Summary
Hypertrophic white adipose tissue (WAT) represents a maladaptive mechanism linked to the risk for developing type 2 diabetes in humans. However, the molecular events that predispose WAT to hypertrophy are poorly defined. Here, we demonstrate that adipocyte hypertrophy is triggered by loss of the corepressor GPS2 during obesity. Adipocyte-specific GPS2 deficiency in mice (GPS2 AKO) causes adipocyte hypertrophy, inflammation, and mitochondrial dysfunction during surplus energy. This phenotype is driven by HIF1A activation that orchestrates inadequate WAT remodeling and disrupts mitochondrial activity, which can be reversed by pharmacological or genetic HIF1A inhibition. Correlation analysis of gene expression in human adipose tissue reveals a negative relationship between GPS2 and HIF1A, adipocyte hypertrophy, and insulin resistance. We propose therefore that the obesity-associated loss of GPS2 in adipocytes predisposes for a maladaptive WAT expansion and a pro-diabetic status in mice and humans.
Keywords: corepressor, transcription, adipose tissue, obesity, type 2 diabetes, insulin resistance, GPS2, HIF1A
Graphical Abstract

Highlights
-
•
Adipose-specific GPS2 deficiency predisposes for adipocyte hypertrophy
-
•
Loss of GPS2 triggers transcriptional activation of HIF1A pathways
-
•
Deregulation of GPS2-HIF1A interplay provokes disrupted mitochondrial activity
-
•
GPS2 and HIF1A levels are negatively correlated in human adipose tissue
Drareni et al. identify a role for the transcriptional corepressor GPS2 in the regulation of adipocyte hypertrophy. They provide evidence that adipocyte-specific loss of GPS2 predisposes toward maladaptive adipose tissue expansion and pro-diabetic status through activation of HIF1A transcriptional activity.
Introduction
Obesity is a highly complex disease and one of the strongest risk factors for the development of insulin resistance (IR) and type 2 diabetes (T2D). The primary cause of T2D is obesity-driven insulin resistance in liver, white adipose tissue (WAT), and skeletal muscle, combined with insufficient secretion of insulin by pancreatic β-cells to overcome this resistance. WAT plasticity has a pivotal role in the development of T2D because it has the capacity to change its dimension in response to nutritional status by remodeling through a series of mechanisms (Klöting et al., 2010, Muir et al., 2016). During positive energy imbalance, adipose tissue expands to store the surplus, but this WAT expansion can become dysfunctional, favoring ectopic fat deposition in other tissues, leading to metabolic dysregulation, progressive insulin resistance, and increased risk for T2D (Blüher, 2010). Maladaptive WAT expansion has been associated with numerous deleterious consequences, including inflammation, fibrosis, hypoxia, and disrupted mitochondrial function (Kusminski et al., 2016, Sun et al., 2011). WAT expansion is achieved through enlargement of pre-existing adipocytes by lipid accumulation (hypertrophy) or by increasing adipocytes number through recruitment of pre-adipocytes from the resident pool of progenitor cells within adipose tissue (hyperplasia). Adipocyte hypertrophy is associated with metabolic alterations such as glucose intolerance and hyperinsulinemia in humans (obese and non-obese) that increases the risk for developing T2D independently of total fat mass (Muir et al., 2016). The balance between hyperplastic and hypertrophic adipose tissue is driven by alterations of adipocyte metabolism that include lipogenesis, lipid catabolism, and adipogenesis. Adiponectin overexpression in genetically obese (Ad-Tg ob/ob) mice showed increased adipogenic capacity (hyperplasia) with a “healthier” phenotype despite a massive obesity (Kim et al., 2007). Moreover, increased lipid catabolism by regulation of mitochondrial function as reported in the MitoNEET model limits adipocyte enlargement and insulin resistance (Kusminski et al., 2012, Wang et al., 2017). However, the molecular mechanisms contributing to the unfavorable sequelae of obese adipose tissue are yet to be fully understood.
A number of recent studies suggest that alterations of the epigenome, driven by transcription factors and coregulators, influence adipose tissue plasticity in mice and humans (Rosen, 2016, Toubal et al., 2013b). Among the candidate coregulators, subunits of the HDAC3 corepressor complex (reviewed in Treuter et al., 2017) have emerged as important regulators of the adipose tissue phenotype in obesity (Cardamone et al., 2014, Cardamone et al., 2018, Cederquist et al., 2016, Fan et al., 2016, Fang et al., 2011, Li et al., 2011, Rohm et al., 2013, Toubal et al., 2013a). Although apparently different aspects of the respective KO phenotypes were characterized, these studies collectively imply that subunit-selective alterations of diverse repression pathways potentially trigger transcriptional reprogramming of adipocytes, which may also affect adipose tissue plasticity favoring hypertrophy.
Of note, earlier work by us and others suggests that a core subunit of the HDAC3 corepressor complex, named G protein pathway suppressor 2 (GPS2), acts as a key regulatory checkpoint in adipose tissue adaptation (Cardamone et al., 2014, Toubal et al., 2013a). By studying human WAT, we previously reported that reduced expression of GPS2 in adipocytes, driven by deregulation of regulatory circuits controlled by the transcription factor TWIST1, coincides with the increased expression of inflammatory genes (Toubal et al., 2013a). These human data are consistent with results obtained from obese and diabetic mouse models that lacked or overexpressed GPS2, both supporting an anti-inflammatory role of GPS2 in adipocytes (Cardamone et al., 2012, Fan et al., 2016, Toubal et al., 2013a). However, the extent to which GPS2 also controls additional pathways that limit metabolic disturbance and the progression of obesity toward T2D is currently unknown.
Here we discover a previously unrecognized role of GPS2 in the transcriptional control of adipocyte hypertrophy. Our study identifies an inflammation-independent mechanism that links alterations of an epigenome regulator to adipose tissue dysfunction and T2D. At the mechanistic level, we find that adipocyte-specific loss of GPS2 in AKO mice triggers transcriptional alterations of WAT remodeling gene signatures linked to the transcription factor HIF1A. Our functional analysis further reveals that GPS2 deregulation and HIF1A over-activation disrupts mitochondrial activity that otherwise limits adipose tissue expansion. Additionally, we demonstrate that pharmacological and genetic inhibition of HIF1A partially reverses the GPS2 AKO phenotype. Finally, correlative analysis in humans revealed a relationship among WAT GPS2 levels, hypertrophic adipocytes, HIF1A levels, and insulin resistance.
Results
Loss of GPS2 in Adipocytes Promotes Adipocyte Hypertrophy and Unhealthy Adipose Tissue Expansion
To investigate whether and how GPS2 controls AT expansion and glucose metabolism during high-fat diet (HFD) feeding, we generated adipocyte-specific Gps2-knockout mice (referred to as GPS2 AKO mice). The reduction of Gps2 mRNA and protein is efficient and was demonstrated in mature isolated and cultured adipocytes of WATs from GPS2 AKO mice compared with wild-type (WT) mice (Figure S1A). WT littermate controls and GPS2 AKO mice were fed an HFD from 7 weeks of age for 1, 4, and 12 weeks. During these periods, there were no significant differences in body weight, food intake, or energy expenditure between the two genotypes (Figures 1A and S1C). Moreover, lean and fat mass and the weight of fat pads were not different between the two genotypes (Figures S1B and S1D). To determine the effects of GPS2 deficiency in mature adipocytes on glucose homeostasis, we compared glucose tolerance and insulin response between GPS2 AKO and WT mice in normal chow diet (CD)-fed and in HFD-fed conditions. GPS2 AKO mice were more glucose intolerant than WT controls after 4 and 12 weeks of HFD (Figure 1B), while no difference was observed in CD-fed mice. The systemic insulin sensitivity tests upon those conditions were similar between the two genotypes (Figure S1E).
Figure 1.
The Loss of GPS2 in Adipocytes Predisposes to Aberrant WAT Remodeling and Glucose Intolerance
(A) Body weight curve during a time course of 12 weeks of CD and 1, 4, and 12 weeks of HFD of WT and GPS2 AKO mice (CD, n = 8; 1 week HFD, n = 7; 4 weeks HFD, n = 12; 12 weeks HFD, n = 13).
(B) Oral glucose tolerance test (OGTT) in WT and GPS2 AKO mice in normal CD and after 4 and 12 weeks of HFD (CD, n = 8; 4 weeks HFD, n = 12; 12 weeks HFD, n = 13).
(C) Representative H&E and perilipin immunofluorescence staining and average of the adipocyte size of eWAT from WT and GPS2 AKO mice upon normal CD and after 1, 4, and 12 weeks of HFD (CD, n = 8; 1 week HFD, n = 7; 4 weeks HFD, n = 12; 12 weeks HFD, n = 13). Scale bars, 100 μm.
(D) Basal or insulin-stimulated phospho-AKT western blotting in eWAT of WT and GPS2 AKO mice after 1 and 4 weeks of HFD (n = 3).
(E) Measurement of basal or insulin-stimulated glucose uptake (using 2-deoxyglucose) on eWAT explants from WT and GPS2 AKO mice after 4 weeks of HFD (n = 3).
(F) Basal or isoproterenol-stimulated phospho-HSL, HSL, and ATGL western blotting on explants of eWAT of WT and GPS2 AKO mice after 4 weeks of HFD (n = 3).
(G) Basal or isoproterenol-stimulated concentration of glycerol and NEFA in the eWAT explant media from WT and GPS2 AKO mice after 4 weeks of HFD (n = 3).
(H) RT-qPCR analysis of Atgl in eWAT and serum concentration of NEFA from WT and GPS2 AKO mice under normal CD and after 1, 4, and 12 weeks of HFD (CD, n = 8; 1 week HFD, n = 7; 4 weeks HFD, n = 12; 12 weeks HFD, n = 13).
All data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figures S1 and S2.
Next, we analyzed adipocyte size in epididymal WAT (eWAT) and inguinal WAT (ingWAT) of WT and GPS2 AKO mice (Figures 1C and S1F). Under normal CD-fed conditions, we did not observe significant differences in eWAT and ingWAT adipocyte size between WT and GPS2 AKO mice (Figures 1C and S1F). However, on the HFD, GPS2 AKO mice were characterized by a significant increase of adipocyte size in eWAT and ingWAT compared with WT control mice. This hypertrophic phenotype of GPS2 AKO mice was seen after only 1 week of HFD, and the difference increased with the duration of the feeding time (Figures 1C and S1F). This increase of adipocyte size of both fat pads in HFD-fed GPS2 AKO mice was corroborated with an impairment of eWAT insulin sensitivity, measured by insulin-stimulated AKT phosphorylation and insulin-stimulated glucose uptake, in the GPS2 AKO mice at 1 and 4 weeks after HFD (Figures 1D and 1E).
Additionally, WAT of AKO mice was characterized by higher macrophage accumulation, adipose tissue inflammation, and adipokine deregulation (Figures S1G–S1I, S2A, and S2B). We also observed increased lipolysis in eWAT and ingWAT of AKO mice compared with WT upon HFD. This was characterized by increased Atgl expression and NEFA concentration and isoproterenol-stimulated lipolysis (Figures 1F–1H, S2B, and S2C). The increase of WAT lipolysis in GPS2 AKO mice was corroborated with a moderate ectopic fat deposition in liver compared with WT controls (Figure S2D). Taken together, these results suggest that the loss of GPS2 in adipocytes predisposes WAT to maladaptation during energy surplus.
GPS2 Deficiency in Adipocytes Potentiates HIF1A-Dependent Pathways
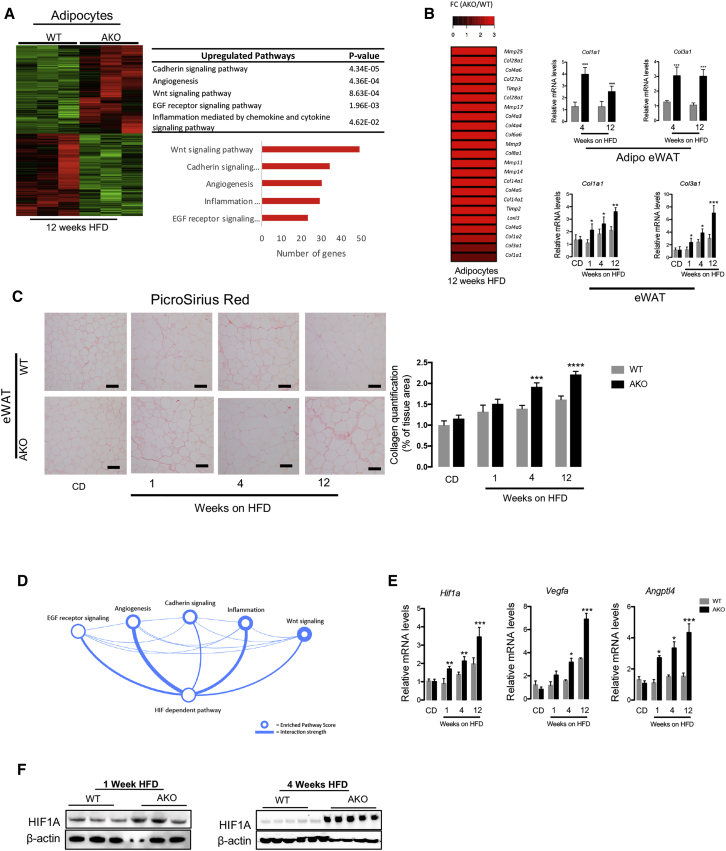
RNA sequencing from isolated adipocytes of eWAT from WT control and GPS2 AKO mice fed an HFD for 12 weeks (Figure 2A) revealed that 2,239 transcripts were induced in isolated adipocytes of eWAT from GPS2 AKO mice relative to WT mice. Among these upregulated transcripts were genes involved in inflammation, as expected, but also genes likely involved in adipose tissue expansion (including cadherin/Wnt signaling pathways and angiogenesis), suggesting a crucial function of GPS2 in adipocyte remodeling (Figure 2A). Importantly, this signature was also seen at the whole-tissue level in GPS2 AKO mice (Figures S3A and S3B).
Figure 2.
HIF1A Signaling Is Enhanced in GPS2-Deficient Adipocytes during HFD Feeding
(A) Heatmap representing global gene expression in eWAT adipocytes of WT and GPS2 AKO mice after 12 weeks of HFD (n = 3). Gene enrichment analyses of the top-upregulated pathways enriched in the datasets.
(B) Heatmap representing the “remodeling” genes upregulated in eWAT adipocytes of the GPS2 AKO compared with WT mice after 12 weeks of HFD (n = 3). RT-qPCR analysis of Col1a1 and Col3a1 in isolated adipocytes and eWAT from WT and GPS2 AKO mice after 4 and 12 weeks of HFD (4 weeks HFD, n = 12; 12 weeks HFD, n = 13).
(C) Representative picrosirius red staining images and collagen quantification in eWAT from WT and GPS2 AKO mice upon normal CD and after 1, 4, and 12 weeks of HFD (CD, n = 8; 1 week HFD, n = 7; 4 weeks HFD, n = 12; 12 weeks HFD, n = 13). Scale bars, 100 μm.
(D) Network analyses of biological processes deregulated in eWAT adipocytes of GPS2 AKO mice after 12 weeks of HFD.
(E) RT-qPCR analysis of Hif1a, Vegfa, and Angptl4 genes in eWAT of WT and GPS2 AKO mice under normal CD and after 1, 4, and 12 weeks of HFD (CD, n = 8; 1 week HFD, n = 7; 4 weeks HFD, n = 12; 12 weeks HFD, n = 13).
(F) HIF1A western blotting in isolated eWAT adipocytes of WT and GPS2 AKO mice after 1 and 4 weeks of HFD (n = 3 and n = 5, respectively).
All data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S3.
Interestingly, our transcriptome analysis revealed that isolated adipocytes of GPS2 AKO mice fed an HFD are characterized by significant increases of genes encoding for collagens and enzymes involved in extracellular matrix remodeling, such as MMPs and TIMPs (Figure 2B). The increased expression of Col1a1 and Col3a1 in isolated adipocytes and eWAT of AKO mice fed an HFD was confirmed by RT-qPCR analyses (Figure 2B). Consistent with this, collagen deposition was gradually increased in eWAT and ingWAT of AKO mice compared with WT mice during the time of regimen (Figures 2C and S3C).
Network analysis of genes significantly increased in eWAT adipocytes of GPS2 AKO mice indicated that HIF1A-dependent pathways could play a central role in the disrupted adipocyte adaptation in GPS2 AKO mice (Figure 2D). To confirm this, we measured the expression levels of Hif1a and its target genes (Vegfa and Angptl4) in eWAT, ingWAT, and eWAT adipocytes after CD and 1, 4, and 12 weeks of HFD (Figures 2E, S4A, and S4B). In both adipose tissue depots and isolated adipocytes, Hif1a and its target genes (Vegfa and Angptl4) were significantly increased in GPS2 AKO mice compared with WT control mice. HIF1A was also strongly upregulated at the protein levels in eWAT of 1 and 4 week HFD-fed GPS2 AKO mice compared with WT mice (Figure 2F).
Next, we executed series of experiments to investigate how GPS2 potentially controls the transcriptional activity of HIF1A. First, we analyzed GPS2 occupancy at promoters of hypoxic and remodeling genes, particularly onto Vegfa, Hif1a, and Angptl4 promoter regions (Figure 3A). Published chromatin immunoprecipitation sequencing (ChIP-seq) data (Cardamone et al., 2014) and our own ChIP-qPCR experiments demonstrate that GPS2 is bound onto the promoters of these genes that could explain their increased expression in GPS2-deficient adipocytes (Figure 3B). This was confirmed by quantifying the histone mark H3K4me3, linked to active transcription, at the Hif1a promoter (Figure 3C). Intriguingly, de novo transcription factor motif analysis of GPS2-bound regions revealed that HIF1A might be the main target transcription factor for GPS2 in adipocytes (Figure 3D). Second, transcription assays using a Vegfa-promoter-luciferase reporter demonstrated that overexpression of GPS2 represses HIF1A-dependent induction of Vegfa promoter activity (Figure 3E). Third, interaction of the two factors was confirmed using co-immunoprecipitation from cellular extracts expressing HA-GPS2 and Flag-tagged HIF1A and further revealed that GPS2 interacts with HIF1A as strongly as with its known direct binding partners in the complex, NCOR and SMRT (Figure 3F). The interaction between overexpressed GPS2 and endogenous HIF1A is confirmed in 3T3-L1 adipocytes (Figure 3F). Forth, we cultured primary adipocytes from WT and GPS2 AKO mice under hypoxic conditions (1% O2 instead of 21% O2) for 6 hr. We then analyzed Hif1a, Vegfa, Angptl4, Col1a1, Col3a1, and Hk2 mRNA levels. Expression of these genes was significantly increased after 6 hr exposure to hypoxia, demonstrating that GPS2-deficient adipocytes are more sensitive to a hypoxic environment (Figure 3G). The higher sensitivity of WAT from GPS2 AKO mice to hypoxia (on the basis of Vegfa expression levels) was also confirmed in vivo, where WT and GPS2 AKO mice were maintained in hypoxic chambers (10% O2 instead of 21% O2) for 1 week (Figure S4C). Importantly, ectopic expression of GPS2 or GFP in primary adipocyte of GPS2 AKO mice cultured upon hypoxia strongly reversed the expression of hypoxia-response genes such as Hif1a, Vegfa, Angptl4, Col1a1, Col3a1, and Hk2 (Figures 3H and S4D). In addition, overexpression of GPS2 in 3T3-L1 mature adipocytes cultured upon hypoxia significantly reduced expression of these hypoxia-sensitive genes (Figures 3I and S4D). Collectively, our data suggest that GPS2 exerts a repressive action on HIF1A.
Figure 3.
GPS2 Directly Controls the Transcriptional Activity of HIF1A
(A) Genome browser tracks of GPS2 peak distribution at Hif1a, Vegfa, and Angptl4 promoters in 3T3-L1 cells (from public database; Cardamone et al., 2014).
(B) ChIP-qPCR analysis of GPS2 recruitment onto Hif1a and Vegfa loci in mature 3T3L1. IgG was used as control.
(C) H3K4me3 measurement by ChIP-qPCR onto Hif1a loci in mature 3T3L1 shGfp (as control) and shGps2. IgG was used as control.
(D) GPS2 binding motif analysis at promoters of GPS2-targeted genes in 3T3-L1 cells.
(E) Repression assays in differentiated 3T3-L1 cells. HIF1A response element from Vegfa promoter upstream a gene encoding for luciferase was transfected into 3T3-L1 cells with HIF1A or GPS2.
(F) Co-immunoprecipitation of HA-GPS2 with Flag-HIF1A in HEK293 cells. Flag-SMRT (aa 2–297), Flag-NCOR (aa 2–320), and HA-GPS2 were used as positive controls. Co-immunoprecipitation of HA-GPS2 with endogenous HIF1A in 3T3-L1 cells.
(G) RT-qPCR analysis of Hif1a, Vegfa, Hk2, Angptl4, Col1a1, and Col3a1 genes in primary adipocytes from ingWAT of WT and GPS2 AKO mice under normoxic or 6 hr hypoxic conditions (n = 4).
(H) RT-qPCR analysis of Hif1a, Vegfa, Hk2, Angptl4, Col1a1, and Col3a1 genes in primary adipocytes from ingWAT of WT and GPS2 AKO mice overexpressing adenovirus encoding GFP (as control) or GPS2 upon normoxic or 6 hr hypoxic conditions (n = 4).
(I) RT-qPCR analysis of Hif1a, Vegfa, Agptl4, Col1a1, and Col3a1 target genes in differentiated 3T3-L1 cells overexpressing adenovirus encoding GFP (as control) or GPS2 upon normoxic or 6 hr hypoxic conditions (n = 6).
All data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S4.
Mitochondrial Dysfunction in GPS2-Deficient Adipocytes Limits Adipose Tissue Remodeling
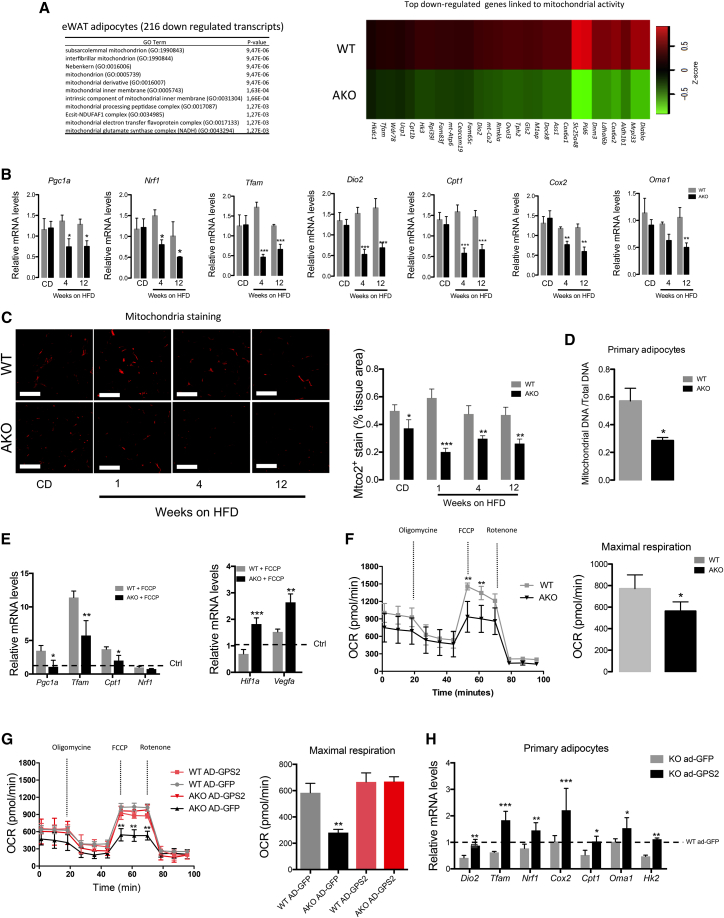
Increased activity of HIF1A in adipocytes is suggested to provoke mitochondrial dysfunction, which restricts fatty acid oxidation and favors adipocyte hypertrophy (Krishnan et al., 2012, Zhang et al., 2010). Interestingly, our adipocyte transcriptome analyses revealed that a significant proportion of the dysregulated genes were linked to mitochondrial activity (GPS2 AKO mice compared with WT littermates upon 12 weeks of HFD) (Figure 4A). Indeed, a significant downregulation of genes involved in mitochondrial function (Cpt1, Cox2, and Oma1) and biogenesis (Pgc1a, Nrf1, Tfam, and Dio2) was observed in eWAT and ingWAT of GPS2 AKO mice (Figures 4B and S5A). These expression data were confirmed by quantifying mitochondria within eWAT of WT and GPS2 AKO mice upon normal CD and HFD for 1, 4, and 12 weeks by mitochondrial staining (Figure 4C). MtDNA was also significantly reduced in primary cultured adipocytes from ingWAT of GPS2 AKO mice compared with WT control mice (Figure 4D), which supports a dysregulation of mitochondrial function in adipocytes from GPS2 AKO mice.
Figure 4.
The Loss of GPS2 in Adipocytes Disrupts Mitochondrial Activity
(A) Cellular component analysis of 216 downregulated genes and heatmap representing the top downregulated genes linked to mitochondrial activity in eWAT adipocytes from WT and GPS2 AKO mice after 12 weeks of HFD (n = 3).
(B) RT-qPCR analysis of mitochondrial genes involved in biogenesis and function in eWAT of WT and GPS2 AKO under normal CD and 4 and 12 weeks of HFD (CD, n = 8; 4 weeks HFD, n = 12; 12 weeks HFD, n = 13).
(C) Representative images of mitochondrial (MTCO2) immunofluorescence staining and quantification in eWAT from WT and GPS2 AKO mice in normal chow diet (CD) and after 1, 4, and 12 weeks of HFD (CD, n = 8; 1 week HFD, n = 7; 4 weeks HFD, n = 12; 12 weeks HFD, n = 3). Scale bars, 100 μm.
(D) Quantification of mtDNA of primary-differentiated adipocytes from ingWAT of WT and GPS2 AKO mice (n = 4)
(E) RT-qPCR analysis of Hif1a, Vegfa, and mitochondrial genes Pgc1a, Tfam, Cpt1, and Nrf1 in differentiated 3T3-L1 cells upon FCCP treatment for 12 hr (n = 6).
(F) Oxygen consumption rate (OCR) of primary-differentiated adipocytes from ingWAT of WT and GPS2 AKO mice (n = 4).
(G) OCR of primary-differentiated adipocytes from ingWAT of WT and GPS2 AKO mice (n = 4) overexpressing adenovirus encoding GFP (as control) or GPS2 (n = 5).
(H) RT-qPCR analysis of mitochondrial genes involved in biogenesis and function in primary-differentiated adipocytes from ingWAT of WT and GPS2 AKO mice (n = 4) overexpressing adenovirus encoding GFP (as control) or GPS2 (n = 5).
All data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S5.
To explore this hypothesis, WT and AKO adipocytes were treated with the uncoupling agent carbonylcyanide-4-(trifluomethoxy)phenylhydrazone (FCCP) to depolarize mitochondria for 12 hr, followed by measuring the expression of genes involved in mitochondrial function. Upon FCCP treatment, we observed significant decreases of Pgc1a, Tfam, and Cpt1 in adipocytes of AKO mice compared with WT (Figure 4E). Interestingly, Hif1a and Vegfa mRNA was significantly increased in AKO adipocytes upon FCCP treatment (Figure 4E). Consistent with this disrupted mitochondrial function, FCCP-induced oxygen consumption rate (OCR) was significantly altered in cultured primary adipocytes from WAT and brown adipose tissue (BAT) of GPS2 AKO mice compared with WT mice as measured by extracellular flux analysis (Figures 4F and S5B). OCR stimulation by palmitate treatment is also impaired in AKO adipocytes compared with WT adipocytes (Figure S5C).
To further support that disrupted mitochondrial function was due to the loss of GPS2, we ectopically rescued GPS2 expression in GPS2 AKO primary adipocytes and then measured OCR. We found that GPS2 re-expression in GPS2-deficient adipocytes restored mitochondrial function by increasing OCR, comparable with WT mice (Figure 4G). This was confirmed at the gene expression level (Figure 4H). A similar phenotype was observed in BAT primary adipocytes (Figure S5D).
Increased mitochondrial activity and biogenesis is an essential biological process involved in WAT remodeling and browning following cold exposure and β-adrenergic receptor stimulation (Inagaki et al., 2016). To visualize the faults in mitochondrial activity and adaptation in vivo, WT and GPS2 AKO mice were subjected either to treatment with an agonist of β3-adrenergic receptor or cold exposure for 5 days. Cold exposure or β3-adrenergic stimulation provoked, in WT mice, increased mitochondrial biogenesis and adipose tissue beiging characterized by increasing MTCO2 and UCP-1 staining, respectively (Figure 5A). In contrast, GPS2 AKO mice upon those conditions did not respond as WT control mice (Figure 5A). Mitochondrial biogenesis and WAT beiging (on the basis of UCP-1 staining) was strongly impaired in GPS2 AKO mice upon both conditions in ingWAT and BAT (Figures 5A and S6A).
Figure 5.
Disrupted Mitochondrial Activity in GPS2 AKO Mice Limits Adipose Tissue Remodeling upon β3-Adrenegic Receptor Agonist and Cold Exposure
(A) Representative images of mitochondrial (MTCO2), UCP1 immunofluorescence staining, and H&E staining of ingWAT from WT and GPS2 AKO control vehicle-treated mice (n = 8), after β3-adrenegic receptor agonist treatment (n = 8) and upon cold exposure during 5 days (n = 12). Scale bars, 100 μm.
(B) Heatmap representing global gene expression pattern analyses of the most dysregulated pathways and heatmap representing the top downregulated in ingWAT of WT and GPS2 AKO mice treated with β3-adrenegic receptor agonist treatment (n = 3).
(C) RT-qPCR analysis of mitochondrial genes involved in biogenesis and function in ingWAT of WT and GPS2 AKO control vehicle-treated mice, after β3-adrenegic receptor agonist treatment (n = 4) or cold exposure (n = 6).
(D) HIF1A western blotting (n = 3) and mRNA levels in ingWAT WT and GPS2 AKO mice after β3-adrenegic receptor agonist treatment (n = 4) or cold exposure (n = 6).
(E) Body temperature of WT and AKO mice after 5 days of cold exposure and measurement of Vo2 consumption during cold exposure (n = 6).
All data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S6.
RNA sequencing of ingWAT from WT and GPS2 AKO mice upon treatment with β3-adrenergic receptor agonist revealed that the 216 downregulated genes were linked to lipid metabolism and nicotinic receptor pathways, which may reflect a deregulation of the mitochondrial activity and adipose tissue browning processes (Figure 5B). This genome-wide signature of ingWAT from GPS2 AKO mice was confirmed by quantifying genes involved in mitochondrial function and WAT browning and BAT (Figures 5C and S6B). Importantly, these inadequate responses to cold exposure or β3-adrenergic receptor agonist treatment observed in GPS2 AKO mice were corroborated with an increased expression of Hif1a in ingWAT of GPS2 AKO mice compared with WT control mice (Figure 5D). Adipose tissue maladaptation to cold exposure of GPS2 AKO mice was characterized at the whole-body level by a significant decrease in body temperature and alteration of O2 consumption (Figure 5E). Collectively, these data suggest that the dysfunctional adipose tissue observed in GPS2 AKO mice could be in part driven by disrupted mitochondrial activity through HIF1A over-activation.
Inhibition of HIF1A in GPS2 AKO Mice Reverses the Pro-diabetic Phenotype
To test above hypothesis, 12 week HFD-fed WT and GPS2 AKO mice were treated with the pharmacologic inhibitor of HIF1A (PX-478) or vehicle (PBS) for 10 days. Subsequent characterization of the metabolic phenotype of these mice did not reveal differences in body weight of the two genotypes after PX-478 or vehicle treatments (Figure S7A). Consistent with earlier findings (Sun et al., 2013), HIF1A inhibition improved glucose tolerance by 15% in WT control mice (Figure 6A). Importantly, the effect of HIF1A inhibition on glucose tolerance was more pronounced (40% improvement) in GPS2 AKO mice, supporting that HIF1A was more active in GPS2 AKO mice than in WT controls (Figure 6A). Unexpectedly, GPS2 AKO mice become more insulin sensitive after PX-478 treatment (Figure S7B). At the adipose tissue level, the pro-diabetogenic phenotype characteristic of GPS2 AKO mice was corrected by HIF1A inhibition. First, we did not observe significant differences between WT controls and GPS2 AKO mice in eWAT and ingWAT phenotypes regarding adipocyte size, number of mitochondria, and WAT inflammation (Figures 6B–6D, S7C, S7D, and S8A). Lipolytic genes, plasma concentration of triglyceride (TG) and hepatic steatosis of GPS2 AKO mice were also normalized to WT conditions after PX-478 treatment (Figures S7E and S7F). Second, the overexpression of HIF1A and its target genes in HFD-fed GPS2 AKO mice was corrected by PX-478 treatment to a similar level of WT control mice in eWAT and ingWAT (Figures 6C and S8B). Third, expression levels of the genes linked to mitochondria activity (Cpt1, Cox2, Oma1, Pgc1a, Nrf1, Tfam, and Dio2) in eWAT and ingWAT of GPS2 AKO mice were similar to WT mice, suggesting that mitochondrial activity of GPS2 AKO was restored upon HIF1A inhibition (Figures 6E and S8C). To further confirm that upon HIF1A inhibition the mitochondrial function was restored in AKO mice, WT and GPS2 AKO mice were subjected to PX-478 treatment followed by of β3-adrenergic receptor stimulation for 5 days. The results show that PX-478 treatment restored the response to β3-adrenergic receptor stimulation by inducing ingWAT browning (characterized by mitochondrial biogenesis and UCP staining) in AKO mice to levels observed in WT mice (Figures 6F and 6G).
Figure 6.
Pharmacologic Inhibition of HIF1A Reverses the GPS2 AKO Phenotype
(A) Oral glucose tolerance test (OGTT) of HFD-fed WT and GPS2 AKO mice after control vehicle (n = 4) or PX-478 (n = 7) treatment for 10 days. Percentage of area under the curve (AUC) of the OGTT after versus before PX-478 treatment (n = 7).
(B) Representative images of H&E staining and average of adipocyte size of eWAT from HFD-fed WT and GPS2 AKO mice after control vehicle (n = 4) or PX-478 (n = 7) treatment for 10 days. Scale bars, 100 μm.
(C) RT-qPCR analysis of Hifa, Vegfa, and Angptl4 genes in eWAT of HFD-fed WT and GPS2 AKO mice after control vehicle (n = 4) or PX-478 (n = 7) treatment for 10 days.
(D) Representative images of mitochondrial (MTCO2) immunofluorescence staining and quantification in eWAT from WT and GPS2 AKO mice after control vehicle (n = 4) or PX-478 (n = 7) treatment for 10 days (n = 7). Scale bars, 100 μm.
(E) RT-qPCR analysis of mitochondrial genes involved in biogenesis and function in eWAT of HFD-fed WT and GPS2 AKO mice after control vehicle (n = 4) or PX-478 (n = 7) treatment for 10 days.
(F) Representative images of mitochondrial (MTCO2), UCP1 immunofluorescence staining, and H&E staining of ingWAT from WT and GPS2 AKO mice after PX-478 treatment for 10 days overlapping with 5 days β3-adrenegic receptor agonist treatment (n = 6). Scale bars, 100 μm.
(G) RT-qPCR analysis of mitochondrial genes involved in biogenesis and function in eWAT of HFD-fed WT and GPS2 AKO mice after 10 days of PX-478 treatment overlapping with 5 days β3-adrenegic receptor agonist treatment (n = 6). Scale bars, 100 μm.
(H) Oxygen consumption rate (OCR) of primary adipocytes from ingWAT of WT and GPS2 AKO mice in presence or absence of 10 μM PX-478 for 16 hr (n = 4) and OCR of primary adipocytes from ingWAT of WT and GPS2 AKO mice treated with siRNA control or siRNA targeting Hif1a for 24 hr (n = 6).
All data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figures S7 and S8.
To substantiate these in vivo observations, pharmacological inhibition and small interfering RNA (siRNA)-mediated depletion of HIF1A were performed in WT and GPS2 AKO adipocytes, and O2 consumption was measured (Figure 6H). Under these conditions, HIF1A inhibition or depletion restored O2 consumption in GPS2 AKO mice to a similar level as the WT controls (Figure 6H).
Altogether, the results support the hypothesis that the loss of GPS2 initially causes HIF1A activation, which then triggers transcriptional re-reprogramming to orchestrate mitochondrial dysfunction, adipocyte hypertrophy, WAT remodeling and insulin resistance, and systemic glucose intolerance.
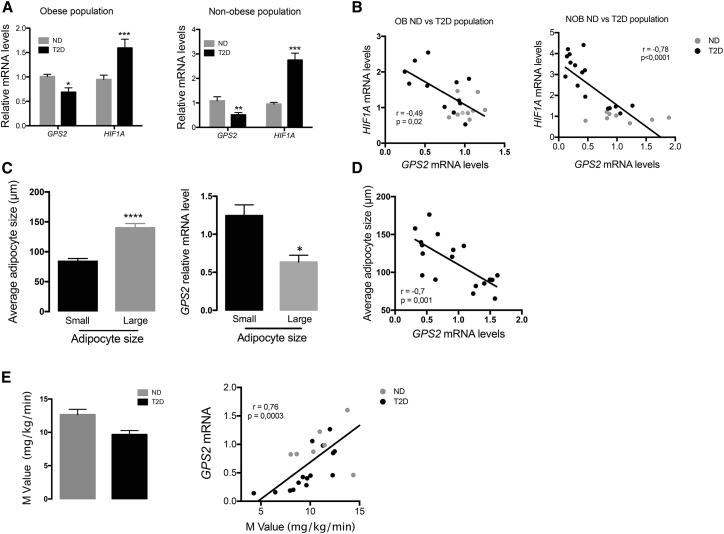
Adipose Tissue Maladaptation and HIF1A Levels Are Negatively Correlated to GPS2 Levels in Humans
To evaluate the human relevance of our experimental mouse data, we measured GPS2 and HIF1A transcript levels in subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT) in two populations of non-obese and obese subjects with or without diagnosed T2D (Table S1, populations 1 and 2). GPS2 expression levels were significantly reduced in non-obese and obese diabetic subjects compared with subjects without T2D, while HIF1A mRNA levels were significantly upregulated in diabetic subjects compared with non-diabetic subjects (Figure 7A). Correlative analysis of GPS2 and HIF1A expression showed significant negative correlation between these two factors in two different populations of non-obese (SAT) and obese (VAT) subjects (Figure 7B). Next, we selected obese subjects with small (diameter < 100 μm) versus large (diameter > 120 μm) adipocytes (Table S1, population 3). Measuring GPS2 expression in the two subgroups revealed that GPS2 expression was higher in the population with smaller adipocytes (Figure 7C). Accordingly, GPS2 expression was significantly correlated with the average adipocyte size of this population (Figure 7D). Of note, we could not find a significant correlation between GPS2 mRNA levels and adipocyte size by stratifying obese patients according to the diabetic status (data not shown).
Figure 7.
GPS2 mRNA Levels in Human Adipose Tissue Are Negatively Correlated to Adipocyte Size, HIF1A mRNA Levels, and Systemic Insulin Resistance
(A) mRNA expression levels of GPS2 and HIF1A in SAT or VAT of non-obese (n = 18) and obese (n = 21) subjects with or without diagnosed T2D.
(B) Correlative analysis of the expression of GPS2 versus HIF1A (B) in SAT and VAT from non-obese (NOB) (n = 18) and obese (OB) (n = 21) populations.
(C) Average adipocytes size from obese subject classified into small (n = 9) and large (n = 9) and GPS2 expression into these two classes of subjects.
(D) Correlative analysis of GPS2 expression with adipocyte size from obese subjects (population 2) and mRNA expression of GPS2 in large versus small adipocytes (n = 9).
(E) Measurement of the M value by hyperinsulinemic-euglycemic clamp in non-obese subjects with our without diagnosed T2D (ND, n = 8; T2D, n = 10). Correlative analysis of the expression of GPS2 versus M value in non-obese subjects with our without diagnosed T2D.
All data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Table S1.
Finally, instead of having the opportunity to measure adipocyte size in the non-obese population 1, we performed a 100 min euglycemic hyperinsulinemic clamp at 80 mU/m2/min insulin infusion to measure insulin-stimulated glucose disposal rate (M value) while blood glucose was clamped at 100 mg/dL (5.56 mol/L) (Nguewa et al., 2017). We found that GPS2 level was negatively correlated to the M value, which indicates that decreased expression of GPS2 in adipose tissue might be linked to insulin resistance even in non-obese humans (Figure 7E).
Collectively, these correlation data point at a conserved key function of GPS2 in influencing adipocyte metabolism and WAT fate in humans.
Discussion
The expansion of WAT significantly influences adipocyte biology and subsequently interferes with whole-body glucose homeostasis. Adipocyte hypertrophy represents a marker and a driver of adipose tissue dysfunction in non-obese and obese populations (Acosta et al., 2016). Quantitative associations have been found between adipocyte size thresholds for increased risk for T2D in obese and non-obese populations (Andersson et al., 2014, Cotillard et al., 2014). Thus, the factors linking obesity and T2D are associated mainly with the hypertrophic WAT phenotype rather than the amount of fat itself. However, the mechanisms underlying the predisposition of WAT to hypertrophy are incompletely understood.
Our findings suggest that the loss of the transcriptional corepressor GPS2 in mature adipocytes is sufficient to predispose WAT to pathological expansion characterized by hypertrophic adipocytes independently of the weight gain. GPS2 AKO mice recapitulate pro-diabetic traits, including WAT insulin resistance, ectopic fat deposition, and systemic glucose intolerance. GPS2 AKO mice are prone to dysfunctional fat depots. We provide evidence that this maladaptive WAT expansion seen in GPS2-deficient adipocytes is a consequence of the uncontrolled activation of the transcription factor HIF1A, thereby identifying a hitherto unknown transcription factor target of the corepressor GPS2.
HIF1A, the master regulator of hypoxia, is well known to be increased during obesity in WAT of humans and mice and has been linked to the pathological changes driving obesity-associated disorders. Overexpression of HIF1A (WT or dominant negative) in WAT accelerates adipocyte hypertrophy and insulin resistance, while HIF1A ablation in WAT leads to metabolic improvements in obese mice (Halberg et al., 2009, Jiang et al., 2011, Lee et al., 2014, Zhang et al., 2010).
Intriguing similarities exist between the phenotype of our GPS2 AKO mice and transgenic HIF1A mice (Halberg et al., 2009, Lee et al., 2014). We have experimentally confirmed the antagonistic interplay between GPS2 and HIF1A by demonstrating that pharmacological or genetic inhibition of HIF1A in GPS2 AKO mice could partially reverse the WAT phenotype. Notably, our data suggest that GPS2 might interfere with HIF1A action in adipocytes at multiple transcriptional levels. GPS2 may restrict the accessibility of HIF1A to promoter regions, and/or GPS2 may directly repress the transcriptional activity of HIF1A. In support of the latter scenario, SMRT and HDAC3, subunits of GPS2 complex, were proposed to control HIF1A activity at the Vegfa promoter (He et al., 2011). These results suggest that GPS2 controls the activity of HIF1A as integral part of an adipocyte corepressor sub-complex containing SMRT but not NCOR. Indeed, the phenotype of adipocyte-specific NCOR KO mice has the opposite phenotype of our GPS2 AKO mice (Li et al., 2011), characterized by adequate WAT expansion and optimal insulin sensitivity during obesity mainly because of the over-activation of PPARγ, the key mediator of adipogenesis.
Up to now, there is no adipose-specific KO model for SMRT, which limits the comparison with our GPS2 AKO mouse model. However, a mutant model for SMRT lacking the nuclear receptor-binding domain 1 (SMRTmRID1) has been described (Fang et al., 2011). Importantly, SMRTmRID1 mice do not develop any metabolic alterations when chow fed, while this SMRTmRID1 model is characterized by hypertrophic adipocytes and systemic glucose intolerance upon diet-induced obesity, as we observed in GPS2 AKO mice. The hypertrophic phenotype of SMRTmRID1 mice is linked to disrupted activity of mitochondria, probably driven by a decrease of mitochondrial biogenesis and fatty acid oxidation. The decreased activity of mitochondrial in SMRTmRID1 mice might be due to decreased expression of PGC1A, key mediator of mitochondrial biogenesis and activity. We also observed a decreased PGC1A expression in adipocytes of GPS2 AKO, which might explain the decreased number of mitochondria.
Interestingly, HIF1A has been reported to interfere with PGC1A action in adipocytes, which provokes alterations of adipocyte fatty acid catabolism (Krishnan et al., 2012). HIF1A over-expression in adipocytes causes the transcriptional inhibition of sirtuin 2, leading to the decreased de-acetylation of PGC1A (the active form), which then suppresses expression of genes involved in fatty acid catabolism in the mitochondria. Of note is also that HDAC3, the enzymatic core subunit of the corepressor complex, has been reported to de-acetylate PGC1A in brown adipocytes (Emmett et al., 2017). In that study, loss of HDCA3 in adipocytes caused deficient WAT remodeling and browning, while the opposite was reported for an independent HDAC3 AKO model (Ferrari et al., 2017). Interestingly, we show here that WAT browning was strongly impaired in GPS2 AKO upon cold exposure or β3-adrenergic receptor stimulation, suggesting that GPS2 might cooperate with HDAC3 to influence PGC1A-dependent browning, in addition to its cooperation with HIF1A (Basse et al., 2017).
TBLR1 is another corepressor complex subunit that controls lipid mobilization in WAT during fasting or energy surplus. Mice lacking TBLR1 in adipocytes have impaired fasting-induced lipolysis, which accelerates adiposity and glucose intolerance. Even if the adipocyte TBLR1 KO phenotype shares similarities with the GPS2 AKO, we believe that the underpinning molecular mechanisms are different. In particular, lipolysis was enhanced in GPS2 AKO mice, opposite to the reduction evident in TBLR1 AKO mice. It is possible that GPS2 cooperates in adipocytes with TBL1 rather than with TBLR1, in agreement with earlier in vitro data (Huang et al., 2015).
Complementary to repressive mechanisms of action, GPS2 may in some contexts act as transcriptional “co-activator” or “pioneer cofactor” via earlier identified mechanisms involving histone demethylases, including KDM4A and H3K9 demethylation (Cardamone et al., 2014, Jakobsson et al., 2009). Notably, GPS2 has been described to positively regulate nuclear-encoded mitochondrial (neMITO) genes and lipolytic PPARγ target genes in the 3T3L1 adipocyte cell line (Cardamone et al., 2014, Cardamone et al., 2018). This is consistent with the downregulation of 216 genes, including neMITO genes, in our GPS2 AKO mice. Although we do not exclude that some downregulated genes may be under direct positive control of GPS2, we believe that our data strongly support the model that the repressive action of GPS2 upon HIF1A is a major mechanism that favors appropriate mitochondrial function upon energy surplus.
We note that the GPS2-HIF1A relationship became not evident in recently reported adipocyte-specific GPS2 KO mice and in studies using GPS2-depleted 3T3-L1 adipocytes in vitro (Cardamone et al., 2014, Cardamone et al., 2018, Cederquist et al., 2016). A main reason could be the surprisingly different, in part opposite, AKO phenotype of the two GPS2 AKO models, generated via different exon targeting strategies and in different genetic mouse backgrounds (e.g., mixed 129sv/C57BL6J versus C57BL6J for our GPS2 AKO mice). Strikingly, AKO mice generated by Perissi and coworkers (Cederquist et al., 2016) become fatter with age on a CD, which was not seen in our GPS2 AKO mice, neither on chow nor on HFD. It appears puzzling that this adiposity phenotype was associated with an increase of insulin sensitivity (Cederquist et al., 2016), opposite to our data and despite disrupted mitochondrial function (Cardamone et al., 2018). Because alterations of mitochondrial biogenesis and activity have been described to be deleterious for glucose homeostasis and insulin signaling in WAT (Choo et al., 2006, Fucho et al., 2017, Ritov et al., 2005), the current discrepancy is unclear. A detailed comparative characterization of the respective GPS2 AKO models would be needed to reveal possible model-specific alterations in the interactions between these key pathways.
The human relevance of our mouse study is supported by correlation data in human populations. Both together allow to propose a conserved role of the epigenome regulator GPS2, along with its corepressor complex, in determining WAT fate and expansion under surplus energy toward a dysmetabolic state and risk for developing T2D. Despite the importance of genetics in obesity and T2D, recent epigenome-wide association studies (EWAS) have suggested a key role of epigenetic modifications in whole-body glucose homeostasis (Morris et al., 2012, Voight et al., 2010). Additionally, DNA methylation of WAT was also altered in diabetic subjects and associated with whole-body glucose homeostasis, inflammation, and metabolism (Andersen et al., 2018, Perfilyev et al., 2017). However, most of the current association-based studies cannot yet answer whether epigenome alterations are a cause, a consequence, or both, of the metabolic alterations during disease progression.
Modulations of histone modifications are attractive targets for future epigenetic therapies because of their reversibility that potentially permits the reprogramming of adipocytes into a “healthier” state. Pharmacological inhibition or genetic deletions of specific histone de-acetylases or DNA methyltransferases have already been shown to exert profound beneficial metabolic effects (e.g., by affecting whole-body glucose homeostasis) (Sharma and Taliyan, 2016). As a consequence, our findings raise the possibility that targeting the adipocyte epigenome via the GPS2-HIF1A axis could be used therapeutically to limit inadequate WAT expansion during obesity.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti Mitochondria antibody [MTC02] | Abcam | Cat# ab3298, RRID:AB_303683 |
| Goat polyclonal anti Perilipin antibody | Abcam | Cat# ab118605, RRID:AB_10901587 |
| Rabbit polyclonal anti UCP-1 antibody | Sigma-Aldrich | Cat# U6382, RRID:AB_261838 |
| Mouse monoclonal anti HIF1a antibody | Novus | Cat# NB100-105G, RRID:AB_1643855 |
| Mouse monoclonal anti beta Actin antibody | Abcam | Cat# ab8226, RRID:AB_306371 |
| Rabbit polyclonal anti Phospho-AKT (Ser21/9) antibody | Cell Signaling Technology | Cat# 9331, RRID:AB_329830 |
| Rabbit polyclonal anti AKT antibody | Cell Signaling Technology | Cat# 9272, RRID:AB_329827 |
| Mouse monoclonal anti flag antibody | Sigma-Aldrich | Cat# F3165, RRID:AB_259529 |
| Rabbit polyclonal anti HSL antibody | Cell Signaling Technology | Cat# 4107, RRID:AB_2296900 |
| Rabbit polyclonal anti Phospho-HSL (Ser660) antibody | Cell Signaling Technology | Cat# 4126, RRID:AB_490997 |
| Rabbit polyclonal anti ATGL Antibody | Cell Signaling Technology | Cat# 2138, RRID:AB_2167955 |
| Biological Samples | ||
| Human Adipose tissue biopsies | Lariboisière hospital and Geoffroy Saint-Hilaire | N/A |
| Mouse adipose tissue explants | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CL316,243 hydrate | Sigma | Cat#C5976 |
| PX-478 HCl - 10 mg | Clinisciences | Cat#A14319-10 |
| PX-478 HCl - 10mM ∗ 1mL in DMSO | Clinisciences | Cat#A14319-10mM-D |
| Insulin (HUMALOG 100 unites/ml) | lilly | N/A |
| Insulin, Human Recombinant | Sigma | Cat#I-9278 |
| Rosiglitazone | Sigma | Cat#R-2408 |
| Iso-butyl-methylxanthine | Sigma | Cat#I5879-SG |
| Dexamethasone | Sigma | Cat#D1756 |
| Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) | Sigma | Cat#C2920 |
| Critical Commercial Assays | ||
| Mouse Adiponectin/Acrp30 DuoSet ELISA | R&D systeme | Cat#DY1119 |
| Mouse Leptin DuoSet ELISA | R&D systeme | Cat# DY498 |
| Randox Triglycerides | RANDOX | Cat#tR213 |
| Randox NEFA (Non-Esterified Fatty Acids) | RANDOX | Cat#FA115 |
| Mitochondrial DNA Isolation | abcam | Cat#ab65321 |
| Dual-Luciferase® Reporter Assay System | Promega | Cat#E1910 |
| Glucose Uptake Fluorometric Assay Kit | SIGMA-ALDRICH | Cat# MAK084-1KT |
| Deposited Data | ||
| Data files for RNA sequencing | NCBI Gene expression | GEO: GSE111647 |
| Experimental Models: Cell Lines | ||
| 3T3L1 | ATCC | Cat#CRL-3242 |
| HEK293 | ATCC | Cat#CRL-1573 |
| Primary pre-adipocytes | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Adipoq-Cre | Jackson Laboratory | 010803 |
| Mouse: Gps2flox/flox mice | Fan et al., 2016 | Generated at Ozgene Pty, Ltd. (Bentley DC, Australia) |
| Oligonucleotides | ||
| See Table S2 for the list of Oligos | N/A | N/A |
| pre-designed Hif1a siRNA, AM16708, Assay ID 158955 | ThermoFisher Scientific | Cat#AM16708, Assay ID 158955 |
| siRNA Control (, Silencer Negative Control) | ThermoFisher Scientific | Please add cat# |
| Recombinant DNA | ||
| pGL3 luciferase vector | This paper | N/A |
| Empty pcDNA3.1 | This paper | N/A |
| Renilla luciferase | This paper | N/A |
| HIF1a plasmide | This paper | N/A |
| GPS2 plasmide | Prof. Eckardt Treuter Departement of Bioscience and Nutrition, Karolinska Institutet | N/A |
| pcDNA3-HA-GPS2 | Prof. Eckardt Treuter Departement of Bioscience and Nutrition, Karolinska Institutet | N/A |
| Flag-tagged HIF1A | Prof. Jorge Ruas at Department of Physiology and Pharmacology, Karolinska Institutet | N/A |
| Flag-tagged SMRT (2–297) and NCOR (2-320) | Prof. Eckardt Treuter Departement of Bioscience and Nutrition, Karolinska Institutet | N/A |
| Ad-GPS2 | Prof. Eckardt Treuter Departement of Bioscience and Nutrition, Karolinska Institutet | N/A |
| Ad-GFP | Prof. Eckardt Treuter Departement of Bioscience and Nutrition, Karolinska Institutet | N/A |
| Software and Algorithms | ||
| FIJI | FIJI Software | https://fiji.sc/ |
| Graphpad 7 | GraphPad Software | https://www.graphpad.com/ |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Nicolas Venteclef (nicolas.venteclef@inserm.fr).
Experimental Model and Subject Details
Human populations
Adipose tissue biopsies (SAT and VAT) were obtained from different populations admitted to the Lariboisière and Geoffroy Saint-Hilaire hospitals (Paris, France). Clinical and anthropometric data are detailed in Populations 1 and 2 summarized in Table S1. The study was conducted in accordance with the Helsinki Declaration and was registered in a public trial registry (Clinicaltrials.gov; NCT02368704) as previously described (Fan et al., 2016). The Ethics Committee of CPP Ile-de-France approved the clinical investigations for all individuals, and written informed consent was obtained from all individuals. SAT and VAT samples from non-obese subjects were obtained after local surgery, while SAT and VAT from obese subjects was obtained during bariatric surgery.
Animals
Gps2flox/flox mice were generated at Ozgene Pty, Ltd. (Bentley DC, Australia) using a targeting construct, which contained loxP sites flanking exons 2 and 5, followed by a FRT site and a neomycin cassette inserted between exons 5 and 6. The targeting vector was electroporated into C57BL/6 Bruce4 embryonic stem (ES) cells. The correctly recombined ES colony was then injected into C57BL/6 blastocysts. Male chimeras were mated with female C57BL/6 mice to get mice with a targeted Gps2 allele. The mice were crossbred with C57BL/6 flp-recombinase mice to remove the neomycin cassette to create heterozygous Gps2flox/+ mice. The mice were then crossbred with C57BL/6 mice for nine generations before being bred with heterozygous Gps2flox/+ mice to get the Gps2flox/flox mice (Fan et al., 2016). To generate adipocyte-specific Gps2 KO mice, Gps2flox/flox mice were crossed with adiponectin-Cre mice (B6;FVB-Tg (Adipoq-Cre) 1Evdr/J; Jackson Laboratory stock no. 010803). KO mice were bred for at least nine generations before the experiments were started. Adipoq-Cre-Gps2flox/flox littermates were used as control. All mice used in the studies were male, between 7– 8 weeks old at the time of the experiment starting point, and randomized before any experiment was started. All animal experiments were approved by the French ethical board (Paris-Sorbonne University, Charles Darwin N°5, 01026.02) and conducted in accordance with the guidelines stated in the International Guiding Principles for Biomedical Research Involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS). All mice strains were bred and maintained at the “Centre exploration fonctionnel (CEF)” at Paris University (UPMC, 91 and 105 boulevard de hospital Paris 13).
Cell cultures
Primary adipocytes isolation and differentiation
Stromal vascular fraction (SVF) was isolated from eWAT, ingWAT and BAT, and primary adipocytes were differentiated in vitro. Briefly, WAT and BAT depots were dissected from euthanized 8-12 weeks old mice. Tissue was digested for 30 min shaking at 37°C with collagenase B (Sigma). Cell suspension was filtered through 200 μm cell strainers. Red Blood cells were lysed and filtered through 70 μm cell strainers. Re-suspended SVF cells were plated in growth medium (DMEM medium supplemented with 10% FCS and 1% Penicillin-Streptomycin) and expanded for 3-5 days. Cells were differentiated in growth medium supplemented with 1 μg/mL insulin, 2 μM rosiglitazone, 0.5 mM iso-butyl-methylxanthine and 0.25 mM dexamethasone. Cells were maintained on differentiation medium for 48-72h, then maintained in growth medium supplemented with only 1 μg/mL insulin for 48-72h. After that, cells were placed in growth medium for 48h.
Cell lines culture
3T3L1 cells were differentiated in growth medium (high-glucose DMEM supplemented with 1 μg/mL insulin, 2 μM rosiglitazone, 0.5 mM iso-butyl-methylxanthine and 0.25 mM dexamethasone. Cells were maintained on differentiation medium for 48 h, then maintained in growth medium supplemented with only 1 μg/mL insulin for 48h. After that, cells were placed in growth medium for 48h.
HEK293FT cells were maintained in growth medium (high-glucose DMEM supplemented with 10% fetal bovine serum and 1% Penicillin–Streptomycin). All cells were maintained in 37°C and 5% CO2 equilibrated with atmospheric O2 in a humidified incubator.
Methods Details
In vivo treatments, metabolic and anthropometric measurements
Diet-induced obesity and insulin resistance
7–8-weeks-old WT and GPS2 AKO mice were fed with a 60%-fat diet (HFD, Research Diets, D12492) for 1, 4 and 12 weeks.
Cold exposure and β3-adrenergic receptor agonist treatment
For cold exposure, mice were subjected to 4°C for 5 days. Mice were sacrificed at day 5. For β3-adrenergic receptor agonist treatment, mice were intraperitoneal (i.p.) injected with CL316,243 hydrate (C5976 Sigma) (1mg/kg) once daily for 5 consecutive days. Mice were sacrificed 24h after the last treatment.
HIF1A inhibitor treatment
For HIF1A inhibitor treatment, 7–8-weeks-old male WT and GPS2 AKO mice (n = 7) were fed with a 60%-fat diet (HFD, Research Diets, D12492). The mice were then dosed by gavage with 5mg/kg of PX-478 (Clinisciences) in PBS or with PBS (as control) every second day. The gavage began 12 weeks after the initial exposure to the HFD and continued for 10 days.
Indirect Calorimetric measurement (metabolic chamber)
For the metabolic cage studies, mice were housed individually in metabolic chambers and maintained on a 12-h dark-light cycle with lights on from 6:00 a.m. to 6:00 p.m. at 4°C or 22°C. Oxygen consumption, CO2 emission, food consumption, movement, and energy expenditure were measured using TSA metabolic chambers (TSA System, Germany) in an open-circuit indirect calorimetric system.
Core body temperature measurements
The measurements of internal core body temperatures were recorded from 12-weeks-old male mice maintained at 4°C using a digital thermometer (Bioseb, BIO-TK8856) and rectal thermocouple probe (Bioseb, RET-3 probe).
Body composition analysis by EchoMRI
Lean tissue and fat mass were measured using on MinispecPlus LF90 (Bruker) according to the manufacturer’s instructions.
Oral glucose tolerance test (OGTT)
Mice were fasted overnight before receiving an oral gavage of glucose (2 g/kg). Blood glucose levels were measured directly from tail vein blood at 0, 15, 30, 45, 60 and 90 min using a glucometer (Accu-Chek Performa, Roche). Blood samples were taken from the tail vein at 0, 15, 30 and 60 min.
Insulin tolerance test (ITT)
Mice were fasted for 5h before receiving an i.p. injection of insulin (0.5 U/kg; Humalog, Lilly). Glycaemia was measured at 0, 15, 30, 45, 60 and 90 min.
Metabolic analyses in vivo were performed on the Functional & Physiological Exploration Platform) in the ICAN PRECLINIC platform (Institute of Cardiometabolism and Nutrition, Pitie-Salpêtrière Hospital, Paris, France), thanks to Amelie Lacombe.
Analysis of different metabolic blood circulating parameters
Adiponectin (Mouse Adiponectin/Acrp30 DuoSet ELISA, R&D Systems) and leptin (Mouse Leptin DuoSet ELISA, R&D Systemes) concentrations were determined by ELISA. Plasma NEFA TG and Glycerol were measured using the colorimetric diagnostic kit, according to the manufacturer’s instructions (Randox Laboratories).
Histology, immunofluorescence and microscopy
WAT and liver tissue samples were fixed in 10% formaldehyde solution overnight and embedded in paraffin. Tissue slides were stained with H&E for the evaluation of the tissue morphology and PicroSirius Red for the quantification of collagen deposition, following standardized protocols. Adipocyte size was measured by the diameters of the adipocytes in light-microscopy images (20X) of AT and liver sections (n = 50 adipocytes per section, three sections per mouse), (1 week HFD n = 7, 4 weeks HFD n = 12, 12 weeks HFD n = 13) and analyzed using ImageJ software. Experiments were done in a blinded manner.
Epitope-specific antibodies were used for IF detection of Perilipin (Abcam, ab118605, 1:200), Mitochondria (Abcam, ab3298, 1:200), UCP1 (Sigma, U6382, 1:500), staining. For IF analysis, cleared and rehydrated sections were quenched with 3% H2O2 for 15 minutes at room temperature then washed twice for 5 minutes each with TBS + 0.1% (vol/vol) Tween-20. Sections were then blocked with TBS + 3% (vol/vol) BSA for 30 minutes at room temperature. Sections were then incubated for one hour at room temperature or overnight at 4°C with diluted primary antibodies, washed 3 times for 5 minutes with TBS + 0.1% (vol/vol) Tween-20, followed by incubation with appropriate flourophore-conjugated secondary antibodies (Invitrogen). Sections were then washed twice for 5 minutes with TBS then mounted with hard-set DAPI-containing mounting media (Vectashield, Vector Labs). Images were acquired on an Axiovert 200M microscope or through whole slide scanning with ZEISS AxioScan.
Quantitative analysis of the collagen deposition, perilipin and mitochondria staining in WAT were performed using positive pixels algorithm (Indica Labs) on digital slides. Results are expressed as percentage of positive pixels.
HIF1A inhibitor treatment
Primary differentiated adipocytes, or mature 3T3L1 cells were treated with 10 mM (in DMSO) of PX-478 (Clinisciences, A14319) or DMSO during 16h.
Hypoxic chamber
Cells were maintained either in normal conditions (37°C and 5% CO2 equilibrated with atmospheric O2 in a humidified incubator) that contains 21% O2 (hereafter referred as normoxia) or in a hypoxia incubator chamber (Stemcell) (1% O2, 5% CO2, balanced with N2 and humidified) that was placed in 37°C (hereafter referred as hypoxia).
Adenoviral infection
For infections, cultured adipocytes were incubated with adenovirus particles for 2 days at MOI 10. Transduction was efficient after 2 days of culture, as determined by western blot analysis and GFP fluorescence.
siRNA treatment
Mouse pre-designed Hif1a siRNA (ThermoFisher Scientific, AM16708, Assay ID 158955) and siRNA Control (ThermoFisher Scientific, Silencer Negative Control) were used. 100 nM siRNAs, along with Lipofectamine 2000 (Life Technologies) were used for the transfection of primary differentiated adipocytes. Transfection was efficient after 24h.
FCCP treatment
Mouse primary differentiated adipocytes from ingWAT were treated with 2,5 μM (in ethanol) of FCCP (Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone) (SIGMA-ALDRICH, C2920) or ethanol (as control) during 12h.
Western blot analysis
Samples were lysed in RIPA buffer supplemented with protease and phosphatase inhibitors and were diluted to a concentration of 20 μg of protein and heated at 99°C for 5 min with a denaturation buffer. Proteins were separated by SDS–PAGE electrophoresis and transferred to PVDF membranes (Amersham International). Membranes were incubated in blocking reagent (PBS with 0.5% Tween 20 and 3% of bovine serum albumin) for 30 min, then in primary antibody (in the blocking solution) overnight at 4°C. The antibodies and their concentrations are the following: anti-HIF1a (Novus, NB100-105; 1:500), anti-β-actin (Abcam, ab8226; 1:30000), anti-phospho-AKT (Cell signaling, 9331S; 1:1000), anti-AKT (Cell signaling, 9272S; 1:1000), anti-Flag (Sigma-Aldrich, F3165; 1:5000), anti-HSL (cell signaling, 4107; 1:1000), anti-phospho-HSL (ser660) (cell signaling, 4126; 1:1000), anti-ATGL (cell signaling, 2138; 1:1000) and custom-made anti-GPS2 N/C ((Fan et al., 2016). After several washes in PBS with 0.5% Tween 20, membranes were incubated in horseradish peroxidase (HRP)-labeled secondary antibodies (1:10000) for 1 h at room temperature in the blocking solution. Membranes were incubated with ECL western-blotting substrate (Pierce, Ref# 32106) and imaged by myECL Imager (ThermoFisher).
Measurement of oxygen consumption
SVF was generated from ingWAT and BAT as described above, and primary adipocytes were differentiated in vitro. For differentiation for Seahorse assay, cells were plated at defined density (20,000 cells/well) into wells of Seahorse XF24 culture plates (Agilent). Oxygen consumption rates in primary adipocytes were measured by Seahorse assay at day 7 of differentiation. Cells were washed once with respiration medium (Seahorse XF base medium with 1 mM pyruvate, 20 mM glucose), supplied with 0.5 mL/well respiration medium, and incubated in CO2-less conditions for 60 min. Equilibrated Seahorse XFe24 cartridges were loaded with port injections to deliver the following drug treatments: oligomycin (1μM final), FCCP (0.5 μM final), rotenone (1μM final). Three measurements were made under basal conditions and after each drug injection. Each measurement cycle had these time parameters: mix 3 min, wait 0 min, measure 3 min. Data are presented as average well OCR (pmol O2/min) at each time point. Error bars represent SEM.
Mitochondrial DNA extraction and quantification
Mitochondrial DNA was extracted with Mitochondrial DNA Isolation Kit (Abcam) according to the manufacturer’s instructions. Total DNA was extracted using AllPrep DNA/RNA Mini Kit (QIAGEN). Mitochondrial and Total DNA were dosed with The Infinite M200 PRO (Tecan) then the ratio Mitochondrial DNA/Total DNA was calculated.
Luciferase activity assay
Luciferase assays were performed by transient transfection of HEK/3T3L1 cells seeded at a density of 105 cells per well (12-well plate) using fugene (3:1 ratio) (Promega). Optimized transfections were performed using 2 ng DNA pGL3 luciferase vector and 2.0 ng Renilla luciferase (for normalization) plus factor expression vectors and/or empty pcDNA3.1. Twenty-four hours after transfection, cell lysates were harvested using a Dual Luciferase kit (Promega) to measure luciferase activities on a Synergy HT plate reader (Biotek).
RT–qPCR analysis
RNA was extracted from tissues or purified cells using the RNeasy RNA Mini Kit (QIAGEN). Complementary DNAs were synthesized using M-MLV Reverse Transcriptase kit (Promega). RT–qPCR was performed using the QuantStudio 3 Real-Time PCR Systems (ThermoFisher Scientific). 18S was used for normalization to quantify relative mRNA expression levels. Relative changes in mRNA expression were calculated using the comparative cycle method (2−ΔΔCt). Primers are listed in Table S2 (see below).
Co-immunoprecipitation of GPS2 and HIF1A
3T3-L1 and HEK293 (ATCC, CRL-1573) cells were co-transfected with pcDNA3-HA-GPS2 (WT) and expression plasmids for a Flag-tagged HIF1A was kindly provided by Prof. Jorge Ruas at Department of Physiology and Pharmacology, Karolinska Institutet. Flag-tagged SMRT (2–297) and NCOR (2-320) served as a positive control for comparison. Cells were lysed 48 h after transfection, and the lysate was incubated with rabbit anti-FLAG (Sigma-Aldrich, F7425)-coupled protein A magnetic beads for 3 h, at 4°C (previously15 μL beads with 2 μg of antibody for 2 h, at 4°C). The beads were washed with lysis buffer five times and eluted at 98°C for 10 min. The eluted samples were loaded in an acrylamide gel by following the western blot protocol, and blotted with anti-FLAG or anti-HA. Whole-cell lysis was used as input.
Explants isolation and treatments
WAT biopsies (0.1 g) were minced and incubated in 1 mL of endothelial cell basal medium (Promocell, Heidelberg, Germany) containing 1% SVF, penicillin (100 units/mL), and streptomycin (100 mg/mL) for 24 h.
Measurement of lipolysis upon isoproterenol stimulation
Explants were washed twice in PBS and stimulated with 10 mM isoproterenol for 30 minutes in ECBM (endothelial cell basal medium) supplemented with 1% SVF and 1% PS. NEFA and glycerol were measured from the supernatants and were calculated relative to tissue weight.
Glucose uptake
To measure the insulin dependent glucose up-take on explants we have used the Glucose up-take Fluorometric Assay Kit from SIGMA-ALDRICH (MAK084) following the manufacturer’s instructions.
RNA sequencing and transcriptomic analysis
The analyses were performed using the Eoulsan pipeline (Jourdren et al., 2012), including read filtering, mapping, alignment filtering, read quantification, normalization, and differential analysis. Before mapping, poly N read tails were trimmed, reads ≤ 40 bases were removed, and reads with quality mean ≤ 30 were discarded. Reads were then aligned against the genome using STAR (version 2.4.0k), and Ensemble annotation version 81 Alignments from reads matching more than once on the reference genome were removed using the Java version of SAMtools. To compute gene expression, GRCm38.p4 GFF3 genome annotation version 81 from the Ensembl database (http://useast.ensembl.org) was used. All overlapping regions between alignments and referenced exons were counted using HTSeqcount 0.5.3 (Anders et al., 2015). The sample counts were normalized using DESeq 1.8.3 (Anders and Huber, 2010). Statistical treatments and differential analyses were also performed using DESeq 1.8.3. For ontological clustering of genes, the database for annotation, visualization, and integrated discovery (EnrichR) was used, and for pathway analyses the Panther classification system was used (Kuleshov et al., 2016). The heatmap were subsequently produced to illustrate the global gene expression for each dataset using heatmap.2 functions from the ggplot2 package of R version 3.3.3. We used an attribute-based layout to visually organize pathways in the network on the basis of their functional attributes. To construct pathway guided network using attributes, Cytoscape (version 3.5.1) was used (Franz et al., 2016). These are (name, value) pairs that map edge and node names to specific data values. RNA sequencing and transcriptome analysis were supported by the France Génomique national infrastructure, funded as part of the “Investissements d’Avenir” program managed by the Agence Nationale de la Recherche (ANR-10-INBS-09).
Lentivirus production and transfection of 3T3L1 cells
For knockdown of Gps2 shRNA oligos targeting Gps2 or GFP, as control, were cloned into pLKO.1 TCR cloning vector (Addgene) and lentivirus particles were produced in human embryonic kidney (HEK) 293T cells. 70% confluent 3T3L1 fibroblasts were transfected with lentivirus in fresh media containing 8 μg/ml Polybrene (Sigma Aldrich). 24 hours after transfection, cells were subject to puromycin selection for 4 days. 3T3L1 shGFP/shGPS2 preadipocytes were differentiate to adipocytes by stimulation with 0.5 mM iso-butyl-methylxanthine, 1 μM dexamethasone and 5 μg/ml insulin for 48 h. Then cells were maintained in growth medium supplement with only 5 μg/ml insulin.
ChIP
ChIP experiments were performed as previously described (Siersbæk et al., 2012) with some minor modifications. Briefly, 3T3-L1 and 3T3-L1 shGFP/shGPS2 adipocytes were double crosslinked with 2 mM disuccinimidyl glutarate (DSG) for 30 min, followed by 1% formaldehyde for 10 min. The reaction was stopped with glycine at final concentration of 0.125 M for 10 min, followed by addition of ChIP nuclear isolation buffer (1% SDS, 50mM Tris-HCl, 20mM EDTA). Isolated nuclei were resuspended in ChIP buffer (0.1% SDS, 1% Triton X-100, 0.15 M NaCl, 1mM EDTA, 20 mM Tris-HCl) and subsequently sonicated 3x10 cycles (30 s ON/OFF) in the UCD-200 Bioruptor (Diagenode), to generate DNA-fragment sizes of 0.2 – 0.5 kb. Approximately chromatin from 10–20 × 106 cells was used for histone marks, 20–30 × 106 cells for GPS2 ChIP. Each lysate was immunoprecipitated with the following antibodies: control rabbit IgG (Santa Cruz, sc-2027, 1-4 μg), anti-H3K4me3 (Abcam, ab8580, 1 μg) and anti-GPS2 (N and C-terminal 4 μg) (Venteclef et al., 2010). Following 3 h rotation at 4°C, 25 μL protein A Dynabeads (Invitrogen) were added and samples were incubated overnight at 4°C with rotation. On the next day, the sample were washed twice with IP wash buffer 1 (1% Triton, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, 20 mM Tris, pH 8.0, and 0.1% sodium deoxycholate [NaDOC]), twice with IP wash buffer 2 (1% Triton, 0.1% SDS, 500 mM NaCl, 1 mM EDTA, 20 mM Tris, pH 8.0, and 0.1% NaDOC), twice with IP wash buffer 3 (0.25 M LiCl, 0.5% NP-40, 1 mM EDTA, 20 mM Tris, pH 8.0, and 0.5% NaDOC), and finally once with IP wash buffer 4 (10 mM EDTA and 200 mM Tris, pH 8), all at 4°C. The immune-bound chromatin was eluted in the elution buffer (1% SDS and 0.1 M NaHCO3) and de-cross-linked by adding NaCl to a final concentration of 0.2 M at 65°C O/N. After RNase A (Fermentas) and proteinase K (Fermentas) treatment, the immunoprecipitated DNA was purified using the QIAquick PCR purification kit (QIAGEN) and analyzed by RT-qPCR.
Quantification and Statistical Analysis
Data are expressed as mean ± SEM. Experiments were performed at least 3 times. Statistical analysis was performed using 2-tailed Student’s t test. Comparisons between more than 2 groups were carried out using one-way ANOVA. P value less than 0.05 was considered statistically significant. The statistical details of experiments can be found in figure legends.
Data and Software Availability
Data Resources
The accession number for the RNA-sequencing data reported in this manuscript is GEO: GSE111647.
Acknowledgments
We acknowledge Ozgene Pty Ltd. for generating floxed GPS2 mice. E.T. was supported by grants from the Swedish Research Council, the Swedish Cancer Society, the Swedish Diabetes Foundation, the Novo Nordisk Foundation, the European Union FP7 Marie Curie ITN NR-NET, and the Center for Innovative Medicine (CIMED) at the Karolinska Institutet. N.V. was supported by grants from the French National Agency of Research (CONRAD and PROVIDE), Region Ile de France (CORDDIM), Paris city (EMERGENCE), the French Foundation for Diabetes (SFD), and the European Union H2020 framework (ERC-EpiFAT 725790). The clamp studies in humans were performed at the Clinical Investigation Center (Groupe Hospitalier Saint-Louis/Lariboisière, Paris) and were supported by Assistance Publique des Hôpitaux de Paris (APHP; PHRC AOR09087; J.-F.G., principal investigator [PI]).
Author Contributions
K.D., N.V., and E.T. conceived the study, interpreted data, and wrote the manuscript, with contributions from F.F., F.A., and J.-F.G. The in vivo experiments were performed by K.D., R.B., F.A., and A.T., with contributions from S.B., C.C., and R.F. S.B. performed ChIP. N.L. conducted the co-immunoprecipitation (co-IP) experiments, and Z.H. produced the virus. M.J.M. performed the majority of genomic analyses, with contributions from M.K., K.D., and N.V. J.-F.G., R.R., A.S., and N.V. collected, analyzed, and interpreted the human data.
Declaration of Interests
The authors declare no competing interests.
Published: September 11, 2018
Footnotes
Supplemental Information includes eight figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.08.032.
Contributor Information
Eckardt Treuter, Email: eckardt.treuter@ki.se.
Nicolas Venteclef, Email: nicolas.venteclef@inserm.fr.
Supplemental Information
References
- Acosta J.R., Douagi I., Andersson D.P., Bäckdahl J., Rydén M., Arner P., Laurencikiene J. Increased fat cell size: a major phenotype of subcutaneous white adipose tissue in non-obese individuals with type 2 diabetes. Diabetologia. 2016;59:560–570. doi: 10.1007/s00125-015-3810-6. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E., Ingerslev L.R., Fabre O., Donkin I., Altıntaş A., Versteyhe S., Bisgaard T., Kristiansen V.B., Simar D., Barrès R. Preadipocytes from obese humans with type 2 diabetes are epigenetically reprogrammed at genes controlling adipose tissue function. Int. J. Obes. 2018 doi: 10.1038/s41366-018-0031-3. Published online February 20, 2018. [DOI] [PubMed] [Google Scholar]
- Andersson D.P., Eriksson Hogling D., Thorell A., Toft E., Qvisth V., Näslund E., Thörne A., Wirén M., Löfgren P., Hoffstedt J. Changes in subcutaneous fat cell volume and insulin sensitivity after weight loss. Diabetes Care. 2014;37:1831–1836. doi: 10.2337/dc13-2395. [DOI] [PubMed] [Google Scholar]
- Basse A.L., Isidor M.S., Winther S., Skjoldborg N.B., Murholm M., Andersen E.S., Pedersen S.B., Wolfrum C., Quistorff B., Hansen J.B. Regulation of glycolysis in brown adipocytes by HIF-1α. Sci. Rep. 2017;7:4052. doi: 10.1038/s41598-017-04246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M. The distinction of metabolically ‘healthy’ from ‘unhealthy’ obese individuals. Curr. Opin. Lipidol. 2010;21:38–43. doi: 10.1097/MOL.0b013e3283346ccc. [DOI] [PubMed] [Google Scholar]
- Cardamone M.D., Krones A., Tanasa B., Taylor H., Ricci L., Ohgi K.A., Glass C.K., Rosenfeld M.G., Perissi V. A protective strategy against hyperinflammatory responses requiring the nontranscriptional actions of GPS2. Mol. Cell. 2012;46:91–104. doi: 10.1016/j.molcel.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardamone M.D., Tanasa B., Chan M., Cederquist C.T., Andricovich J., Rosenfeld M.G., Perissi V. GPS2/KDM4A pioneering activity regulates promoter-specific recruitment of PPARγ. Cell Rep. 2014;8:163–176. doi: 10.1016/j.celrep.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardamone M.D., Tanasa B., Cederquist C.T., Huang J., Mahdaviani K., Li W., Rosenfeld M.G., Liesa M., Perissi V. Mitochondrial retrograde signaling in mammals is mediated by the transcriptional cofactor GPS2 via direct mitochondria-to-nucleus translocation. Mol. Cell. 2018;69:757–772.e7. doi: 10.1016/j.molcel.2018.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist C.T., Lentucci C., Martinez-Calejman C., Hayashi V., Orofino J., Guertin D., Fried S.K., Lee M.J., Cardamone M.D., Perissi V. Systemic insulin sensitivity is regulated by GPS2 inhibition of AKT ubiquitination and activation in adipose tissue. Mol. Metab. 2016;6:125–137. doi: 10.1016/j.molmet.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo H.J., Kim J.H., Kwon O.B., Lee C.S., Mun J.Y., Han S.S., Yoon Y.S., Yoon G., Choi K.M., Ko Y.G. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia. 2006;49:784–791. doi: 10.1007/s00125-006-0170-2. [DOI] [PubMed] [Google Scholar]
- Cotillard A., Poitou C., Torcivia A., Bouillot J.L., Dietrich A., Klöting N., Grégoire C., Lolmede K., Blüher M., Clément K. Adipocyte size threshold matters: link with risk of type 2 diabetes and improved insulin resistance after gastric bypass. J. Clin. Endocrinol. Metab. 2014;99:E1466–E1470. doi: 10.1210/jc.2014-1074. [DOI] [PubMed] [Google Scholar]
- Emmett M.J., Lim H.W., Jager J., Richter H.J., Adlanmerini M., Peed L.C., Briggs E.R., Steger D.J., Ma T., Sims C.A. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature. 2017;546:544–548. doi: 10.1038/nature22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R., Toubal A., Goñi S., Drareni K., Huang Z., Alzaid F., Ballaire R., Ancel P., Liang N., Damdimopoulos A. Loss of the co-repressor GPS2 sensitizes macrophage activation upon metabolic stress induced by obesity and type 2 diabetes. Nat. Med. 2016;22:780–791. doi: 10.1038/nm.4114. [DOI] [PubMed] [Google Scholar]
- Fang S., Suh J.M., Atkins A.R., Hong S.H., Leblanc M., Nofsinger R.R., Yu R.T., Downes M., Evans R.M. Corepressor SMRT promotes oxidative phosphorylation in adipose tissue and protects against diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. U S A. 2011;108:3412–3417. doi: 10.1073/pnas.1017707108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari A., Longo R., Fiorino E., Silva R., Mitro N., Cermenati G., Gilardi F., Desvergne B., Andolfo A., Magagnotti C. HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat. Commun. 2017;8:93. doi: 10.1038/s41467-017-00182-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–311. doi: 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucho R., Casals N., Serra D., Herrero L. Ceramides and mitochondrial fatty acid oxidation in obesity. FASEB J. 2017;31:1263–1272. doi: 10.1096/fj.201601156R. [DOI] [PubMed] [Google Scholar]
- Halberg N., Khan T., Trujillo M.E., Wernstedt-Asterholm I., Attie A.D., Sherwani S., Wang Z.V., Landskroner-Eiger S., Dineen S., Magalang U.J. Hypoxia-inducible factor 1alpha induces fibrosis and insulin resistance in white adipose tissue. Mol. Cell. Biol. 2009;29:4467–4483. doi: 10.1128/MCB.00192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Gao Z., Yin J., Zhang J., Yun Z., Ye J. Regulation of HIF-1alpha activity in adipose tissue by obesity-associated factors: adipogenesis, insulin, and hypoxia. Am. J. Physiol. Endocrinol. Metab. 2011;300:E877–E885. doi: 10.1152/ajpendo.00626.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Cardamone M.D., Johnson H.E., Neault M., Chan M., Floyd Z.E., Mallette F.A., Perissi V. Exchange factor TBL1 and arginine methyltransferase PRMT6 cooperate in protecting G protein pathway suppressor 2 (GPS2) from proteasomal degradation. J. Biol. Chem. 2015;290:19044–19054. doi: 10.1074/jbc.M115.637660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T., Sakai J., Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. Cell Biol. 2016;17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson T., Venteclef N., Toresson G., Damdimopoulos A.E., Ehrlund A., Lou X., Sanyal S., Steffensen K.R., Gustafsson J.A., Treuter E. GPS2 is required for cholesterol efflux by triggering histone demethylation, LXR recruitment, and coregulator assembly at the ABCG1 locus. Mol. Cell. 2009;34:510–518. doi: 10.1016/j.molcel.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Jiang C., Qu A., Matsubara T., Chanturiya T., Jou W., Gavrilova O., Shah Y.M., Gonzalez F.J. Disruption of hypoxia-inducible factor 1 in adipocytes improves insulin sensitivity and decreases adiposity in high-fat diet-fed mice. Diabetes. 2011;60:2484–2495. doi: 10.2337/db11-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdren L., Bernard M., Dillies M.A., Le Crom S. Eoulsan: a cloud computing-based framework facilitating high throughput sequencing analyses. Bioinformatics. 2012;28:1542–1543. doi: 10.1093/bioinformatics/bts165. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., van de Wall E., Laplante M., Azzara A., Trujillo M.E., Hofmann S.M., Schraw T., Durand J.L., Li H., Li G. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Invest. 2007;117:2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöting N., Fasshauer M., Dietrich A., Kovacs P., Schön M.R., Kern M., Stumvoll M., Blüher M. Insulin-sensitive obesity. Am. J. Physiol. Endocrinol. Metab. 2010;299:E506–E515. doi: 10.1152/ajpendo.00586.2009. [DOI] [PubMed] [Google Scholar]
- Krishnan J., Danzer C., Simka T., Ukropec J., Walter K.M., Kumpf S., Mirtschink P., Ukropcova B., Gasperikova D., Pedrazzini T., Krek W. Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 2012;26:259–270. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski C.M., Holland W.L., Sun K., Park J., Spurgin S.B., Lin Y., Askew G.R., Simcox J.A., McClain D.A., Li C., Scherer P.E. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat. Med. 2012;18:1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski C.M., Bickel P.E., Scherer P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat. Rev. Drug Discov. 2016;15:639–660. doi: 10.1038/nrd.2016.75. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Kim J.W., Osborne O., Oh D.Y., Sasik R., Schenk S., Chen A., Chung H., Murphy A., Watkins S.M. Increased adipocyte O2 consumption triggers HIF-1α, causing inflammation and insulin resistance in obesity. Cell. 2014;157:1339–1352. doi: 10.1016/j.cell.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Fan W., Xu J., Lu M., Yamamoto H., Auwerx J., Sears D.D., Talukdar S., Oh D., Chen A. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell. 2011;147:815–826. doi: 10.1016/j.cell.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir L.A., Neeley C.K., Meyer K.A., Baker N.A., Brosius A.M., Washabaugh A.R., Varban O.A., Finks J.F., Zamarron B.F., Flesher C.G. Adipose tissue fibrosis, hypertrophy, and hyperplasia: Correlations with diabetes in human obesity. Obesity (Silver Spring) 2016;24:597–605. doi: 10.1002/oby.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguewa J.L., Lontchi-Yimagou E., Agbelika F., AitDjoudi M., Boudou P., Choukem S., Sobngwi E., Gautier J.F. Relationship between HHV8 infection markers and insulin sensitivity in ketosis-prone diabetes. Diabetes Metab. 2017;43:79–82. doi: 10.1016/j.diabet.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Perfilyev A., Dahlman I., Gillberg L., Rosqvist F., Iggman D., Volkov P., Nilsson E., Risérus U., Ling C. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am. J. Clin. Nutr. 2017;105:991–1000. doi: 10.3945/ajcn.116.143164. [DOI] [PubMed] [Google Scholar]
- Ritov V.B., Menshikova E.V., He J., Ferrell R.E., Goodpaster B.H., Kelley D.E. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes. 2005;54:8–14. doi: 10.2337/diabetes.54.1.8. [DOI] [PubMed] [Google Scholar]
- Rohm M., Sommerfeld A., Strzoda D., Jones A., Sijmonsma T.P., Rudofsky G., Wolfrum C., Sticht C., Gretz N., Zeyda M. Transcriptional cofactor TBLR1 controls lipid mobilization in white adipose tissue. Cell Metab. 2013;17:575–585. doi: 10.1016/j.cmet.2013.02.010. [DOI] [PubMed] [Google Scholar]
- Rosen E.D. Epigenomic and transcriptional control of insulin resistance. J. Intern. Med. 2016;280:443–456. doi: 10.1111/joim.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Taliyan R. Histone deacetylase inhibitors: Future therapeutics for insulin resistance and type 2 diabetes. Pharmacol. Res. 2016;113(Pt A):320–326. doi: 10.1016/j.phrs.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Siersbæk M.S., Loft A., Aagaard M.M., Nielsen R., Schmidt S.F., Petrovic N., Nedergaard J., Mandrup S. Genome-wide profiling of peroxisome proliferator-activated receptor γ in primary epididymal, inguinal, and brown adipocytes reveals depot-selective binding correlated with gene expression. Mol. Cell. Biol. 2012;32:3452–3463. doi: 10.1128/MCB.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Kusminski C.M., Scherer P.E. Adipose tissue remodeling and obesity. J. Clin. Invest. 2011;121:2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Halberg N., Khan M., Magalang U.J., Scherer P.E. Selective inhibition of hypoxia-inducible factor 1α ameliorates adipose tissue dysfunction. Mol. Cell. Biol. 2013;33:904–917. doi: 10.1128/MCB.00951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubal A., Clément K., Fan R., Ancel P., Pelloux V., Rouault C., Veyrie N., Hartemann A., Treuter E., Venteclef N. SMRT-GPS2 corepressor pathway dysregulation coincides with obesity-linked adipocyte inflammation. J. Clin. Invest. 2013;123:362–379. doi: 10.1172/JCI64052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubal A., Treuter E., Clément K., Venteclef N. Genomic and epigenomic regulation of adipose tissue inflammation in obesity. Trends Endocrinol. Metab. 2013;24:625–634. doi: 10.1016/j.tem.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Treuter E., Fan R., Huang Z., Jakobsson T., Venteclef N. Transcriptional repression in macrophages-basic mechanisms and alterations in metabolic inflammatory diseases. FEBS Lett. 2017;591:2959–2977. doi: 10.1002/1873-3468.12850. [DOI] [PubMed] [Google Scholar]
- Venteclef N., Jakobsson T., Ehrlund A., Damdimopoulos A., Mikkonen L., Ellis E., Nilsson L.M., Parini P., Jänne O.A., Gustafsson J.A., Steffensen K.R., Treuter E. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXR beta in the hepatic acute phase response. Genes Dev. 2010;24 doi: 10.1101/gad.545110. 381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., MAGIC investigators. GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Landry A.P., Ding H. The mitochondrial outer membrane protein mitoNEET is a redox enzyme catalyzing electron transfer from FMNH2 to oxygen or ubiquinone. J. Biol. Chem. 2017;292:10061–10067. doi: 10.1074/jbc.M117.789800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lam K.S., Ye H., Chung S.K., Zhou M., Wang Y., Xu A. Adipose tissue-specific inhibition of hypoxia-inducible factor 1alpha induces obesity and glucose intolerance by impeding energy expenditure in mice. J. Biol. Chem. 2010;285:32869–32877. doi: 10.1074/jbc.M110.135509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.