Abstract
Analogous to the c-Myc (Myc)/Max family of bHLH-ZIP transcription factors, there exists a parallel regulatory network of structurally and functionally related proteins with Myc-like functions. Two related Myc-like paralogs, termed MondoA and MondoB/carbohydrate response element–binding protein (ChREBP), up-regulate gene expression in heterodimeric association with the bHLH-ZIP Max-like factor Mlx. Myc is necessary to support liver cancer growth, but not for normal hepatocyte proliferation. Here, we investigated ChREBP's role in these processes and its relationship to Myc. Unlike Myc loss, ChREBP loss conferred a proliferative disadvantage to normal murine hepatocytes, as did the combined loss of ChREBP and Myc. Moreover, hepatoblastomas (HBs) originating in myc−/−, chrebp−/−, or myc−/−/chrebp−/− backgrounds grew significantly more slowly. Metabolic studies on livers and HBs in all three genetic backgrounds revealed marked differences in oxidative phosphorylation, fatty acid β-oxidation (FAO), and pyruvate dehydrogenase activity. RNA-Seq of livers and HBs suggested seven distinct mechanisms of Myc–ChREBP target gene regulation. Gene ontology analysis indicated that many transcripts deregulated in the chrebp−/− background encode enzymes functioning in glycolysis, the TCA cycle, and β- and ω-FAO, whereas those dysregulated in the myc−/− background encode enzymes functioning in glycolysis, glutaminolysis, and sterol biosynthesis. In the myc−/−/chrebp−/− background, additional deregulated transcripts included those involved in peroxisomal β- and α-FAO. Finally, we observed that Myc and ChREBP cooperatively up-regulated virtually all ribosomal protein genes. Our findings define the individual and cooperative proliferative, metabolic, and transcriptional roles for the “Extended Myc Network” under both normal and neoplastic conditions.
Keywords: Myc (c-Myc), hepatocellular carcinoma, Warburg effect, mitochondria, fatty acid oxidation, liver cancer, hepatoblastoma, MondoA, MYC proto-oncogene bHLH transcription factor, Oxphos
Introduction
The c-Myc (Myc) oncoprotein is deregulated in many human tumors while also playing roles in normal proliferation, differentiation, and metabolism (1–5). Myc coordinates the transcription of thousands of protein-coding genes, rRNAs and tRNAs, microRNAs, and long-noncoding RNAs (6–15). However, its role in maintaining growth and survival of normal or neoplastic cells depends on the timing and level of its expression and its cellular context. For example, whole-body depletion of Myc in the embryo is lethal but is compatible with postnatal survival (16–18). Indeed, total-body dominant-negative inhibition of endogenous Myc in older mice is associated with mild and reversible intestinal epithelial and hematopoietic hypoplasia and long-term survival (19), yet these mice demonstrate regression of Ras oncogene-induced lung adenocarcinomas, thereby demonstrating the differential Myc dependence of normal and transformed tissues. Myc hypomorphism is associated with smaller organs resulting from a reduction in cell number but not size (18).
The consequences of Myc knockout in the liver have been controversial, particularly with regard to its role in regeneration following partial hepatectomy (PH) 3 (20–23). We recently reexamined this in a novel mouse model of Type I hereditary tyrosinemia in which the replacement of diseased hepatocytes by transplanted nontyrosinemic hepatocytes can be monitored over several months (24–26). Using competitive transplants, we demonstrated that WT and myc−/− (Myc-KO) hepatocyte populations possessed similar long-term repopulation potential. In contrast, hepatoblastoma (HB) development driven by mutant forms of β-catenin and yes-associated protein (YAP) was markedly delayed in Myc-KO livers (13, 27), indicating that Myc plays distinct roles in normal and neoplastic proliferation. Alternatively, Myc-KO hepatocytes could be rescued by factors that functionally substitute for Myc. Indeed, Myc-KO HBs up-regulated expression of the weakly transforming Myc paralog L-Myc (13, 28, 29).
Myc, L-Myc, and another paralog, N-Myc, are members of a group of related bHLH-ZIP transcription factors that bind canonical E-box elements (CA(C/T)GTG) and regulate gene expression. The central member of this group is Max, whose dimerization with Myc is necessary for E-box binding. In contrast, heterodimers formed between Max and the four bHLH-ZIP members of the Mxd family compete with Myc-Max heterodimers for DNA binding and transcriptional modulation (30, 31). Hereafter, we refer to these proteins as the “Myc-Max-Mxd” family.
Following the discovery of the Myc-Max-Mxd family, it was appreciated that a related bHLH-ZIP family with Myc-like functions exists. Analogously, this also consists of a central, Max-like bHLH-ZIP protein, Mlx; two closely related Myc-like factors termed MondoA/Mlxip and MondoB/ChREBP/Mlxipl (hereafter ChREBP) that positively regulate gene expression; and a negative regulator termed Mnt (32–34). We refer to this as the “ChREBP-Mlx-Mnt” family and to the two families collectively as the “Extended Myc Network” (Fig. S1A). Cross-talk occurs by virtue of the fact that Mxd1 and Mxd4 also interact with Mnt. The canonical binding site for the ChREBP-Mlx-Mnt family, termed the carbohydrate response element (ChoRE), consists of two Myc-like E-box elements separated by 5 nucleotides (CAYGYGnnnnnCRCRTG) (35–38). Whereas the two families regulate common genes (14, 39, 40), those under ChREBP-Mlx-Mnt family control comprise a subset of those regulated by the Myc-Max-Mxd family, particularly those involved in carbohydrate and lipid metabolism (14, 32, 34, 36, 41–45). Moreover, unlike nuclearly localized Myc proteins, MondoA and ChREBP shuttle between the nucleus and cytoplasm in response to their binding of glucose 6-phosphate, thereby suggesting that they serve as nutrient sensors and regulate target gene expression in accord with metabolic status (34, 40, 46, 47).
We have now compared both the repopulation and tumorigenic potential of Myc-KO (or MKO) hepatocytes with those lacking ChREBP (ChREBP-KO or CKO) and both Myc and ChREBP (Double-KO or DKO). We chose for study ChREBP rather than the related MondoA because, even though both are widely distributed, ChREBP expression is more prominent in liver and appears to play a more important role (32, 48, 49). Our findings demonstrate that, in contrast to Myc, ChREBP's absence leads to a profound proliferative disadvantage in competitive hepatocyte repopulation assays and that Double-KO hepatocytes are even more impaired. HB growth rates were also markedly slowed, although tumor initiation remained unaffected. A combination of metabolic, biochemical, and transcriptional profiling better defined the abnormal phenotypes. Our findings indicate that the inhibition of normal and neoplastic proliferation resulting from Myc and/or ChREBP loss originates from multiple and in some cases overlapping and cooperative effects on key metabolic processes, primarily centered around carbohydrate and lipid metabolism and protein synthesis.
Results
Myc-KO, ChREBP-KO, and Double-KO livers accumulate neutral lipid
Livers from young Myc-KO mice appear relatively normal but gradually accumulate neutral lipids (25). We confirmed this by demonstrating more prominent vacuolization in hematoxylin and eosin–stained Myc-KO livers (Fig. S1B). ChREBP-KO livers from comparably aged mice were even more abnormal, with lipids tending to be particularly prominent in subcapsular and periendothelial regions. Similar changes were seen in Double-KO livers, with certain areas being more noticeably affected. Hepatic triglyceride content largely reflected these findings (Fig. 1C). Thus, ChREBP appears to play a more important role than Myc in maintaining hepatic triglyceride content.
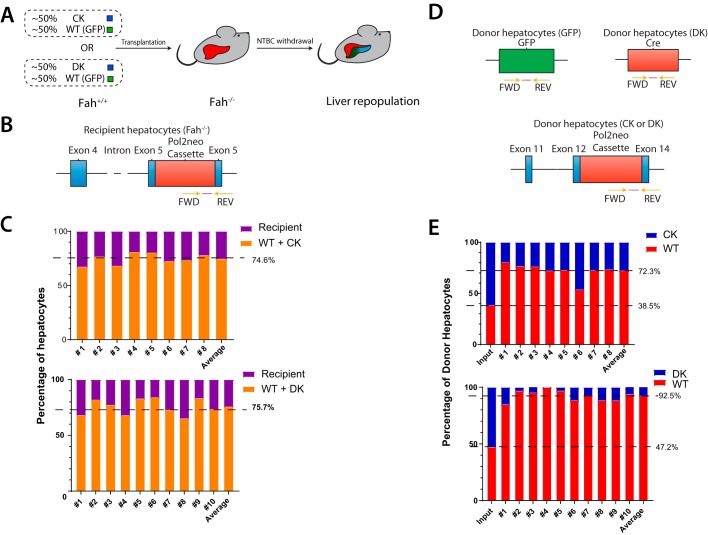
Figure 1.
Competitive disadvantage of ChREBP-KO and Double-KO fah+/+ donor hepatocytes. A, basis of the FAH competitive repopulation assay. GFP+ fah+/+ hepatocytes were used as the control, WT donor population. They were mixed in approximately equal numbers with ChREBP-KO or Double-KO hepatocytes (both GFP− and fah+/+). Following intrasplenic injection of the mixed hepatocyte population into fah−/− mice, animals were maintained on NTBC for 4 days and then periodically cycled off and on the drug until weights stabilized. Hepatocytes were then isolated, and the relative proportions of donor and recipient populations were determined. B, TaqMan primer set used to quantify the proportion of total fah−/− recipient, post-transplant hepatocyte population. Primers were designed to amplify the flanking region of the neo cassette-exon 5 region of the fah−/− recipient population. C, reconstitution of livers following transplantation of a donor population composed of WT + ChREBP-KO hepatocytes (top) or WT + Double-KO hepatocytes (bottom). Total donor and recipient populations from 8–10 transplanted mice were quantified as described in B. Numbers to the right indicate the final mean percentage repopulation by the total donor population. D, TaqMan primers used to distinguish each of the two donor populations. The primer set used to amplify GFP+ WT donor hepatocytes amplified a region of the GFP cassette. The primer set used to amplify the ChREBP-KO donor population amplified the region flanking the neo cassette and exon 14 of the chrebp gene. The primer set used to detect the Double-KO population amplified a region of Cre recombinase, which was present only in the Myc-KO population. E, hepatocyte DNAs from C were used as templates for the TaqMan primers depicted in D to allow a determination of the relative proportion of WT and ChREBP-KO donor hepatocytes (top) or WT + Double-KO donor hepatocytes (bottom). The first bar (input) indicates the percentage composition of the initial input population. Numbers to the left indicate the percentage of WT hepatocytes in input or transplanted hepatocyte populations. The values for C and E were obtained from reconstitution experiments using eight different known ratios of each of the two DNA populations being quantified as described previously (51). Correlation coefficients in each case were >0.99 (not shown).
In contrast, HBs were largely devoid of neutral lipids, although occasional areas of vacuolization were observed. However, the adjacent regions of nontransformed parenchyma largely retained the patterns of neutral lipid staining described above (Fig. S1B). Tumor triglyceride content was also uniformly low (Fig. S1B).
ChREBP-KO and Double-KO hepatocytes function poorly in repopulation assays
Liver repopulation by WT and Myc-KO hepatocytes occurs at similar rates (25). To assess this, we employed the hereditary tyrosinemia model described above. Donor mice lack the enzyme fumaryl acetoacetate hydrolase (FAH) (50) and succumb to the progressive accumulation of toxic tyrosine catabolites unless rescued with the drug 2-(2-nitro-4-trifluoro-methyl-benzoyl)-1,3-cyclohexanedione (NTBC), which blocks the upstream enzyme p-hydroxyphenylpyruvate dioxygenase. Mice can also be rescued by the intrasplenic injection of fah+/+ hepatocytes (25, 26, 51, 52). During iterative periods of NTBC discontinuation, donor cells gradually replace their fah−/− counterparts and, upon reaching an equilibrium state, comprise as much as 80% of the recipient's hepatocytes. This assay is superior to PH for assessing donor hepatocyte proliferative potential in that it imposes greater replicative demand that can reveal subtle differences in repopulation potential not appreciable with the PH model. Additionally, whereas 30–40% of post-PH hepatocytes do not replicate and about an equal fraction undergo hypertrophy rather than actual division (53, 54), repopulation following transplant assesses only proliferation.
We initially performed transplants using single populations of hepatocytes and noted the time needed to achieve NTBC independence, as defined by the ability of recipients to discontinue the drug and maintain >80% of their pretransplant weight (25). Consistent with previous findings, we found this time to be 9.5 ± 0.3 and 10.1 ± 0.4 weeks following transplantation with WT and Myc-KO hepatocytes, respectively (not significant as determined by Student's two-tailed t test) (25). In contrast, transplantation with ChREBP-KO hepatocytes required 11.8 ± 0.4 weeks to achieve NTBC independence (p = 5.18 × 10−3, compared with WT). Double-KO hepatocytes required 18.9 ± 1.0 weeks (p = 1.40 × 10−8, compared with WT). Additionally, the time to NTBC independence was significantly prolonged in Double-KO mice versus ChREBP-KO mice (p = 1.91 × 10−5). Thus, both ChREBP-KO and Double-KO hepatocytes are replication-impaired.
We next performed competitive repopulation assays with ChREBP-KO + WT or Double-KO + WT hepatocytes (Fig. 1A) (25, 51). Upon attaining NTBC independence, a TaqMan-based approach was used to quantify total donor and recipient populations (Fig. 1). In both cases, ∼75% of hepatocytes were found to be of donor origin (Fig. 1C). Further investigation of the ChREBP-KO + WT transplants showed that ChREBP-KO hepatocytes comprised 61.5 and 27.7% of the pre- and post-transplant donor population, respectively (Fig. 1 (D and E), p = 5.2 × 10−10). Even more striking were the results obtained with Double-KO + WT transplants, where 52.8% of the pretransplant input donor population but only 7.5% of the post-transplant hepatocytes were Double-KO cells (p = 5.2 × 10−10). The difference in repopulation efficiencies between ChREBP-KO and Double-KO was highly significant (p = 5.0 × 10−6). Thus, both ChREBP-KO and even more so Double-KO hepatocytes demonstrated a profound competitive disadvantage.
Growth inhibition of Myc-KO, ChREBP-KO, and Double-KO HBs
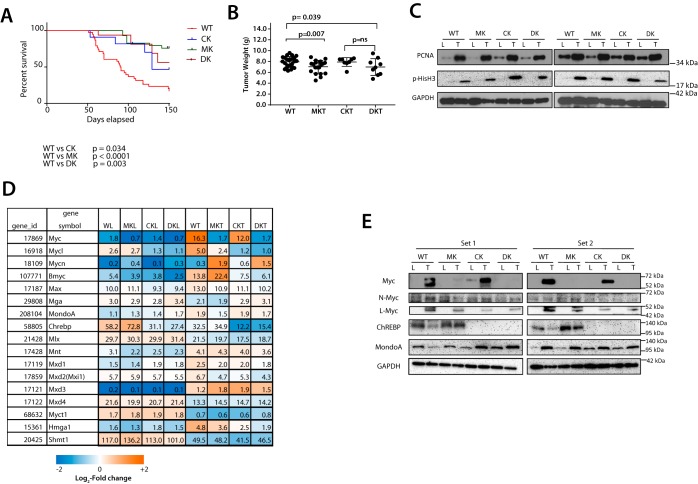
We next examined HB tumorigenesis in response to liver expression of oncogenic forms of β-catenin and YAP (13, 27, 51). WT animals succumbed with a mean survival of ∼13 weeks (Fig. 2A) (13, 27, 51), whereas survival in all knockout groups was delayed, and tumor sizes were somewhat smaller (Fig. 2B). Proliferating cell nuclear antigen and phosphohistone H3 levels as markers of proliferation (55, 56) were indistinguishable among the tumor groups (Fig. 2C). Thus, ChREBP loss, or combined Myc + ChREBP loss, affects tumor growth rates but not initiation.
Figure 2.
Behaviors and properties of WT and KO tumors. A, Kaplan–Meir survival curves of the indicated groups of mice following hydrodynamic tail vein injection of plasmids encoding mutant forms of β-catenin and YAP (13, 27) (n = 10–15 mice/group). B, tumor weights at the time of sacrifice in each group. Note that tumor weights in all three KO groups (Myc-KO (MKT), ChREBP-KO (CKT), and Double-KO (DKT)) were assessed much later than those of WT tumors. C, expression of proliferating cell nuclear antigen (PCNA) and phosphohistone H3 in livers (L) and tumors (T) from the indicated tissues. D, transcript levels of members of the Extended Myc Network. Also included is the expression of genes (myct1/mt-mc1, hmga1, and shmt) that can rescue the growth defect of myc−/− fibroblasts (57, 59). Relative mRNA expression levels of the indicated genes were determined directly from RNA-Seq results (n = 5 samples/group). The numbers within boxes indicate the relative expression of each transcript. Myc transcript levels in Myc-KO and Double-KO livers and tumors and ChREBP transcripts in ChREBP-KO and Double-KO livers and tumors arise almost exclusively from transcripts upstream of the neo cassettes that disrupted the coding region of the genes (not shown). E, expression of select members of the Extended Myc Network in representative livers and HBs from each of the four cohorts. Two sets of immunoblots (Set 1 and Set 2) are depicted (see Fig. S2 for two additional sets). Error bars, S.D.
We examined expression of other members of the Extended Myc Network as well as other Myc target genes capable of restoring myc−/− cell growth, including myct1, hmga1, and shmt (57–60) (Fig. 2D). Aside from confirming that the three knockout tissues lacked expression of their respective full-length transcripts, we showed variable degrees of change in the livers and tumors of mice with regard to a number of these transcripts, including those for L-Myc, N-Myc, Mxd3, Mnt, Myct1, and Shmt.
Members of the Extended Myc Network possess varying degrees of structural and functional redundancy (14, 28, 36, 62–65), suggesting that they might be able to rescue the proliferative defects of KO hepatocytes. As expected, Myc-KO livers and tumors did not express Myc, ChREBP-KO livers and tumors did not express ChREBP, and Double-KO livers and tumors did not express either protein (Fig. 2E and Fig. S2). Regardless of genotype, all livers and tumors expressed low or undetectable levels of N-Myc. L-Myc levels were variably elevated in all tumors. Similarly, livers expressed low levels of MondoA, which was increased modestly in all tumors. ChREBP was expressed at similar levels in WT and Myc-KO livers and tumors. From these studies and the fact that L-Myc and MondoA are about equally up-regulated in all tumors regardless of genotype, we conclude that neither of these is likely to be expressed at levels sufficient to support nontransformed hepatocyte proliferation. The poor correlation between transcript and protein levels for some of these factors (Fig. 2 (D and E) and Fig. S2) indicates that much of their control was post-transcriptional.
Metabolic alterations in livers and tumors
WT and Myc-KO liver and tumor metabolism differ in several critical ways that impact TCA cycle function, FAO, and levels of critical substrates, such as ATP and acetyl-CoA (AcCoA) (13, 25). Because of ChREBP's known influence on carbohydrate and lipid metabolism and its cooperation with Myc in regulating these activities (40, 41, 44, 45, 49, 66–69), we reevaluated some of these behaviors in our four liver and HB cohorts.
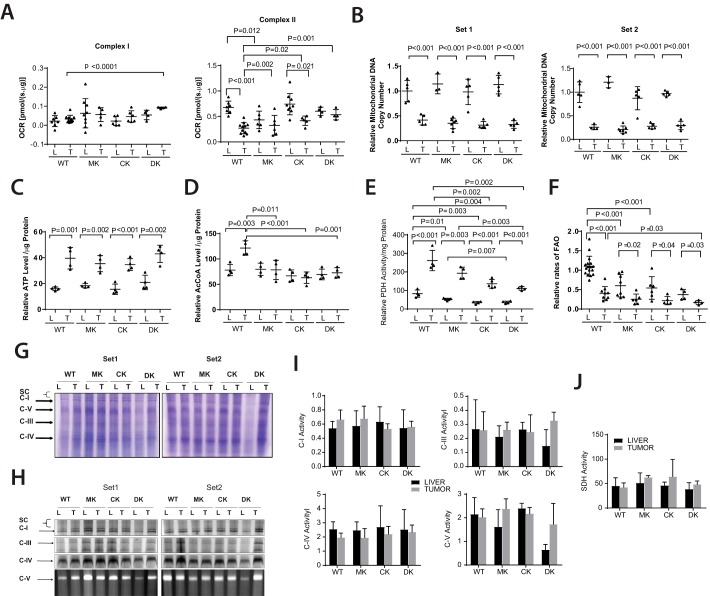
Oxygen consumption rates (OCRs) of WT and Myc-KO livers were similar, as reported previously (Fig. 3A) (13). In contrast, ChREBP-KO liver OCRs were lower than those of WT or Myc-KO livers. This discrepancy was corrected in Double-KO livers, suggesting that, in this largely quiescent population, Myc and ChREBP affected readout oppositely. These differences were largely attributable to alterations in Complex II function, which accounts for the majority of Oxphos in liver (Fig. 3A) (13, 25, 51).
Figure 3.
Metabolic differences distinguish WT, Myc-KO, ChREBP-KO, and Double-KO livers and tumors. A, liver and tumor OCRs. The indicated tissues were dispersed, and their maximal OCRs were determined by respirometry following the addition of the TCA cycle substrates pyruvate, malate, and succinate. Complex I and Complex II responses were distinguished by measuring OCR before and after the addition of the Complex I inhibitor rotenone. B, reduction of mtDNA content in HBs is independent of tumor growth rates and the presence of Myc and/or ChREBP. Two different sets of TaqMan probes (Set 1 and Set 2) were used to independently amplify different regions of the mitochondrial genome. C, ATP levels in livers and tumors. D, AcCoA levels in livers and tumors. E, PDH activity in fresh livers and tumors. PDH was assessed by measuring the release of 14CO2 from 14C-labeled pyruvate (13, 51). F, FAO rates in fresh livers and tumors. FAO was assessed by measuring the release of water-soluble products from 3H-labeled palmitate-BSA (13, 51). G, representative Coomassie Blue–stained nondenaturing gels of ETC complexes from livers (L) and tumors (T) isolated from the indicated groups. The panel shows two sets of results. H, typical appearances of developed nondenaturing gels following in situ enzymatic assays for the ETC complexes I, III, IV, and V. I, quantification of ETC complex activities. Four sets of gels similar to those shown in H were scanned and normalized to the activity of Complex IV, which showed little variation among the different tissues (13, 25, 105). J, complex II activities were assayed in vitro on tissue lysates because the in situ assay is unreliable. All results were normalized to the mean activity of each complex measured in WT livers. Error bars, S.D.
Also, as shown previously, WT and Myc-KO tumor OCRs were significantly lower than their corresponding normal livers, indicating greater reliance on glycolysis (13, 24, 51). In contrast, both ChREBP-KO tumors and Double-KO tumors were indistinguishable from their corresponding livers, and this was also reflected in their or in vitro activities
The reduction of WT and Myc-KO tumor OCRs was an expected consequence of the Warburg effect, which diverts glycolytic substrates away from mitochondria and into anabolic pathways that abet tumor proliferation (70–73). We recently reported that, like many human cancers (74), both HBs and Myc-induced hepatocellular carcinomas have a reduced mitochondrial DNA (mtDNA) content relative to livers, thus providing a potential physical basis for their reduced Oxphos and the Warburg effect (13, 75). Indeed, this was true for all four sets of HBs (Fig. 3B). Our results indicate that certain ETC functions are under differential control by ChREBP and Myc but that neither is responsible for the maintaining mtDNA.
Despite their reduced mtDNA content, WT and Myc-KO HBs had higher ATP levels than their corresponding livers (13). We confirmed this and further showed that these persisted in ChREBP-KO and Double-KO HBs (Fig. 3C). This correlated with a higher AcCoA content in WT HBs but not in Myc-KO, ChREBP-KO, or Double-KO tumors (Fig. 3D). Thus, the impaired growth of knockout tumors cannot be attributed to ATP deficiencies but could be due to an AcCoA deficit.
Increased pyruvate dehydrogenase (PDH) activity accompanies HB and hepatocellular carcinoma tumorigenesis and may reflect a reduced reliance on FAO and/or an attempt to maximize the conversion of limiting amounts of pyruvate to AcCoA (76). We confirmed this but also showed that Myc-KO, ChREBP-KO, and Double-KO livers and tumors had progressively larger reductions in PDH activity. Most strikingly, PDH activity in Double-KO tumors, although still elevated relative to its corresponding liver, was not significantly higher than the activity in WT livers (Fig. 3E).
The relatively high FAO rate of normal liver is Myc-dependent and is reduced by ∼50% in its absence. A further ∼50% decline in FAO rates then occurs in both WT and Myc-KO HBs, with the latter reflecting the already reduced activity in Myc-KO livers (13). FAO rates were equally dependent on ChREBP, and Double-KO livers and HBs had the lowest FAO rates as well as the lowest PDH activities among their respective groups (Fig. 3F). These results reveal that FAO is positively regulated by the combined actions of Myc and ChREBP. Tumors and their corresponding livers therefore progressively suppress PDH and FAO activities that normally function reciprocally and serve as major sources of ATP and AcCoA.
Myc's loss is associated with significant ETC dysfunction in both livers and HBs (Fig. 3A) (13, 25). To determine how ChREBP and Myc + ChREBP affected these processes, we resolved ETC complexes using blue native gel electrophoresis (Fig. 3G) and performed functional assays for all complexes. As seen in Fig. 3 (H–J), the only consistent abnormality was a loss of Complex V (ATP synthase) activity in Double-KO livers that was lower than the corresponding activity in Double-KO tumors or other liver or tumor groups.
Transcriptional profiling of livers
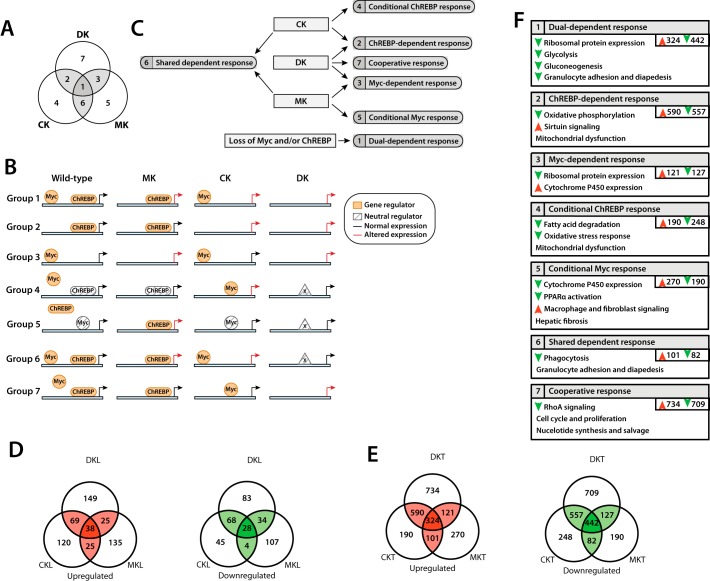
We next conducted RNA-Seq on five tissues from each group and identified distinct transcript expression profiles. The relationships of these to one another and the ways in which Myc and ChREBP may potentially interact to differentially affect target gene expression are depicted in Fig. 4 (A–C).
Figure 4.
Summary of observed gene expression changes in WT, Myc-KO, ChREBP-KO, and Double-KO livers and tumors. A, Venn diagram summarizing the relationships of transcriptional profile differences among livers and HBs. All transcripts could be grouped into one of seven categories (1–7) based on which gene expression differences were shared between or among different groups. B, potential mechanistic explanations for the types of gene expression changes in the groups described in A, assuming Myc and/or ChREBP act as direct positive or negative regulators. The models outlined here are meant to be heuristic in nature and not necessarily inclusive of all members of the indicated group. Group 1 (Dual-dependent response) is composed of genes that are regulated by both Myc and ChREBP (Table S8). The elimination of either one, or both, leads to altered expression (either up or down). Group 2 (ChREBP-dependent response) is composed of genes regulated only by ChREBP and whose expression is not altered by Myc-KO. Group 3 (Myc-dependent response) is composed of genes only regulated by Myc and whose expression is not affected by ChREBP-KO. Group 4 (Conditional ChREBP response) is composed of genes whose expression is not altered by ChREBP binding; rather, ChREBP binding precludes Myc binding at the same or a different site. In ChREBP's absence, Myc now binds and alters expression. In the Double-KO state, a WT level of expression is restored, potentially through a third factor (X), which can only bind and/or activate its cognate gene in the absence of both Myc and ChREBP. This factor may or may not bind to E-box or ChoRE elements. Group 5 (Conditional Myc response) is related to the Group 4 response and is composed of genes that, in WT cells, are bound but not directly regulated by Myc. Rather, Myc insulates the gene from ChREBP, which, when bound, alters expression. As with Group 4, the restoration of normal target gene expression may be mediated via a third factor. Group 6 (Shared dependent response) is related to the Group 1 response and is composed of genes regulated by both Myc and ChREBP, either one of which is sufficient to insulate the gene from a third factor, which, when bound, restores a WT level of expression. Group 7 (Cooperative response) is composed of genes that may be bound by Myc and/or ChREBP but with equivalent responses under either condition. Only when both factors are absent is there a change in expression. C, flowchart representing the types of gene expression changes occurring in response to the knockout of Myc, ChREBP, or both, according to the terminology used in A and B. D, summary of the number of significant gene expression changes observed in ChREBP-KO, Myc-KO, and Double-KO livers when compared with WT liver, according to the Venn diagram layout shown in A (q < 0.05). E, summary of shared gene expression changes observed in tumors (q < 0.05). F, description of types of gene expression changes observed in ChREBP-KO, Myc-KO, and Double-KO tumors. Genes falling into each of the Venn diagram groups shown in A were analyzed using gene ontology and pathway analysis. Numbers in the top left corner of each box correspond to each of the seven groups. Up (red) and down (green) arrows signify general up- or down-regulation of genes belonging to a given pathway. Pathways without arrows contain dysregulated genes but could not be assigned directionality with any degree of confidence. An analysis for livers is not shown, as the number of dysregulated genes in each Venn diagram group were generally too low to reliably predict pathway directionality.
In ChREBP-KO livers, 397 genes were significantly dysregulated compared with normal liver (q value < 0.05), corresponding to groups 1, 2, 4, and 6 of Fig. 4 (A–C). 252 were up-regulated, and 145 were down-regulated (Fig. 4D). Many could be grouped into several pathways, including those involved in retinoid X receptor (RXR)/liver X receptor and farnesoid X receptor/RXR signaling and lipid metabolism (Fig. S3A and http://prochownik.pitt.edu/chrebp). Noteworthy genes included previously described positive ChREBP targets, such as liver-type pyruvate kinase (Pklr; down-regulated 3.2-fold), fatty acid synthase (Fasn; down-regulated 1.8-fold), and the fatty acid elongase Elovl6 (down-regulated 2.2-fold) (40, 68, 77), as well as other functionally related genes, such as Elovl2 (down-regulated 1.6-fold), and the acyl-CoA thioesterases Acot1, Acot2, and Acot4 (down-regulated 2.5-, 2.0-, and 1.7-fold, respectively). These are most commonly involved in overlapping functions related to cholesterol and bile acid biosynthesis and transport, fatty acid biosynthesis, and triglyceride hydrolysis (78–80).
165 of the above 397 genes were dysregulated only in ChREBP-KO livers and demonstrated no further dysregulation compared with WT liver following additional knockout of Myc. These are defined as being “conditional ChREBP-KO–responsive” genes, because their dysregulation is conditional upon Myc's presence (Fig. 4D, Group 4). They included the pH-sensitive phosphate transporter Slc34a2 (2.6-fold down-regulation), which is overexpressed in some cancers and positively regulates Myc plus five down-regulated cytochrome P450 (Cyp) genes (1.6–2.8-fold) (Table S2).
137 genes were dysregulated in both ChREBP-KO and Double-KO livers and were classified as being “ChREBP-dependent” (Fig. 4 (B–D), Group 2). These included down-regulated metabolic genes, such as the previously mentioned Fasn and Pklr, as well as peroxisome proliferator activator receptor δ (Ppard); several acyl-CoA thioesterases; Pdk1, a negative regulator of PDH (81); and numerous solute transporters (Table S2).
In Myc-KO livers, 396 genes were significantly dysregulated compared with normal liver, corresponding to Groups 1, 3, 5, and 6 (Fig. 4, B–D) (223 up-regulated and 173 down-regulated). These showed significant involvement in functions previously identified in ChREBP-KO livers, including steroid synthesis and other metabolic pathways (Fig. S3B). Notably up-regulated (1.5–84-fold) targets included nine Cyp genes, which participate in steroid metabolism and fatty acid ω-oxidation (82). Down-regulated genes included lysine demethylase Kdm5d, which has been linked to malignant transformation (down-regulated 5.6-fold) (83, 84).
242 of the above 396 genes were dysregulated only in Myc-KO livers and not in Double-KO livers (Conditional Myc- response; Fig. 4, B–D, Group 5). These included the majority of the aforementioned up-regulated Cyp450 and sulfotransferase genes plus three additional down-regulated Cyp450 genes and Kdm5d (Table S3).
59 genes (25 up-regulated and 34 down-regulated) were strictly Myc-dependent, being similarly dysregulated in Myc-KO and Double-KO livers but not in ChREBP-KO livers (Myc-dependent response; Fig. 4B, Group 3). This group included four Cyp450 genes (up-regulated 3.5–24-fold) and Pvt1 (up-regulated 2-fold), a long noncoding RNA whose gene resides adjacent to MYC, often co-amplifies with it, and stabilizes the Myc protein (Table S4) (85).
29 genes were dysregulated following the loss of ChREBP or Myc, but not both (Shared dependent response; Fig. 4 (B–D), Group 6). Of these, 25 were up-regulated and four were down-regulated. Among these were the estrogen sulfotransferase Sult1e1 (up-regulated 11-fold) and three MHC Class II genes (Table S5). Members of this group were too few in number to permit any further functional classification using gene ontology analysis.
494 genes were dysregulated in Double-KO livers compared with normal liver and included genes contained within Groups 1, 2, 3, and 7 (281 up-regulated and 213 down-regulated; Fig. 4, B–D). Similar to Myc-KO livers, and more so ChREBP-KO livers, pathway analysis revealed roles for many of these genes in the metabolism of steroids and other lipids (Fig. S3C and http://prochownik.pitt.edu/chrebp/).
Dysregulated genes falling into groups 1–3 have been discussed above. Because 232 of these were only dysregulated in Double-KO livers, we classified them as showing a “cooperative response” indicating that loss of either ChREBP or Myc fully compensated for the other's absence (Fig. 4 (B–D), Group 7). Members included 16 down-regulated ribosomal protein (RP) genes (average down-regulation = 1.43-fold) and the gene encoding acetoacetyl-CoA synthetase (Aacs, down-regulated 2.63-fold) (Table S6).
66 genes were dysregulated in all three liver groups (Dual-dependent response; Fig. 4 (B–D), Group 1 and Table S8). Of these, 38 were up-regulated, and 28 were down-regulated. The latter included genes encoding the leptin receptor (Lepr; down-regulated 4-fold) and the rate-limiting enzyme in cholesterol synthesis, HMG-CoA reductase (Hmgcr; down-regulated 1.6-fold) (Table S7).
Transcriptional profiling of tumors
WT and ChREBP-KO tumors differentially expressed 2534 genes, with 1,205 being up-regulated and 1,329 down-regulated in the latter group (Fig. 4, E and F). Many are involved in growth and cancer signaling pathways, metabolism (including glycolysis, gluconeogenesis, sucrose degradation, and acyl-CoA hydrolysis), intracellular signaling (STAT3, Wnt, and protein kinase A signaling), and the immune response (granulocyte and macrophage, interleukin-8, and NFAT signaling) (Fig. S4 and http://prochownik.pitt.edu/chrebp/).
Common to both ChREBP-KO and Double-KO tumors and strictly ChREBP-dependent response (Group 2) were 590 up-regulated and 557 down-regulated transcripts (Fig. 4 (A, B, E, and F), Groups 1 and 2). These involved pathways regulating Oxphos and mitochondrial function, including 20 down-regulated NADH:ubiquinone oxidase genes and four ATP synthase genes (Fig. S5A). Genes involved in Wnt/β-catenin signaling were also among this group (Fig. S5B), as were those encoding the mitochondrially localized members of the sirtuin family, sirt3–5 (Fig. S5C). Sirtuins function as lysine deacylase, ADP-ribosyltransferase, and desuccinylase proteins, and Sirt1 and Sirt3 also serve as tumor suppressors (86, 87). Additionally, NRF2-mediated signaling was predicted to be decreased due to altered expression of 21 genes in this pathway (Fig. S5D). NRF2 is a leucine zipper transcription factor whose own targets include numerous genes involved in cytoprotective and antioxidant responses (88, 89). Other notable genes in this group include Igf2 (up-regulated 5.6-fold) and the previously noted Pklr (down-regulated 3.5-fold).
438 genes (190 up-regulated and 248 down-regulated) were dysregulated only in ChREBP-KO tumors compared with WT tumors, (Fig. 4E, Group 4, Conditional ChREBP response). Many of the latter are involved in lipid metabolism, including triacylglycerol degradation (Abhd6, Ces1e, Ce2a, Lipc, Mgll, and Pnpla7; 1.6-fold average down-regulation), mitochondrial function, Oxphos, and the NRF2-mediated oxidative stress response (Fig. 4F). Other notable genes include six Cyp450 genes (1.8–2.8-fold down-regulation) and the aforementioned solute carrier Slc34a2 (1.8-fold down-regulation).
1,657 genes were differentially expressed in Myc-KO tumors compared with WT tumors (816 up-regulated and 841 down-regulated), with many of the involved pathways being the same as described in ChREBP-KO tumors (Fig. 4, E and F). These included various growth and cancer signaling pathways, metabolic and lipid metabolism pathways, and intracellular signaling (Fig. S6).
The subset of 248 genes dysregulated in both Myc-KO and Double-KO tumors consisted of 121 up-regulated and 127 down-regulated genes (Myc-dependent response; Fig. 4E, Group 3). Notably down-regulated were 12 genes encoding ribosomal proteins and two encoding eukaryotic initiation factors (Fig. S7A). Elevated expression of seven Cyp450 genes was also observed, as well as variable dysregulation of several genes involved in RXR signaling and lipid metabolism, including three phospholipases, carnitine palmitoyltransferase 1B (Cpt1b; down-regulated 2.8-fold) and others (Fig. S7B). Other notable genes in this group included the previously discussed Pvt1 (up-regulated 1.6-fold) and N-Myc (Mycn; up-regulated 5.7-fold).
The 460 genes dysregulated only in Myc-KO tumors but not Double-KO tumors included 270 up-regulated genes and 190 down-regulated genes (Conditional Myc response; Fig. 4 (E and F), Group 5). These were involved in hepatic fibrosis, macrophage and fibroblast signaling, the PPARα/RXRα pathway, and STAT3 signaling. Others included several Cyp450 genes, the putative tumor suppressor Kdm5d (down-regulated 3.0-fold) and the STAT3-regulated oncogene Bcl-3 (Bcl3; up-regulated 1.8-fold) (80, 83).
Compared with WT tumors, 183 genes (101 up-regulated and 82 down-regulated) were differentially expressed when either ChREBP or Myc, but not both, was knocked out (Shared dependent response; Fig. 4E, Group 6). This group was largely concerned with immune signaling, including pathways involving granulocyte adhesion, cellular movement, phagocytosis, and T-cell function.
Double-KO tumors differentially expressed 3,604 transcripts compared with WT tumors (1,769 up-regulated and 1,835 down-regulated). These most commonly functioned in growth and cancer signaling, lipid metabolism and RXR signaling, the immune response, and intracellular signaling pathways (Fig. S8). Dysregulated genes belonging to Groups 1, 2, and 3 (Fig. 4A) have been discussed previously. 1,443 genes were only dysregulated in Double-KO tumors and comprised the largest of the seven groups (Cooperative response, Group 7). 734 were up-regulated, and 709 were down-regulated (Fig. 4, E and F). Analysis of this subset was notable for the predicted down-regulation of the RhoA signaling pathway (Fig. S9A) and revealed genes involved in nucleotide synthesis and salvage pathways. Additionally, up-regulation of Kdm5d (3.5-fold) and eight cytochromes P450 (average up-regulation = 2.4-fold) was observed, as well as the down-regulation of two additional CYP450 genes (Fig. S9B).
The 766 genes deregulated in all experimental groups comprised the “dual-dependent response” (Fig. 4, A, D, E, and F, Group 1). This contained 324 up-regulated and 442 down-regulated genes and was most notable for 64 strongly down-regulated RP genes and six eukaryotic initiation factors, which were most prominent in the Double-KO group (Fig. S9C). This included the down-regulation of 10 genes involved in glycolysis and gluconeogenesis (Fig. S9D) and numerous others involved in granulocyte adhesion and diapedesis (Fig. S9E).
To obtain an approximation of the fraction of transcripts that were products of direct Myc target genes, we queried the ChiP Atlas database with the above 1,657 transcripts whose expression differed between WT and Myc-KO tumors (q < 0.05). 71.2% of these were contained in this database, indicating that a substantial proportion of our differentially expressed transcript population originates from direct Myc target genes.
Myc and ChREBP cooperatively regulate ribosomal protein transcript (RPT) expression
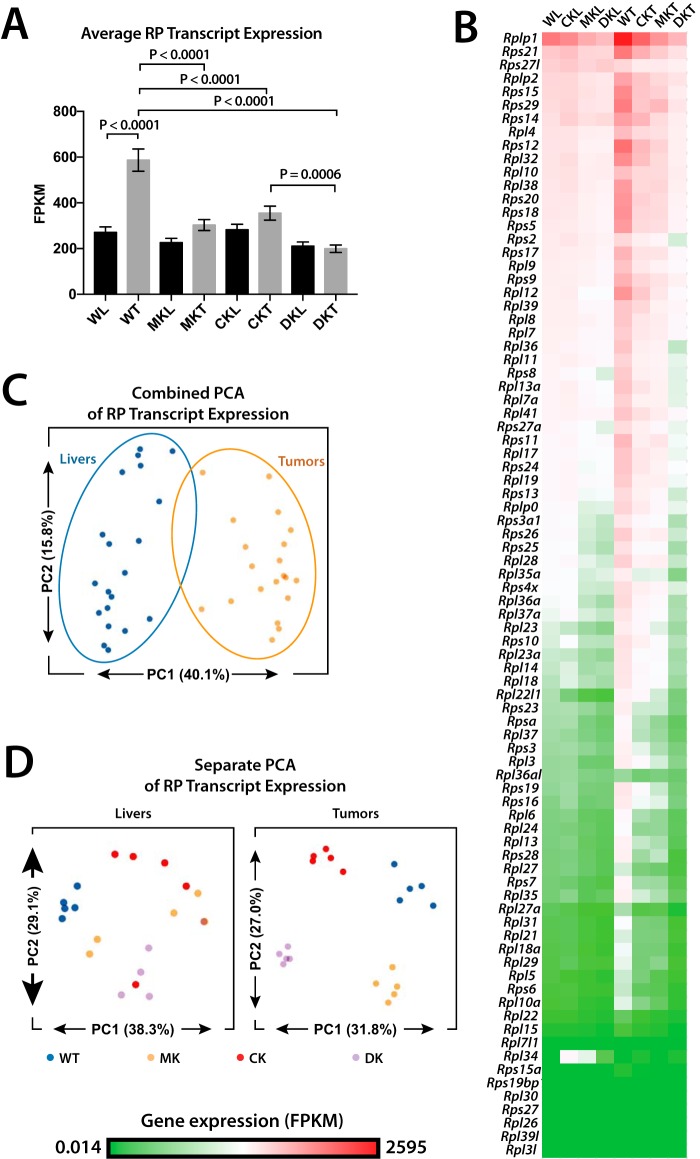
As a group, genes encoding RPTs were up-regulated 2.2-fold in WT tumors relative to WT livers, but only 1.3-fold in Myc-KO and ChREBP-KO tumors and not at all in Double-KO tumors (Fig. 5 (A and B), p < 0.0001). Myc-KO tumors down-regulated 73 of 80 RPTs relative to WT tumors, with the average expression of these being 52% of WT tumors. Similarly, 62 RPTs were down-regulated in ChREBP-KO tumors (average expression = 62% of WT). Double-KO tumors not only down-regulated a larger number of RPTs (79 of 80) but did so to a greater degree (average expression = 36% of WT) (Fig. 5B). This indicated that Myc and ChREBP, either individually or together, regulate virtually all RPTs in tumors and, to a lesser extent, in livers.
Figure 5.
RPT expression in Myc-KO, ChREBP-KO, and Double-KO livers and tumors. A, average expression of all 80 RPTs across all experimental groups. p values were calculated using t tests corrected for multiple comparisons. B, raw expression (FPKM) of all 80 RPTs across all experimental groups displayed as a heat map. The order of transcripts is based on the most to the least abundant in WT livers (WL). C, PCA of RPT relative expression in livers and tumors. Livers and tumors cluster distinctly from one another, indicating that each group possesses distinct patterns of RPT expression. D, PCA of RPT expression performed separately in livers and tumors. WT livers cluster distinctly from knockout livers, but none of the knockout liver groups can be distinguished from one another. All tumor groups cluster distinctly from one another, however, indicating that knockout of Myc, ChREBP, or both leads to distinct expression patterns of RPTs.
RPT expression patterns can distinguish tumor types and normal tissues in ways that are independent of absolute RPT levels (90, 91). Indeed, principal component analysis (PCA) showed that liver and tumor RPT expression patterns were distinct (Fig. 5C). Additionally, RPT patterns of knockout livers were distinct from those of WT livers, indicating that any knockout leads to a signature pattern of RPT expression (Fig. 5D). Whereas RPT expression patterns of all three KO liver groups displayed significant overlap, their cognate tumor groups clustered separately. Thus, RPT expression patterns from normal and transformed tissues are distinct and readily discernible from one another. Myc and ChREBP, therefore, in addition to dictating the absolute levels of tumor RPTs (and to a lesser extent liver RPTs), also regulate their expression patterns.
To determine which RPTs best distinguished WT from KO livers, a random forest classifier feature selection algorithm was used (Fig. S10A). This identified Rplp1, Rpl22l1, Rpl23, and Rps7 as the best discriminators (Fig. S10B). Similarly, Rpl27a, Rps11, Rps28, and Rps7 were the best discriminators of the tumor groups (Fig. S10C).
Finally, RP gene proximal promoters were analyzed for the presence of putative canonical E-boxes and ChoREs. Although 60 RP gene promoters contained at least one E-box within the regions analyzed, putative ChoRE sequences were found only in the promoters of Rps12 and Rpl13a (Fig. S10D). The lack of conserved E-boxes in 25% of RP genes plus the virtual absence of ChoREs argues that a significant amount of Myc regulation and virtually all of the ChREBP regulation of the RP gene family are likely to be indirect. However, it remains possible that less conserved E-boxes and ChoREs located in these promoters or outside the regions examined are involved in Myc- and/or ChREBP-dependent regulation.
Metabolic pathway dysregulation in livers and cancers mediated by the losses of Myc and ChREBP
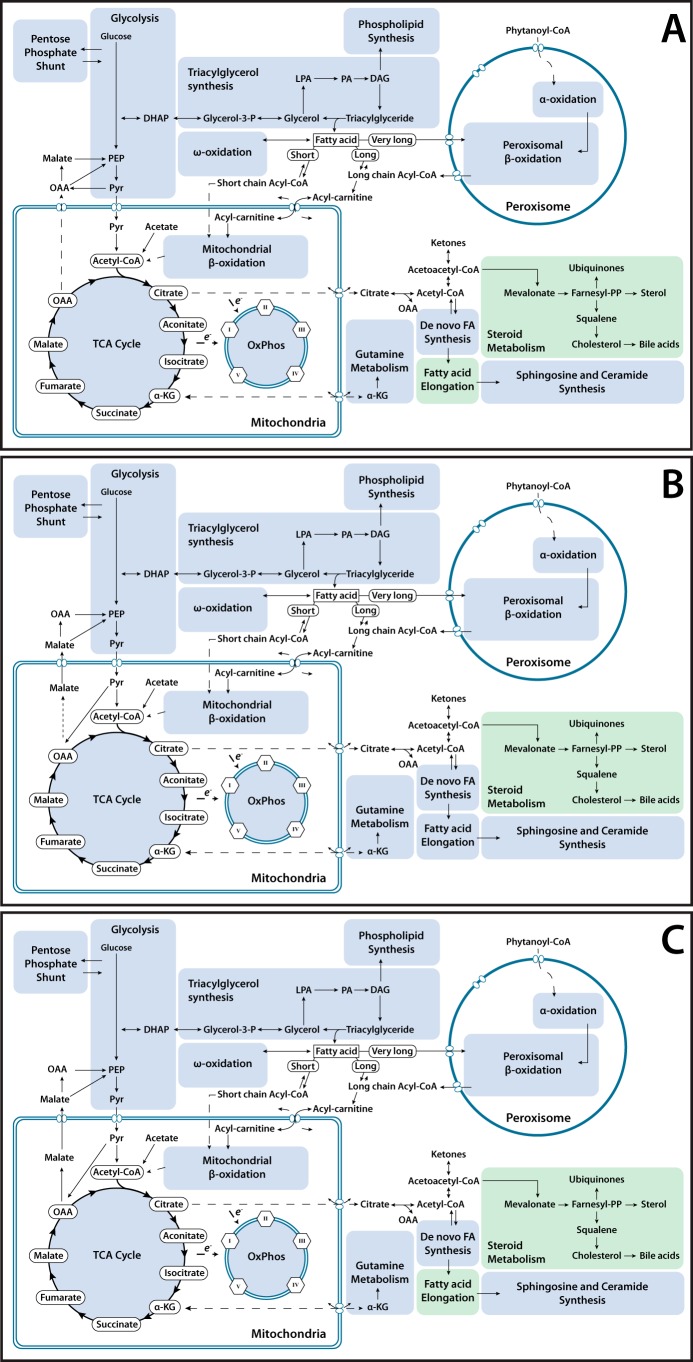
The loss of ChREBP in livers resulted in an overall down-regulation of transcripts encoding fatty acid elongation genes and HMG-CoA reductase, which encodes the rate-limiting enzyme in cholesterol synthesis (Fig. 6A). Myc-KO livers similarly down-regulated HMGCR transcripts but did not demonstrate strong down-regulation of those encoding enzymes involved in fatty acid elongation as did ChREBP-KO livers (Fig. 6B). Double-KO livers demonstrated down-regulation of transcripts involved in fatty acid elongation and cholesterol synthesis and a small number of genes involved in various other metabolic processes (Fig. 6C).
Figure 6.
Metabolic map of transcriptional changes in ChREBP-KO, Myc-KO, and Double-KO livers. Genes involved in direct metabolic processing of carbohydrates and lipids were mapped and grouped into general categories. Metabolic categories likely to be down-regulated relative to WT liver, either by significant down-regulation of genes encoding key rate-limiting enzymes or significant down-regulation of a large portion of many genes in the category, are colored green. Metabolic categories likely to be up-regulated in an experimental group relative to WT liver are colored red. A, summary of transcriptional changes in metabolic pathways in ChREBP-KO livers compared with WT livers, as described above. B, transcriptional changes in metabolic pathways in Myc-KO livers compared with WT livers. C, transcriptional changes in metabolic pathways in Double-KO livers compared with WT livers.
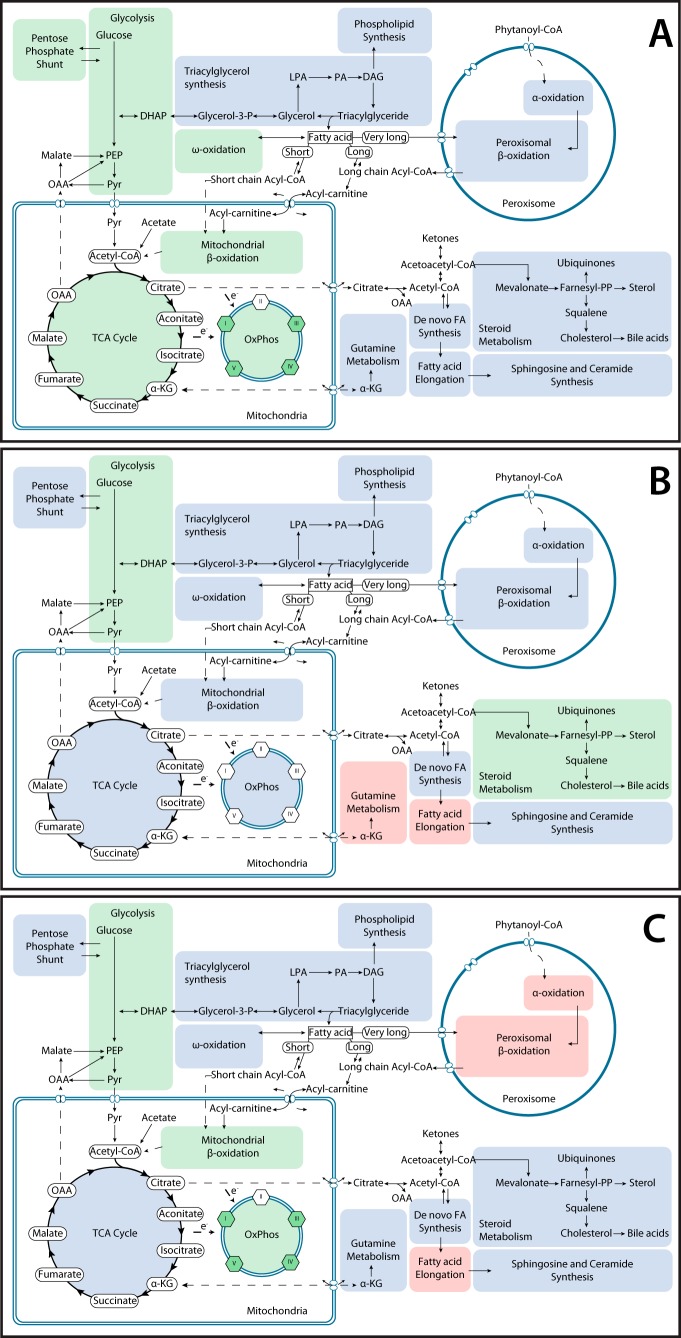
ChREBP-KO tumors strongly down-regulated the genes involved in glycolysis, the pentose phosphate shunt, the TCA cycle, Oxphos, and β- and ω-oxidation. ω-Oxidation is an alternative nonenergy-generating cytosolic and microsomal FAO pathway that becomes activated in response to inefficient β-FAO and is mediated largely by Cyp450 members and alcohol and aldehyde dehydrogenases (Fig. 7A) (82, 92). Myc-KO tumors also down-regulated transcripts for glycolysis and HMGCR and up-regulated those for glutamine metabolism and fatty acid elongation (Fig. 7B). Double-KO tumors also down-regulated many glycolytic genes and mitochondrial β-oxidation and Oxphos genes and up-regulated numerous genes involved in fatty acid elongation and peroxisomal α- and β-FAO (Fig. 7C).
Figure 7.
Metabolic map of transcriptional changes in ChREBP-KO, Myc-KO, and Double-KO tumors. A, summary of transcriptional changes in metabolic pathways in ChREBP-KO tumors compared with WT tumors, as described in Fig. 6. B, transcriptional changes in metabolic pathways in Myc-KO tumors compared with WT tumors. C, transcriptional changes in metabolic pathways in Double-KO tumors compared with WT tumors.
Additional evidence supporting cooperation between Myc and ChREBP in the regulation of lipid metabolism was the finding that 26 of the 102 known murine Cyp450 genes in livers (25%) and 51 of 102 in tumors (50%) were deregulated in response to knockout of ChREBP and/or Myc (Tables S9 and S10). In the latter case, 34 (56%) were members of the Cyp2, Cyp3, or Cyp4 families, which play particularly relevant roles in steroid and eicosanoid metabolism (93–95). In keeping with previous findings that Cyp450 genes are Myc targets both in untransformed hepatocytes and hepatoblastomas (13, 25), we found 35 Cyp450 genes to be exclusive targets of Myc, 15 to be exclusive targets of ChREBP, and six to be deregulated only when both Myc and ChREBP were eliminated. These results indicate that, as a gene family with important roles in fatty acid, sterol metabolism, and eicosanoid metabolism, Cyp450 genes are subject to extensive regulation by Myc and/or ChREBP.
Finally, to validate RNA-Seq results, we selected a small number of representative transcripts from the various families discussed above and performed real-time qRT-PCR analyses (Table S11). We found these results to be in excellent overall agreement with those obtained by RNA-Seq (Fig. S11).
Discussion
Cross-talk among select members of the Extended Myc Network presumably underlies the responsiveness of its members to growth and differentiation signals and helps to coordinate the timing and robustness of these responses with nutrient availability, proliferative needs, and metabolic preparedness (14, 32, 34, 36, 46, 47, 63, 96).
The hepatocyte-specific loss of Myc is associated with mild and age-dependent neutral lipid accumulation (25). Even more neutral lipids accumulate in the livers of ChREBP-KO and Double-KO mice (Fig. S1, B and C). Similar observations have been made in other cell types following Myc or N-Myc inhibition (97–99). This likely represents a generalized loss of coordination between exogenous lipid uptake, FAO, and de novo lipid biosynthesis with the rate of the former exceeding the rate of the latter two. Our finding that all KO livers were defective in FAO, coupled with previous observations that Myc inhibition enhances exogenous fatty acid uptake (13, 97–99), is consistent with this idea. Mitochondrial dysfunction in KO livers and tumors (Fig. 3, A, E, and F), together with the inability to properly up-regulate Oxphos-related genes (Fig. S5A), is consistent with other abnormalities in Myc-KO livers and HBs (13) and in cell lines with MondoA knockdown (63).
We previously found there to be no discernible differences in the rates at which WT and Myc-KO hepatocytes repopulate the fah−/− liver despite the latter's lipid processing defects (25, 26, 50). In contrast, ChREBP-KO and Double-KO hepatocytes, with more severe lipid abnormalities, were even more impaired (Fig. 1, D and E) (37). This suggests that ChREBP-regulated genes are just as important for sustained hepatocyte proliferation as Myc-regulated genes, if not more so. We cannot currently state with certainty whether this represents a direct effect on proliferation or an indirect one resulting, for example, from an inimical intracellular environment generated by accumulated lipids. In addition, the collaboration between the losses of Myc and ChREBP to produce a more severely impaired proliferative defect where none existed for the knockout of Myc alone suggests that this represents a form of synthetic lethality (100).
Despite differences in hepatocyte proliferative rates (Fig. 1E) (25), tumor growth rates were equally impaired (Fig. 2, A and B) (13). Tumors may rely more on the products of certain Myc and/or ChREBP target genes because of their more rapid growth. The pronounced AcCoA deficit in all KO tumors and their failure to increase RPT expression might be two such examples (Figs. 3D and 5 (A and B)). Additionally, Myc and ChREBP target genes are distinct in hepatocytes and tumors and could perhaps account for differential effects on tumor growth (Fig. 4) (13). Finally, β-catenin target gene products other than Myc could alter tumor cell sensitivity to Myc and/or ChREBP in ways that are distinct from those of hepatocytes. For example, Myc and ChREBP regulate smaller subsets of RPTs in livers than in tumors and do so less robustly (Fig. 5, A and B) (90). The defective RPT response of hepatocytes might not be fully appreciated until they are subject to the more demanding proliferative signals associated with transformation. Normal and transformed hepatocytes also have distinct metabolic properties, which may be direct or indirect consequences of Myc, ChREBP, and β-catenin expression (Fig. 3, A, E, and F).
WT and all KO tumor groups up-regulated L-Myc equally, which could potentially rescue the Myc-KO phenotypes if not the ChREBP and Double-KO phenotypes (25). L-Myc serves this function in myc−/− fibroblasts, although its transcriptional activation potential and transforming activity are markedly weaker than Myc's (28, 29, 101). Similarly, elevated MondoA in tumors might partially rescue ChREBP-KO tumor growth. MondoA enhances N-Myc transcriptional activity by altering chromatin conformation on some metabolic target gene promoters and by preventing Mlx's heterodimerization with Mxd and Mnt (60, 90, 91). Whether this occurs with L-Myc as well, however, is unknown.
MondoA and ChREBP target genes have been identified in a variety of cell lines, and ChREBP-bound genes have been cataloged in liver and white adipose tissue (37, 40, 43, 63, 67, 69). However, it remains unclear how many of the nearly 6,000 ChREBP-binding sites actually alter gene expression (37). The gene expression profiles of our three KO liver and tumor groups suggest that at least seven distinct mechanisms account for these transcriptional responses (Fig. 4) and underscore the interplay between Myc and ChREBP.
In addition to the gene targets mentioned above, we unexpectedly found that ChREBP also controls RP genes, particularly in collaboration with Myc (Fig. 5 and Figs. S9C and S10). Whereas numerous canonical E-boxes were identified in the proximal promoters of these genes, ChoRE elements were detected in only two (Fig. S10D). This suggests either that ChREBP regulation of most RP genes is indirect, that some ChoREs are more degenerate than those we defined as bona fide, that ChoREs reside outside the proximal promoter regions we analyzed, and/or that ChREBP binds to E-boxes as well as ChoREs. Regardless of the nature of the regulation, ChREBP's cooperation with Myc can be quite strong, as seen by the complete unresponsiveness of RP genes in tumors. This suggests that Myc and ChREBP are the major determinants of RPT expression in normal and neoplastic stimuli and cannot be replaced by other factors. It is difficult to escape the conclusion that a major proliferative defect in hepatocytes and livers lacking Myc and/or ChREBP is their inability to match translational rates with proliferative demands (102–104).
Given the variability in E-box binding affinities for Myc and a likely similar variability for ChREBP (105–108), it is not surprising that the patterns of RPT expression in different tumor types vary (Fig. 5, C and D). These likely reflect both the relative intrinsic affinities of binding sites within their gene promoters and the levels of Myc, ChREBP, and other members of the Extended Myc Network. Conceivably, the presumed resulting changes in ribosomal subunit composition can impact the efficiency with which different mRNAs are selected and translated, thereby achieving a distinct level of translational control independent of the transcriptional control imparted by Myc and ChREBP themselves (109).
Experimental procedures
Animals, hepatocyte transplants, and tumorigenesis studies
All animal work was performed in compliance with the Public Health Service Policy on Humane Care and Use of Laboratory Animal Research (DLAR) Guide for Care and Use of Laboratory Animals. All procedures were reviewed and approved by the University of Pittsburgh's institutional animal care and use committee. Mice were maintained in a pathogen-free facility under standard conditions with free access to food and water. UBI-GFP/B6-EGFP mice were purchased from Jackson Laboratories (Bar Harbor, ME). These mice, which express enhanced GFP in all tissues under the control of the ubiquitin C promoter, were used as a source of WT hepatocytes for all transplant studies. Mice bearing a conditional, hepatocyte-specific deletion of myc (Myc-KO mice) were generated by crossing c-Mycflox/flox (WT) mice (2, 25) with Alb-Cre recombinase-positive mice (25) and were genotyped as described previously (25). chrebp−/− knockout (ChREBP-KO) mice (B6.129S6-Mlxipltm1Kuy/J mice, strain 010537) were purchased from Jackson Laboratories and were genotyped as recommended by the vendor.
Immunocompromised FRG-NOD fah−/− mice (Yecuris, Inc., Tualatin, OR) were used as hepatocyte transplant recipients. They were maintained on 8 mg/liter NTBC (Yecuris) in their drinking water. Hepatocyte isolation was performed using a two-step collagenase perfusion method (24, 25, 51). For competitive transplant studies, WT and KO hepatocytes (>80% viability) were combined in approximately equal numbers, and 3.0 × 105 were injected intrasplenically into 6–8-week-old recipient FRG-NOD mice. Four days after transplantation, NTBC was discontinued and then reinstated when mice lost 20% of their body weight. NTBC cycling was continued until body weights stabilized. Mice were then maintained off NTBC for 4–5 additional weeks to ensure maximal hepatic replacement and body weight normalization. Hepatocytes were then isolated as described above (24, 25, 51), washed twice in PBS, and snap-frozen for subsequent PCR-based analysis.
In some cases, mice were reconstituted noncompetitively using only Myc-KO, ChREBP-KO, or Double-KO donor hepatocytes. In these instances, the time to NTBC independence was noted, and entire livers were removed for histological assessment.
Determining the extent to which recipient fah−/− mice were reconstituted with the total donor population used a two-step TaqMan-based assay. PCRs were performed with a Kapa Probe Fast qPCR kit (Kapa Biosystems, Wilmington, MA) in a volume of 12 μl with 10 ng of genomic DNA as described previously (51). Conditions for amplification were as follows: 95 °C for 10 s, 40 cycles at 95 °C for 15 s, 60 °C for 60 s. Repopulation was calculated with primers that amplified the Pol2neo cassette inserted in exon 5 of fah−/− mice (forward, 5′-GGGAGGATTGGGAAGACAATAG-3′; reverse, 5′-ATTCTCCTTGCCTCTGAACATAA-3′). The product was detected and quantified in real time with the probe, 5′/56-FAM/CTTCTGAGG/ZEN/CGGAAAGAA/3IABkFQ/-3′. A standard curve with known amounts of fah−/− and fah+/+ hepatocyte DNA was generated to allow the fractional contribution of each transplanted sample to be determined. To determine the contributions of Myc-KO and ChREBP-KO hepatocytes relative to WT hepatocytes or to one another, we used the approach depicted in Fig. 1A.
Induction of HBs
Tumorigenesis was induced in 6–8-week-old mice using the hydrodynamic tail vein injection–mediated delivery of Sleeping Beauty plasmids (13, 27, 51). Mice received 10 μg each of plasmids encoding a mutant, patient-derived form of human β-catenin (Δ90) and YAP (S27A) along with 2 μg of a vector encoding Sleeping Beauty transposase (13, 51). Plasmids were purified using Qiagen tip-500 columns (Qiagen, Inc., Valencia, CA) and were delivered in a total volume of 2 ml in 0.9% NaCl solution over ∼5–9 s. Mice were evaluated three times weekly and euthanized when tumors reached ∼2 cm in maximal diameter. Tumor weights and survival were recorded in all instances.
Metabolic studies
Livers or HBs were removed, immediately placed in a sterile tissue culture plate, and maintained on ice while pieces were apportioned for each of the assays described below or immediately snap-frozen in liquid nitrogen. Additional tissues were used for the isolation of mitochondria (25), which were snap-frozen in liquid nitrogen and stored at −80 ºC. Remaining tissues were apportioned in small aliquots, snap-frozen, and stored at −80º C for studies not requiring fresh tissue.
For FAO assays, ∼50 mg of fresh tissue was minced in 350 μl of SET buffer at 4 °C (25, 51, 75) and disrupted by five passes in a Potter–Elvehjem homogenizer; 5 μl of the homogenate was added to 195 μl of FAO reaction buffer containing 100 mm sucrose, 10 mm Tris-HCl, pH 7.4, 0.2 mm EDTA, 80 mm KCl, 5 mm KH2PO4, 1 mm MgCl2, 0.1 mm malate, 0.05 mm CoA, 2 mm l-carnitine, 2 mm ATP, 1 mm DTT, 0.3% fatty acid-free BSA, pH 8.0, and 0.5 μCi/ml 3H-labeled palmitate conjugated to BSA (PerkinElmer Life Sciences). Reactions were incubated at 37 °C for 2 h and stopped with the addition of 40 μl of 1 m KOH. They were further incubated at 60 °C for 1 h to hydrolyze newly synthesized acyl-carnitine esters. 40 μl of 4 m perchloric acid was added, and incubations were continued on ice for an additional 60 min. After organic extraction, water-soluble 3H-labeled products were measured by scintillation counting.
PDH complex activity was determined as described previously (25, 51, 75). Briefly, 50–100 mg of fresh tissue was minced in ice-cold glutamine-, glucose-, and pyruvate-free Dulbecco's-modified minimal essential medium and gently homogenized by several passages through an 18-gauge needle. 0.5-ml aliquots were placed into 0.5 ml of 2× assay buffer containing 0.075 μCi of [1-14C]pyruvate (PerkinElmer Life Sciences). Reactions were incubated at 37 °C for 40 min with continuous shaking and terminated by injecting 0.5 ml of 4.0 m perchloric acid through the rubber stopper, which contained a central hanging basket (Kimble-Chase Life Science and Research Products, Rockwood, TN) into which had been placed a 25-mm glass microfiber filter (Whatman/GE Healthcare) saturated with 0.5 m KOH. 14CO2 was collected for 40 min at 37 °C onto the filters and quantified by scintillation counting.
Respirometry was performed as described previously (25, 51, 75). Briefly, ∼50 mg of fresh tissue was disrupted in ice-cold MiR05 buffer. ∼300 μg of the lysate was used to determine OCRs using an Oroboros Oxygraph 2k instrument (Oroboros Instruments, Inc., Innsbruck, Austria). Each reaction contained cytochrome c (10 μm) (25), malate (2.0 mm), pyruvate (5 mm), ADP (5 mm), and glutamate (10 mm) to initiate electron flow via Complex I, followed by the addition of succinate to a final concentration of (10 mm) to measure the combined activities of ETC Complexes I + II. Finally, rotenone was added (0.5 μm final concentration) to inhibit Complex I and to allow for Complex II quantification. Activities were normalized to total protein content.
To quantify mtDNA content, two probe sets were used to amplify unique regions of mtDNA from livers and tumors using a TaqMan-based approach as described previously (25, 51, 75). The amplified segments included 101 bp of the cytochrome c oxidase I gene (probe set 1) and 90 bp of the D-loop region (probe set 2). Results were normalized to the product of a third TaqMan reaction that amplified a 73-bp segment of the nuclear apolipoprotein B gene. All reactions contained 10 ng of total DNA and were performed simultaneously on a CFX96 TouchTM real-time PCR detection system (Bio-Rad) using the following conditions: 95 °C for 10 s, 40 cycles at 95 °C for 15 s, and 60 °C for 60 s.
ATP assays were performed on previously snap-frozen tissues using the ATP Lite detection system (PerkinElmer Life Sciences). AcCoA assays were also performed on frozen tissues using an assay kit (MAK039, Sigma-Aldrich). Both assays were performed according to the suppliers' directions. Triglyceride content was measured as described previously (110).
Purified mitochondria were used in nondenaturing blue native gel electrophoresis to separate each of the multisubunit complexes of the electron transport chain (ETC), followed by Coomassie Blue staining (25, 51, 75). In situ assays for ETC Complexes I, III, IV, and V were performed as described previously, whereas Complex II (succinate dehydrogenase) assays were performed on separately prepared lysates (13, 25, 111). Enzymatic activity for each sample was quantified by scanning the bands corresponding to enzymatic activity in gels using a Fluorchem M system (Protein Simple, San Jose, CA), and relative activity was compared with that Complex IV, which showed the least sample-to-sample or group-to-group variability.
Western blotting
This was performed as described (13, 51, 75). Frozen tissues were disrupted in lysis buffer containing protease and phosphatase inhibitors, sonicated, boiled, and stored in small aliquots at −80 °C. The antibodies used, their dilutions, and the vendors from which they were purchased are provided in Table S1.
Transcriptional profiling of livers and HBs
All procedures were performed as described previously (13, 75). Briefly, RNA purification from five representative tissues of each group under study was performed using Qiagen RNeasy columns (Qiagen, Inc., Valencia, CA) followed by DNase digestion. Final sample integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Forster City, CA), and only samples with RNA Integrity Number (RIN) values >8 were processed further. RNA-Seq was performed at the Children's Hospital of Pittsburgh of UPMC Core Genomics Facility. Single-end single-indexed sequencing (∼75 bp) was performed with an Illumina NextSeq 500 sequencer (Illumina, Inc., San Diego, CA). Gene expression was calculated using Cufflinks and expressed as fragments per kilobase million (FPKM). Gene expression counts were normalized across samples and statistically compared between experimental groups using Cuffdiff, with a false discovery rate–adjusted p value (or q value) cutoff of 0.05. Raw and processed original data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (61) and are accessible through GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114634) at accession number GSE114634. Ingenuity Pathway Analysis (IPA, Qiagen) was used to categorize differentially expressed genes into pathways, with pathway significance was adjusted for false discovery using Bonferonni–Hochberg correction and a p value cutoff of 0.05. For each pathway, the following were recorded for display: overall significance (p value), ratios of dysregulated genes compared with all possible genes associated with that pathway (ratio), and predicted pathway activation or inhibition (Z-score, where applicable). Not all IPA pathways included the ability to estimate a Z-score. Significant pathways were then grouped into “superpathways” according to function using IPA's internal category scheme. These and their component transcripts can be accessed in an interactive format at https://prochownik.pitt.edu/chrebp/.
For select transcripts (Fig. S11), the same RNAs used for the above-described RNA-Seq experiments were used in qRT-PCRs to validate RNA-Seq results. Each qRT-PCR (20 μl) was performed with a Power SYBR Green RNA-to-CTTM 1-Step kit (Applied Biosystems, Inc., Waltham, MA) according to the manufacturer's instructions. Real-time reactions were monitored on a StepOne Plus Real PCR system (Applied Biosystem). Each reaction was performed in duplicate with <10% variation between samples. A list of PCR primers is shown in Table S11.
Author contributions
H. W. and E. V. P. conceptualization; H. W. and J. M. D. data curation; H. W. and J. M. D. software; H. W., J. M. D., S. K., J. L., L. E. J., and E. V. P. formal analysis; H. W., J. M. D., S. K., J. L., J. M., L. E. J., F. A., A. W. D., and E. V. P. investigation; H. W. and J. L. visualization; H. W., J. L., A. W. D., and E. V. P. writing-review and editing; J. M. D. and E. V. P. methodology; J. L., F. A., A. W. D., and E. V. P. resources; A. W. D. and E. V. P. supervision; A. W. D. and E. V. P. funding acquisition; A. W. D. and E. V. P. writing-original draft; E. V. P. validation; E. V. P. project administration.
Supplementary Material
This work was supported by National Institutes of Health RO1 Grants DK103645 (to A. W. D.) and CA174713 (to E. V. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S11 and Tables S1–S11.
Raw and processed original data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under accession number GSE114634.
- PH
- partial hepatectomy
- AcCoA
- acetyl coenzyme A
- bHLH-ZIP
- basic helix-loop-helix leucine zipper
- ChoRE
- carbohydrate response element
- ChREBP
- carbohydrate response element–binding protein
- KO
- knockout
- CKO
- ChREBP knockout or chrebp−/−
- Cyp
- cytochrome P450
- DKO
- ChREBP-KO + Myc-KO or chrebp−/−/myc−/−
- ETC
- electron transport chain
- FAH
- fumaryl acetoacetate hydrolase
- FAO
- fatty acid oxidation
- HB
- hepatoblastoma
- HMG
- hydroxymethylglutaryl
- HMGCR
- HMG coenzyme A reductase
- MKO
- Myc KO or myc−/−
- mtDNA
- mitochondrial DNA
- NTBC
- 2-(2-nitro-4-trifluoro-methyl-benzoyl)-1,3-cyclo-hexanedione
- OCR
- oxygen consumption rate
- Oxphos
- oxidative phosphorylation
- PCA
- principle component analysis
- PDH
- pyruvate dehyydrogenase
- RP
- ribosomal protein
- RPT
- ribosomal protein transcript
- RXR
- retinoid X receptor
- YAP
- Yes-associated protein
- qRT-PCR
- quantitative RT-PCR
- FPKM
- fragments per kilobase million
- IPA
- Ingenuity Pathway Analysis
- TCA
- tricarboxylic acid.
References
- 1. Anantharaman A., Lin I. J., Barrow J., Liang S. Y., Masannat J., Strouboulis J., Huang S., and Bungert J. (2011) Role of helix-loop-helix proteins during differentiation of erythroid cells. Mol. Cell. Biol. 31, 1332–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fernandez D., Sanchez-Arevalo V. J., and de Alboran I. M. (2012) The role of the proto-oncogene c-myc in B lymphocyte differentiation. Crit. Rev. Immunol. 32, 321–334 10.1615/CritRevImmunol.v32.i4.30 [DOI] [PubMed] [Google Scholar]
- 3. Kalkat M., De Melo J., Hickman K. A., Lourenco C., Redel C., Resetca D., Tamachi A., Tu W. B., and Penn L. Z. (2017) MYC deregulation in primary human cancers. Genes 8, E151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Müller M., Hermann P. C., Liebau S., Weidgang C., Seufferlein T., Kleger A., and Perkhofer L. (2016) The role of pluripotency factors to drive stemness in gastrointestinal cancer. Stem Cell Res. 16, 349–357 10.1016/j.scr.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 5. Nesbit C. E., Tersak J. M., and Prochownik E. V. (1999) MYC oncogenes and human neoplastic disease. Oncogene 18, 3004–3016 10.1038/sj.onc.1202746 [DOI] [PubMed] [Google Scholar]
- 6. Hamilton M. J., Young M. D., Sauer S., and Martinez E. (2015) The interplay of long non-coding RNAs and MYC in cancer. AIMS Biophys. 2, 794–809 10.3934/biophy.2015.4.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hart J. R., Roberts T. C., Weinberg M. S., Morris K. V., and Vogt P. K. (2014) MYC regulates the non-coding transcriptome. Oncotarget 5, 12543–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iaccarino I. (2017) lncRNAs and MYC: an intricate relationship. Int. J. Mol. Sci. 18, E1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jackstadt R., and Hermeking H. (2015) MicroRNAs as regulators and mediators of c-MYC function. Biochim. Biophys. Acta 1849, 544–553 10.1016/j.bbagrm.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 10. Levens D. (2013) Cellular MYCro economics: balancing MYC function with MYC expression. Cold Spring Harb. Perspect. Med. 3, a014233 10.1101/cshperspect.a014233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Psathas J. N., and Thomas-Tikhonenko A. (2014) MYC and the art of microRNA maintenance. Cold Spring Harb. Perspect. Med. 4, a014233 10.1101/cshperspect.a014175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soga T. (2013) Cancer metabolism: key players in metabolic reprogramming. Cancer Sci. 104, 275–281 10.1111/cas.12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang H., Lu J., Edmunds L. R., Kulkarni S., Dolezal J., Tao J., Ranganathan S., Jackson L., Fromherz M., Beer-Stolz D., Uppala R., Bharathi S., Monga S. P., Goetzman E. S., and Prochownik E. V. (2016) Coordinated activities of multiple Myc-dependent and Myc-independent biosynthetic pathways in hepatoblastoma. J. Biol. Chem. 291, 26241–26251 10.1074/jbc.M116.754218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilde B. R., and Ayer D. E. (2015) Interactions between Myc and MondoA transcription factors in metabolism and tumourigenesis. Br. J. Cancer 113, 1529–1533 10.1038/bjc.2015.360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaytseva O., and Quinn L. M. (2017) Controlling the master: chromatin dynamics at the MYC promoter integrate developmental signaling. Genes 8, E118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis A. C., Wims M., Spotts G. D., Hann S. R., and Bradley A. (1993) A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7, 671–682 10.1101/gad.7.4.671 [DOI] [PubMed] [Google Scholar]
- 17. Dubois N. C., Adolphe C., Ehninger A., Wang R. A., Robertson E. J., and Trumpp A. (2008) Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development 135, 2455–2465 10.1242/dev.022707 [DOI] [PubMed] [Google Scholar]
- 18. Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G. R., and Bishop J. M. (2001) c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414, 768–773 10.1038/414768a [DOI] [PubMed] [Google Scholar]
- 19. Soucek L., Whitfield J., Martins C. P., Finch A. J., Murphy D. J., Sodir N. M., Karnezis A. N., Swigart L. B., Nasi S., and Evan G. I. (2008) Modelling Myc inhibition as a cancer therapy. Nature 455, 679–683 10.1038/nature07260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baena E., Gandarillas A., Vallespinós M., Zanet J., Bachs O., Redondo C., Fabregat I., Martinez-A C., and de Alborán I. M. (2005) c-Myc regulates cell size and ploidy but is not essential for postnatal proliferation in liver. Proc. Natl. Acad. Sci. U.S.A. 102, 7286–7291 10.1073/pnas.0409260102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li F., Xiang Y., Potter J., Dinavahi R., Dang C. V., and Lee L. A. (2006) Conditional deletion of c-myc does not impair liver regeneration. Cancer Res. 66, 5608–5612 10.1158/0008-5472.CAN-05-4242 [DOI] [PubMed] [Google Scholar]
- 22. Qu A., Jiang C., Cai Y., Kim J. H., Tanaka N., Ward J. M., Shah Y. M., and Gonzalez F. J. (2014) Role of Myc in hepatocellular proliferation and hepatocarcinogenesis. J. Hepatol. 60, 331–338 10.1016/j.jhep.2013.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanders J. A., Schorl C., Patel A., Sedivy J. M., and Gruppuso P. A. (2012) Postnatal liver growth and regeneration are independent of c-myc in a mouse model of conditional hepatic c-myc deletion. BMC Physiol. 12, 1 10.1186/1472-6793-12-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duncan A. W., Hickey R. D., Paulk N. K., Culberson A. J., Olson S. B., Finegold M. J., and Grompe M. (2009) Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet. 5, e1000385 10.1371/journal.pgen.1000385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edmunds L. R., Otero P. A., Sharma L., D'Souza S., Dolezal J. M., David S., Lu J., Lamm L., Basantani M., Zhang P., Sipula I. J., Li L., Zeng X., Ding Y., Ding F., et al. (2016) Abnormal lipid processing but normal long-term repopulation potential of myc−/− hepatocytes. Oncotarget 7, 30379–30395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grompe M., Lindstedt S., al-Dhalimy M., Kennaway N. G., Papaconstantinou J., Torres-Ramos C. A., Ou C. N., and Finegold M. (1995) Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat. Genet. 10, 453–460 10.1038/ng0895-453 [DOI] [PubMed] [Google Scholar]
- 27. Tao J., Calvisi D. F., Ranganathan S., Cigliano A., Zhou L., Singh S., Jiang L., Fan B., Terracciano L., Armeanu-Ebinger S., Ribback S., Dombrowski F., Evert M., Chen X., and Monga S. P. S. (2014) Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 147, 690–701 10.1053/j.gastro.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Birrer M. J., Segal S., DeGreve J. S., Kaye F., Sausville E. A., and Minna J. D. (1988) L-Myc cooperates with Ras to transform primary rat embryo fibroblasts. Mol. Cell. Biol. 8, 2668–2673 10.1128/MCB.8.6.2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barrett J., Birrer M. J., Kato G. J., Dosaka-Akita H., and Dang C. V. (1992) Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol. Cell. Biol. 12, 3130–3137 10.1128/MCB.12.7.3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baudino T. A., and Cleveland J. L. (2001) The Max network gone mad. Mol. Cell. Biol. 21, 691–702 10.1128/MCB.21.3.691-702.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grandori C., Cowley S. M., James L. P., and Eisenman R. N. (2000) The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16, 653–699 10.1146/annurev.cellbio.16.1.653 [DOI] [PubMed] [Google Scholar]
- 32. Billin A. N., and Ayer D. E. (2006) The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism. Curr. Top. Microbiol. Immunol. 302, 255–278 [DOI] [PubMed] [Google Scholar]
- 33. McFerrin L. G., and Atchley W. R. (2011) Evolution of the Max and Mlx networks in animals. Genome Biol. Evol. 3, 915–937 10.1093/gbe/evr082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Shea J. M., and Ayer D. E. (2013) Coordination of nutrient availability and utilization by MAX- and MLX-centered transcription networks. Cold Spring Harb. Perspect. Med. 3, a014258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaadige M. R., Looper R. E., Kamalanaadhan S., and Ayer D. E. (2009) Glutamine-dependent anapleurosis dictates glucose uptake and cell growth by regulating MondoA transcriptional activity. Proc. Natl. Acad. Sci. U.S.A. 106, 14878–14883 10.1073/pnas.0901221106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peterson C. W., and Ayer D. E. (2011) An extended Myc network contributes to glucose homeostasis in cancer and diabetes. Front. Biosci. (Landmark Ed.) 16, 2206–2223 10.2741/3848 [DOI] [PubMed] [Google Scholar]
- 37. Poungvarin N., Chang B., Imamura M., Chen J., Moolsuwan K., Sae-Lee C., Li W., and Chan L. (2015) Genome-wide analysis of ChREBP binding sites on male mouse liver and white adipose chromatin. Endocrinology 156, 1982–1994 10.1210/en.2014-1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shih H. M., Liu Z., and Towle H. C. (1995) Two CACGTG motifs with proper spacing dictate the carbohydrate regulation of hepatic gene transcription. J. Biol. Chem. 270, 21991–21997 10.1074/jbc.270.37.21991 [DOI] [PubMed] [Google Scholar]
- 39. Diolaiti D., McFerrin L., Carroll P. A., and Eisenman R. N. (2015) Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis. Biochim. Biophys. Acta 1849, 484–500 10.1016/j.bbagrm.2014.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang P., Metukuri M. R., Bindom S. M., Prochownik E. V., O'Doherty R. M., and Scott D. K. (2010) c-Myc is required for the CHREBP-dependent activation of glucose-responsive genes. Mol. Endocrinol. 24, 1274–1286 10.1210/me.2009-0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Havula E., and Hietakangas V. (2018) Sugar sensing by ChREBP/Mondo-Mlx-new insight into downstream regulatory networks and integration of nutrient-derived signals. Curr. Opin. Cell Biol. 51, 89–96 10.1016/j.ceb.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 42. Ishii S., Iizuka K., Miller B. C., and Uyeda K. (2004) Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc. Natl. Acad. Sci. U.S.A. 101, 15597–15602 10.1073/pnas.0405238101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jeong Y. S., Kim D., Lee Y. S., Kim H. J., Han J. Y., Im S. S., Chong H. K., Kwon J. K., Cho Y. H., Kim W. K., Osborne T. F., Horton J. D., Jun H. S., Ahn Y. H., Ahn S. M., and Cha J. Y. (2011) Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PLoS One 6, e22544 10.1371/journal.pone.0022544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma L., Tsatsos N. G., and Towle H. C. (2005) Direct role of ChREBP: Mlx in regulating hepatic glucose-responsive genes. J. Biol. Chem. 280, 12019–12027 10.1074/jbc.M413063200 [DOI] [PubMed] [Google Scholar]
- 45. Ma L., Robinson L. N., and Towle H. C. (2006) ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J. Biol. Chem. 281, 28721–28730 10.1074/jbc.M601576200 [DOI] [PubMed] [Google Scholar]
- 46. Sans C. L., Satterwhite D. J., Stoltzman C. A., Breen K. T., and Ayer D. E. (2006) MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol. Cell. Biol. 26, 4863–4871 10.1128/MCB.00657-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stoltzman C. A., Peterson C. W., Breen K. T., Muoio D. M., Billin A. N., and Ayer D. E. (2008) Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc. Natl. Acad. Sci. U.S.A. 105, 6912–6917 10.1073/pnas.0712199105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Postic C., Dentin R., Denechaud P. D., and Girard J. (2007) ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu. Rev. Nutr. 27, 179–192 10.1146/annurev.nutr.27.061406.093618 [DOI] [PubMed] [Google Scholar]
- 49. Uyeda K., and Repa J. J. (2006) Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 4, 107–110 10.1016/j.cmet.2006.06.008 [DOI] [PubMed] [Google Scholar]
- 50. Kvittingen E. A. (1995) Tyrosinaemia–treatment and outcome. J. Inherit. Metab. Dis. 18, 375–379 10.1007/BF00710049 [DOI] [PubMed] [Google Scholar]
- 51. Jackson L. E., Kulkarni S., Wang H., Lu J., Dolezal J. M., Bharathi S. S., Ranganathan S., Patel M. S., Deshpande R., Alencastro F., Wendell S. G., Goetzman E. S., Duncan A. W., and Prochownik E. V. (2017) Genetic dissociation of glycolysis and the TCA cycle affects neither normal nor neoplastic proliferation. Cancer Res. 77, 5795–5807 10.1158/0008-5472.CAN-17-1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Overturf K., Al-Dhalimy M., Tanguay R., Brantly M., Ou C. N., Finegold M., and Grompe M. (1996) Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat. Genet. 12, 266–273 10.1038/ng0396-266 [DOI] [PubMed] [Google Scholar]
- 53. Marongiu F., Serra M. P., Sini M., Marongiu M., Contini A., and Laconi E. (2014) Cell turnover in the repopulated rat liver: distinct lineages for hepatocytes and the biliary epithelium. Cell Tissue Res. 356, 333–340 10.1007/s00441-014-1800-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miyaoka Y., Ebato K., Kato H., Arakawa S., Shimizu S., and Miyajima A. (2012) Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr. Biol. 22, 1166–1175 10.1016/j.cub.2012.05.016 [DOI] [PubMed] [Google Scholar]
- 55. Mehta R. (1995) The potential for the use of cell proliferation and oncogene expression as intermediate markers during liver carcinogenesis. Cancer Lett. 93, 85–102 10.1016/0304-3835(95)03790-4 [DOI] [PubMed] [Google Scholar]
- 56. Tapia C., Kutzner H., Mentzel T., Savic S., Baumhoer D., and Glatz K. (2006) Two mitosis-specific antibodies, MPM-2 and phospho-histone H3 (Ser28), allow rapid and precise determination of mitotic activity. Am. J. Surg. Pathol. 30, 83–89 10.1097/01.pas.0000183572.94140.43 [DOI] [PubMed] [Google Scholar]
- 57. Nikiforov M. A., Chandriani S., O'Connell B., Petrenko O., Kotenko I., Beavis A., Sedivy J. M., and Cole M. D. (2002) A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell. Biol. 22, 5793–5800 10.1128/MCB.22.16.5793-5800.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rogulski K. R., Cohen D. E., Corcoran D. L., Benos P. V., and Prochownik E. V. (2005) Deregulation of common genes by c-Myc and its direct target, MT-MC1. Proc. Natl. Acad. Sci. U.S.A. 102, 18968–18973 10.1073/pnas.0507902102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rothermund K., Rogulski K., Fernandes E., Whiting A., Sedivy J., Pu L., and Prochownik E. V. (2005) C-Myc-independent restoration of multiple phenotypes by two C-Myc target genes with overlapping functions. Cancer Res. 65, 2097–2107 10.1158/0008-5472.CAN-04-2928 [DOI] [PubMed] [Google Scholar]
- 60. Yin X., Grove L., Rogulski K., and Prochownik E. V. (2002) Myc target in myeloid cells-1, a novel c-Myc target, recapitulates multiple c-Myc phenotypes. J. Biol. Chem. 277, 19998–20010 10.1074/jbc.M200860200 [DOI] [PubMed] [Google Scholar]
- 61. Edgar R., Domrachev M., and Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybidization array data repository. Nucleic Acids Res. 30, 207–210 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Birrer M. J., Raveh L., Dosaka H., and Segal S. (1989) A transfected L-myc gene can substitute for c-myc in blocking murine erythroleukemia differentiation. Mol. Cell. Biol. 9, 2734–2737 10.1128/MCB.9.6.2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carroll P. A., Diolaiti D., McFerrin L., Gu H., Djukovic D., Du J., Cheng P. F., Anderson S., Ulrich M., Hurley J. B., Raftery D., Ayer D. E., and Eisenman R. N. (2015) Deregulated Myc requires MondoA/Mlx for metabolic reprogramming and tumorigenesis. Cancer Cell 27, 271–285 10.1016/j.ccell.2014.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cole M. D., and McMahon S. B. (1999) The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18, 2916–2924 10.1038/sj.onc.1202748 [DOI] [PubMed] [Google Scholar]
- 65. Malynn B. A., de Alboran I. M., O'Hagan R. C., Bronson R., Davidson L., DePinho R. A., and Alt F. W. (2000) N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 14, 1390–1399 [PMC free article] [PubMed] [Google Scholar]
- 66. Iizuka K., and Horikawa Y. (2008) ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr. J. 55, 617–624 10.1507/endocrj.K07E-110 [DOI] [PubMed] [Google Scholar]
- 67. Sae-Lee C., Moolsuwan K., Chan L., and Poungvarin N. (2016) ChREBP regulates itself and metabolic genes implicated in lipid accumulation in β-cell line. PLoS One 11, e0147411 10.1371/journal.pone.0147411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Suzuki T., Muramatsu T., Morioka K., Goda T., and Mochizuki K. (2015) ChREBP binding and histone modifications modulate hepatic expression of the Fasn gene in a metabolic syndrome rat model. Nutrition 31, 877–883 10.1016/j.nut.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 69. Witte N., Muenzner M., Rietscher J., Knauer M., Heidenreich S., Nuotio-Antar A. M., Graef F. A., Fedders R., Tolkachov A., Goehring I., and Schupp M. (2015) The glucose sensor ChREBP links de novo lipogenesis to PPARγ activity and adipocyte differentiation. Endocrinology 156, 4008–4019 10.1210/EN.2015-1209 [DOI] [PubMed] [Google Scholar]
- 70. Dang C. V., Le A., and Gao P. (2009) MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15, 6479–6483 10.1158/1078-0432.CCR-09-0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Goetzman E. S., and Prochownik E. V. (2018) The role for Myc in coordinating glycolysis, oxidative phosphorylation, glutaminolysis, and fatty acid metabolism in normal and neoplastic tissues. Front. Endocrinol. (Lausanne) 9, 1–25 10.3389/fendo.2018.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mikawa T., LLeonart M. E., Takaori-Kondo A, Inagaki N., Yokode M., and Kondoh H. (2015) Dysregulated glycolysis as an oncogenic event. Cell. Mol. Life Sci. 72, 1881–1892 10.1007/s00018-015-1840-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ward P. S., and Thompson C. B. (2012) Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 10.1016/j.ccr.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Reznik E., Miller M. L., Şenbabaoğlu Y., Riaz N., Sarungbam J., Tickoo S. K., Al-Ahmadie H. A., Lee W., Seshan V. E., Hakimi A. A., and Sander C. (2016) Mitochondrial DNA copy number variation across human cancers. ELife 5, e10769 10.7554/eLife.10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dolezal J. M., Wang H., Kulkarni S., Jackson L., Lu J., Ranganathan S., Goetzman E. S., Bharathi S. S., Beezhold K., Byersdorfer C. A., and Prochownik E. V. (2017) Sequential adaptive changes in a c-Myc-driven model of hepatocellular carcinoma. J. Biol. Chem. 292, 10068–10086 10.1074/jbc.M117.782052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Delarue J., and Magnan C. (2007) Free fatty acids and insulin resistance. Curr. Opin. Clin. Nutr. Metab. Care 10, 142–148 10.1097/MCO.0b013e328042ba90 [DOI] [PubMed] [Google Scholar]
- 77. Bae J. S., Oh A. R., Lee H. J., Ahn Y. H., and Cha J. Y. (2016) Hepatic Elovl6 gene expression is regulated by the synergistic action of ChREBP and SREBP-1c. Biochem. Biophys. Res. Commun. 478, 1060–1066 10.1016/j.bbrc.2016.08.061 [DOI] [PubMed] [Google Scholar]
- 78. Kassam A., Miao B., Young P. R., and Mukherjee R. (2003) Retinoid X receptor (RXR) agonist-induced antagonism of farnesoid X receptor (FXR) activity due to absence of coactivator recruitment and decreased DNA binding. J. Biol. Chem. 278, 10028–10032 10.1074/jbc.M208312200 [DOI] [PubMed] [Google Scholar]
- 79. Massafra V., and van Mil S. W. C. (2018) Farnesoid X receptor: a “homeostat” for hepatic nutrient metabolism. Biochim. Biophys. Acta 1864, 45–59 10.1016/j.bbadis.2017.10.003 [DOI] [PubMed] [Google Scholar]
- 80. Zhao C., and Dahlman-Wright K. (2010) Liver X receptor in cholesterol metabolism. J. Endocrinol. 204, 233–240 10.1677/JOE-09-0271 [DOI] [PubMed] [Google Scholar]
- 81. Patel M. S., Nemeria N. S., Furey W., and Jordan F. (2014) The pyruvate dehydrogenase complexes: structure-based function and regulation. J. Biol. Chem. 289, 16615–16623 10.1074/jbc.R114.563148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wanders R. J., and Tager J. M. (1998) Lipid metabolism in peroxisomes in relation to human disease. Mol. Aspects Med. 19, 69–154 [DOI] [PubMed] [Google Scholar]
- 83. Arseneault M., Monlong J., Vasudev N. S., Laskar R. S., Safisamghabadi M., Harnden P., Egevad L., Nourbehesht N., Panichnantakul P., Holcatova I., Brisuda A., Janout V., Kollarova H., Foretova L., Navratilova M., et al. (2017) Loss of chromosome Y leads to down regulation of KDM5D and KDM6C epigenetic modifiers in clear cell renal cell carcinoma. Sci. Rep. 7, 44876 10.1038/srep44876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Komura K., Jeong S. H., Hinohara K., Qu F., Wang X., Hiraki M., Azuma H., Lee G. S., Kantoff P. W., and Sweeney C. J. (2016) Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc. Natl. Acad. Sci. U.S.A. 113, 6259–6264 10.1073/pnas.1600420113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tseng Y. Y., and Bagchi A. (2015) The PVT1-MYC duet in cancer. Mol. Cell. Oncol. 2, e974467 10.4161/23723556.2014.974467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. O'Callaghan C., and Vassilopoulos A. (2017) Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell 16, 1208–1218 10.1111/acel.12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. van de Ven R. A. H., Santos D., and Haigis M. C. (2017) Mitochondrial sirtuins and molecular mechanisms of aging. Trends Mol. Med. 23, 320–331 10.1016/j.molmed.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Loboda A., Damulewicz M., Pyza E., Jozkowicz A., and Dulak J. (2016) Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell. Mol. Life Sci. 73, 3221–3247 10.1007/s00018-016-2223-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rushworth S. A., and Macewan D. J. (2011) The role of nrf2 and cytoprotection in regulating chemotherapy resistance of human leukemia cells. Cancers 3, 1605–1621 10.3390/cancers3021605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kulkarni S., Dolezal J. M., Wang H., Jackson L., Lu J., Frodey B. P., Dosunmu-Ogunbi A., Li Y., Fromherz M., Kang A., Santana-Santos L., Benos P. V., and Prochownik E. V. (2017) Ribosomopathy-like properties of murine and human cancers. PLoS One 12, e0182705 10.1371/journal.pone.0182705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dolezal J. M., Dash A. P., and Prochownik E. V. (2018) Diagnostic and prognostic implications of ribosomal protein transcript expression patterns in human cancers. BMC Cancer 18, 275 10.1186/s12885-018-4178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yu S., Rao S., and Reddy J. K. (2003) Peroxisome proliferator-activated receptors, fatty acid oxidation, steatohepatitis and hepatocarcinogenesis. Curr. Mol. Med. 3, 561–572 10.2174/1566524033479537 [DOI] [PubMed] [Google Scholar]
- 93. Nelson D. R., Koymans L., Kamataki T., Stegeman J. J., Feyereisen R., Waxman D. J., Waterman M. R., Gotoh O., Coon M. J., Estabrook R. W., Gunsalus I. C., and Nebert D. W. (1996) P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6, 1–42 10.1097/00008571-199602000-00002 [DOI] [PubMed] [Google Scholar]
- 94. Nelson D. R., Zeldin D. C., Hoffman S. M., Maltais L. J., Wain H. M., and Nebert D. W. (2004) Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 14, 1–18 10.1097/00008571-200401000-00001 [DOI] [PubMed] [Google Scholar]
- 95. Nebert D. W., Wikvall K., and Miller W. L. (2013) Human cytochromes P450 in health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120431 10.1098/rstb.2012.0431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D. R., Tessarollo L., Casellas R., Zhao K., and Levens D. (2012) c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 10.1016/j.cell.2012.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Müller I., Larsson K., Frenzel A., Oliynyk G., Zirath H., Prochownik E. V., Westwood N. J., and Henriksson M. A. (2014) Targeting of the MYCN protein with small molecule c-MYC inhibitors. PLoS One 9, e97285 10.1371/journal.pone.0097285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang H., Sharma L., Lu J., Finch P., Fletcher S., and Prochownik E. V. (2015) Structurally diverse c-Myc inhibitors share a common mechanism of action involving ATP depletion. Oncotarget 6, 15857–15870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zirath H., Frenzel A., Oliynyk G., Segerström L., Westermark U. K., Larsson K., Munksgaard Persson M., Hultenby K., Lehtiö J., Einvik C., Påhlman S., Kogner P., Jakobsson P. J., and Henriksson M. A. (2013) MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. U.S.A. 110, 10258–10263 10.1073/pnas.1222404110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. O'Neil N. J., Bailey M. L., and Hieter P. (2017) Synthetic lethality and cancer. Nat. Rev. Genet. 18, 613–623 10.1038/nrg.2017.47 [DOI] [PubMed] [Google Scholar]
- 101. Landay M., Oster S. K., Khosravi F., Grove L. E., Yin X., Sedivy J., Penn L. Z., and Prochownik E. V. (2000) Promotion of growth and apoptosis in c-myc nullizygous fibroblasts by other members of the myc oncoprotein family. Cell Death Differ. 7, 697–705 10.1038/sj.cdd.4400701 [DOI] [PubMed] [Google Scholar]
- 102. Kafri M., Metzl-Raz E., Jonas F., and Barkai N. (2016) Rethinking cell growth models. FEMS Yeast Res. 16, fow081 10.1093/femsyr/fow081 [DOI] [PubMed] [Google Scholar]
- 103. Li G. W., Burkhardt D., Gross C., and Weissman J. S. (2014) Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 10.1016/j.cell.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Marguerat S., and Bähler J. (2012) Coordinating genome expression with cell size. Trends Genet. 28, 560–565 10.1016/j.tig.2012.07.003 [DOI] [PubMed] [Google Scholar]
- 105. Prochownik E. V., and VanAntwerp M. E. (1993) Differential patterns of DNA binding by myc and max proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 960–964 10.1073/pnas.90.3.960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Desbarats L., Gaubatz S., and Eilers M. (1996) Discrimination between different E-box-binding proteins at an endogenous target gene of c-myc. Genes Dev. 10, 447–460 10.1101/gad.10.4.447 [DOI] [PubMed] [Google Scholar]
- 107. Fernandez P. C., Frank S. R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., and Amati B. (2003) Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 10.1101/gad.1067003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Allevato M., Bolotin E., Grossman M., Mane-Padros D., Sladek F. M., and Martinez E. (2017) Sequence-specific DNA binding by MYC/MAX to low-affinity non-E-box motifs. PLoS One 12, e0180147 10.1371/journal.pone.0180147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shi Z., and Barna M. (2015) Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 31, 31–54 10.1146/annurev-cellbio-100814-125346 [DOI] [PubMed] [Google Scholar]
- 110. Jouihan H. (2012) Measurement of liver triglyceride content. Bio-protocol 2, e223 10.21769/BioProtoc.223 [DOI] [Google Scholar]
- 111. Graves J. A., Rothermund K., Wang T., Qian W., Van Houten B., and Prochownik E. V. (2010) Point mutations in c-Myc uncouple neoplastic transformation from multiple other phenotypes in rat fibroblasts. PLoS One 5, e13717 10.1371/journal.pone.0013717 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.