Abstract
Bacterial type III secretion systems (T3SS) are used to inject proteins into mammalian cells to subvert cellular functions. The Shigella T3SS apparatus (T3SA) is comprised of a basal body, cytoplasmic sorting platform and exposed needle with needle “tip complex” (TC). TC maturation occurs when the translocator protein IpaB is recruited to the needle tip where both IpaD and IpaB control secretion induction. IpaB insertion into the host membrane is the first step of translocon pore formation and secretion induction. We employed disruptive insertional mutagenesis, using bacteriophage T4 lysozyme (T4L), within predicted IpaB loops to show how topological features affect TC functions (secretion control, translocon formation and effector secretion). Insertions within the N‐terminal half of IpaB were most likely to result in a loss of steady‐state secretion control, however, all but the two that were not recognized by the T3SA retained nearly wild‐type hemolysis (translocon formation) and invasiveness levels (effector secretion). In contrast, all but one insertion in the C‐terminal half of IpaB maintained secretion control but were impaired for hemolysis and invasion. These nature of the data suggest the latter mutants are defective in a post‐secretion event, most likely due to impaired interactions with the second translocator protein IpaC. Intriguingly, only two insertion mutants displayed readily detectable T4L on the bacterial surface. The data create a picture in which the makeup and structure of a functional T3SA TC is highly amenable to physical perturbation, indicating that the tertiary structure of IpaB within the TC is more plastic than previously realized.
Keywords: Shigella, type III secretion, IpaB, translocon
Abbreviations
- T3SS
type III secretion system
- T3SA
type III secretion apparatus
- TC
tip complex
- Ipa
invasion plasmid antigen
- LDAO
N,N‐dimethyldodecylamine N‐oxide
- OPOE
n‐octyl‐poly‐oxyethylene
- T4L
bacteriophage T4 lysozyme
- SRB
sulforhodamine B
- IMAC
Immobilized Metal Affinity Chromatography
- DOPC
dioleoylphosphatidylcholine
- DOPG
dioleoylphosphatidylglycerol
- RBC
red blood cells.
Introduction
Shigella flexneri is a Gram‐negative, enteric pathogen that causes shigellosis (bacillary dysentery). Shigellosis is a severe bloody diarrhea that leads to dehydration, ulcerative colitis, and other complications. In a 1999 report, there were estimated to be >160 million cases of shigellosis with 1.1 million deaths annually.1 While the mortality associated with shigellosis has dropped since that time, the global burden of disease caused by Shigella continues to be high with devastating effects on children under the age of five.2, 3 Because as few as 10–100 organisms can cause disease, regional epidemics are common in all parts of the world.4 Shigella uses its type III secretion system (T3SS)1 to translocate effector proteins into host cells to alter their normal functions for the benefit of the pathogen.5 Many important Gram‐negative pathogens use the T3SS as essential virulence factors and despite pathogen‐specific differences in T3SS effector functions, the T3SS apparatus (T3SA) maintains significant structural homology across diverse species boundaries.6 The T3SA is anchored by a basal body that spans the entire bacterial envelope and supports an external needle composed of multiple copies of a small, polymerized needle protein terminating at a needle tip complex (TC) that maintains control of secretion.7
Shigella is the only pathogen for which the individual steps in T3SA needle TC maturation have been described.8, 9, 10 As a first step in TC maturation in Shigella, invasion plasmid antigen D (IpaD) forms a pentamer at the tip of the nascent MxiH needle.8, 10, 11 In the absence of IpaD, type III secretion in Shigella is constitutive. As an environmental sensor, IpaD can detect small molecules in the environment to promote recruitment and maintenance of IpaB, the first translocator protein, to the mature needle TC.10, 12, 13 This does not lead to induced secretion, but it does promote changes in the pathogen that enhance invasiveness, probably contributed to by increased adherence to host cells.14
From the T3SA needle tip, IpaB inserts into cholesterol‐ and sphingolipid‐rich host cell membranes—a behavior that can be mimicked in vitro using liposomes.9, 15 The interaction of IpaB with host cells or cholesterol/sphingolipid‐rich liposomes then promotes recruitment of IpaC, the second translocator protein, to the T3SA needle TC to give rise to formation of the “translocon pore” in the host cytoplasmic membrane. This completes formation of the conduit between Shigella and its host cell and induces effector protein secretion and translocation into the host cell.9 Because of the insight it has provided on secretion induction, the Shigella T3SA is arguably the best model available for studying the temporal assembly of the T3SA needle TC and the molecular events leading to type III secretion induction. The Shigella T3SA also provides a needed model for characterizing the interactions occurring between the needle TC proteins, especially as they relate to their roles in translocon formation and function.
When made in the Shigella cytoplasm, hydrophobic IpaB is stabilized by association with its chaperone IpgC.16 While no high‐resolution structures exist for full‐length IpaB, the structure of a stable N‐terminal core (residues 74–224) has been solved at a 2.1 Å resolution and found to be an elongated coiled‐coil.17 This structure represents a quarter of the overall protein and consists of an elongated (>100 Å) coiled‐coil. The fragment containing this structure (residues 28–226) binds to IpaD in vitro, suggesting it is involved in anchoring IpaB to the T3SA needle tip.13 IpaB can be purified after co‐expression with IpgC followed by removal of the chaperone using mild detergents,18, 19 however, the behavior of IpaB following separation from IpgC is greatly affected by the detergent used. In the presence of N,N‐dimethyldodecylamine N‐oxide (LDAO), IpaB forms a monomer in solution that binds to but does not lyse liposomes. In contrast, IpaB prepared using n‐octyl‐poly‐oxyethylene (OPOE) forms a stable tetramer in solution that penetrate liposomes to form a pore with a diameter of approximately 3 nm.18 Thus, it is likely this state (tetramer) that resides at tip of the mature Shigella T3SA needle tip, anchored by the N‐terminal coiled‐coil.13 This tetramer would then insert into the target cell membrane and initiate translocon pore formation.
We generated a series of bacteriophage T4 lysozyme (T4L) insertion mutants within IpaB to explore the functional effects of this added density in an ipaB‐null strain of Shigella. T4L possesses N‐ and C‐termini that are spatially very close together (∼9 Å) and by inserting it at predicted loops, the likelihood of IpaB misfolding was minimalized and this was confirmed by spectroscopic analysis. The added densities within IpaB were expected to be disruptive and potentially interfere at protein‐protein interfaces for interactions involving IpgC (in the cytoplasm), IpaD (within the TC), itself (within the TC or translocon), IpaC (within the translocon) and/or MxiH (within the TC). Likewise, insertions that masked the hydrophobic region of IpaB were expected to eliminate the ability of the protein to insert into host cell membranes. Surprisingly, almost all of the T4L insertion mutants were active in one or more activities related to wild‐type IpaB function. Based upon the acquired data, we present a model that summarizes the functional topology of IpaB. This model is based upon functions already attributed different regions of the protein and how these functions are influenced by disruptive insertional mutagenesis.
Results
Expression of IpaB T4L‐insertion mutants in a Shigella flexneri ipaB‐null strain
To generate a library of IpaB insertion mutants, predicted loops (random coil regions) were identified for inserting bacteriophage T4 lysozyme (T4L). T4L was selected because of its stability, high solubility20 and the small distance (∼9 Å) between its N‐ and C‐termini (Fig. S1). This last property was anticipated to minimize disruption of IpaB folding and negated the need for including a large peptide linker (GS‐T4L‐GS). Twelve T4L insertion mutants were generated [Fig. 1(A)] and cloned into an expression plasmid for complementing the S. flexneri ipaB‐null strain SF620.21 Some of the general regions selected were previously identified as having important roles in IpaB function (reviewed in ref. 22). For example, recognition of the IpaB N‐terminus is known to be important in targeting it to the T3SA. Additionally, the N‐terminal third of the protein is critical for interaction with IpgC through a canonical P/VxLxxP chaperone binding motif located between residues 65 and 70.23, 24, 25 Disruption of this interaction could destabilize IpaB within the Shigella cytoplasm. Other insertions in the N‐terminal half of IpaB lie within or near the IpaB coiled‐coil motif [residues 74–224, Fig. 1(B)]. Biochemical13, 23 and genetic26 data support the idea that the IpaB coiled‐coil and adjacent regions are important for interactions with IpgC in the cytoplasm and/or IpaD within the TC. Finally, we predicted that insertions at or near the hydrophobic region (approximately residues 300 to 420) and IpaB C‐terminus could lead to defects in pathogenesis potentially involving interactions with IpaC, the host membrane and/or with itself as part of the TC.9, 27, 28 When expressed in S. flexneri SF620, all of the mutant IpaB proteins were found in whole cell extracts prepared from equal numbers of bacteria, however, IpaB‐T4L‐60 was degraded to a lower molecular weight form (intermediate between the insertion mutant and wild‐type IpaB), IpaB‐T4L‐C expression levels appeared to be reduced and IpaB‐T4L‐171 and IpaB‐T4L‐435 were present at barely detectable levels (Fig. S2). It is possible that the proximity of position 60 to the known IpgC binding site reduced the stability of IpaB‐60‐T4L, however, IpaB‐T4L‐N, 52 and 91 all accumulated at near wild‐type levels, suggesting that IpgC association was unaffected in these mutants despite their relative proximity to the chaperone binding region(s). Interestingly, another mutant with reduced protein expression was IpaB‐T4L‐171 (Fig. S2). This insertion lies within a hairpin loop near one end of the N‐terminal IpaB coiled‐coil for which a high‐resolution structure has been solved [Fig. 1(B)]. This low level of expression is curious because it is not indicative of a loss of important virulence functions as shown below. Additionally, IpaB‐T4L‐435 had reduced protein levels in Shigella, which is consistent with a previous deletion mutagenesis study where residues 410–417 were truncated.29
Figure 1.

Generation of IpaB‐T4 lysozyme (T4L) insertion mutants. (A) The primary sequence of IpaB from S. flexneri is shown with the following bioinformatics predictions: secondary structure (α‐helix, red text; random coil, black text; coiled‐coil, bold), regions of disorder (underlined), and transmembrane helices (green highlight). The IpaB (residues 74–224) crystal structure is represented with solid green bars above the amino acid numbering. Locations of T4L insertions are depicted with blue squares (pegs), more information can be found in Table SI. The figure was generated using XtalPred.33 (B) The crystal structure of IpaB (residues 74–224) from S. flexneri (PDB ID: 3U0C) is shown with colors from blue (N‐terminus) to red (C‐terminus). The location of T4L insertions 91, 171, and 226 are indicated in the context of the structure.
Recombinant co‐expression of the IpaB‐T4L insertion mutants with IpgC in E. coli was carried out to allow for examination of their in vitro biophysical properties (see Fig. S3). These co‐expression experiments appeared to confirm that IpaB‐T4L‐60 was destabilized with regard to IpgC binding because it could not be purified from the E. coli lysates despite the fact that it could be detected at reduced levels in Shigella. Likewise, while IpaB‐T4L‐435 could be detected at reduced levels in Shigella whole cell extracts, it could not be co‐purified with IpgC from E. coli. Once again, this could be due to a destabilization of the chaperone complex that leads to the IpaB's degradation when expressed outside the context of Shigella, possibly because residue 435 is located within a long stretch of predicted α‐helical secondary structure [Fig. 1(A)]. In complete contrast, IpaB‐T4L‐C and IpaB‐T4L‐171 were both seen at greatly reduced levels in Shigella, but they accumulated at levels similar to that seen for the other constructs when co‐expressed with IpgC in E. coli (Fig. S3), thus suggesting that chaperone binding is unaffected.
It is worth noting that when co‐expressed with IpgC in E. coli, every IpaB‐T4L insertion mutant that could be expressed maintained a stable α‐helical structure content as determined by circular dichroism (CD) spectroscopy (Fig. S4). While this provided prima facie evidence that the T4L insertions had caused minimal disruption of IpaB's overall folding, it should be noted that T4L also is an α‐helical protein and is contributing to the overall CD signal. Nevertheless, when the CD signal at 222 nm was monitored as a function of temperature, all of the purified protein complexes had a thermal unfolding temperature midpoint similar to that of wild‐type IpaB (Fig. S5). This was surprising because it suggests that, at least for IpaB in complex with IpgC, none of the added T4L densities compromised IpaB's behavior in solution. To test this more rigorously, IpaB's function within the context of Shigella T3SS functions was explored.
Secretion of the IpaB‐T4L insertion mutants by S. flexneri
The Shigella T3SS is known to display low‐level background levels of secretion, which can be observed by examining the protein profiles in culture supernatant fractions of overnight cultures. In ipaD and ipaB null strains, overnight type III secretion is elevated relative to wild‐type Shigella or S. flexneri SF620 complemented with a plasmid to express ipaB. Thus, S. flexneri SF620 was complemented with the genes for the IpaB‐T4L insertion mutants so that control of overnight secretion could be monitored. There was no elevated secretion in wild‐type Shigella, SF620 expressing ipaB, or an ipaC‐null strain of Shigella [Fig. 2(A) and Table 1]. When the IpaB‐T4L insertion mutants were examined, a range of overnight secretion profiles was observed [Fig. 2(A)]. Loss of secretion control was seen for IpaB having T4L insertions at the N‐terminus or at residues 60, 226, 238 and 435. The fact that IpaB‐T4L‐60 failed to control secretion could be accounted for by an inability to associate with IpgC, which fits with the lack of co‐expression with the chaperone in E. coli. This would also prevent it from being recognized for its own secretion which is shown to be the case by immunoblot analysis [Fig. 2(B)]. The inability to control secretion for IpaB‐T4L‐N would also most likely be due to an inability for the T3SA to recognize it for secretion, though degraded forms of this mutant do appear in Shigella overnight supernatants [Fig. 2(B)]. It's not yet clear why the other T4L insertion mutants fail to control secretion, though for IpaB‐T4L‐435 it is possible that instability is an issue since it cannot be co‐expressed with IpgC in E. coli. Perhaps there is something unique about this region that also contributes to chaperone binding in the bacterial cytoplasm.
Figure 2.
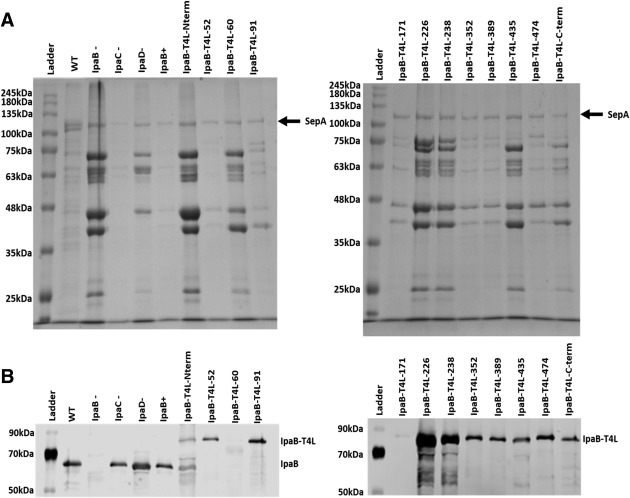
Secretion by Shigella producing the IpaB‐T4L insertion mutants. (A) Overnight protein secretion was analyzed by SDS‐PAGE (10% polyacrylamide gel) with Coomassie blue staining after precipitation of culture supernatants. (B) Immunoblot of IpaB and the IpaB‐T4L insertion mutants present in the overnight culture supernatants (same samples as used in “A”) after probing with anti‐IpaB rabbit antibodies. Molecular mass markers (labeled Ladder) are indicated in each panel and the position of Shigella SepA (not secreted by the T3SA) is shown by an arrow in “A.”
Table 1.
Summary of IpaB‐T4L Insertion Mutant Characteristics
| Expressiona | T3SS Activity | |||||
|---|---|---|---|---|---|---|
| Site | Shigella | E. coli | O/N Secretion Controlb | Hemolysisc | Invasiond | Liposome Lysise |
| N | 1.1 | + | 46.3 ± 21.3 | 0.5 ± 0.3%** | 2.1 ± 0.5%** | 177 ± 7% |
| 52 | 1.2 | + | 1.5 ± 0.1 | 70 ± 5%* | 129 ± 35% | 185 ± 12% |
| 60 | 1.8* | – | 13.0 ± 10.4 | 0.1 ± 0.0%** | 0.3 ± 0.3%** | n/a |
| 91 | 1.5 | + | 3.8 ± 3.2 | 92 ± 10% | 103 ± 21% | 197 ± 13% |
| 171 | 0.1 | + | 3.4 ± 1.1 | 53 ± 8%* | 103 ± 8% | 125 ± 4% |
| 226 | 2.2 | + | 22.5 ± 5.8 | 75 ± 12%* | 68 ± 9% | 192 ± 12% |
| 238 | 3.3 | + | 19.4 ± 9.9 | 57 ± 12%* | 85 ± 15% | 207 ± 10% |
| 352 | 2.2 | + | 3.0 ± 0.8 | 8.7 ± 1.2%** | 12 ± 5%** | 184 ± 13% |
| 389 | 2.0 | + | 3.0 ± 0.7 | 20 ± 2%** | 10 ± 4%** | 175 ± 8% |
| 435 | 0.2 | – | 14.0 ± 7.9 | 0.1 ± 0.0%** | 0.5 ± 0.3%** | n/a |
| 474 | 2.5 | + | 3.8 ± 1.0 | 9.6 ± 1.0%** | 0.0 ± 0.0%** | 145 ± 11% |
| C | 0.7 | + | 8.5 ± 4.1 | 7.4 ± 1.1%** | 1.2 ± 0.4%** | 232 ± 20% |
Expression in Shigella was determined by observation of a band of the appropriate size detected by immunoblots of whole cell extracts using rabbit anti‐IpaB antiserum (see Fig. S2). The values shown are relative to IpaB present in S. flexneri SF620 expressing wild‐type IpaB after a densitometer scan of a representative immunoblot. *IpaB‐T4L‐60 was observed but it had degraded to a number of smaller molecular weight species and the value given in the approximate relative density of all the sized fragments combined. In E. coli, IpaB was co‐expressed with IpgC and the presence of IpaB determined by SDS‐PAGE with Coomassie staining after IMAC purification based on a His6 tag on the IpaB. A positive result indicated that the IpaB‐IpgC complex could be purified (see Fig. S3).
Overnight secretion was determined by separating the proteins from overnight culture supernatants from equal numbers of bacteria by SDS‐PAGE followed by Coomassie blue staining (see Fig. 2). Samples were then scanned using densitometry for the amounts of IpaC and IpaD present relative to that for S. flexneri SF620 expressing wild‐type IpaB (normalized value of 1.0) ±SD in representative gels.
% Hemolysis is relative to B+ [Fig. 3(A)] with * indicating P‐value ≤0.05 and ** indicating P‐values ≤0.01 using a standard Student's t‐test.
% Invasion is relative to B+ [see Fig. 3(B)] with * indicating P‐value ≤0.05 and ** indicating P‐values ≤0.01 using a standard Student's t‐test.
% Dye release from liposomes is relative to wild‐type IpaB (see Fig. 4). All IpaB‐T4L mutants that could be purified were considered to be fully functional for liposome lysis.
Perhaps the most interesting observation is the number of mutants able to restore control of secretion to Shigella SF620. T4L insertions at 52, 91, 171, 352, 389, 474 and at the C‐terminus all secrete at normal levels in overnight cultures. This occurs despite the fact that IpaB‐T4L‐171 is significantly reduced in expression in S. flexneri. In fact, even when secretion of IpaB is specifically examined by immunoblotting, very little IpaB‐T4L‐171 can be observed in the culture supernatants. Meanwhile, the insertions at 226 and 238 result in similar levels of IpaB in overnight culture supernatants and the remainder have similar levels of IpaB in the cell supernatant as does wild‐type IpaB. These findings show that expression and stability of these insertion mutants cannot be specifically correlated with IpaB secretion or secretion control. Therefore, a more rigorous assessment of IpaB functionality for these mutants was to test for translocon pore formation and invasion of cultured cells (proper effector secretion).
Shigella contact‐mediated hemolysis and cellular invasion as a measure of translocon pore formation and effector translocation
To test for the IpaB‐T4L insertion mutants' ability to form translocon pores, a standard contact‐mediated hemolysis assay was performed. It was at this point that the most profound IpaB functional defects were expected. Surprisingly, all but three of the mutants restored some level of hemolytic activity to SF620. Mutants completely devoid of hemolytic activity were IpaB‐T4L‐N, IpaB‐T4L‐60 and IpaB‐T4L‐435, even though the insertion at 435 behaved normally in restoring secretion control [Fig. 3(A) and Table 1]. The absence of hemolysis for Shigella expressing IpaB‐T4L‐N and IpaB‐T4L‐60 may be understandable since each has an apparent secretion defect, however, the absence of hemolytic activity for IpaB‐T4L‐435 is less clear. This mutant is secreted reasonably well though its expression level is somewhat reduced and it appears to be unstable when co‐expressed with IpgC in E. coli.
Figure 3.

The ability for IpaB‐T4L insertion mutants to restore virulence to S. flexneri SF620. (A) Contact‐mediated hemolysis activity was tested in sheep red blood cells for 30 min at 37°C. (B) Invasion of HeLa cells after a 1 hr incubation with the different IpaB‐expressing Shigella strains. The values shown in each panel are relative to S. flexneri SF620 expressing wild‐type IpaB (set at 100%).
Of the remaining insertion mutants, those in the N‐terminal half of IpaB restored nearly normal levels of contact hemolysis while those in the C‐terminal half restored a basal level (about 10%) of contact hemolysis [Fig. 3(A)]. The latter observation is worth noting, because this is the level of contact hemolysis seen for ipaC null mutants.27 This may suggest that the T4L insertions in the C‐terminal half of IpaB may form a fully functional TC but that upon insertion into a host cell membrane, they are unable to properly interact with secreted IpaC to form a functional translocon pore. We would attribute this to the formation of a “pre‐translocon pore” formed by IpaB alone.18 On the other hand, two of the T4L insertions (at residues 226 and 338) were unable to restore secretion control but were nearly normal with regard to contact hemolysis. These are located immediately downstream from the identified coiled‐coil of IpaB, suggesting that they are located in a region of transition between the N‐terminal TC anchoring region and the C‐terminal hydrophobic portion of IpaB.
The ultimate test of IpaB function within the context of the Shigella T3SS is its ability to direct invasion of cultured cells, which requires translocon pore formation and the coordinated translocation of effector proteins into host cells. Consistent with the contact‐mediated hemolysis results, T4L insertions at the N‐terminus and residues 60 and 435 were completely unable to direct host cell invasion [Fig. 3(B) and Table 1]. On the other hand, there is a complete loss of function for T4L insertions at the C‐terminus and position 474. Meanwhile, the remainder of the T4L insertions within the N‐terminal half of the protein had nearly wild‐type invasiveness and this is despite the fact that insertions at 226 and 238 display completely uncontrolled type III secretion [Figs. 2 and 3(B)].
Release of small molecules from liposomes by IpaB‐T4L insertion mutants
Proper function (i.e., pore formation) of IpaB in the context of the Shigella T3SS TC requires multiple interactions (e.g., MxiH, IpaD, IpaB, and/or IpaC) that could potentially be disrupted by the extra density provided by T4L insertion mutagenesis. Because T4L insertions in the C‐terminal half of IpaB do not affect secretion control, but do appear to affect translocon formation and/or function, it is possible that they are impaired in their ability to insert into the host membrane. To test for this, the release of sulforhodamine B (SRB) from liposomes was monitored using recombinant IpaB separated from IpgC using OPOE (Fig. 4). We have previously shown that IpaB in OPOE forms an oligomer that behaves like a pore following insertion into liposomes.18, 30 In this assay, the release of SRB from the liposomes relieves auto‐quenching of fluorescence, which can be monitored spectroscopically [Fig. 4(A)]. Each of the 10 purified IpaB‐T4L insertion mutants was capable of lysing SRB‐loaded liposomes at levels equal to or higher than wild‐type IpaB [Fig. 4(B) and Table 1]. This includes the N‐terminal insertion mutant, further supporting the hypothesis that this mutant is attenuated in Shigella simply due to its inability to be recognized for transport. In particular, none of the insertions in the C‐terminal half of IpaB showed any reduced ability to interact with liposomes. These data suggest that T4L insertions within the C‐terminal half of IpaB (352, 389, 435, 474, and C) reduce translocon formation and invasiveness at steps downstream of IpaB membrane contact, potentially through disrupted interactions with IpaC, which is thought to interact with a region of IpaB spanning residues 367–458.31
Figure 4.

Ability of IpaB‐T4L insertion mutants to release SRB from liposomes. (A) Release of SRB from liposomes results in relief of auto‐quenching and is measured as an increase in SRB fluorescence emission. Addition of IpaB (prepared in OPOE) results in a time‐dependent release of SRB. Addition of Triton X‐100 results in 100% liposome lysis. (B) Percentage of SRB release relative to total dye release was calculated and is shown as a relative percent of release caused by wild‐type IpaB. All samples contained PBS + 0.5% (v/v) OPOE with the exception of IpgC/IpaB, which was in PBS lacking detergent. OPOE alone caused no release of SRB (column 2).
Surface localization of the IpaB‐T4L insertion mutant strains
Surface localization of the TC components IpaD and IpaB was analyzed for the series of T4L insertion mutants using immunofluorescence labeling and flow cytometry. IpaD surface levels were not substantially affected by any of the IpaB T4L insertion mutants although it was mildly reduced for IpaB‐T4L‐N [Fig. S6(A)]. The presence of IpaB on the surface of Shigella is indicative of a functional interaction between IpaD and IpaB within the mature TC. When the degree of IpaB surface labeling was examined after growing the bacteria to log phase, modest labeling of wild‐type Shigella was observed and this was completely lost in the ipaB‐null strain SF620 [Fig. 5(A) and Table 1]. Consistent with the data presented thus far, no surface labeling was observed for IpaB‐T4L‐N and IpaB‐T4L‐60, which are poorly secreted, if at all, by Shigella [Fig. 5(A)]. It is noteworthy that IpaB‐labeling with T4L insertion at positions 52, 352, 389, 435, 474 and at the C‐terminus all appear to be elevated relative to the levels seen in wild‐type bacteria [Fig. 5(A)]. All of these mutants except for the C‐terminal fusion were able to prevent uncontrolled secretion [Fig. 2(A)]. Taken together, these data are consistent with the formation of a mature, uninduced T3SA TC, however, of these only IpaB‐T4L‐52 restored substantial levels of contact‐mediated hemolysis and invasiveness (Fig. 3). T4L insertion at positions 91, 171, 226 and 238 displayed low levels of IpaB surface labeling (Fig. 5). This labeling was somewhat lower than that of wild‐type Shigella but still readily detectable. Once again, no clear pattern exists between secretion control and surface localization of IpaB.
Figure 5.
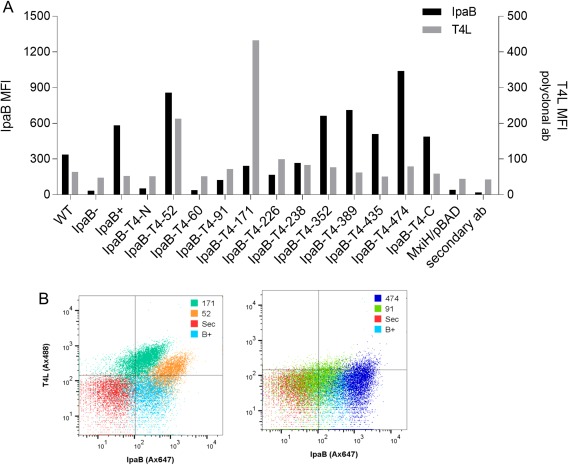
Mean fluorescence intensity (MFI) of specific markers detected by flow cytometry at the surface of IpaB‐T4L insertion mutant strains grown to mid‐log phase. (A) MFI of IpaB (black bars) and T4L (grey bars) were detected using polyclonal antibodies previously generated against the purified recombinant proteins. Additional negative controls include a mxiH null mutant and a sample lacking the secondary antibody. (B) Flow cytometry analysis for IpaB and T4L labeling on the Shigella surface. Sec represents bacteria for which the secondary antibody was not added (staining negative control).
Surface localization of T4L
T4L has a molecular weight of 18.6 kDa, which represents a substantial addition to the density of IpaB within the T3SA needle TC. Such added density would be expected to interfere with IpaB packing within the TC and in many cases lead to the presence of detectable levels of T4L on the Shigella. The elevated surface presence of IpaB for mutants within the C‐terminal half of IpaB were considered the best candidates for having detectable T4L exposed on the bacterial surface. Surprisingly, none of the C‐terminal T4L insertions that displayed elevated IpaB on the surface had detectable levels of T4L on the bacterial surface using either polyclonal rabbit serum [Fig. 5(A) and Table 1] or a commercial monoclonal antibody (Fig. S6) with cell sorting. In fact, only two IpaB‐T4L insertion mutants clearly had T4L on the surface and it could be detected using either of the antibodies [Fig. 5(A) and Fig. S6]. Of these two, only one, IpaB‐T4L‐52, also displayed elevated IpaB on the surface. The other was the IpaB‐T4L‐171 that is not only expressed at very low levels, but is also secreted at very low levels, yet it is able to control steady‐state secretion.
These results indicate that the tertiary structure of IpaB displays a high degree of plasticity and that positions 52 and 171 are the most solvent exposed as part of the TC. One potential caveat of this approach, however, is that we have not defined which T4L epitopes are contained within the generated polyclonal sera, which could potentially bias IpaB/T4L surface labeling towards specific regions of the antigen. Nevertheless, essentially all aspects of IpaB function appear to be quite accommodating to the added density of T4L at most positions within the protein without accompanying exposure of the T4L moiety on the bacterial surface. This suggests that the multiple functions of IpaB are accommodated by an unusually high degree of structural plasticity, which could explain why high‐resolution structures of full‐length IpaB have been difficult to obtain.
Crystal structure of IpaB 74–242
Almost all of the IpaB‐T4L insertion mutants retained some aspect of normal IpaB function. With the exception of T4L insertions that affect IpaB's secretion (at the N‐terminus or after residue 60), the majority of the insertions upstream or at residue 238 seem to retain most or all of IpaB's ability to lyse red blood cells (RBCs) and to invade cultured cells. Despite this, only the insertions at positions 52, 91, and 171 were able to control overnight secretion. The added T4L density at position 226 and 238 caused IpaB to lose its ability to control steady‐state secretion but it still retained its other virulence related activities. Interestingly, both of these insertions are near one end of the only known high‐resolution structure for IpaB, which only previously extended from residue 74 to residue 224 [Fig. 1(B)]. It is clear that a region just to the C‐terminal side of this structure is critical for properly controlling background secretion, but doesn't appear to otherwise influence IpaB functions. To address this, we were able to determine the crystal structure of an IpaB construct spanning residues 74–242 (Table 2). The structure of IpaB74–242 (Fig. 6) is similar to IpaB74–224 with respect to topology with both proteins being comprised of an extended coiled‐coil, however, the structures only align with an RMSD of 2.58 Å (149/149 Cα atoms) (Fig. S7). This structural difference arises from an alteration in how the coiled‐coil twists as it approaches the hairpin region, although this difference is likely a result of the different crystal packing between each structure (P21 vs. I4 space groups).
Table 2.
Diffraction Data and Refinement Statistics
| IpaB 74–242 | |
|---|---|
| Data Collection | |
| Unit‐cell parameters (Å) | a = b = 171.43, c = 40.59 |
| Space group | I4 |
| Resolution (Å)a | 42.86–2.10 (2.16–2.10) |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Observed reflections | 233,850 (18,814) |
| Unique reflections | 34,988 (2862) |
| <I/σ(I)>a | 11.1 (2.0) |
| Completeness (%)a | 99.8 (100) |
| Multiplicitya | 6.7 (6.6) |
| R merge (%)a, b | 9.1 (98.1) |
| R meas (%)a, e | 9.9 (106.6) |
| R pim (%)a, e | 3.8 (41.4) |
| CC1/2 a, c | 0.997 (0.802) |
| Refinement | |
| Resolution (Å) | 35.87–2.10 |
| Reflections (working/test) | 33,279/1683 |
| R factor/R free (%)d | 20.1/24.5 |
| No. of atoms (Protein/Water) | 2475/174 |
| Model Quality | |
| R.m.s deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 0.921 |
| Average B‐factor (Å2) | |
| All Atoms | 52.5 |
| Protein | 52.9 |
| Water | 48.6 |
|
Coordinate error (maximum likelihood) (Å) |
0.27 |
| Ramachandran Plot | |
| Most favored (%) | 97.8 |
| Additionally allowed (%) | 1.3 |
| Outliers (%) | 0.9 |
Values in parenthesis are for the highest resolution shell.
R merge = ΣhklΣi |I i(hkl) – <I(hkl)>|/ΣhklΣi I i(hkl), where I i(hkl) is the intensity.
measured for the ith reflection and <I(hkl)> is the average intensity of all reflections with indices hkl.
R meas = redundancy‐independent (multiplicity‐weighted) R merge.37, 48 R pim = precision‐indicating (multiplicity‐weighted) R merge.49, 50
CC1/2 is the correlation coefficient of the mean intensities between two random half‐sets of data.51, 52
R factor = Σhkl ǁF obs (hkl) | – |F calc (hkl) ǁ/Σhkl |F obs (hkl)|; Rfree is calculated in an.
identical manner using 5% of randomly selected reflections that were not included in the refinement.
Figure 6.

Crystal structure of IpaB74–242 at 2.10 Å. The crystal structure of S. flexneri IpaB (residues 74–239) is shown in cartoon ribbon format, colored blue (N‐terminus) to red (C‐terminus). Two copies of each polypeptide are found within the asymmetric unit with a single copy shown for clarity. The inset shows the representative model‐to‐map correlation for residues 225–239. The 2Fo – Fc weighted electron density map (contoured at 2.0 σ) is drawn as a blue cage.
The structure of IpaB74–242 also reveals that residues 231–233 (Ala‐Ser‐Ala) adopt a random coil structure, creating a turn that then transitions back into an α‐helical structure. Further structural characterization will be required to assess whether this turn is adopted in the context of the complete IpaB polypeptide, however, it may be providing a flexible linker between what are largely functionally distinct N‐ and C‐termini. In any event, it is intriguing that the addition of significant new density near this apparent linker between the N‐ and C‐terminal domains of IpaB has such a dramatic effect on steady‐state secretion rates, but none of the downstream activities of IpaB. Furthermore, neither of the T4L insertions here, nor any of those C‐terminal to this point, result in exposure of T4L on the bacterial surface. It is clear that there is still much to learn about the structure‐function relationships within IpaB and disruptive insertional mutagenesis has now provided a foundation for initiating such studies.
Discussion
There remains a great deal to be learned about the structure and particularly the structural dynamics occurring within the T3SA needle TC. To understand the topology and spatial relationships involving IpaB in the Shigella T3SA TC, we generated a series of T4L insertion mutants to determine the disruptive effects the added densities have on IpaB function in Shigella (summarized in Table 1). T4L properties allowed us to minimize the likelihood of IpaB misfolding and this appeared to be the case since all but two of the insertion mutants retained wild‐type secondary structure content based on CD spectroscopy. Indeed, only two of the 12 insertion mutants (T4L‐60 and T4L‐435) could not be purified as recombinant proteins in a complex with IpgC. In all cases, the insertion mutants described here were expected to introduce new densities into IpaB that would disrupt important protein‐protein interactions in the cytoplasm, within the TC or within the translocon. Likewise, insertions masking the hydrophobic region of IpaB might be expected to eliminate IpaB's ability to insert into phospholipid membranes. Surprisingly, nearly all of the insertion mutants were active in one or more IpaB functions (chaperone binding, secretion control, translocon formation, membrane interaction and translocation of effectors). A model of IpaB showing the protein's known and proposed structural features based on these T4L insertional mutagenesis data is shown in Figure 7.
Figure 7.
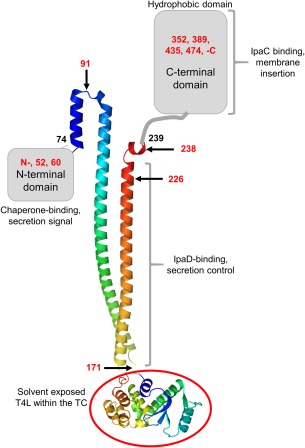
Summary of the functional landscape of IpaB based on T4L insertion mutagenesis. The positions of T4L insertion sites are indicated by red numbers and key functions associated with each regions are described. Structures have not been determined for the N‐terminal domain (residues 1–73) or the C‐terminal domain which possesses a major hydrophobic component (∼residues 300–420) which contains two predicted transmembrane helices [see Fig. 1(A)]. At the bottom, the structure of T4L is shown (red circle) as it is proposed to occur for insertion at IpaB position 171. T4L is recognized by T4L anti‐sera on the surface of Shigella when inserted at positions 52 and 171 (see Fig. 5).
The acquired data reveal patterns of behavior that allow an updated assessment of IpaB spatiotemporal functions. Two mutants gave rise to functional defects that might be expected based on the literature. T4L at the IpaB N‐terminus had no effect on association with IpgC, however, only very small amounts of this protein (mostly degraded) could be found in culture supernatants. This was not unexpected since IpaB's immediate N‐terminal sequence is required for it to be recognized for secretion. The added density at the immediate N‐terminus most likely alters the context of this sequence. IpaB with T4L at residue 60 could not be generated as a recombinant protein and in Shigella it was degraded in the cytoplasm as would be expected if the added density impaired IpgC binding. Indeed, a canonical chaperone binding motif has been described involving residues 65–70 in IpaB.23, 24, 25 Thus, this is the only insertion within the N‐terminal half of IpaB that was defective for all IpaB functions, likely a result of impaired chaperone binding.
One other mutant, IpaB‐T4L‐435, was impaired for all IpaB functions despite the fact that it could be found in Shigella culture supernatants. This mutant could not be purified after co‐expression with IpgC in E. coli and was found at greatly reduced levels in the Shigella cytoplasm—hallmarks of impaired chaperone binding. However, its presence at only slightly reduced levels in Shigella culture supernatants is perplexing. We would propose that this region influences chaperone binding, but that the dominant binding within the N‐terminus of IpaB allows for aberrant secretion of IpaB that is not otherwise productive. All three of the mutants just described are unable to control type III secretion in Shigella (Fig. 2 and Fig. S2). Thus, it appears that some facet of IpaB secretion is required for the control of type III secretion (IpaB‐T4L‐N and IpaB‐T4L‐60), however, secretion alone is not sufficient for secretion control (IpaB‐T4L‐435). This would argue that IpaB secretion and an activity involving IpgC work in a coordinated fashion to control overnight secretion. Perhaps an interaction between IpgC and a component of the sorting platform occurs during IpaB presentation influences secretion control.
Recent studies have demonstrated that the IpaB coiled‐coil motif [Fig. 1(B), residues 74–224] and the region immediately upstream of it can allow for interaction with IpaD as part of the TC.13, 26 Intriguingly, multiple T4L insertion mutants within or flanking this region (52, 91, 171, 226, and 238) complement a Shigella ipaB‐null strain at near wild‐type levels for both contact hemolysis and invasion. Strikingly, the ability to control secretion does not directly correlate with retention of these functions. T4L insertions at position 52, 91 and 171 restore control of overnight secretion, but insertions at residues 226 and 238 do not. Interestingly, T4L at position 171 restored most of IpaB's invasion functions, however, the protein was poorly expressed and secreted at very low levels while still providing secretion control. Relative to the coiled‐coil, position 171 is at the opposite end from 226 and 228, which may be significant with regard to secretion control, but not other virulence functions. Position at a point relative to the coiled‐coil, however, does not strictly dictate the level of secretion control since IpaB‐T4L‐91 is located the same end as residues 226 and 238, but it is fully capable of controlling overnight secretion. Two key features that seem to stand out with regard to loss of secretion control are being in the N‐terminal half of the protein and being located near a sharp turn located on the C‐terminal side of the IpaB coiled‐coil (see Figs. 6 and 7). Future structural work needs to focus on this portion of IpaB to clarify what may be occurring at the structural level.
An unexpected observation was the ability for all of the T4L insertions in the C‐terminal half of IpaB (except for the insertion at position 435 as described above) to maintain the ability to control overnight type III secretion. This seems to correlate with elevated IpaB detected on the Shigella surface (although no T4L is detected there). While all of the N‐terminal insertions are considered relative to the coiled‐coil, the C‐terminal insertions are better considered relative to the known hydrophobic regions contained here. Despite controlling secretion, none of these mutants restored substantial levels of contact hemolysis or invasion activities, even though all recombinant proteins were able to insert into liposomes in vitro at wild‐type levels or better. Two of them, T4L at 352 and 389 did restore background levels of contact hemolysis, but not invasiveness. The other two (T4L position 474 and the C‐terminus) were defective for both activities. Because Shigella ipaC null mutants are noninvasive but retain ∼10% of their hemolysis activity, we have proposed that IpaB is able to insert into target cell membranes (possibly as a tetramer) to form a “pre‐translocon pore.” This may be what occurs with the insertions at residues 352 and 389. Thus, these mutants may contribute to the first step of translocon formation, but cannot complete the process, suggesting they are defective for recruitment of or perhaps association with IpaC. Meanwhile, IpaB with insertions at 474 and the C‐terminus may not form the pre‐translocon pore. Unfortunately, this is not entirely consistent with their ability to lyse liposomes in vitro. Future studies warrant determining how T4L insertion at these sites affects IpaB oligomerization, which has been linked with its ability to form pore‐like structures in liposomes.
In our hands, this is the first time we have seen secretion control so thoroughly uncoupled from other essential IpaB functions, but it seems that secretion control is an activity that is largely associated with structures at the N‐terminus of IpaB. It should be noted that deletions within the extreme C‐terminus of IpaB32 have been reported to result in loss of overnight secretion control, so it cannot be ruled out that all control mechanisms are localized to the N‐terminus, however, T4L at the C‐terminus did not seem to impair secretion control. In these studies, added densities within the C‐terminal half of IpaB seem to be more fundamentally impaired at post secretion control steps, suggesting the heart of IpaB's function as a translocator protein reside here. A remarkable aspect that stands out, however, is that so many of the insertion mutants retain one or more of their normal functions despite harboring an added density of ∼19 kDa. On top of this, only insertions at residues 52 and 171 appear to actually display T4L on the bacterial surface, regardless of the amount of IpaB detected there.
The data presented here indicate that there is a great deal of resilience within the Shigella T3SA TC against the disruptive addition of additional protein densities. Perhaps the ability to incorporate significant new densities into IpaB while maintaining specific functional features will allow for future studies on the TC using protein‐based probes. Likewise, this finding suggests that IpaB can be targeted for the addition of cargo that allows it to be tracked inside macrophages prior to the induction of pyroptosis. In any event, by generating this panel of T4L insertion mutants, we now know that IpaB harbors essentially three major functional hot spots including: (a) the immediate N‐terminus which is needed for recognition by the secretion apparatus and chaperone binding; (b) an N‐terminal coiled‐coil within a soluble N‐terminal fragment that is involved in anchoring IpaB within the TC and for important aspects of secretion control; and (c) the C‐terminal half which is key to the activities needed for formation of an active translocon and possibly for translocation itself. Additionally, the structure of IpaB74–242 reveals that the N‐terminal coiled‐coil and C‐terminal hydrophobic domain are connected by a turn that could be a mediator of the high level of structural plasticity that has not been fully appreciated until now. Future studies using these insertions mutants will be targeting these individual regions for their structural and functional dynamics with respect to translocon assembly and secretion‐state regulation.
Materials and Methods
Generation of IpaB T4L insertion mutants in Shigella
Secondary structure elements within IpaB were predicted using the XtalPred server,33 which coordinates data analysis with the following external servers PSI‐BLAST, PSIPRED, DISOPRED2, COILS, and TMHMM, among others. This information was used to guide the selection of Bacteriophage T4 lysozyme (T4L) insertion sites. The T4L coding sequence (catalytically inactive mutant E11A) was inserted at the N‐ and C‐terminus of IpaB, as well as after the following amino acids: 52, 60, 91, 171, 226, 238, 352, 389, 435, and 474 using ligation independent cloning with wild‐type IpaB in the pT7HMT vector as a template.34 A Gly‐Ser linker was added in between IpaB and T4L coding sequences during this step as well. Each of the resulting insertion mutants were then used as templates for cloning into NdeI/BamHI‐digested pWPsf4 vector27 and electroporated into the Shigella flexneri ipaB‐null strain SF62021 for phenotypic analyses. DNA oligo information for cloning can be found in Table SI.
Recombinant protein expression
Wild‐type IpaB and the T4L insertion mutants described above were expressed as previously described.35 Briefly, all IpaB derivatives were cloned into pT7HMT and co‐expressed in E. coli BL21(DE3) with its cognate chaperone IpgC in pACYCDuet‐1 and the protein present in the cytosolic fraction was purified by Immobilized Metal Affinity Chromatography (IMAC). The chaperone was removed by the addition of 0.5% (v/v) OPOE and then separating the IpaB by a second round of IMAC in the presence of the detergent. IpaB74–242 was generated by amplifying a region of the opening reading frame (residues 74–242) of S. flexneri ipaB via PCR and subcloned into BamHI/NotI‐digested pT7HMT.34 This vector was transformed into Tuner(DE3) E. coli competent cells and expressed and purified as previously described for IpaB28–226.17
Crystallization
IpaB74–242 (10 mg/mL in 10 mM Tris–HCl pH 7.5, 50 mM NaCl) was crystallized by vapor diffusion in Compact Jr. (Rigaku Reagents) sitting drop plates at 20°C by mixing 0.75 µL of protein solution with 0.75 µL of reservoir solution. Crystals were obtained in Molecular Dimensions ProPlex B7 (0.2 M ammonium acetate, 0.1 M sodium acetate (pH 4.0) and 15% (w/v) PEG 4000) and flash‐cooled in a cryoprotectant solution consisting of mother liquor supplemented with 20% (v/v) glycerol.
Diffraction data collection, structure determination, refinement, and analysis
X‐ray diffraction data were collected on IpaB74–242 crystals at 1.0000 Å at 100 K using a Dectris Pilatus 6M pixel array detector at IMCA‐CAT beamline 17ID at the APS (Table 2). Following data collection, individual reflections from each dataset were integrated with XDS36 and scaled with Aimless.37 Experimental phase information was obtained for the IpaB74–242 structure using molecular replacement (PHASER; search model obtained from PDB entry 3U0C17) within the Phenix suite.38, 39
Structure refinement for the IpaB74–242 structure was carried out using Phenix.38, 39 One round of individual coordinates and isotropic atomic displacement factor refinement was conducted, and the refined model was used to calculate both 2Fo‐Fc and Fo‐Fc difference maps. Manual building within Coot40, 41 was used to iteratively improve the model, followed by subsequent refinement cycles. TLS refinement42 was incorporated in the final stages to model anisotropic atomic displacement parameters. Hydrogen atoms were included during all rounds of refinement. Ordered solvent molecules were added according to the default criteria of Phenix and inspected manually within Coot prior to model completion. The following residues within the IpaB74–242 structure were not modeled as a result of poor map quality: residues 241–242 (chain A) and residues 231–242 (chain B).
T3SS protein overnight secretion
Overnight protein secretion as a measure of leaky secretion was assessed as previously reported.43 Briefly, Shigella flexneri strains were grown overnight and culture supernatants collected after centrifugation. Supernatants were then precipitated with subsequent trichloroacetic acid and ice‐cold acetone treatments. SDS‐PAGE was then used to separate the protein content of precipitated samples. Gels were stained with Coomassie Brilliant blue dye.
Contact‐mediated hemolysis and invasion
Contact‐mediated hemolysis using sheep's erythrocytes has been previously reported.27 Briefly, S. flexneri strains were grown in culture at 37°C to log‐phase and 6.25E8 CFUs incubated with 50 µl of RBCs for 1 h at 37°C. RBCs were then extruded with cold PBS to obtain maximum hemoglobin release, a direct measure of pore formation ability by Shigella. Hemoglobin was measured with a plate reader set for absorbance at 545 nm. Invasion of HeLa cells was measured with a modified gentamicin protection assay.43 Briefly, log‐phase S. flexneri cultures were used to infect HeLa cell monolayers grown in 24‐well plates at a multiplicity of infection (MOI) of 1:100 and invasion allowed to proceed for 30 min at 37°C in a humidified 5% CO2 environment. Then, gentamicin was used to remove bacteria in the extracellular environment. Cells were extruded with 0.2% Triton X‐100 in PBS and serially diluted. Dilutions were grown on filter plates overnight and imaged with a CTL ELISpot reader as described before.44
Liposome dye release assay
To determine whether recombinantly expressed IpaB‐T4L insertion mutants retained the ability to cause the release of small molecules from liposomes, a dye release assay was performed as previously described.30 Unilamellar liposomes comprised of DOPC (dioleoylphosphatidylcholine) and DOPG (dioleoylphosphatidylglycerol) and cholesterol (66.8, 23.5, and 9.7 mol%, respectively) were prepared in chloroform and dried under nitrogen to create lipid films. The films were hydrated in PBS alone containing 100 mM SRB, sonicated and extruded through 100‐nm pore membranes at 42°C. Excess dye was separated from the SRB liposomes by size exclusion chromatography on Sephadex G‐50. SRB release was measured as an increase in SRB fluorescence as a function of time in the presence of added IpaB or the corresponding T4L insertion mutant (100 nM). The amount of dye release was quantified relative to total release seen following the addition of Triton X‐100 to a final concentration of 0.1% (v/v).
Immunofluorescence and flow cytometry
S. flexneri cultures were grown at 37°C until log‐phase, washed and 5 × 108 CFU resuspended in PBS. Bacteria were immunolabeled as previously reported and analyzed by flow cytometry.9, 18 Briefly, cells were fixed with 2% of paraformaldehyde and blocked with a solution 1% BSA 50% PBS‐based Odyssey blocking buffer in PBS. Cells were then labeled with polyclonal anti‐IpaB, anti‐IpaD rabbit serum, or anti‐T4L guinea pig serum; followed by labeling with appropriate secondary antibodies coupled to Alexa dyes. Cells were analyzed by flow cytometry (collection of 1 × 106 events) in a BD FACSAria Fusion cell sorter.
Generation of anti‐T4L serum
Recombinantly expressed and purified T4L was used to immunize guinea pigs (Pocono Rabbit Farm & Laboratory). Target recognition under native conditions was confirmed by surface plasmon resonance (data not shown). Recombinant T4L protein was immobilized through surface exposed amine groups on a CMD200 surface (Xantec Bioanalytics). Dilutions of immune serum were prepared in HBS‐T buffer (20 mM Hepes (pH 7.4), 140 mM NaCl, 0.005% Tween‐20) and injected over the T4L and reference surfaces using a single‐cycle kinetics approach. The reference‐subtracted sensorgrams were analyzed by BiaCore T‐200 Evaluation Software (v3.0) (GE Life Sciences) and fit to a 1:1 binding model.
Multiple sequence alignments and figure modeling
Multiple sequence alignments were carried out using ClustalW45 and aligned with secondary structure elements using ESPRIPT.46 Representations of all structures were generated using PyMol.47
Statistical analysis
All statistical analyses were performed with GraphPad Prism 7. Ordinary one‐way ANOVA with a post hoc Dunnett test was done by setting the IpaB complemented strain as the control sample. An alpha value of 0.05 was used.
Conflicts of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author Contributions
M.L.B., S.T., M.M., O.A., C.E.V., and K.X.R. performed and analyzed experiments in this study. W.D.P., W.L.P., and B.V.G. designed and coordinated the study. All authors reviewed the results and approved the final version of the manuscript.
Supporting information
Supplementary Figure S1. Crystal structure of T4 lysozyme. The crystal structure of lysozyme from bacteriophage T4 (PDB ID: 2LZM) is shown colored from blue (N‐terminus) to red (C‐terminus). The distance between the N‐ and C‐termini is ∼9.1 Å.
Supplemental Figure S2. Production of IpaB and the IpaB‐T4L insertion mutants by Shigella flexneri. Bacteria were grown to an absorbance at 600 nm of 2.5 and then 1 ml of the culture was centrifuged to collect the bacteria which were then solubilized using SDS‐sample buffer. 10 µl of each was then separated by SDS‐PAGE (10% polyacrylamide gel) and the IpaB detected by immunoblot using rabbit anti‐IpaB antibodies. IpaB could be detected in all samples except for the ipaB null strain SF620 (IpaB‐) though the levels were reduced for IpaB‐T4L‐171 and IpaB‐T4L‐435. IpaB‐T4L60 appeared to undergo degradation.
Supplementary Figure S3. Recombinant expression and purification of IpaB‐T4L insertion mutants. Each wild‐type or T4L mutant was co‐expressed with IpgC and purified. This figure shows a Coomassie blue‐stained SDS PAGE gel of those protein pairs that could be detected after expression in E. coli BL21 (DE3). IpaB‐T4L‐60 and 435 mutants were not expressed and thus are not included in this figure. Some loss of chemical stability was observed for T4L insertions at positions 352 and 389, however, IpgC expressed well in all cases (even for the insertions at positions 60 and 435) when E. coli whole cell extracts were examined (data not shown). The same amount of protein (∼5 µg) was separated in each lane based upon the absorbance at 280 nm.
Supplementary Figure S4. Circular dichroism (CD) analysis of IpaB (wild‐type and T4L insertion mutants) in complex with IpgC after IMAC purification.
Only IpaB‐T4L‐60 and IpaB‐T4L‐435 could not be purified. For all the IpaB‐IpgC complexes that could be purified, the CD spectra indicated stable folding into the expected α‐helical structure (see WT) for this complex. The orange spectra are after heating the protein to 90°C (see Bottom panel) and then cooling back to 10°C. The final panel is recombinant T4L alone. (The CD spectrum of IpgC alone can be found in Birket SE, et al. 2007. Biochemistry 46:8128‐8137.)
Supplementary Figure S5. Thermal unfolding of the purified IpaB‐T4L insertion mutants based on CD analyses. The molar ellipticity at 222 nm was monitored as a function of temperature for all of the recombinant IpaB‐T4L insertion mutants complexed with IpgC. The thermal unfolding curve for IpgC alone can be found in Birket SE, et al. 2007. Biochemistry 46:8128‐8137.)
Supplementary Figure S6. Surface presentation of IpaD and T4L. A) The Mean Fluorescence Intensity (MFI) of IpaD was detected by flow cytometry using IpaD‐specific rabbit polyclonal serum. The only cases where substantial surface labeling was not observed were for the ipaD null strain S. flexneri SF621 (IpaD‐) and a strain that does not express the needle protein MxiH (MxiH/pBAD). B) The MFI of T4L was similarly detected using a commercial monoclonal antibody against T4L. Only in two cases was any level of T4L surface localization observed with this monoclonal antibody and that labeling was very weak.
Figure S7. Structural superposition of IpaB74‐224 and IpaB74‐242. Structural superposition of the coiled‐coil domains from IpaB74‐224 (cyan) and IpaB74‐242 (green) is depicted in cartoon ribbon format, with an overall RMSD of 2.58 Å over 149/149 Cα atoms.
Figure S8. Characterization of Guinea Pig anti‐rT4L antisera by Surface Plasmon Resonance. Recombinant T4L (rT4L) was immobilized to a final ligand density of 830 RU on an NHS‐EDC‐activated surface via amide bond formation using T4L surface amine groups. Sequential four‐fold dilutions of immune serum were injected over rT4L surface using a single cycle approach, followed by a single dissociation phase. The reference subtracted sensorgram is shown in black, while the outcome of fitting the response data to a binding model is shown in red. Due to the polyclonal nature of the anti‐rT4L immune serum, accurate determination of binding constants was not possible. Nevertheless, these data show clear evidence of dose‐dependent binding of immune serum‐derived IgGs to the immobilized rT4L that is characterized by the slow off‐rate (10−4 to 10−5/s) characteristic of high‐affinity antibodies.
Acknowledgments
Flow cytometry analysis was performed at the Kansas Vaccine Institute Immunology Core Laboratory. We also acknowledge the assistance provided by the Protein Structure (X‐ray) Laboratory, which is supported in part by the Protein Structure and Function Center of Biomedical Research Excellence (NIH P30 GM110761). This research was funded by awards from the National Institutes of Health (NIH R01 AI099489 and AI123351) to W.D.P. and the Kansas Agriculture Experiment Station to B.V.G.
Broader Impact: Bacterial type III secretion systems enable pathogens to communicate with mammalian cells. A critical step in this process is the bacterium's ability to recognize contact with the host, which promotes the coordinated delivery of effector proteins. This study explores proteins on the Shigella surface that allow this process to proceed.
References
- 1. Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM (1999) Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ 77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 2. Bardhan P, Faruque AS, Naheed A, Sack DA (2010) Decrease in shigellosis‐related deaths without Shigella spp.‐specific interventions, Asia. Emerg Infect Dis 16:1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque ASG, Zaidi AKM, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins‐Browne RM, Levine MM (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case‐control study. Lancet 382:209–222. [DOI] [PubMed] [Google Scholar]
- 4. DuPont HL, Levine MM, Hornick RB, Formal SB (1989) Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis 159:1126–1128. [DOI] [PubMed] [Google Scholar]
- 5. Killackey SA, Sorbara MT, Girardin SE (2016) Cellular aspects of Shigella pathogenesis: Focus on the manipulation of host cell processes. Front Cell Infect Microbiol 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schroeder GN, Hilbi H (2008) Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 21:134–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, Strynadka NCJ, Finlay BB (2017) Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol 15:323–337. [DOI] [PubMed] [Google Scholar]
- 8. Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL (2006) IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri . Infect Immun 74:4391–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Epler CR, Dickenson NE, Olive AJ, Picking WL, Picking WD (2009) Liposomes recruit IpaC to the Shigella flexneri type III secretion apparatus needle as a final step in secretion induction. Infect Immun 77:2754–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD (2007) Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun 75:2626–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheung M, Shen DK, Makino F, Kato T, Roehrich AD, Martinez‐Argudo I, Walker ML, Murillo I, Liu X, Pain M, Brown J, Frazer G, Mantell J, Mina P, Todd T, Sessions RB, Namba K, Blocker AJ (2015) Three‐dimensional electron microscopy reconstruction and cysteine‐mediated crosslinking provide a model of the type III secretion system needle tip complex. Mol Microbiol 95:31–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stensrud KF, Adam PR, La Mar CD, Olive AJ, Lushington GH, Sudharsan R, Shelton NL, Givens RS, Picking WL, Picking WD (2008) Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem 283:18646–18654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dickenson NE, Arizmendi O, Patil MK, Toth RTt, Middaugh CR, Picking WD, Picking WL (2013) N‐terminus of IpaB provides a potential anchor to the Shigella type III secretion system tip complex protein IpaD. Biochemistry 52:8790–8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faherty CS, Redman JC, Rasko DA, Barry EM, Nataro JP (2012) Shigella flexneri effectors OspE1 and OspE2 mediate induced adherence to the colonic epithelium following bile salts exposure. Mol Microbiol 85:107–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adam PR, Dickenson NE, Greenwood JC 2nd, Picking WL, Picking WD (2014) Influence of oligomerization state on the structural properties of invasion plasmid antigen B from Shigella flexneri in the presence and absence of phospholipid membranes. Proteins 82:3013–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Menard R, Sansonetti P, Parsot C, Vasselon T (1994) Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri . Cell 79:515–525. [DOI] [PubMed] [Google Scholar]
- 17. Barta ML, Dickenson NE, Patil M, Keightley A, Wyckoff GJ, Picking WD, Picking WL, Geisbrecht BV (2012) The structures of coiled‐coil domains from type III secretion system translocators reveal homology to pore‐forming toxins. J Mol Biol 417:395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dickenson NE, Choudhari SP, Adam PR, Kramer RM, Joshi SB, Middaugh CR, Picking WL, Picking WD (2013) Oligomeric states of the Shigella translocator protein IpaB provide structural insights into formation of the type III secretion translocon. Protein Sci 22:614–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Birket SE, Harrington AT, Espina M, Smith ND, Terry CM, Darboe N, Markham AP, Middaugh CR, Picking WL, Picking WD (2007) Preparation and characterization of translocator/chaperone complexes and their component proteins from Shigella flexneri . Biochemistry 46:8128–8137. [DOI] [PubMed] [Google Scholar]
- 20. Weaver LH, Matthews BW (1987) Structure of bacteriophage T4 lysozyme refined at 1.7 A resolution. J Mol Biol 193:189–199. [DOI] [PubMed] [Google Scholar]
- 21. Menard R, Sansonetti PJ, Parsot C (1993) Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol 175:5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Picking WL, Picking WD (2016) The many faces of IpaB. Front Cell Infect Microbiol 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adam PR, Patil MK, Dickenson NE, Choudhari S, Barta M, Geisbrecht BV, Picking WL, Picking WD (2012) Binding affects the tertiary and quaternary structures of the Shigella translocator protein IpaB and its chaperone IpgC. Biochemistry 51:4062–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lunelli M, Lokareddy RK, Zychlinsky A, Kolbe M (2009) IpaB‐IpgC interaction defines binding motif for type III secretion translocator. Proc Natl Acad Sci USA 106:9661–9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lokareddy RK, Lunelli M, Eilers B, Wolter V, Kolbe M (2010) Combination of two separate binding domains defines stoichiometry between type III secretion system chaperone IpgC and translocator protein IpaB. J Biol Chem 285:39965–39975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murillo I, Martinez‐Argudo I, Blocker AJ (2016) Genetic dissection of the signaling cascade that controls activation of the Shigella type III secretion system from the needle tip. Sci Rep 6:27649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD (2005) IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun 73:1432–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Veenendaal AK, Hodgkinson JL, Schwarzer L, Stabat D, Zenk SF, Blocker AJ (2007) The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol 63:1719–1730. [DOI] [PubMed] [Google Scholar]
- 29. Guichon A, Hersh D, Smith MR, Zychlinsky A (2001) Structure‐function analysis of the Shigella virulence factor IpaB. J Bacteriol 183:1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adam PR, Barta ML, Dickenson NE (2017) Characterization of type three secretion system translocator interactions with phospholipid membranes. Methods Mol Biol 1531:81–91. [DOI] [PubMed] [Google Scholar]
- 31. Page AL, Fromont‐Racine M, Sansonetti P, Legrain P, Parsot C (2001) Characterization of the interaction partners of secreted proteins and chaperones of Shigella flexneri . Mol Microbiol 42:1133–1145. [DOI] [PubMed] [Google Scholar]
- 32. Roehrich AD, Martinez‐Argudo I, Johnson S, Blocker AJ, Veenendaal AK (2010) The extreme C terminus of Shigella flexneri IpaB is required for regulation of type III secretion, needle tip composition, and binding. Infect Immun 78:1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slabinski L, Jaroszewski L, Rychlewski L, Wilson IA, Lesley SA, Godzik A (2007) XtalPred: a web server for prediction of protein crystallizability. Bioinformatics 23:3403–3405. [DOI] [PubMed] [Google Scholar]
- 34. Geisbrecht BV, Bouyain S, Pop M (2006) An optimized system for expression and purification of secreted bacterial proteins. Prot Expr Purif 46:23–32. [DOI] [PubMed] [Google Scholar]
- 35. Barta ML, Adam PR, Dickenson NE (2017) Recombinant expression and purification of the Shigella translocator IpaB. Methods Mol Biol 1531:173–181. [DOI] [PubMed] [Google Scholar]
- 36. Kabsch W (2010) Xds. Acta Cryst D 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Evans PR (2011) An introduction to data reduction: space‐group determination, scaling and intensity statistics. Acta Cryst D 67:282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L‐W, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Cryst D 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams PD, Grosse‐Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Cryst D 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 40. Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Cryst D 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 41. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Cryst D 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Painter J, Merritt EA (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Cryst D 62:439–450. [DOI] [PubMed] [Google Scholar]
- 43. Osiecki JC, Barker J, Picking WL, Serfis AB, Berring E, Shah S, Harrington A, Picking WD (2001) IpaC from Shigella and SipC from Salmonella possess similar biochemical properties but are functionally distinct. Mol Microbiol 42:469–481. [DOI] [PubMed] [Google Scholar]
- 44. Liu X, Wang S, Sendi L, Caulfield MJ (2004) High‐throughput imaging of bacterial colonies grown on filter plates with application to serum bactericidal assays. J Immunol Methods 292:187–193. [DOI] [PubMed] [Google Scholar]
- 45. Thompson J, Higgins D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gouet P, Courcelle E, Stuart D, Métoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305–308. [DOI] [PubMed] [Google Scholar]
- 47. DeLano WL. The PyMOL Molecular Graphics System. 2002;2009. http://www.pymol.org.
- 48. Evans P (2006) Scaling and assessment of data quality. Acta Cryst D 62:72–82. [DOI] [PubMed] [Google Scholar]
- 49. Diederichs K, Karplus PA (1997) Improved R‐factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol 4:269–275. [DOI] [PubMed] [Google Scholar]
- 50. Weiss MS (2001) Global indicators of X‐ray data quality. J Appl Cryst 34:130–135. [Google Scholar]
- 51. Karplus PA, Diederichs K (2012) Linking crystallographic model and data quality. Science 336:1030–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Evans P (2012) Biochemistry. Resolving some old problems in protein crystallography. Science 336:986–987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Crystal structure of T4 lysozyme. The crystal structure of lysozyme from bacteriophage T4 (PDB ID: 2LZM) is shown colored from blue (N‐terminus) to red (C‐terminus). The distance between the N‐ and C‐termini is ∼9.1 Å.
Supplemental Figure S2. Production of IpaB and the IpaB‐T4L insertion mutants by Shigella flexneri. Bacteria were grown to an absorbance at 600 nm of 2.5 and then 1 ml of the culture was centrifuged to collect the bacteria which were then solubilized using SDS‐sample buffer. 10 µl of each was then separated by SDS‐PAGE (10% polyacrylamide gel) and the IpaB detected by immunoblot using rabbit anti‐IpaB antibodies. IpaB could be detected in all samples except for the ipaB null strain SF620 (IpaB‐) though the levels were reduced for IpaB‐T4L‐171 and IpaB‐T4L‐435. IpaB‐T4L60 appeared to undergo degradation.
Supplementary Figure S3. Recombinant expression and purification of IpaB‐T4L insertion mutants. Each wild‐type or T4L mutant was co‐expressed with IpgC and purified. This figure shows a Coomassie blue‐stained SDS PAGE gel of those protein pairs that could be detected after expression in E. coli BL21 (DE3). IpaB‐T4L‐60 and 435 mutants were not expressed and thus are not included in this figure. Some loss of chemical stability was observed for T4L insertions at positions 352 and 389, however, IpgC expressed well in all cases (even for the insertions at positions 60 and 435) when E. coli whole cell extracts were examined (data not shown). The same amount of protein (∼5 µg) was separated in each lane based upon the absorbance at 280 nm.
Supplementary Figure S4. Circular dichroism (CD) analysis of IpaB (wild‐type and T4L insertion mutants) in complex with IpgC after IMAC purification.
Only IpaB‐T4L‐60 and IpaB‐T4L‐435 could not be purified. For all the IpaB‐IpgC complexes that could be purified, the CD spectra indicated stable folding into the expected α‐helical structure (see WT) for this complex. The orange spectra are after heating the protein to 90°C (see Bottom panel) and then cooling back to 10°C. The final panel is recombinant T4L alone. (The CD spectrum of IpgC alone can be found in Birket SE, et al. 2007. Biochemistry 46:8128‐8137.)
Supplementary Figure S5. Thermal unfolding of the purified IpaB‐T4L insertion mutants based on CD analyses. The molar ellipticity at 222 nm was monitored as a function of temperature for all of the recombinant IpaB‐T4L insertion mutants complexed with IpgC. The thermal unfolding curve for IpgC alone can be found in Birket SE, et al. 2007. Biochemistry 46:8128‐8137.)
Supplementary Figure S6. Surface presentation of IpaD and T4L. A) The Mean Fluorescence Intensity (MFI) of IpaD was detected by flow cytometry using IpaD‐specific rabbit polyclonal serum. The only cases where substantial surface labeling was not observed were for the ipaD null strain S. flexneri SF621 (IpaD‐) and a strain that does not express the needle protein MxiH (MxiH/pBAD). B) The MFI of T4L was similarly detected using a commercial monoclonal antibody against T4L. Only in two cases was any level of T4L surface localization observed with this monoclonal antibody and that labeling was very weak.
Figure S7. Structural superposition of IpaB74‐224 and IpaB74‐242. Structural superposition of the coiled‐coil domains from IpaB74‐224 (cyan) and IpaB74‐242 (green) is depicted in cartoon ribbon format, with an overall RMSD of 2.58 Å over 149/149 Cα atoms.
Figure S8. Characterization of Guinea Pig anti‐rT4L antisera by Surface Plasmon Resonance. Recombinant T4L (rT4L) was immobilized to a final ligand density of 830 RU on an NHS‐EDC‐activated surface via amide bond formation using T4L surface amine groups. Sequential four‐fold dilutions of immune serum were injected over rT4L surface using a single cycle approach, followed by a single dissociation phase. The reference subtracted sensorgram is shown in black, while the outcome of fitting the response data to a binding model is shown in red. Due to the polyclonal nature of the anti‐rT4L immune serum, accurate determination of binding constants was not possible. Nevertheless, these data show clear evidence of dose‐dependent binding of immune serum‐derived IgGs to the immobilized rT4L that is characterized by the slow off‐rate (10−4 to 10−5/s) characteristic of high‐affinity antibodies.


