Abstract
The ancient and pluripotent progranulins contain multiple repeats of a cysteine‐rich sequence motif of ∼60 amino acids, called the granulin/epithelin module (GEM) with a prototypic structure of four β‐hairpins zipped together by six inter‐hairpin disulfide bonds. Prevalence of this disulfide‐enforced structure is assessed here by an expression screening of 19 unique GEM sequences of the four progranulins in the zebrafish genome, progranulins 1, 2, A and B. While a majority of the expressed GEM peptides did not exhibit uniquely folded conformations, module AaE from progranulin A and AbB from progranulin B were found to fold into the protopypic 4‐hairpin structure along with disulfide formation. Module AaE has the most‐rigid three‐dimensional structure with all four β‐hairpins defined using high‐resolution (H–15N) NMR spectroscopy, including 492 inter‐proton nuclear Overhauser effects, 23 3J(HN,Hα) coupling constants, 22 hydrogen bonds as well as 45 residual dipolar coupling constants. Three‐dimensional structure of AaE and the partially folded AbB re‐iterate the conformational stability of the N‐terminal stack of two beta‐hairpins and varying degrees of structural flexibility for the C‐terminal half of the 4‐hairpin global fold of the GEM repeat. A cell‐based assay demonstrated a functional activity for the zebrafish granulin AaE in promoting the survival of neuronal cells, similarly to what has been found for the corresponding granulin E module in human progranulin. Finally, this work highlights the remaining challenges in structure‐activity studies of proteins containing the GEM repeats, due to the apparent prevalence of structural disorder in GEM motifs despite potentially a high density of intramolecular disulfide bonds.
Keywords: 3D structure, granulin/epithelin module, progranulin, zebrafish, hairpin stack fold, NMR spectroscopy
Short abstract
PDB Code(s): http://firstglance.jmol.org/fg.htm?mol=6CKU
Abbreviations
- ARIA
ambigous restraints for iterative assignment
- GEM
granulin/epithelin module
- GEP
granulin/epithelin precursor
- HPLC
high‐performance liquid chromatography
- HSQC
heteronuclear single‐quantum correlation
- IPTG
isopropyl β‐D‐thiogalactoside
- MJ‐HSQC
J‐mutiplied HSQC
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser effect spectrosscopy
- NMR
nuclear magnetic resonance
- PCDGF
PC cell‐derived growth factor
- PCR
polymerase chain reaction
- 2QF‐COSY
double‐quantum filtered correlation spectrosocpy
- RDC
residual dipolar coupling
- RMSD
root mean square difference
- TFA
trifluoacetic acid
- TOCSY
total correlation spectroscopy
- TOPCAT
TOpology and PreCalculation Assignment.
Introduction
Proteins harboring the granulin or granulin/epithelin modules (GEM) have recently emerged as an important class of structurally stable molecules with such diverse biological functions as wound repair, angiogenesis and related inflammatory processes as well as in embryogenesis, neurobiology and cancer development.1, 2, 3, 4, 5 Precursors carrying the GEM repeat are among the most ancient extracellular regulatory proteins still employed in lower organisms and mammals ranging from metazoa, teleost fishes, elephant sharks to man.3 Mammalian GEM proteins were originally purified from human peripheral leukocytes,6 human bone marrow,7 rat kidneys,8 and equine neutrophils.9 They are products of the proteolytic processing of progranulin, the granulin/epithelin precursor which in the case of humans contain seven and a half tandem GEM repeats.7 The precursor has also been identified as the PCDGF10 and acrogranin.11 A structural study of all GEM repeats in human progranulin12 confirmed the structural modularity and stability of the hairpin stack fold proposed for this unique class of cysteine‐rich protein repeats.13
Prototypic forms of GEMs, the granulins, were also first isolated from the hematopoietic organs of a teleost fish cyprinus carpio14 and later from goldfish intestines.15 These individual granulins have an approximate molecular weight of 6 kDa, with a unique and highly conserved pattern of 10 or 12 cysteine residues that is found in all other GEM proteins.2, 3 Curiously, teleost granulins may be derived from two distinct classes of progranulins, one including progranulins 1 and 2, which contain one and a half GEM repeats, and the second the progranulins A and B, which contain 10 and 9 GEM repeats, as shown by an analysis of the zebrafish progranulin gene family.16 Isolated carp granulins are essentially identical in sequence to the full GEM of zebrafish progranulins 1 and 2.14, 16 It is not known to this day, however, whether individual GEM proteins can be generated from the GEM repeats of the larger zebrafish progranulins A and B (also referred to as progranulins Aa and Ab in the following text).
In the current study, we report a structural dissection of all four zebrafish granulin precursors, namely progranulin A (referred to as Aa in the form of a DNA clone), progranulin B (or the Ab clone) and progranulins 1 and 2, using bacterial expression to generate GEM proteins and high‐resolution NMR spectroscopy for structural characterizations. Both homonuclear and heteronuclear NMR techniques were applied for three‐dimensional (3D) structure screening and for high‐resolution 3D structure determination, especially by incorporation of NMR residual dipolar coupling constraints. Detailed structure studies of relatievely well‐folded zebrafish GEM proteins revealed a conformational diversity of the C‐terminal subdomains of the hairpin stack fold in GEM proteins. Very importantly, we identified a well‐structured GEM protein AaE from progranulin Aa that also exhibits functional activities in promoting the survival of neuronal cells.
Results
Protein expression and purification
A total of 19 distinct GEM sequences (Fig. 1) were selected from the four granulin precursors in the zebrafish genome, the progranulin A, progranulin B, progranulin 1 and progranulin 2.16 The genomic sequences of progranulins A and B each contain 10 and 9 GEM sequences, respectively [Fig. 1(A)]. However, a partial clone of the progranulin A gene, referred to as progranulin Aa [Fig. 1(B) and Table S1(A)] was used here to derive eight GEM sequences from a total of 10 potential GEM repeats in progranulin A. These eight GEM sequences include those of the first four granulins, referred to as AaH (or the 1st module), AaG (2nd module), AaF (3rd module), AaB (4th module) and of the last four granulins, named as AaA (the 7th module), AaC (8th module), AaD (9th module), and AaE (10th module) [Fig. 1(A)]. It was found that all zebrafish granulins have a similarity of 42–50% with the seven granulin modules in human progranulin, which does not generate an unambiguous correspondence with granulins G, F, B, A, C, D, and E that appear sequentially along the human progranulin sequence.7, 16 Therefore, module 2 of the progranulin Aa clone was named here as the zebrafish AaG module based on an identical pattern of cysteine residues as granulin G in human progranulin, that is, missing two cysteine residues [Fig. 1(B)]. Modules 5 and 6 were not considered further in this study since they exhibit a high degree of sequence similarity with modules 4 and 7,3, 16 that is, with AaB and AaA, respectively [Fig. 1(B)]. The GEM motifs of zebrafish progranulins 1 and 2 are grouped together with the first GEM repeats of progranulin A [or the Aa clone, Table SI(A)] and progranulin B [or the progranulin Ab clone, Table SI(A)], which are named as AaH and AbH, respectively. Consequently, the last granulin module in progranulin Ab is identified as AbI, which stands alone from the rest of the granulin sequences (Fig. 1). Progranulins 1 and 2 are the precursors of teleost granulin‐1 and granulin‐2,16 first isolated from the head kidney of carp fishes.14 Both progranulins 1 and 2 contain one complete GEM followed by a short sequence resembling a half GEM or para‐granulin in human progranulin,16 which are grouped together here in the sequence alignment [Fig. 1(B)].
Figure 1.
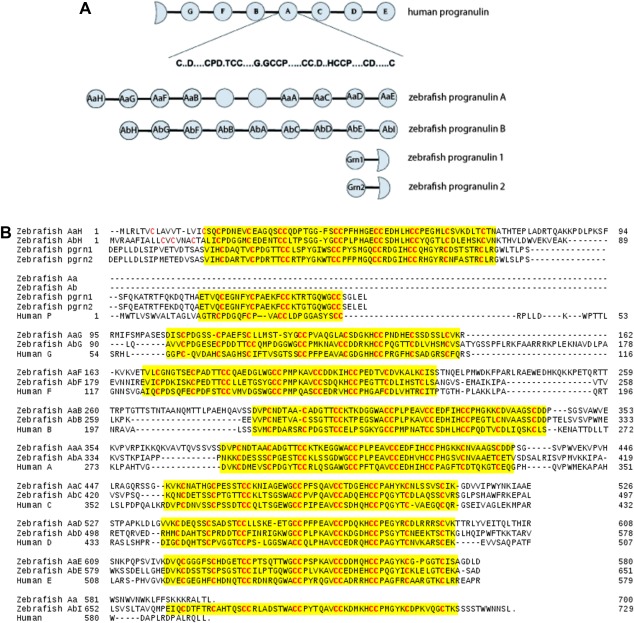
(A) A schematic illustration for the numenclature of granulin modules in zebrafish progranulins Aa, Ab, 1 and 2 as compared to human progranulin. Human P, G, F, B, A, C, D, and E identify granulin modules that appear sequentially in the human progranulin gene.7 (B) Sequence alignment of progranulin clones used in this study. Granulin modules 5 and 6 are missing from the partial clone, progranulin Aa, of zebrafish progranulin A. Progranulin Ab, on the other hand, is a full‐length clone spanning the entire sequence of zebrafish progranulin B.16 Both zebrafish granulin AaG and human granulin G have 10 cysteine residues instead of the canonical 12 cysteines for a GEM repeat [Fig. 1(A)] with a pair of cysteine residues missing (replaced) in two (the first and the third) of the four characteristic CC or double‐cysteine repeats.
Table 1.
Amino Acid Residues at Equivalent Positions of 18 and 24 in Human, Carp and Zebrafish Granulins
| Residue 18 | Residue 24 | |
|---|---|---|
| Ala | ||
| Val | hF | |
| Leu | Grn1,AaG,AaD,AbF,AbH | |
| Ile | cGrn1,AbE | |
| Met | ||
| Cys | ||
| Phe | AbD | AaH |
| Trp | hA,hC,hF,cGrn1,Grn1,Grn2,AaA,AaB,AaC,AaE,AaF,AbA,AbB,AbC,AbD,AbE,AbG,AbI | |
| Gly | ||
| Pro | AaE | |
| Ser | ||
| Thr | AaD | |
| Tyr | hB,AaG,AbF,AbH | |
| Asn | ||
| Gln | hC,AaF,AaH,AbG | |
| Asp | ||
| Glu | hB | |
| Lys | AaA,AaB,AaC,AbA,AbB,AbC | |
| Arg | hA,Grn2,AbI | |
| His |
Note: cGrn1 and hA, hC, hF indicate carp granulin‐1 and human granulins A, C and F, respectively. Other GEMs are named according to the conventions used in this work (see Fig. 1), for example, AaE—GEM “E” from zebrafish progranluin Aa and AbB—GEM B from zebrafish progranulin Ab, etc.
Following a previous study with human progranulin,12 a high‐throughput approach was utilized to assess the structural diversity of the 19 selected zebrafish GEM domains and to determine the high‐resolution three‐dimensional (3D) structures of purified and well‐folded GEM peptides. Therefore, 19 DNA fragments encoding all 19 GEM sequences [Fig. 1(B) and Table SI(B)] were subcloned into the pET32b plasmid vector. After DNA sequencing, a total of 14 plasmids had the correct DNA sequences [Table SI(C)] and were found to be inducible by IPTG with a high level of fusion protein production in an oxidizing intracellular environment and assisted by the thioredoxin tag to facilitate disulfide bond formation, see the Methods Section. These included AaA, AaC, AaD, AaE, AaF, AaG, and AaH from the progranulin Aa clone and AbA, AbB, AbE, AbF, and AbG from progranulin Ab as well as the Grn1 and Grn2 modules from progranulin 1 and progranulin 2. Each of the 14 fusion proteins was purified from the cell culture, subjected to enterokinase cleavage, and further purified by use of reversed‐phase HPLC. Levels of fusion protein expression were uniformly high: ∼100 mg from 2 L of cell cultures for all fusion proteins. With AaE, for example, 25 mg of the fusion protein protein was recovered after purification, which generated 6 mg of the released AaE fragment and 2.27 mg of folded AaE after further HPLC purification. So the yield of each GEM peptide depended only on the efficiency of fusion protein digestion by enterokinase and subsequent HPLC purification. Very noticeably, the HPLC profiles of all 14 granulin peptides (Fig. 2) show that the first fraction of zebrafish AaE eluted the earliest (at ∼19% acetonitrile), much earlier than the other 13 modules, even earlier than zebrafish Grn1. This elution property indicates the formation of a well‐folded structure by AaE as also found in the purification of naturally occurring carp granulins 1 and 2.13, 14 For this study, HPLC fractions of all the GEM peptides (Fig. 2) were therefore prioritized only according to early elution times, quantity of purified peptide materials as well as the quality of proton NMR spectra for the selected fractions.
Figure 2.
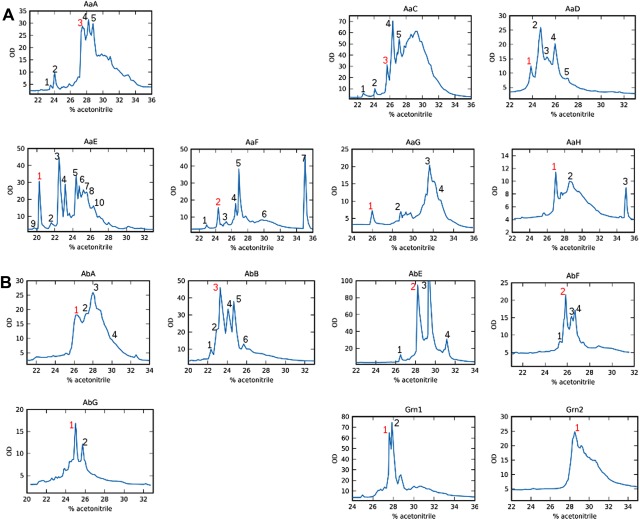
HPLC profiles of the expressed and cleaved fusion proteins carrying 14 of the 19 zebrafish GEM peptides (Fig. 1). Each fusion protein contains a thioredoxin tag and a His‐tag, which are separated from the GEM peptide after proteolytic cleavage (see the Experimental Section). The numbered peaks indicate the collected HPLC fractions and the red‐colored peak was chosen for further NMR analyses (Fig. 3) after a preliminary screening of collected fractions. (A) HPLC elution profiles for the purification of seven progranulin Aa GEMs: AaA, AaC, AaD, AaE, AaF, AaG, and AaH. Fractions for NMR spectra screening included 3 and 5 of AaA, 3 of AaC, 1 and 2 of AaD, 1, 3, and 4 of AaE, 2, 4, 5, and 7 of AaF, 1 of AaG, and 1 and 3 of AaH. (B) Elution profiles of five GEMs from progranulin Ab, AbA, AbB, AbE, AbF, and AbG and the two GEMs, Grn1 and Grn2, from progranulins 1 and 2, respectively. Fractions for NMR screening included 1 of AbA, 3, 4, and 5 of AaB, 2, 3, and 4 of AbE, 2 and 4 of AbF, 1 of AbG, 1 of Grn1 and Grn2.
Folding properties of zebrafish GEM proteins
Simple one‐dimensional proton NMR spectra were then used to assess the extent of structuring for the expressed GEM peptides since the proton chemical shift is a sensitive measure of three‐dimensional molecular structure/conformations and environmental differences such as proximity to polar or aromatic groups.17 For an unstructured protein, for example, the amide region in proton NMR spectra has a characteristically large and broad signal at approximately 8.3 ppm. On the other hand, a folded or well‐structured protein has a typical proton NMR spectrum with a large dispersion of amide proton resonances, for example, ranging from ∼6.5 to 10 ppm. Based on their one‐dimensional proton NMR spectra (Fig. 3), most purified zebrafish GEMs appear to be partially unfolded with a rather intense “random‐coil” peak admixed with some downfield‐shifted signals, indicating that every module also has a certain population of folded species. One clear exception is the proton NMR spectrum of zebrafish AaE, with not only a good dispersion of amide proton signals, but also a notable absence of “random‐coil” peaks. Furthermore, existence of a few resonances between 5 and 6 ppm demonstrates the formation of a β‐sheet structure characteristic of GEM proteins.13
Figure 3.
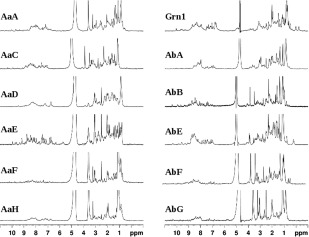
500 MHz proton NMR spectra of HPLC fractions selected for NMR analyses (see Fig. 2). Most spectra contain a broad spectral feature in between 7.5 and 8.5 ppm, with the exception of AaE and AbB which have well‐dispersed and sharp proton resonances indicating the formation of folded three‐dimensional structures (see the text descriptions for details). Samples of AaG and Grn2 did not produce NMR spectra of sufficient S/N ratio, indicating either aggregation or insufficient amount of purified materials.
Figure 4 is a comparison of the two‐dimensional (2D) proton‐proton TOCSY spectra of four GEMs, from that of AbE, AbB, zebrafish granulin‐1 (Grn1), to AaE. Resonance peaks in the TOCSY spectrum of AaE were impressively sharp and relatively uniform in intensities and well‐dispersed (Fig. 4). In comparison, AbB also exhibits well‐dispersed TOCSY peaks, but in addition to less‐dispersed and broader signals. The granulin module AbB was therefore also selected for a detailed structural analyses and comparison.
Figure 4.
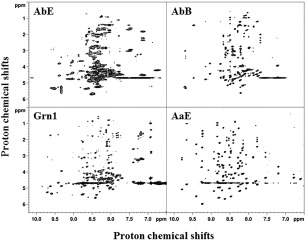
Two‐dimensional NMR TOCSY spectra showing relatively well‐resolved NH‐to‐sidechain proton connectivies of four GEM peptides, AbE, AbB, Grn1, and AaE. The TOCSY spectra are arranged in the order of increasing number of peaks and increasing resonance dispersion, for example, from 6.5 to 10 ppm for AaE covering >3.5 ppm for the NH proton resonances. Broader spectra features are also evident in the TOCSY spectra of AbE, as already seen in the one‐dimensional proton NMR spectrum of this GEM peptide (Fig. 3), which indicates either conformational flexibility and/or sample aggregation.
All proton NMR signals of the AbB and AaE modules could be assigned (Table SII) using their 2D TOCSY and NOESY spectra. NH chemical shift assignments, in particular, show that both AbB and AaE exhibit a large dispersion of amide proton resonances in the N‐terminal half [of almost 2.5 ppm and 3.3 ppm, respectively, Table SII(A) and SII(B)], demonstrating good folding in the N‐terminal subunits of these two GEMs. On the other hand, AaE also shows a high NH signal dispersion in the C‐terminal half [of 2.88 ppm, Table SII(B)], indicating a well‐structured C‐terminal subdomain, while AbB appears to be devoid of an ordered structure in the C‐terminal region, that is, with a significantly reduced NH signal dispersion [of only ∼1 ppm, Table SII(A)]. The differing structural properties of AbB and AaE modules are more evident in the diagonal plots of the NOE connectivities (Fig. 5). The typical pattern of a β‐hairpin structure can be easily identified with four β‐strands in the entire structure of AaE [Fig. 5(B)], but only two β‐strands/hairpins in the N‐terminal region of AbB [Fig. 5(A)].
Figure 5.
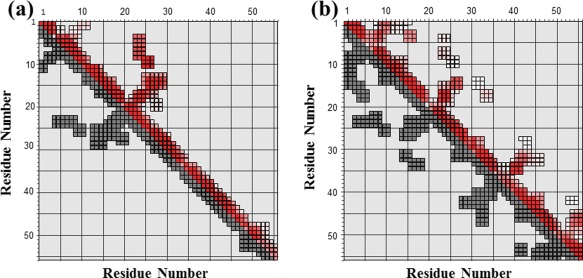
Diagonal plots of NOE connectivities observed for GEM peptides AbB and AaE. (A) NOE connectivites of AbB indicate medium‐range folding and the formation of two β‐hairpins. (B) NOE connectivities of AaE show the formation of four β‐hairpins and signs of two structural subdomains in the N‐ and C‐terminal region. Input NOE peak lists with chemical shift assignments (lower triangle in black) are plotted (using the Perl script ariaoverview.pl, see Vranken et al.22) against assigned NOE constaints (upper triangle in color). Both axes of the plots represent residue numbers of AbB in (A) and AaE in (B).
The better‐folded AaE module was further characterized by use of the NMR heteronuclear single‐quantum coherence (HSQC) experiment with preparations of 15N‐labeled AaE peptide samples. The HSQC spectrum of AaE shows crosspeaks of rather uniform intensities [Fig. 6(A)]. Using three‐dimensional (3D) 15N‐HSQC‐TOCSY and 15N‐HSQC‐NOESY spectra, all 51 cross‐peaks in the HSQC spectrum of AaE were assigned, corresponding to expected backbone amide protons, based on the amino acid sequence of AaE [Table SI(C)], along with crosspeaks of sidechain amide protons. The remaining five residues, all prolines devoid of a proton‐bound nitrogen, cannot be observed in the 2D 1H–15N HSQC spectrum. In all, zebrafish AaE is a stably folded and well‐structured GEM protein as further indicated by a rather uniform distribution of the steady‐state 1H–15N NOEs [Fig. 6(B)].
Figure 6.
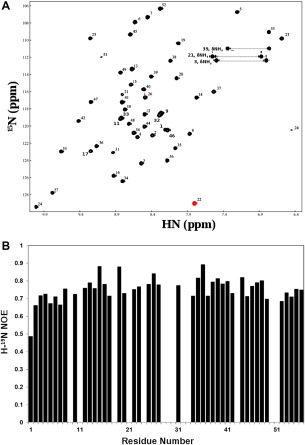
Heteronuclear (H‐15N) NMR data showing a uniquely folded three‐dimensional structure for the GEM peptide AaE. (A) Proton (H)‐nitrogen (15N) correlation (HSQC) spectra of 15N‐labeled AaE (see the Experimental Section for sample conditions). A widespread dispersion of HSQC peaks mirrors similar characteristics of the TOCSY spectrum for AaE (Fig. 4). The HSQC peaks also have rather uniform intensities, which further indicates good folding and conformational rigidity for the three‐dimensional structure of AaE. (B) Heteronuclear (H‐15N) NOEs of AaE displaying uniformity, with the exception of terminal residue Asp1, of the atomic motions in AaE, again indicating stable folding for AaE (see text descriptions for details). Proline residues, that is, Pro18, Pro28, Pro30, Pro43, and Pro50, do not have amide protons for H‐15N NOE measurements, and heteronuclear NOE data were not quantitated for residues Ser9, His11, Ser29, Val33, Ala32, and Gly51 due to resonance overlaps and for Gln21, Trp24, and Gly51 because of diminished intensities of their H‐15N HSQC peaks.
The three‐dimensional structure of zebrafish AaE
For structure determination, proton chemical shift assignments for AaE were established with a single 15N‐labeled AaE sample using 2D 1H‐1H TOCSY, 2D 1H‐1H NOESY spectra and re‐confirmed by 3D 15N HSQC‐TOCSY and 3D 15N HSQC‐NOESY spectra. NOE connectivities were identified in 2D NOESY recorded in both D2O and H2O and 3D 15N NOESY‐HSQC spectra in H2O. NOE intensities were estimated by measuring peak heights and were used for assigning interproton distances to the NOE restraints as described in the Materials and Methods section. The final list of NOE data consists of 492 uniquely assigned distance restraints, including 197 sequential, 190 medium‐range (1 < | i – j | < 5), and 105 long‐range (| i – j | ≥ 5) restraints. The structural restraints also included 23 3J(HN,Hα) coupling constants measured using an MJ‐HSQC experiment with the 15N‐labeled AaE sample [Table SII(B)] and converted into dihedral angle restraints. Hydrogen bond distance restraints were applied for 22 candidate hydrogen bonds identified on the basis of NOE patterns characteristic of regular (beta‐sheet) secondary structures and with reduced amide hydrogen/deuterium exchange rates [Table SII(B)].
For a small protein with a nearly spherical shape, the NOE‐derived distances, hydrogen bond and dihedral angle constraints are normally sufficient for defining the 3D structure at a good level of atomic resolution. However, AaE, like other GEM proteins, has a brick‐shaped folding with four parallel β‐sheets/hairpins giving rise to long‐range NOEs mainly within the individual β‐hairpins. Consequently, each β‐hairpin unit can be defined locally by hydrogen bonds and characteristic medium‐range NOEs, but their relative orientation may remain under‐defined. However, 15N relaxation data [Fig. 6(B)] show similar amplitudes across all residues for local backbone motions on the nanosecond‐to‐picosecond time scale, indicating inherent structural rigidity across the entire molecule. Such a structural rigidity allows the use of the dipolar couplings to establish the spatial orientations between each β‐hairpin unit.
1H–15N residual dipolar couplings were measured for AaE in the liquid crystal media of Pf1 phage with a phage concentration of 12 mg/ml. Residual dipolar coupling constants were obtained for 45 residues [Table SII(B)] but measurement was not possible for the other 11 residues as a result of peak overlaps, absence of HN signals for Pro residues and the disappearance of peaks at pH 6.5 resulting from fast exchange with the solvent water. Using the approach of Clore et al.18 an initial value of Da = 0.32 and R = 9.08 was estimated from the distribution of residual dipolar couplings, then optimized by running a series of structure calculations with Da values in the range of (0.26 ∼ 0.46) in steps of 0.02 and R values varying from 7.6 to 10 in steps of 0.2. The values of parameters that gave the lowest energy structures were selected, that is, Da = 9.48 and R = 0.40. A superposition of the 16 final simulated annealing structures is shown in Figure 7B. The quality of the RDC‐refined structure [Fig. 7(B)] is significantly improved compared with the structure calculated only with NOE, dihedral angle and hydrogen bond constraints [Fig. 7(A)]. Mean backbone RMSD to the average structure for the whole molecule is 0.697, compared with 1.223 for the structures without RDC. There is also a slight improvement in the stereochemical quality shown using PROCHECK calculations: 67.2% torsion angles of all residues in the most favored regions of the Ramachandran plot compared with 63.9% for the structures without RDC.
Figure 7.
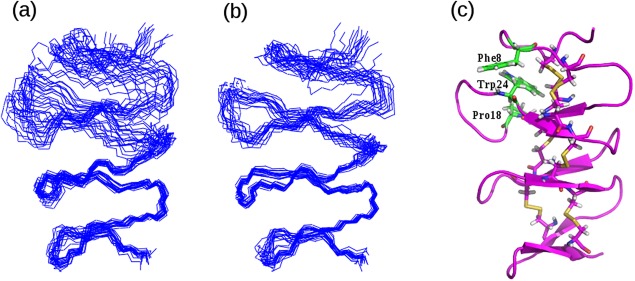
Three‐dimensional structure of AaE showing structural subdomains and critical/packing interactions among selected residues. (A) Backbone superposition of a set of converged structures of AaE calculated using NOE distances, dihedral angle and hydrogen‐bonding constraints. (B) Superposition of a set of refined structures of AaE after inclusion of residual (long‐range) dipolar coupling constants. Subdomain flexibility is still discernable, especially when the structure models are superimposed using the backbone atoms of the C‐terminal subdomain. (C) A ribbon diagram of the refined 3D structure of AaE rendered using the set of structures shown in (B). Three unique residues, Phe8, Pro18, and Trp24 are shown here with close‐contact side‐chain packing potentially stablizing the 3D structure of the N‐terminal subdomain.
Experimental identification of disulfide bonds
All 14 zebrafish granulin modules were expressed in the cytoplasm of the Origami bacterial strain, which is favorable for the spontaneous formation of stable disulfide bonds. In addition, all granulin DNA fragments were subcloned into the pET32b bacterial vector containing an N‐terminal thioredoxin tag, which promotes formation of disulfide bonds in the cytoplasmic space for the over‐expressed proteins. Indeed, with the well‐structured AaE module, disulphide links were identified by the observed long‐range NOEs between the CαH/CβH and CβH/CβH proton pairs of residues Cys4–Cys16, Cys10–Cys26, Cys17–‐Cys34, Cys27–Cys41, Cys35–Cys48, and Cys42–Cys55. These same SS bridges were also evident in the calculated 3D structures [Fig. 7(B)] using NOE‐derived distance, dihedral angle, hydrogen bond and residual dipolar coupling restraints.
Cell survival assay results
NSC 34 cells were cultured at different concentrations of the well‐structured AaE peptide, ranging from 10 nM to 10 µM. Slight differences were noticed on day 1 for cell cultures with different concentrations of AaE, but the differences were no longer significant on day 2 (Fig. 8). Very interestingly, cells cultured in the presence of higher concentrations of AaE (either 1 or 10 µM) had increased survival compared to control cultures and to lower concentrations of AaE (either 10 nM or 100 nM) over the entire period of 8 days, particularly from day 6 to day 8. It therefore appears that AaE at higher concentrations (1 to a few µM) promotes the growth and survival of differentiated neuronal cells represented by the NSC‐34 cell line developed from combining embryonic spinal cord cells with neuroblastoma cells.19, 20
Figure 8.
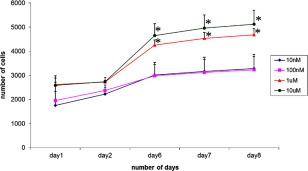
Effects of varying concentrations of the GEM peptide AaE on the survival of a neuronal cell line, NSC34. Cells that are cultured in the presence of 1 µM (or higher) of AaE had significantly increased survival compared to controls and to the presence of lower concentrations of AaE. Asterisks (*) indicate experimentally determined cell survival rates as significatly different (P < 0.050) from those of control cell cultures without added granulin. Statistical analysis was carried out using ANOVA available in the SigmaPlot4 software program.
Discussions and Conclusion
Structural characteristics of the GEM protein fold was first determined using the prototypic carp granulin‐1,13 then investigated through granulin‐derived peptides21, 22, 23, 24 and more recently via a dissection of granulin modules in human progranulin.12 Despite being a small scaffold (of ∼60 residues) reinforced by a large number (6) of disulfide bonds, well‐folded GEM proteins still appear to be composed of two distinct structural and, possibly, functional subdomains. The N‐terminal subdomain forms a well‐structured stack of two β‐hairpins, directing the formation of consensus disulfide pairing patterns found in the intact stack of four β‐hairpins.21, 22, 23 Three‐dimensional structure of the N‐terminal stack of two β‐hairpins has been shown to be quite tolerant of the loop size in the second β‐hairpin: ranging from 8 to 9 residues in a typical granulin module (Fig. 1) to 13 residues in a plant GEM motif.24 On the other hand, the C‐terminal subdomain appears to be more diverse and structurally more flexible, which is unique for each GEM protein and which may be related to distinct biological functions. The zebrafish GEM proteins again exhibit substantial structural diversity, as exemplified by their different patterns of spontaneous refolding during bacterial expression and purification (Figs. 2 and 3). The relatively autonomous folding of the N‐terminal stack of two β ‐hairpins is again evident in the NMR signatures of the two better‐structured zebrafish GEMs, AaE and AbB (Fig. 4 and descriptions in the Results section). Specifically, the NH proton chemical shift dispersion of AbB are 1.2 and 2.48 ppm, for the first and the second beta‐hairpins, respectively [Table SII(A)], while its entire C‐terminal region has a greatly reduced NH shift dispersion of only 0.6–1 ppm. As well, even the best‐folded AaE has a smaller shift dispersion (of 2.88 ppm) for the C‐terminal region than the N‐terminal subdomain (up to 3.29 ppm), indicating a certain degree of internal structural flexibility in between the two subdomains.
The full three‐dimensional structure of AaE is represented in Figure 7, which shows two clearly defined subdomains. The N‐terminal subdomain of AaE is formed by the two well‐folded antiparallel β‐sheets similar to previously studied GEM proteins and peptide fragments. The connecting structure between the two β‐hairpins has a conformation of a type‐II beta‐turn, which is stabilized by a hydrogen bond between the amide proton of Glu14 and the carbonyl oxygen of His11. Most notably, the C‐terminal subdomain was also found to be well structured in the AaE module. Several observed hydrogen bonds [Table SII(B)], typical of antiparallel beta‐sheets, may strongly stabilize the two C‐terminal β‐hairpins and there is a similar type‐II turn as observed in the N‐terminal subunit, with the carbonyl oxygen of Pro43 accepting a hydrogen bond from Tyr46 HN. The entire 3D structure of AaE is again dominated by six unique disulfide bonds, with several specific hydrophobic interactions contributing to the overall fold stabilization. For example, it has the typical contacts between the aromatic side chain of Trp24 and the side chains of residues Phe8 and Pro18 [Fig. 7(C)], all stacked against the two disulfide bonds as in other granulin modules and fragments.12, 21, 22, 23, 24 The structurally equivalent residues in carp granulin‐1 are Trp24, Thr8, and Ile18, respectively21, 22 and Trp24, Val8, and Arg18 in human granulin A.12, 23 This subdomain nature of yet another well‐structured GEM protein, zebrafish AaE, is perhaps the manifestation of the prevalent two‐exon coding of GEM repeats, where, for each GEM, one exon encodes the N‐terminal stack of two hairpins and a second exon encodes the C‐terminal stack of two hairpins,25 leading to unique patterns of duplication of the GEM repeats during evolution.3
Table 1 shows the amino acid preferences at position 18 and 24 of all GEM repeats including carp granulin‐113 and human granulin A, C, F,12 whose 3D structures have been determined, and the zebrafish granulin repeats studied here (Fig. 1). A Trp residue at position 24 is strongly correlated with the folding into a stack of β ‐hairpins, as in human granulins A, C and F12 and the appearance of well‐structured species in purified zebrafish GEMs, especially Grn1, AaA, AaC, AaE, AaF, AbA, AbB, and AbE (Fig. 3). This same Trp residue had previously been highlighted as a conserved element in the evolution of granulin repeats.3 On the other hand, other residues are found at this position for the rest of the zebrafish GEMs: Phe for AaH, Thr for AaD, Tyr for AaG, and AbF and AbH, which appears to be related to a predominant lack of three‐dimensional folding (Fig. 3) or very low yield of the intact peptide [e.g., Fig. 2(A)—for AaG]. In this regard, human granulin B also has a Tyr residue in place of Trp24, a negatively charged Glu at position 18 in place of positively charged (e.g., Arg and Lys) and more hydrophobic residues such as Leu, Ile or Phe for other folded GEM peptides (Table 1). Lack of favorable packing interactions among more hydrophobic sidechains may be the origin of alternative disulfide pairing observed in recombinantly expressed human granulin B (also referred to as granulin‐3).26 Very interestingly, position 18 in zebrafish granulin AaE turns out to be a unique proline (Table 1), which is next to Cys17 of the Cys17‐Cys34 disulfide pair, where other GEMs generally have a hydrophobic or polar residue with a long side chain, for example, Leu for human granulin F and Arg for human granulin A, zebrafish Grn2 and zebrafish AbI. This Pro residue in AaE may therefore also have important positive effects on the folding, on the formation of consensus disulfide bonds and on the three‐dimensional structure stability of the prototypic stack of four beta‐hairpins (Fig. 7).
Progranulins, progranulin‐derived GEM proteins and other GEM peptides have emerged as very important cell‐growth modulators as well as mediators of lysosomal function,27, 28 playing a significant role in many biological processes.1, 3, 4, 5, 29 Recent interests have also focused on GEM proteins in the infectious and carcinogenic human liver parasite fluke;30, 31 particularly as inducers of organ fibrosis and potentiators of metastatic carcinomas of the human liver,32, 33 and cardiovascular systems,33 which can be targets for the development of anti‐cancer therapies. Like human progranulin and GEM proteins,34 parasitic GEM‐like proteins may also find therapeutic applications as novel agents for wound healing.35 In the context of neurobiology, neuronal NSC34 cells cultured with AaE indicate that the well‐structured AaE may promote neuronal survival (Fig. 8), but at the high concentrations above 100 nM. This finding is consistent with earlier reports that (human) progranulin increases neuronal cell survival36, 37, 38, 39, 40, 41 and a human granulin peptide, granulin‐E, increased cell survival in both cortical neurons and spinal cord motor neurons.39, 42 Curiously, rather high concentrations (>800 nM) of fluke GEM peptides were also found to be required for promoting the proliferation of human cholangiocytes.35 A disulfide‐reinforced peptide mimicking the fluke N‐terminal stack of two hairpins had greatly increased cell proliferative activity, but its effectiveness remained the same for wound healing. Taken together, all these observations outline the remaining challenges for structure‐activity studies of this important class of growth‐modulating GEM proteins.
In a structure dissection of human progranulin, relatively well‐defined three‐dimensional structures were found only for three, that is, human granulins A, C, and F, of the seven full GEM repeats.12 All three human granulins A, C and F contain a well‐folded N‐terminal stack of two beta‐hairpins and all three had significant cell growth‐modulating activities: strong growth inhibition of the MDA‐MB‐468 cancer cell line by granulin A, weak inhibition of cell growth by granulin C and strong growth promotion by granulin F, albeit at relatively high concentrations (>5 μM). Granulin B did not emerge as a folded protein in that study, even though the unfolded granulin B had some growth‐stimulating activities and it was demonstrated previously to exert proinflammatory effects.43 Very interestingly, human granulin B without disulfide bond formation was shown to be a intrinsically disordered protein, or an IDP, even with the native Tyr28 residue replaced by the structure‐reinforcing residue Trp.44 In fact, a high degree of disorder has been uncovered for human granulin modules 1 (or G), 3 (or B), 5 (or C), and 7 (or E), with granulin B exhibiting the highest degree of structure disorder.26 Lack of unique folding for an individual GEM motif makes structure‐activity studies of granulin B and other individual human granulin modules a challenging task12 and tandem repeats incorporating a more structured module may be needed26 for restoring the potency of GEM fragments to the level observed for progranulins. It is also possible that conformational disorder may be a prerequisite for full functional activities for some GEM proteins. In this regard, it is interesting to notice that a structurally constrained analogue of the fluke GEM protein did not offer specific advantages to the wound‐healing activities of fluke‐derived GEM peptides.35
Materials and Methods
Cloning and expression
A total of 19 distinct granulin fragments (Fig. 1) were amplified using PCR from the cDNA clones of zebrafish progranulins [Table SI(A)], identified and maintained by Bennett's research laboratory at the Research Institute of the McGill University Health Center (MUHC), Montreal. After double digestion with the restriction enzymes Nco I and EcoR I, the PCR products were subcloned using synthetic primers [Table SI(B)] into the pET32b bacterial plamid vector, which expresses proteins with a thioredoxin and hexa‐histidine tags attached to the N‐terminus followed by a specific enterokinase cleavage site. Positive clones were confirmed by colony PCR screening with the Taq DNA polymerase and by DNA sequencing [Table SI(C)].
All fusion proteins were expressed in the E. coli strain Origami (DE3), which favors disulfide bond formation in progranulins through an oxidizing intracellular environment together with the thioredoxin tag.12 Cells were grown at room temperature on LB medium to produce the encoded proteins. At an OD600 of 0.6 ∼ 0.8, cells were induced to express the His‐tagged fusion proteins by addition of isopropyl β‐D‐thiogalactoside. (IPTG) to a final concentration of 0.4 mM. Ampicillin was added to a concentration of 100 μg/ml. Four hours later, the cells were harvested and cell pellets were stored at −20°C prior to further processing.
For an 15N‐labeled protein sample (of the AaE module), a single colony was first grown at 37°C in l mL LB medium for 8 h, then transferred to 60 mL of diluted 15N‐labeled rich medium Bio‐Express‐1000 (Cambridge Isotope Labs) and grown overnight. On the second day, the overnight culture was inoculated with the same diluted 15N‐labeled Bio‐Express medium containing 100 μg/mL ampicillin in a total volume of 225 mL, and allowed to grow until an OD600 of 0.6 ∼ 0.8 was obtained, at which time protein expression was induced overnight with 0.1 mM IPTG. In the third day, the cells were harvested for further processing.
Protein purification and NMR sample preparation
Frozen cell pellets were thawed by adding 50 mL of 100 mM sodium phosphate at pH 7.0 with 10 mM Tris–HCl, 300 mM sodium chloride. Lysozoyme was added to a concentration of 0.8 mg/ml, and cell suspension was shaken over ice for 30 min. Before sonication, the protease inhibitor, phenylmethylsulfonyl fluoride (PMSF), was added to a final concentration of 100 μg/mL and the cell debris was spun down after sonication. An amount of 12 g of solid urea was added to a final concentration of 4M into the supernatant, which was followed by gentle shaking with 10 mL (total volume) of suspended Ni‐NTA resin for 1 h. His‐tagged fusion protein was purified on a Ni‐NTA column under denaturing conditions. The Ni2+ affinity beads were washed three times with 5‐column volumes of 100 mM sodium phosphate, 10 mM Tris–HCl, 300 mM sodium chloride and 8M urea at pH 6.3. Proteins were eluted with five column volumes of 100 mM sodium phosphate, 10 mM Tris–HCl and 300 mM sodium chloride and 8M urea at pH 5.5.
The protein was eluted from Ni‐NTA resin in the presence of 8M urea and was refolded by first dialyzing into 2.66M urea (1:3 dilution) at pH 7.0 at room temperature, and further dialyzing into 0.88M urea (another 1:3 dilution) at pH 7.0 and 4°C. Finally, the fusion protein with a concentration of 1 mg/mL was dissolved in the refolding buffer containing 10 mM sodium phosphate at pH 7.0 with 10 mM Tris–Cl, 150 mM sodium chloride, 0.01% azide, and 2 mM CaCl2. The folded fusion protein was subjected to enterkinase cleavage for 24 h at 37°C, which produced the GEM peptides [Table SI(C)] preceeded by a two‐residue (Ala‐Met or AM) extension derived from the enterokinase specificity site.12
The cleavage reaction was stopped by addition of trifluoroacetic acid (TFA) to a final concentration of 0.3% (v/v), followed by loading the solution onto C18 Sep‐Pak cartridges. After repeat loading until the OD280 of the flow‐through was very low, the cartridge was rinsed by 0.1% TFA (v/v), and the cleaved fusion proteins were eluted by use of aqueous acetonitrile at 35% (v/v) containing 0.1% TFA (v/v). Purified protein materials were lyophilized before final purification using high‐performance liquid chromatography (HPLC).
The Sep‐Pak eluates were lyophilized and redissolved in 0.1% TFA and the GEM peptides were subjected to reversed‐phase chromatography using a C18 Vydac column. After equilibration in 10% acetonitrile (v/v), Sep‐Pak eluates were loaded by injection and the column eluted with a linear gradient of 10%‐50% acetonitrile (v/v) containing 0.1% TFA (v/v). The column eluate was monitored by UV absorbance at 280 nm. Selected peak fractions were dried and characterized initially by mass spectroscopy and in further details by nuclear magnetic resonance (NMR).
The purified and lyophilized peptides were redissolved in 450 μL of 10% D2O and 90% of an NMR buffer that contains 10 mM sodium phosphate, 150 mM sodium chloride and 0.2 mM EDTA at pH 6.8. For NMR experiments in D2O, the samples were first lyophilized and then redissolved in pure D2O. All samples were buffered at pH 5–6.5 and NMR experiments were performed at 30°C.
NMR measurements
Standard one‐dimensional (1D) 1H spectra and two‐dimensional (2D) TOCSY and NOESY spectra were collected on a Bruker Avance 500 MHz spectrometer equipped with a pulse‐field gradient unit that was used to suppress the solvent (water) signal. The NMR data were processed by use of the XWINNMR software package.
Sequential assignment of NMR signals was obtained initially using unlabeled protein samples. The following 2D homonuclear spectra were recorded in 90% H2O/10%D2O using standard pulse sequences and phase cycles: 2D TOCSY (with a mixing time of 70 ms), and 2D NOESY (mixing times of 100 and 200 ms). Assignments were confirmed and extended using 3D heteronuclear spectra recorded using an 15N‐labeled peptide sample. The following heteronuclear NMR spectra were recorded in 90% H2O/10% D2O: 2D 1H–15N HSQC, 3D 15N TOCSY‐HSQC (with a mixing time of 70 ms), and 3D 15N NOESY‐HSQC (mixing times of 100 and 200 ms). For the HSQC and NOESY‐HSQC spectra, solvent suppression was achieved using pulsed field gradients and water flip‐back pulses. The 15N‐labeled sample was also used to measure amide hydrogen/deuterium exchange kinetics via 2D 1H–15N HSQC spectra and for subsequent acquisition of NMR spectra in pure D2O. The following spectra were recorded in 99.9% D2O: 2QF‐COSY, TOCSY (with a mixing time of 70 ms) and NOESY (mixing times of 100 and 200 ms).
Steady‐state 1H–15N NOEs were measured using pulse sequences from Farrow et al.45 with a recycle delay of 6 s. The spectra were acquired with or without saturation of the proton nuclei during the recycle delay, corresponding to the presence or absence of the 1H–15N NOEs, respectively. The steady‐state NOEs were calculated as the ratios of peak heights in the spectra recorded with and without proton saturation.
The 3J (HNHα) coupling constants were determined by use of the MJ‐HSQC experiment.46 A series of MJ‐HSQC spectra were acquired with increasing multiplication factor 2N = 3, 5, 7. The observable signals are split by 2N*3J(HNHα) in the F1 dimension. The J coupling constant can be accurately obtained by least‐squares fitting of the apparent doublets in ω1 with respective multiplication factors.
Residual dipolar couplings and their use for structure refinement
Residual dipolar coupling constants were measured using an 15N labeled peptide sample with partial alignment achieved by diluting into a liquid‐crystalline Pf1 phage medium (profos AG). The concentration of the Pf1 phage medium was increased stepwise from 7 mg/ml to 12 mg/ml and to 16 mg/ml to achieve a maximum alignment while minimizing non‐specific interactions between the peptide molecules and the orienting media. The 1H–15N residual dipolar couplings were measured from the 1JNH splitting appearing in the 15N dimension of an HSQC experiment that incorporated a semi‐TROSY effect.47 The experiment was therefore carried out at 800 MHz and processed to give a final digital resolution of 1 Hz/pt (along the F2 1H dimension) and 5 Hz/pt (F1–15N dimension). The residual dipolar coupling was taken as the difference between the splitting observed in the oriented Pf1 phage medium and the isotropic medium.
3D structure calculations and refinement
Interproton distance restraints were derived from the NOE cross peaks in 3D 1H–15N NOESY‐HSQC (with a mixing time of 200 ms) spectrum of the 15N‐labeled sample, and 2D homonuclear NOESY (mixing time of 200 ms) spectra of an unlabeled sample in 90% H2O/10% D2O and 99.9% D2O, respectively. The NOE peaks were calibrated automatically during the structure calculation by use of ARIA, a method that combines an iterative NOE interpretation scheme with dynamic assignment of ambiguous NOE cross peaks.48
Hydrogen bond restraints were based on the observation of reduced amide hydrogen/deuterium exchange rates and characteristic NOEs involving 1HN and 1Hα resonances. Each hydrogen bond thus identified was defined by two distance restraints: rNH–O =1.8–2.4 A and rN–O = 2.7–3.5 A. The phi dihedral angle restraints were derived from 3JHN‐Hα coupling constants. The phi angle was restrained to the range −150°C to −90°C for 3JHN–Hα > 8 Hz, and −90°C to −30°C for 3J HN–Hα < 5 Hz.
Residual dipolar couplings contain non‐local (global) structural constraints that can be used for structure refinement in combination with NOE distances, dihedral angle, and hydrogen bond restraints. Structure refinement is achieved by incorporating the residual dipolar couplings into the structure calculation by means of a penalty function.49
Here “calc” and “obs” refer to the calculated and observed values of the residual dipolar couplings and k dip is a force constant. The inclusion of the residual dipolar couplings in the structure calculations also requires the values of Da, the axial component of the alignment tensor, and R, the rhombicity defined by the method of Clore et al.18 from a histogram displaying the distributions of residual dipolar couplings. From the high and low extreme values and from the most populated value, the Da and R was estimated, and subsequently refined by conducting a grid search to find values that yield the lowest energy structures.
Structural calculation was performed with the software package ARIA48 interfaced with the CNS program. Starting from an extended polypeptide chain, a first set of 200 structures was calculated on the basis of restraints as follows: 22 hydrogen bond restraints, 23 dihedral angle restraints, six disulfide bond restraints, 432 NOE‐based distance restraints derived from manual assignment of different NOESY spectra, and 88 ambiguous NOEs assigned using a frequency window tolerance around their chemical shift values of 0.02 ppm along the 1H acquisition dimension, 0.04 ppm along the 1H indirect dimension and 0.5 ppm along the heteronuclear 15N direction (as deposited with the BioMagResBank, BMRB accession No. 30422). During the following eight iterations, ARIA performs an automatic assignment of NOESY spectra peaks based on the chemical shift list and its compatibility with the interproton distances measured in the initial structures. Conversion of peak volumes into distance restraints was accomplished through the structure calculation. In each iteration, out of all the 200 calculated structures, half (i.e., 100) were retained for further refinement, and 35% of the structures with lower energy were used for assignment purposes. Also, from the third iteration on, residual dipolar couplings were introduced into the refinement process, with orientation of the alignment tensor as a floating variable. During the entire eight ARIA iterations, the schematic parameters were modified as follows: the assignment parameter (p) from 1.01 to 0.80, the violation threshold (Tv) from 1000 to 0.1, and the violation ratio (Nv) held constant at 0.8. Every run was monitored and analyzed by home‐built (Perl) scripts collectively called TopCat, including ariaview.pl.22
This iterative structure calculation/refinement process led to 16 structures of low energy, which were used for the final analysis and for structure deposition (PDB accession No. 6CKU). The stereochemical quality of the 16 structures was analyzed using the program PROCHECK.50 The structure was visualized using the MOLMOL software program.51
NSC‐34 cell culture and survival assay
The NSC‐34 cell line was originally developed by fusing embryonic spinal cord cells with neuroblastoma cells and found to exhibit motor neuronal properties in the differentiated state.19, 20 For the survival assay,38 the NSC‐34 cells were maintained in the DMEM media supplemented with 10% fetal bovine serum. For stimulation with the progranulin module AaE, NSC‐34 cells were cultured in 96 well plates at an initial density of 2000 cells per well. After 24 h, the cultures were treated with or without AaE under serum‐free conditions in DMEM for several days. Cell survival/viability was assessed using the AlamarBlue (Biosource, Camarillo, CA) colorimetric assay, which provides a quantitative assessment of the metabolic activities, leading to a chemical reduction of AlamarBlue from the oxidized form (blue) to the reduced form (pink). Plates were read in an ELISA plate reader at twodifferent wavelengths, 570 and 630 nm, 24 h after addition of AlamarBlue.
Supporting information
Supporting Information
Table SI(A): Sequences of the cDNA clones of zerbrafish progranulins Aa, Ab, 1 and 2
Table SI(B): Synthetic primers for subcloning of all (19) zebrafish granulin modules from the cDNAs of progranulins Aa, Ab, 1 and 2.
Table SI(C): DNA and predicted peptide sequences of 14 subcloned zebrafish granulin modules.
Table SII(A): 1H chemical shifts of zebrafish granulin AbB at pH 6.8 and 303 K
Table SII(B): 15N and 1H chemical shifts of zebrafish granulin AaE at pH 6.8 and 303 K plus 3J(HN‐Ha) coupling constants, residual dipolar constants (RDCs) and hydrogen‐bond pairs from reduced hydrogen‐deuterium exchange rates of the NH protons.
Acknowledgments
This work was supported in part by the Natural Sciences and Engineering Research Council of Canada (RGPIN 141019‐97 to Feng Ni and RGPIN 327185‐11 to Hugh Bennett) and by the Canadian Cancer Society Research Institute (CCSRI 700379 to Andrew Bateman). Work at the National Research Council of Canada was supported by the Biologics and Biomanufacturing Program and by a Genonomics and Health Initiatives (GHI) Program, both sponsored by the Government of Canada. This is NRCC Publication No. NRC‐HHT_53379.
References
- 1. Bateman A, Bennett HP (1998) Granulins: the structure and function of an emerging family of growth factors. J Endocrinol 158:145–151. [DOI] [PubMed] [Google Scholar]
- 2. Bateman A, Bennett HP (2009) The granulin gene family: from cancer to dementia. Bioessays 31:1245–1254. [DOI] [PubMed] [Google Scholar]
- 3. Palfree RG, Bennett HP, Bateman A (2015) The evolution of the secreted regulatory protein progranulin. PLoS One 10:e0133749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chitramuthu BP, Bennett HPJ, Bateman A (2017) Progranulin: a new avenue towards the understanding and treatment of neurodegenerative disease. Brain 140:3081–3104. [DOI] [PubMed] [Google Scholar]
- 5. Abella V, Pino J, Scotece M, Conde J, Lago F, Gonzalez‐Gay MA, Mera A, Gómez R, Mobasheri A, Gualillo O (2017) Progranulin as a biomarker and potential therapeutic agent. Drug Discov Today 22:1557–1564. [DOI] [PubMed] [Google Scholar]
- 6. Bateman A, Belcourt D, Bennett H, Lazure C, Solomon S (1990) Granulins, a novel class of peptide from leukocytes. Biochem Biophys Res Commun 173:1161–1168. [DOI] [PubMed] [Google Scholar]
- 7. Bhandari V, Palfree RG, Bateman A (1992) Isolation and sequence of the granulin precursor cDNA from human bone marrow reveals tandem cysteine‐rich granulin domains. Proc Natl Acad Sci USA 89:1715–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shoyab M, McDonald VL, Byles C, Todaro GJ, Plowman GD (1990) Epithelins 1 and 2: isolation and characterization of two cysteine‐rich growth‐modulating proteins. Proc Natl Acad Sci USA 87:7912–7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Couto MA, Harwig SS, Cullor JS, Hughes JP, Lehrer RI (1992) Identification of eNAP‐1, an antimicrobial peptide from equine neutrophils. Infect Immun 60:3065–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou J, Gao G, Crabb JW, Serrero G (1993) Purification of an autocrine growth factor homologous with mouse epithelin precursor from a highly tumorigenic cell line. J Biol Chem 268:10863–10869. [PubMed] [Google Scholar]
- 11. Baba T, Hoff HB III, Nemoto H, Lee H, Orth J, Arai Y, Gerton GL (1993) Acrogranin, an acrosomal cysteine‐rich glycoprotein, is the precursor of the growth‐modulating peptides, granulins, and epithelins, and is expressed in somatic as well as male germ cells. Mol Reprod Dev 34:233–243. [DOI] [PubMed] [Google Scholar]
- 12. Tolkatchev D, Malik S, Vinogradova A, Wang P, Chen Z, Xu P, Bennett HP, Bateman A, Ni F (2008) Structure dissection of human progranulin identifies well‐folded granulin/epithelin modules with unique functional activities. Protein Sci 17:711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hrabal R, Chen Z, James S, Bennett HP, Ni F (1996) The hairpin stack fold, a novel protein architecture for a new family of protein growth factors. Nat Struct Biol 3:747–752. [DOI] [PubMed] [Google Scholar]
- 14. Belcourt DR, Lazure C, Bennett HP (1993) Isolation and primary structure of the three major forms of granulin‐ like peptides from hematopoietic tissues of a teleost fish (Cyprinus carpio). J Biol Chem 268:9230–9237. [PubMed] [Google Scholar]
- 15. Uesaka T, Yano K, Yamasaki M, Ando M (1995) Somatostatin‐, vasoactive intestinal peptide‐, and granulin‐like peptides isolated from intestinal extracts of goldfish, Carassius auratus. Gen Comp Endocrinol 99:298–306. [DOI] [PubMed] [Google Scholar]
- 16. Cadieux B, Chitramuthu BP, Baranowski D, Bennett HP (2005) The zebrafish progranulin gene family and antisense transcripts. BMC Genom 6:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rehm T, Huber R, Holak TA (2002) Application of NMR in structural proteomics: screening for proteins amenable to structural analysis. Structure 10:1613–1618. [DOI] [PubMed] [Google Scholar]
- 18. Clore GM, Gronenborn AM, Bax A (1998) A robust method for determining the magnitude of fully asymmetric alignment tensor of oriented macromolecules in the absence of structural information. J Magn Reson 133:216–221. [DOI] [PubMed] [Google Scholar]
- 19. Cashman NR, Durham HD, Blusztajn JK, Oda K, Tabira T, Shaw IT, Dahrouge S, Antel JP (1992) Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn 194:209–221. [DOI] [PubMed] [Google Scholar]
- 20. Durham HD, Dahrouge S, Cashman NR (1993) Evaluation of the spinal cord neuron X neuroblastoma hybrid cell line NSC‐34 as a model for neurotoxicity testing. Neurotoxicology 14:387–395. [PubMed] [Google Scholar]
- 21. Vranken WF, Chen ZG, Xu P, James S, Bennett HP, Ni F (1999) A 30‐residue fragment of the carp granulin‐1 protein folds into a stack of two beta‐hairpins similar to that found in the native protein. J Pept Res 53:590–597. [DOI] [PubMed] [Google Scholar]
- 22. Vranken WF, James S, Bennett HP, Ni F (2002) Solution structures of a 30‐residue amino‐terminal domain of the carp granulin‐1 protein and its amino‐terminally truncated 3–30 subfragment: implications for the conformational stability of the stack of two beta‐hairpins. Proteins 47:14–24. [PubMed] [Google Scholar]
- 23. Tolkatchev D, Ng A, Vranken W, Ni F (2000) Design and solution structure of a well‐folded stack of two beta‐hairpins based on the amino‐terminal fragment of human granulin A. Biochemistry 39:2878–2886. [DOI] [PubMed] [Google Scholar]
- 24. Tolkatchev D, Xu P, Ni F (2001) A peptide derived from the C‐terminal part of a plant cysteine protease folds into a stack of two beta‐hairpins, a scaffold present in the emerging family of granulin‐like growth factors. J Pept Res 57:227–233. [DOI] [PubMed] [Google Scholar]
- 25. Bhandari V, Bateman A (1992) Structure and chromosomal location of the human granulin gene. Biochem Biophys Res Commun 188:57–63. [DOI] [PubMed] [Google Scholar]
- 26. Ghag G, Holler CJ, Taylor G, Kukar TL, Uversky VN, Rangachari V (2017) Disulfide bonds and disorder in granulin‐3: An unusual handshake between structural stability and plasticity. Protein Sci 26:1759–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holler CJ, Taylor G, Deng Q, Kukar T (2017) Intracellular proteolysis of progranulin generates stable, lysosomal granulins that are haploinsufficient in patients with frontotemporal dementia caused by GRN mutations. eNeuro 4:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evers BM, Rodriguez‐Navas C, Tesla RJ, Prange‐Kiel J, Wasser CR, Yoo KS, McDonald J, Cenik B, Ravenscroft TA, Plattner F, Rademakers R, Yu G, White CL, Herz J (2017) Lipidomic and transcriptomic basis of lysosomal dysfunction in progranulin deficiency. Cell Rep 20:2565–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kao AW, McKay A, Singh PP, Brunet A, Huang EJ (2017) Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neurosci 18:325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smout MJ, Mulvenna JP, Jones MK, Loukas A (2011) Expression, refolding and purification of Ov‐GRN‐1, a granulin‐like growth factor from the carcinogenic liver fluke, that causes proliferation of mammalian host cells. Protein Expr Purif 79:263–270. [DOI] [PubMed] [Google Scholar]
- 31. Machicado C, Marcos LA, Zimic M (2016) Hypothetical granulin‐like molecule from Fasciola hepatica identified by bioinformatics analysis. Springerplus 5:773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smout MJ, Sotillo J, Laha T, Papatpremsiri A, Rinaldi G, Pimenta RN, Chan LY, Johnson MS, Turnbull L, Whitchurch CB, Giacomin PR, Moran CS, Golledge J, Daly N, Sripa B, Mulvenna JP, Brindley PJ, Loukas A (2015) Carcinogenic parasite secretes growth factor that accelerates wound healing and potentially promotes Neoplasia. PLoS Pathog 11:e1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang C, Lei H, Tian Y, Shang M, Wu Y, Li Y, Zhao L, Shi M, Tang X, Chen T, Lv Z, Huang Y, Tang X, Yu X, Li X (2017) Clonorchis sinensis granulin: identification, immunolocalization, and function in promoting the metastasis of cholangiocarcinoma and hepatocellular carcinoma. Parasit Vectors 10:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He Z, Ong CH, Halper J, Bateman A (2003) Progranulin is a mediator of the wound response. Nat Med 9:225–229. [DOI] [PubMed] [Google Scholar]
- 35. Bansal PS, Smout MJ, Wilson D, Cobos Caceres C, Dastpeyman M, Sotillo J, Seifert J, Brindley PJ, Loukas A, Daly NL (2017) Development of a potent wound healing agent based on the liver fluke granulin structural fold. J Med Chem 60:4258–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gao X, Joselin AP, Wang L, Kar A, Ray P, Bateman A, Goate AM, Wu JY (2010) Progranulin promotes neurite outgrowth and neuronal differentiation by regulating GSK‐3β. Protein Cell 1:552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo A, Tapia L, Bamji SX, Cynader MS, Jia W (2010) Progranulin deficiency leads to enhanced cell vulnerability and TDP‐43 translocation in primary neuronal cultures. Brain Res 1366:1–8. [DOI] [PubMed] [Google Scholar]
- 38. Ryan CL, Baranowski DC, Chitramuthu BP, Malik S, Li Z, Cao M, Minotti S, Durham HD, Kay DG, Shaw CA, Bennett HP, Bateman A (2009) Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci 10:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Damme P, Van Hoecke A, Lambrechts D, Vanacker P, Bogaert E, van Swieten J, Carmeliet P, Van Den Bosch L, Robberecht W (2008) Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J Cell Biol 181:37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kleinberger G, Wils H, Ponsaerts P, Joris G, Timmermans JP, Van Broeckhoven C, Kumar‐Singh S (2010) Increased caspase activation and decreased TDP‐43 solubility in progranulin knockout cortical cultures. J Neurochem 115:735–747. [DOI] [PubMed] [Google Scholar]
- 41. Xu J, Xilouri M, Bruban J, Shioi J, Shao Z, Papazoglou I, Vekrellis K, Robakis NK (2011) Extracellular progranulin protects cortical neurons from toxic insults by activating survival signaling. Neurobiol Aging 32:2326.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Muynck L, Herdewyn S, Beel S, Scheveneels W, Van Den Bosch L, Robberecht W, Van Damme P (2013) The neurotrophic properties of progranulin depend on the granulin E domain but do not require sortilin binding. Neurobiol Aging 34:2541–2547. [DOI] [PubMed] [Google Scholar]
- 43. Zhu J, Nathan C, Jin W, Sim D, Ashcroft GS, Wahl SM, Lacomis L, Erdjument‐Bromage H, Tempst P, Wright CD, Ding A (2002) Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell 111:867–878. [DOI] [PubMed] [Google Scholar]
- 44. Ghag G, Wolf LM, Reed RG, Van Der Munnik NP, Mundoma C, Moss MA, Rangachari V (2016) Fully reduced granulin‐B is intrinsically disordered and displays concentration‐dependent dynamics. Protein Eng Des Sel 29:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman‐Kay JD, Kay LE (1994) Backbone dynamics of a free and phosphopeptide‐complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33:5984–6003. [DOI] [PubMed] [Google Scholar]
- 46. Heikkinen S, Aitio H, Permi P, Folmer R, Lappalainen K, Kilpelainen L (1999) J‐multiplied HSQC (MJ‐HSQC): a new method for measuring 3J(HNHalpha) couplings in 15N‐labeled proteins. J Magn Reson 137:243–246. [DOI] [PubMed] [Google Scholar]
- 47. Lerche MH, Meissner A, Poulsen FM, Sorensen OW (1999) Pulse sequences for measurement of one‐bond 15N‐1H coupling constants in the protein backbone. J Magn Reson 140:259–263. [DOI] [PubMed] [Google Scholar]
- 48. Nilges M, Donoghue SI (1998) Ambiguous NOEs and automated NOE assignment. Prog NMR Spectrosc 32:107–139. [Google Scholar]
- 49. Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A (1997) Use of dipolar 1H‐15N and 1H‐13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nat Struct Biol 4:732–738. [DOI] [PubMed] [Google Scholar]
- 50. Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) Procheck: A program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291. [Google Scholar]
- 51. Koradi R, Billeter M, Wüthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics 14:51–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Table SI(A): Sequences of the cDNA clones of zerbrafish progranulins Aa, Ab, 1 and 2
Table SI(B): Synthetic primers for subcloning of all (19) zebrafish granulin modules from the cDNAs of progranulins Aa, Ab, 1 and 2.
Table SI(C): DNA and predicted peptide sequences of 14 subcloned zebrafish granulin modules.
Table SII(A): 1H chemical shifts of zebrafish granulin AbB at pH 6.8 and 303 K
Table SII(B): 15N and 1H chemical shifts of zebrafish granulin AaE at pH 6.8 and 303 K plus 3J(HN‐Ha) coupling constants, residual dipolar constants (RDCs) and hydrogen‐bond pairs from reduced hydrogen‐deuterium exchange rates of the NH protons.


