.2% glucose to an A600 nm of 0.5. Expression of MBP-GlpQ was induced with 0.3 mm isopropylthiogalactoside, and cells were further grown for 16 h at 30 °C. Cells were lysed with 0.4 mg/ml lysozyme and by sonication. The lysate was purified by affinity chromatography using amylose resin (Qiagen) and 20 mm Tris-HCl (pH 7.6) and 200 mm NaCl as washing buffer. MBP-GlpQ was eluted with the same buffer but using 20 mm NaCl and 10 mm maltose. The purity of the protein was confirmed by SDS-PAGE, and protein concentration was measured using the Bradford protein assay (Bio-Rad) using BSA as the standard.
Extraction of WTA and cell wall
Extraction of WTA from S. aureus cells was performed as described previously (22). In brief, 2 liters of overnight culture in BM were harvested and washed with 20 mm ammonium acetate buffer (pH 4.8). Lysis was performed mechanically using a cell mill (Euler). Samples were incubated with DNase and RNase overnight at 37 °C. SDS was added to 2%, and samples were treated with sonication (output 30%, cycle 50%, 15 min) and subsequent incubation at 65 °C for 1 h. SDS was removed by extensive washing with ammonium acetate buffer, and samples were incubated with 5% TCA for 4 h at 60 °C to remove WTA from peptidoglycan before finally dialyzing against bidistilled water over 2 days at 4 °C with a membrane of 3.5 kDa molecular weight cut-off (MWCO). Samples were quantified by their phosphate amount as described previously (22), concentrated to 50 mm Pi, and stored at −20 °C. Cell wall samples were spared from TCA treatment, dialyzed against water, and quantified by their dry weight.
Enzyme activity assay
Enzyme activity was performed essentially as described previously (10) by measurement of GroP formation in a spectrometric assay coupled to an NAD-dependent enzymatic reaction with glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Sigma). The assay mixture was performed in a 96-well plate in a final volume of 50 μl. The reaction buffer contained 0.9 m glycine in 1 m hydrazine (pH 9), 5 mm CaCl2, 0.5 m NAD+, and 10 units/ml GAPDH. 2.5 mm extracted WTA sample was incubated with 5 μg of MBP-GlpQ or MBP alone. The kinetics of the reaction were measured until oxidation of Gro-3P by GAPDH was complete, and the Gro-3P concentration was calculated from the absorbance change at 340 nm using an NADH standard curve. The kinetics data were analyzed using Prism 6 software (GraphPad).
Activity of recombinant MBP-GlpQ was evaluated on cell wall extracts washed previously with 20 mm Tris-HCl (pH 7.8) and 20 mm NaCl. An identical sample was dried in a SpeedVac to determine its dry weight. Cell wall extracts were incubated in the presence of 5 mm CaCl2 and 10 μg MBP-GlpQ or MBP alone for 24 h at 37 °C and 700 rpm shaking. After centrifugation, the supernatant was analyzed for phosphate content using the Biomol GREEN kit according to the manufacturer's instructions. As a positive control, mutanolysin (Sigma-Aldrich), an N-acetylmuramidase that solubilizes cell walls by specifically cleaving the β-N-acetylmuramyl-(1→4)–GlcNAc linkage of the peptidoglycan backbone, was used to release fragments of peptidoglycan with WTA attached. The mutanolysin-treated cell wall led to a strong release of soluble phosphate (Fig. S3).
End point enzyme activity was analyzed via LC/MS as described below. Reaction mixtures contained 2.5 mm WTA sample in 20 mm Tris-HCl (pH 8) with 5 mm CaCl2 and 5 μg MBP-GlpQ or MBP. The samples were incubated overnight at 37 °C and analyzed for GroP formation. Supernatants from GlpQ-treated cell wall extracts prepared as described above were also analyzed by LC/MS.
LC/MS analysis of GlpQ substrates
Identification of products formed in the reaction of GlpQ with WTA and cell walls was performed using an Ultimate 3000 HPLC system (Dionex) coupled to a MicrOTOF II detector (Bruker). For HPLC, a Gemini C18 column (150 × 4.6 mm, 110 Å, 5 μm, Phenomenex) was used at 37 °C with a flow rate of 0.2 ml/min. A 5-min washing step with 96% buffer A (0.1% formic acid and 0.05% ammonium formate) was applied, followed by a linear gradient of 0% to 40% acetonitrile in buffer A for 30 min. A final washing step with 40% buffer B for 5 min and a re-equilibration step (100% buffer A) for 5 min completed the method. Samples were ionized via electron spray ionization in positive ion mode. Exact masses in positive ion mode for GroP (m/z+1 173.01) and RboP (m/z+1 153.07) were presented as extracted ion chromatograms with Data Analysis (Bruker). Peak areas were extracted and analyzed, and GroP was quantified using serial dilutions of a GroP standard.
Quantitative real-time qPCR
1 ml of a 16-h culture of S. aureus USA300 grown in MHB was washed four times with 1 ml of RPMI-Pi, reinoculated in 3 to 5 ml of fresh RPMI-Pi to an initial A600 nm of 0.02, and grown for 20 h at 37 °C at 160 rpm. For a 30-min phosphate stress condition, a 16-h culture of S. aureus USA300 grown in MHB was reinoculated in MHB and grown at 37 °C and 160 rpm to an A600 nm of 1. The cells were then washed four times with RPMI-Pi, resuspended in the initial amount of RPMI-Pi, and grown for 30 min at 37 °C and 160 rpm.
Prior to harvesting, RNAprotect bacterial reagent (twice the culture volume, Qiagen) was added to the culture, and the mixture was immediately vortexed for 10 s. The cells were harvested, and the pellet was stored at −80 °C overnight. The next day, the cells were resuspended with 1 ml of TRIzol (Ambion, Life Technologies), followed by mechanical disruption with 0.5 ml of zirconia–silica beads (Carl Roth, Karlsruhe, Germany; 0.1 mm in diameter) using a FastPrep 24 homogenizer (MP Biomedicals) (1 cycle; 20 s; speed, 6.5 m/s). RNA was extracted with 200 μl of chloroform, recovered by precipitation with isopropyl alcohol, washed with 80% ethanol, dried under a vacuum, and resuspended in 30 to 50 μl of bidistilled water. The unlikely presence of DNA was measured with a Qubit 3.0 Fluorometer (Invitrogen). RNA was quantified using a Nanodrop ND-100 spectrophotometer, and 10 μg of RNA was digested with DNase I using the TURBO DNA-freeTM kit (Thermo Fisher Scientific) and stored at −80 °C.
RNA (20 ng) was transcribed into complementary DNA and used directly for real-time qPCR using the Power SYBR® Green RNA-to-CTTM 1-Step Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Real-time qPCR was performed in a QuantStudio 3 System (Thermo Fisher Scientific) using the following conditions: 30 min at 48 °C, 10 min at 95 °C and 40 cycles of 15 s at 95 °C, and 1 min at 60 °C. Quantification of glpQ expression was achieved using a standard curve method for relative quantification. gyrB was used as the endogenous control. The primers used for each gene were as follows: glpQ_P13 (5′-gactatgactaactcttcgaaaag-3′) and glpQ_P14 (5′-gaatagcctgaggtttatttgca-3′); gyr297_F (5′-ttagtgtgggaaattgtcgataat-3′) and gyr_P1_R (5′-tccgttactttaatccagttatc-3′). The specificity of the amplified products was verified by analysis of the dissociation curves generated by the QuantStudioTM Design & Analysis software based on the specific melting temperature for each amplicon.
Phage and GlpQ susceptibility assays
The phage K susceptibility of S. lugdunensis or PS187 secreting S. aureus GlpQ was tested by transforming the bacterial strains with the pTX-Q plasmid so that expression of heterologous S. aureus GlpQ was controlled by a xylose-inducible promoter. The pTX-Q plasmid was constructed by inserting a PCR fragment of GlpQ (including the Shine-Dalgarno sequence region) and using primer pair 5′-cgcGGATCCgagatgaaaggataaagactatg-3′ and 5′-ctCCCGGGctacttaatgacttctttatatttatcagcg-3′ into the BamHI/SmaI restriction site of the Staphylococcus-specific expression vector pTX15 (35). Phage K susceptibility was tested by growing pTX-Q-transformed (or empty vector pTX15-transformed) strains overnight in BM liquid medium with 1% xylose with aeration at 37 °C. 10 μl of phage K lysate dilutions (10−3, 10−4, and 10−5) was spotted in triplicate on tryptic soy broth agar (TSA) supplemented or not with 0.5% or 1% xylose as indicated. Plates were grown for 16 h at 37 °C.
To test the susceptibility of S. lugdunensis to exogenous S. aureus GlpQ, a stationary phase culture of S. lugdunensis was swabbed on TSA, and paper discs soaked with 0.1, 1, or 10 μg of recombinant purified MBP (negative control) or GlpQ were applied on top of the bacterial lawn. Plates were grown for 16 h at 37 °C.
Statistical methods
Statistical analyses were performed with the Prism 6.0 package (GraphPad Software), and the between-group differences were analyzed for significance with a two-tailed Student's t test. p ≤ 0.05 was considered statistically significant.
Author contributions
A. M. J., G. X., and A. P. conceptualization; A. M. J., S. U., G. X., and A. P. data curation; A. M. J., J. S., C. M., and A. P. formal analysis; A. M. J., G. X., C. M., and A. P. supervision; A. M. J. and A. P. funding acquisition; A. M. J., C. M., and A. P. validation; A. M. J., J. S., and S. U. investigation; A. M. J., J. S., S. U., and C. M. methodology; A. M. J. and J. S. writing-original draft; A. M. J. and A. P. project administration; A. M. J., G. X., C. M., and A. P. writing-review and editing; C. M. resources.
Supplementary Material
Acknowledgments
We thank David Gerlach for helpful discussions and Gabriele Hornig and Madeleine Spahmann for technical support.
This work was financed by grants from the Medical Faculty of the University of Tübingen (F1433261 to A. M. J.) and from the German Research Council (TRR34 and TRR156 to A. P. and SFB766 and GRK1708 to C. M. and A. P.) and from the German Center for Infection Research (to A. J. and A. P.). The authors declare that they have no conflicts of interest with the contents of this article. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
This article contains Figs. S1–S3, Tables S1–S3, and references.
- CoNS
- coagulase-negative staphylococci
- GroP
- glycerol-3-Pi
- WTA
- wall teichoic acid
- RboP
- ribitol-5-Pi
- MBP
- maltose-binding protein
- LB
- Luria broth
- MHB
- Müller–Hinton broth
- BM
- basic medium
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- qPCR
- quantitative PCR
- TSA
- tryptic soy broth agar.
References
- 1. Wertheim H. F., Melles D. C., Vos M. C., van Leeuwen W., van Belkum A., Verbrugh H. A., and Nouwen J. L. (2005) The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762 10.1016/S1473-3099(05)70295-4 [DOI] [PubMed] [Google Scholar]
- 2. Byrd A. L., Belkaid Y., and Segre J. A. (2018) The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155 10.1038/nrmicro.2017.157 [DOI] [PubMed] [Google Scholar]
- 3. Zipperer A., Konnerth M. C., Laux C., Berscheid A., Janek D., Weidenmaier C., Burian M., Schilling N. A., Slavetinsky C., Marschal M., Willmann M., Kalbacher H., Schittek B., Brötz-Oesterhelt H., Grond S., et al. (2016) Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 10.1038/nature18634 [DOI] [PubMed] [Google Scholar]
- 4. Krismer B., Weidenmaier C., Zipperer A., and Peschel A. (2017) The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 15, 675–687 10.1038/nrmicro.2017.104 [DOI] [PubMed] [Google Scholar]
- 5. Krismer B., Liebeke M., Janek D., Nega M., Rautenberg M., Hornig G., Unger C., Weidenmaier C., Lalk M., and Peschel A. (2014) Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 10, e1003862 10.1371/journal.ppat.1003862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Portale A. A., Halloran B. P., and Morris R. C. Jr. (1987) Dietary intake of phosphorus modulates the circadian rhythm in serum concentration of phosphorus: implications for the renal production of 1,25-dihydroxyvitamin D. J. Clin. Invest. 80, 1147–1154 10.1172/JCI113172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park Y. D., Jang J. H., Oh Y. J., and Kwon H. J. (2014) Analyses of organic acids and inorganic anions and their relationship in human saliva before and after glucose intake. Arch. Oral. Biol. 59, 1–11 10.1016/j.archoralbio.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 8. Lorin M. I., Gaerlan P. F., and Mandel I. D. (1972) Quantitative composition of nasal secretions in normal subjects. J. Lab. Clin. Med. 80, 275–281 [PubMed] [Google Scholar]
- 9. Burke W. (2014) The ionic composition of nasal fluid and its function. Health 6, 720–728 10.4236/health.2014.68093 [DOI] [Google Scholar]
- 10. Jorge A. M., Schneider J., Unsleber S., Göhring N., Mayer C., and Peschel A. (2017) Utilization of glycerophosphodiesters by Staphylococcus aureus. Mol. Microbiol. 103, 229–241 10.1111/mmi.13552 [DOI] [PubMed] [Google Scholar]
- 11. Kolata J., Bode L. G., Holtfreter S., Steil L., Kusch H., Holtfreter B., Albrecht D., Hecker M., Engelmann S., van Belkum A., Völker U., and Bröker B. M. (2011) Distinctive patterns in the human antibody response to Staphylococcus aureus bacteremia in carriers and non-carriers. Proteomics 11, 3914–3927 10.1002/pmic.201000760 [DOI] [PubMed] [Google Scholar]
- 12. Stentzel S., Sundaramoorthy N., Michalik S., Nordengrün M., Schulz S., Kolata J., Kloppot P., Engelmann S., Steil L., Hecker M., Schmidt F., Völker U., Roghmann M. C., and Bröker B. M. (2015) Specific serum IgG at diagnosis of Staphylococcus aureus bloodstream invasion is correlated with disease progression. J. Proteomics 128, 1–7 10.1016/j.jprot.2015.06.018 [DOI] [PubMed] [Google Scholar]
- 13. Brown S., Santa Maria J. P. Jr., and Walker S. (2013) Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336 10.1146/annurev-micro-092412-155620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winstel V., Xia G., and Peschel A. (2014) Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int. J. Med. Microbiol. 304, 215–221 10.1016/j.ijmm.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 15. Myers C. L., Li F. K., Koo B. M., El-Halfawy O. M., French S., Gross C. A., Strynadka N. C., and Brown E. D. (2016) Identification of two phosphate starvation-induced wall teichoic acid hydrolases provides first insights into the degradative pathway of a key bacterial cell wall component. J. Biol. Chem. 291, 26066–26082 10.1074/jbc.M116.760447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Corda D., Mosca M. G., Ohshima N., Grauso L., Yanaka N., and Mariggiò S. (2014) The emerging physiological roles of the glycerophosphodiesterase family. FEBS J. 281, 998–1016 10.1111/febs.12699 [DOI] [PubMed] [Google Scholar]
- 17. Wise E. M. Jr, Glickman R. S., and Teimer E. (1972) Teichoic acid hydrolase activity in soil bacteria (Bacillus subtilis-sporulation-phosphodiesterase-polyamines-concanavalin A). Proc. Natl. Acad. Sci. U.S.A. 69, 233–237 10.1073/pnas.69.1.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kusser W., and Fiedler F. (1984) A novel glycerophosphodiesterase from Bacillus pumilus. FEBS Lett. 166, 301–306 10.1016/0014-5793(84)80100-3 [DOI] [PubMed] [Google Scholar]
- 19. Cornelissen A., Sadovskaya I., Vinogradov E., Blangy S., Spinelli S., Casey E., Mahony J., Noben J. P., Dal Bello F., Cambillau C., and van Sinderen D. (2016) The baseplate of Lactobacillus delbrueckii bacteriophage Ld17 harbors a glycerophosphodiesterase. J. Biol. Chem. 291, 16816–16827 10.1074/jbc.M116.728279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Myers C. L., Ireland R. G., Garrett T. A., and Brown E. D. (2015) Characterization of wall teichoic acid degradation by the bacteriophage varphi29 appendage protein GP12 using synthetic substrate analogs. J. Biol. Chem. 290, 19133–19145 10.1074/jbc.M115.662866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiang Y., Leiman P. G., Li L., Grimes S., Anderson D. L., and Rossmann M. G. (2009) Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol. Cell 34, 375–386 10.1016/j.molcel.2009.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Winstel V., Sanchez-Carballo P., Holst O., Xia G., and Peschel A. (2014) Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. mBio 5, e00869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Endl J., Seidl H. P., Fiedler F., and Schleifer K. H. (1983) Chemical composition and structure of cell wall teichoic acids of staphylococci. Arch. Microbiol. 135, 215–223 10.1007/BF00414483 [DOI] [PubMed] [Google Scholar]
- 24. Endl J., Seidl P. H., Fiedler F., and Schleifer K. H. (1984) Determination of cell wall teichoic acid structure of staphylococci by rapid chemical and serological screening methods. Arch. Microbiol. 137, 272–280 10.1007/BF00414557 [DOI] [PubMed] [Google Scholar]
- 25. Brown S., Xia G., Luhachack L. G., Campbell J., Meredith T. C., Chen C., Winstel V., Gekeler C., Irazoqui J. E., Peschel A., and Walker S. (2012) Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. U.S.A. 109, 18909–18914 10.1073/pnas.1209126109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Winstel V., Kühner P., Salomon F., Larsen J., Skov R., Hoffmann W., Peschel A., and Weidenmaier C. (2015) Wall teichoic acid glycosylation governs Staphylococcus aureus nasal colonization. mBio 6, e00632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grant W. D. (1979) Cell wall teichoic acid as a reserve phosphate source in Bacillus subtilis. J. Bacteriol. 137, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prompt C. A., Quinton P. M., and Kleeman C. R. (1978) High concentration of sweat calcium, magnesium and phosphate in chronic renal failure. Nephron 20, 4–9 10.1159/000181189 [DOI] [PubMed] [Google Scholar]
- 29. Winstel V., Liang C., Sanchez-Carballo P., Steglich M., Munar M., Bröker B. M., Penadés J. R., Nübel U., Holst O., Dandekar T., Peschel A., and Xia G. (2013) Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat. Commun. 4, 2345 10.1038/ncomms3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schade J., and Weidenmaier C. (2016) Cell wall glycopolymers of Firmicutes and their role as nonprotein adhesins. FEBS Lett. 590, 3758–3771 10.1002/1873-3468.12288 [DOI] [PubMed] [Google Scholar]
- 31. Watson S. P., Clements M. O., and Foster S. J. (1998) Characterization of the starvation-survival response of Staphylococcus aureus. J. Bacteriol. 180, 1750–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Santos-Beneit F. (2015) The Pho regulon: a huge regulatory network in bacteria. Front. Microbiol. 6, 402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelliher J. L., Radin J. N., Grim K. P., Párraga Solórzano P. K., Degnan P. H., and Kehl-Fie T. E. (2018) Acquisition of the phosphate transporter NptA enhances Staphylococcus aureus pathogenesis by improving phosphate uptake in divergent environments. Infect. Immun. 86, e00631–17 10.1128/IAI.00631-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lamarche M. G., Wanner B. L., Crépin S., and Harel J. (2008) The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32, 461–473 10.1111/j.1574-6976.2008.00101.x [DOI] [PubMed] [Google Scholar]
- 35. Peschel A., Ottenwälder B., and Götz F. (1996) Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 137, 279–284 10.1111/j.1574-6968.1996.tb08119.x [DOI] [PubMed] [Google Scholar]
- 36. Sadovskaya I., Vinogradov E., Li J., and Jabbouri S. (2004) Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus epidermidis RP62A, a reference biofilm-positive strain. Carbohydr. Res. 339, 1467–1473 10.1016/j.carres.2004.03.017 [DOI] [PubMed] [Google Scholar]
- 37. Jenni R., and Berger-Bächi B. (1998) Teichoic acid content in different lineages of Staphylococcus aureus NCTC8325. Arch. Microbiol. 170, 171–178 10.1007/s002030050630 [DOI] [PubMed] [Google Scholar]
- 38. Xia G., Maier L., Sanchez-Carballo P., Li M., Otto M., Holst O., and Peschel A. (2010) Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J. Biol. Chem. 285, 13405–13415 10.1074/jbc.M109.096172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.