Abstract
DNA methyltransferase 3A (DNMT3A) is mutated in various myeloid neoplasms including acute myeloid leukemia (AML), especially at the Arg882 and associated with inferior outcomes. Here, we report that the DNMT3A-Arg882His/Cys (R882H/C) mutations led to inactivation of apoptosis through DNA damage signaling following the impairment of differentiation of myeloid leukemia cells. Gene expression profiling analysis revealed aberrant expression of several cell-cycle and apoptosis-related genes, and the DNA methylation assay identified both hypo- and hypermethylation features in different regions throughout the whole genome of DNMT3A mutants-transduced myeloid cells. We found that DNMT3A-R882H/C mutations upregulated the expression of an antioxidant protein, pyroxiredoxin-2 (PRDX2), at the mRNA and protein levels with decreased accumulation of reactive oxygen species (ROS). Augmentation of ROS generation by ROS accumulating agent or by knockdown of PRDX2 from myeloid cells effectively increased drug sensitivity and apoptosis as a consequence of reduced cell proliferation. DNMT3A-R882C/H mutations decreased apoptosis induction in part by increasing the antioxidant capacity of the cell owing to upregulation of PRDX2. Molecularly, both DNMT3A-WT and R882H/C mutants interacted with PRDX2; and R882C/H mutation-induced hypomethylation increased PRDX2 expression which enhanced cell proliferation and growth with impairment of apoptosis, thereby contributing to leukemogenesis.
Introduction
Recent studies have shown that epigenetics plays an important role in cancer biology including leukemia [1], [2]. Acute myeloid leukemia (AML) is a genetically heterogeneous malignancy. Whole genome sequencing found DNMT3A as one of the most frequently mutated genes across a range of hematological malignancies including AML [3], [4]. DNA methylation of CpG dinucleotides represents key epigenetic modifications that control the regulation of gene expression. In mammals, CpG methylation is catalyzed by a family of DNA methyltransferase enzymes including DNMT1, DNMT3A, and DNMT3B [5]. DNMT3A and DNMT3B are the main enzymes to initiate de novo DNA methylation, whereas DNMT1 maintains methyltransferase activity [6]. Gene mutation studies identified somatic mutations of DNMT3A in about 20% of patients with AML, mostly in cases with monocytic lineage (AML-M5 or -M4), and were associated with poor prognosis [7], [8]. Although various DNMT3A mutations have been identified in AML, Arg882His (R882H) is the most frequent, accounting for 70%-80% of cases, and R882C is the next [9]. It also has been reported that DNMT3A mutations caused loss of tetramerization and thereby exert reduced de novo methyl transferase activity and focal hypomethylation [10]. Although Dnmt3a-null hematopoietic stem cells (HSCs) showed both increased and decreased methylation at distinct loci in vivo [11], it has been also reported that Dnmt3a loss from HSCs in conditional Dnmt3a knockout mouse causes impairment of HSC-differentiation and upregulation of self-renewal genes [11]. It has recently been reported that DNMT3A-R882 mutants interacted with polycomb proteins and block HSCs and leukemia cell differentiation [9]. More recent report revealed that DNMT3A-R882H mutation cooperated with NRAS mutation to transform HSC and induced AML development [12].
It has been suggested that DNMT3A-R882H increased the protein level of CDK1, a cell-cycle regulator protein, and enhanced cell-cycle activity [13]. DNMT3A interacts with many other proteins including histone modifiers to suppress their target gene expression [14], [15]. Previous studies have suggested DNMT3A mutations as the fundamental genetic event at the initiation of AML pathogenesis [16], [17]. Despite the current progress of functional role of DNMT3A mutations, the molecular pathogenesis of myeloid malignancies remains poorly understood. The mechanisms of AML transformation and functional role of DNMT3A mutations through its target genes in the leukemogenesis remain to be explored. In this study, we show that DNMT3A mutants impaired apoptosis through DNA damage signaling and target epigenetically augmented PRDX2, an antioxidant protein which may contribute to malignant transformation.
Materials and Methods
Cell Culture, Drug Treatments, Staining, and Cell Proliferation
The human leukemia cell lines K562, HL-60, U937, and THP-1 were cultured in RPMI-1640 medium; HEK293T cells were cultured in DMEM according to standard conditions. HL-60 cells were obtained from ATCC (November 2015), and U937, K562, and THP-1 were obtained from our own stocks. All cell lines were authenticated by cellular morphology and STR analysis at Chang Gung Memorial Hospital (January-February 2017). Murine myeloid leukemia 32Dcl3 (32D) cells were cultured in the presence of 1 ng/ml murine-IL-3 under similar conditions. Phorbol 12-myristate 13-acetate (PMA)–mediated myelomonocytic differentiation of U937 cells and megakaryocytic differentiation of K562 cells were induced by applying 40 nM PMA (Sigma chemicals) dissolved in dimethyl sulfoxide. To induce granulocytic differentiation, U937 cells were treated with 300 nM all-trans retinoic acid (ATRA) for 96 hours. Oxidative stress was induced by tertiary-butyl hydrogen peroxide (TBHP) treatment performed on cells cultured in 12-well or 6-well microplates. For colonogenic growth assays, cells were cultured in 12-well plate at 1-2 × 103 cells/well in Methocult H4435 (StemCell Technologies) medium for 7 days. Photograph was taken by phase contrast microscope (Nikon Eclipse TS100, Japan). For morphological studies, cytospined (Thermo) smears were stained with modified Wright-Giemsa (Sigma). Digital images were acquired using Olympus (model no. U-TV0.5XC-3) microscope equipped with a digital camera. Cell proliferation and survival in the presence of drugs or no drugs were determined at different time points assessed by manual counting using a hemocytometer followed with trypan blue staining.
Plasmid Construction, Lentiviral Preparation, and Infection
The full-length cDNA of human DNMT3A-WT was constructed into the NheI/NotI multiple cloning sites of lentiviral vector pCDH-CMV-MCS-EF1-Puro according to the standard method and verified by sequencing. Point mutants R882C and R882H and deletion mutant Q515X of DNMT3A gene were generated from WT using site-directed mutagenesis (KAPA HiFi HotStart, Kapa Biosystems) and confirmed by full-length DNA sequencing. Wild-type and mutant-DNMT3A with N-terminal 2 × FLAG tagged was then subcloned into pIRES2-EGFP-vector. Lentiviruses production, cell infection, and selection of puromycin-resistant cells were performed as our previous description [18]. The pLKO.1-puro plasmid-based shRNAs, including shLuc (luciferase shRNA, TRCN231719), human PRDX2-sh1 (TRCN0000064906), PRDX2-sh2 (TRCN0000064907), mouse Prdx2-sh1 (TRCN0000120694), Prdx2-sh2 (TRCN0000322525), and human cMYC-sh1 (TRCN0000010389) and cMYC-sh2 (TRCN0000010390), were obtained from the National RNAi Core laboratory, Taiwan. For shRNA-mediated knockdown of PRDX2 or Prdx2, cells were infected with pLKO.1-puro plasmid-based shRNAs lentivector and selected with puromycin.
Flow Cytometry Analysis
Cells were stained with anti-CD11b PE/CD14 APC (eBioscience; Cat. 12-0118/Cat. 17-0149) or anti–CD61-PerCP-Ab (BD Pharmingen), and assays were performed by flow cytometry (BD AriaIII) using the BD Cell-Quest Pro version 4.0.1 software. The apoptosis was detected on a FACSCalibur (BD Biosciences) flow cytometry by Annexin V and propidium iodide (PI) staining according to the manufacturer instructions (Annexin V-FITC kit, Beckman Coulter, Marseille, France).
Immunofluorescence Analysis
PBS-washed cytospin cells were fixed with chilled 100% methanol at room temperature for 5 minutes, washed thrice with cold PBS, blocked with 5% BSA in PBS containing 0.1% Tween 20, and then incubated with the primary antibodies at 4°C for overnight. Cells were subsequently washed thrice, incubated with Rhodamine and FITC-conjugated anti-rabbit or anti-mouse secondary antibodies (Jackson ImunoResearch), and counterstained with 4,6-diamidino-2- phenylindole (DAPI) (Sigma). Cells were washed and mounted, morphological changes of cells were examined under the fluorescence microscope (Olympus BX61), and images were acquired using a digital color camera with Case Data Manager EXPO 6.0 software system. All the images were taken with fixed contrast and exposure time for DAPI of 80 milliseconds, 150 milliseconds for red fluorescence, and 1000 milliseconds for green fluorescence.
Intracellular Reactive Oxygen Species (ROS) Generation Assay
Live green ROS assay kit (Molecular Probes) was used to measure intracellular ROS according to the manufacturer's instructions. DNMT3A-WT and mutants expressing cells including control were incubated in 25 μM 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA) containing PBS at 37°C for 30 minutes. Cells were then washed thrice with PBS to remove the extra reagent that had not entered the cells, and the fluorescence intensity of the cell suspension was immediately measured by flow cytometry. Similar cells were cytospined for microscopic analysis.
Isolation of RNA, Reverse Transcription, and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
Total RNA was extracted from frozen or cultured cells using Trizol total RNA isolation reagent (Invitrogen). Sample was reversely transcribed to cDNA with random hexamers using the Superscript III First-Strand Synthesis System for RT-PCR Kit (Invitrogen). The cDNA was used for quantitative PCR with iQTM SYBR Green Supermix (BIO-Rad) according to manufacturer's protocol. The sequences of oligonucleotides used for qPCR are listed in supplementary Table S1. Samples were run in duplicate, transcript levels were calculated as 2(−∆∆Ct), and the threshold cycle number for different genes was normalized to the expression of GAPDH.
Immunoblotting and Immunoprecipitation Analysis
Immunoblot was performed with the following antibodies: anti-DNMT3A (#3598), anti-γH2AX (#2577), p-cMYC (#13748), and anti-MYC (#5605) obtained from Cell Signaling Technology; anti-PRDX2 (ab133481) and anti-BCL2 (ab692) from AbCam; anti-CDK1 (Cat.GTX20018) and anti-PARP-1 (Cat. GTX100573) from GeneTex; and anti-Flag (F3165) and anti–β-Actin (Cat. A5441) from Sigma-Aldrich. Cells were homogenized and lysed in a buffer containing 50 mM Tris-HCl pH 8.0, 120 mM NaCl, 0.5% NP-40, 0.25% Na deoxycholate, 1 mM DTT, 1 mM PMSF, 1 mM EDTA, 1 mM NAF, 1 mM Na3VO4, and protease inhibitor (all chemicals purchased from Sigma Chemicals, St Louis, MO). A total of 20-40 μg proteins were subjected to electrophoresis followed by immunoblotting analysis using antibodies against the target proteins. A loading control of detection of β-Actin was included for all immunoblots. The protein lysate was resolved in 6-12% SDS polyacrylamide gels according to the protein size, transferred to a nylon membrane (PVDF, Schleiche & Schuell, Einbeck, Germany), and then incubated with primary antibodies. After washing, the membranes were incubated with HRP-conjugated anti-mouse IgG or anti rabbit antibodies (1:5000-10,000). Detection was completed using enhanced chemiluminescence (Amersham Pharmacia). For immunoprecipitation reaction, FLAG-DNMT3A-WT/mutants-expressed HEK293T cells were washed with ice-cold PBS twice and lysed in lysis buffer (150 mM NaCl, 50 mM Tris HCl pH 7.4, 0.5% NP-40) supplemented with protease and phosphatase inhibitors. For DNAse treatments, cell lysates were incubated with an excess DNase I in presence of 2.5 mM MgCl2 for 45 minutes at 37°C prior to immunoprecipitation. At the end of reaction, 5 mM final concentration of EDTA was added. The efficiency of DNAse treatment was checked using plasmidic DNA (Supplementary Figure S1). Lysates containing equal amount of proteins (500-1000 μg) were subjected to immunoprecipitation with anti-FLAG M2 affinity gel (Sigma-Aldrich, Cat. A2220) at 4°C for overnight according to the manufacturer's instructions, washed, and eluted by boiling in Laemmli buffer. Immunoblotting was analysis as described method.
Gene Expression Microarray Analysis
Gene expression analysis was carried out using Affymetrix Human Gene U133 Plus 2. First, RNA was extracted from stably transduced U937 cells using Trizol reagent method. RNA molecules were then reversely transcribed into cDNA by using an enzyme reverse transcriptase. The cDNA product was purified and quantified using standard protocol. Amplification and biotin labeling of fragmented cDNA were carried out using standard Affymetrix protocol. Labeled probes were hybridized to the Affemetrix GeneChip Hybridization Oven 645 and GeneChip Fluidiecs Station 450 and scanned according to manufacturer's protocol. The relative expression level for each gene was stored as an image. Expression data were extracted from image files produced on GeneChip Scanner 3000 7G. The scanned images were analyzed with Standard Affymetrix protocol. Genechip analysis data were normalized with RMA by Affymetrix Expression Consol Ver 1.4 (EC 1.4) software, and fold change was calculated compared to both empty vector control and DNMT3A-WT. DNA microarrays had a lower limit of detection where signal for the probe set was no longer detectable above the background noise of the microarray due to their fixed protocol of hybridization. The differentially expressed genes were selected using the criteria of undergoing a ≥1.5-fold (upregulation) and <0.6-fold (downregulation) change in gene expression. A part of differentially expressed genes from microarray data was validated using qRT-PCR analyses.
Bisulfite Sequencing
Genomic DNA was isolated from U937 cells stably transduced with DNMT3A-WT/MT including control and subjected to bisulfite conversion with the EZ DNA methylation gold kit (Zymo Research) according to the manufacturer's instructions. Bisulfite- converted DNA was amplified by PCR using gene-specific primers. PCR products were cloned using TA-cloning vector (Yeastern Biotech C. Ltd.) and checked CpG methylation pattern after bisulfite sequencing. Sequencing primers used for bisulfite sequencing analysis of PRDX2 gene is: forward primer: GGGGGTTTTAGTTTTTAGTTATGGT and reverse primer: TTAACTACAACCTCCCTCTTTAACC.
DNA Methylation Microarray Analysis
Genome-wide DNA methylation was assessed using the Illumina Infinium Human MethylationEPIC Beadchip (Illumina Inc., CA) according to manufacturer's instructions. The resulting raw data were normalized (control normalization) and background corrected by manufacturer to generate methylation β-values. Genomic methylation pattern (hypo- or hypermethylation) of different samples was analyzed after the variance filter (STD >0.2), and raw data (without filter) were used for the analyses of specific gene methylation pattern.
The data, including all raw gene expression and genomic-methylation data, have been deposited in the Gene Expression Omnibus database with accession number GSE90934.
Statistical Analysis
Data represented here are mean ± S.D. as indicated. The significance of the differences between groups was determined using Student t test (2-tailed). A P value of <.05 was considered significant for all analyses.
Results
DNMT3A-R882 Mutants Lead to Inactivation of Apoptosis Following Impairment of Differentiation of Myeloid Leukemia Cells
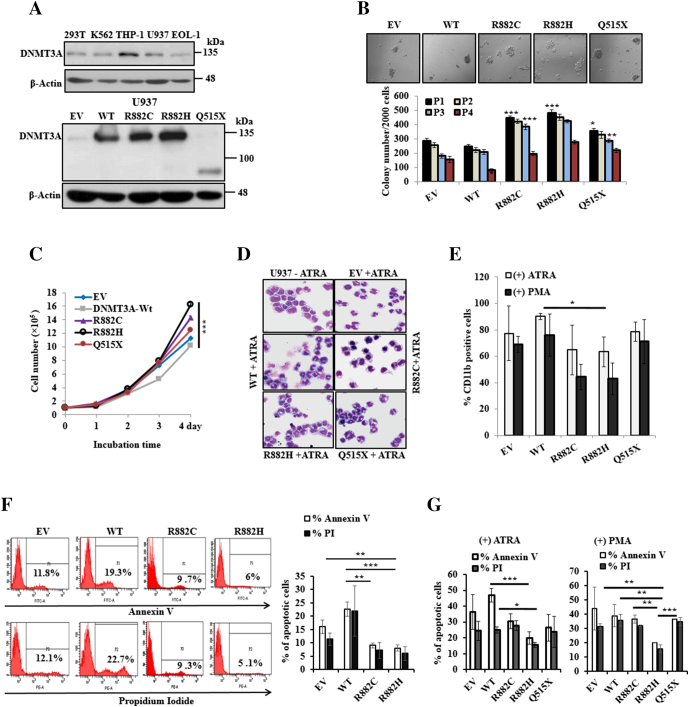
To elucidate the functional role of DNMT3A mutants, we stably expressed WT and mutant- DNMT3A-WT including empty vector (EV) in U937 cells (Figure 1A). The results showed that colony formation ability, self-renewal activity, and cell proliferation were enhanced in cells with DNMT3A-R882H/C mutants compared to EV or WT, whereas C-terminal deletion mutant DNMT3A-Q515X showed a moderate effect (Figure 1, B and C). After treatment with 300 nM ATRA or 40 nM PMA for 96 hours, U937 cells induced morphological change, and cells endured differentiation accompanied by apoptosis. Wright-Giemsa–stained smears showed a greater fraction of more mature granulomonocytes in U937 cells transduced with WT-DNMT3A compared to R882H/C-expressing cells (Figure 1D). Flow cytometry results showed that cellular surface differential antigen CD11b, a marker of both granulocytic and monocytic cell differentiation, decreased in U937 cells expressing R882H/C compared to WT or EV with treatment of ATRA or PMA (Figure 1E). In contrast, cell morphology and cellular surface differential antigen markers in U937 cells transduced with Q515X have no difference compared to EV or DNMT3A-WT expressing cells, indicating different biological effects of Q515X from R882H/C. U937 cells transduced with DNMT3A-WT showed more Annexin V– and PI-positive cells compared to R882H-expressing cells (Figure 1F). On the treatment of ATRA or PMA, DNMT3A-R882H/C–transduced U937 cells showed lesser extent of apoptotic-induced cells compared to EV or WT-expressing cells (Figure 1G).
Figure 1.
DNMT3A-R882H/C mutations impair stimulant-induced differentiation with inactivation of apoptosis of myeloid leukemia cells. (A) Immunoblot of endogenous DNMT3A expression in different leukemic and human fibroblast cell lines (upper panel), and overexpressed WT-, mutant-DNMT3A into U937 cells (lower panel). (B) Colony formation ability in Methocult medium after stable overexpression of WT and mutant-DNMT3A into U937 cells (original magnification: ×100). Columns P1-P4 represent the number of serially replated colonies. (C) Growth curves of U937 cells stably transduced with WT- and mutant-DNMT3A. Representative results from three independent replicates are shown. (D) U937 cells were treated with 300 nM ATRA for 96 hours; cell morphology with Wright-Giemsa–stained smears (original magnification: ×400). (E) Percentage of CD11b-positive cells after treatment with 300 nM ATRA and 40 nM PMA for 96 hours. (F) U937 cells stably transduced with EV-, DNMT3A-WT, and mutants used for apoptosis analysis. Representative flow cytometry analysis (left panel) and the % of Annexin V– and PI-positive cells are shown (right panel). (G) Percentage of apoptotic cells after treatment with 300 nM ATRA and 40 nM PMA for 96 hours. Error bars represent ± S.D. of the mean of two separate experiments. *P < .05, **P < .03, ***P < .01. P value compared to empty vector control in colony formation and self-renewal activity was calculated.
DNMT3A-R882 Mutants Impair Apoptosis Through Attenuation of DNA Damage Signaling of Myeloid Leukemia Cells
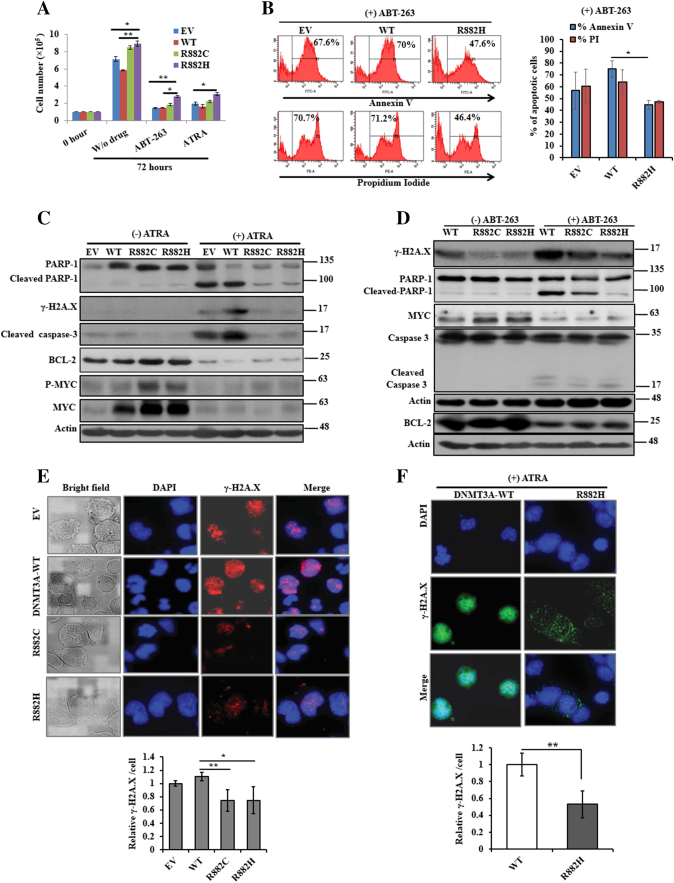
We found that though drugs inhibited significantly cell growth and proliferation, drug-induced cell growth inhibition was lower on R882H mutant cells compared to other mutant-carrying cells (Figure 2A and Supplementary Figure S2A). As shown in Figure 2B, treatment with ABT-263, a potent apoptosis-inducing drug, for 72 hours on U937 cells transduced with EV or WT resulted in marked increase in apoptosis compared to R882H transduced cells. We also found that on the treatment of U937 cells by ATRA, BCL-2 and CDK1 expression was inhibited in a dose- and time-dependent manner (Supplementary Figure S2B). In addition, expression of those proteins was reduced in U937 cells transduced with DNMT3A-WT/MTs in the presence of ATRA (Figure 2C and Supplementary Figure S2C). Moreover, the phosphorylation of histone H2A.X (γ-H2A.X), cleaved PARP-1, and cleaved caspase-3 expression was attenuated in DNMT3A-R882H/C-transduced U937 cells in response to either ATRA or ABT-263 as compared to DNMT3A-WT cells (Figure 2, C and D). Similarly, immunoblot data showed that MYC and BCL-2 expression was reduced on the treatment of both drugs. Immunofluorescence microscopy assays also revealed that DNA-damage signaling nuclear protein H2A.X was more phosphorylated in EV or DNMT3A-WT–expressing cells compared to R882H/C-expressing cells (Figure 2, E and F), indicating that DNMT3A-mutant cells impaired apoptosis through attenuation of DNA damage signaling.
Figure 2.
DNMT3A R882H/C mutants impair apoptosis through attenuation of DNA damage signaling. (A) Cell proliferation of stably transduced U937 cells treated with 300 nM ATRA and 300 nM ABT-263 for 72 hours or no drug. Data presented were the average of at least two replicates. (B) Representative flow cytometry analysis (left panel) and the % of Annexin V– and PI-positive cells (right panel) of mutant and WT-DNMT3A U937 cells including EV on the treatment of 300 nM ABT-263 for 72 hours are shown. (C and D) DNA damage signaling protein levels including c-MYC were examined with or without treatment of ATRA (C) or ABT-263 (D) by immunoblot analyses. β-Actin was used as a control for equal loading. (E and F) Phosphorylation of H2A.X (γ-H2A.X) levels was verified in transformed U937 cells without drug (E) and in the presence of 300 nM ATRA (F) by immunofluorescence microscopy (original magnification: ×1000). All the images were taken with same contrast and exposure time. Quantitation of the intensity of γ-H2A.X per cell was measured using ImageJ software (NIH, USA). Each data point represents the mean ± S.D. of three different microscopic field. All studies were repeated at least once; *P < .05, **P < .03.
DNMT3A-R882 Mutants Deregulate Cell Cycle and Apoptosis-Associated Genes
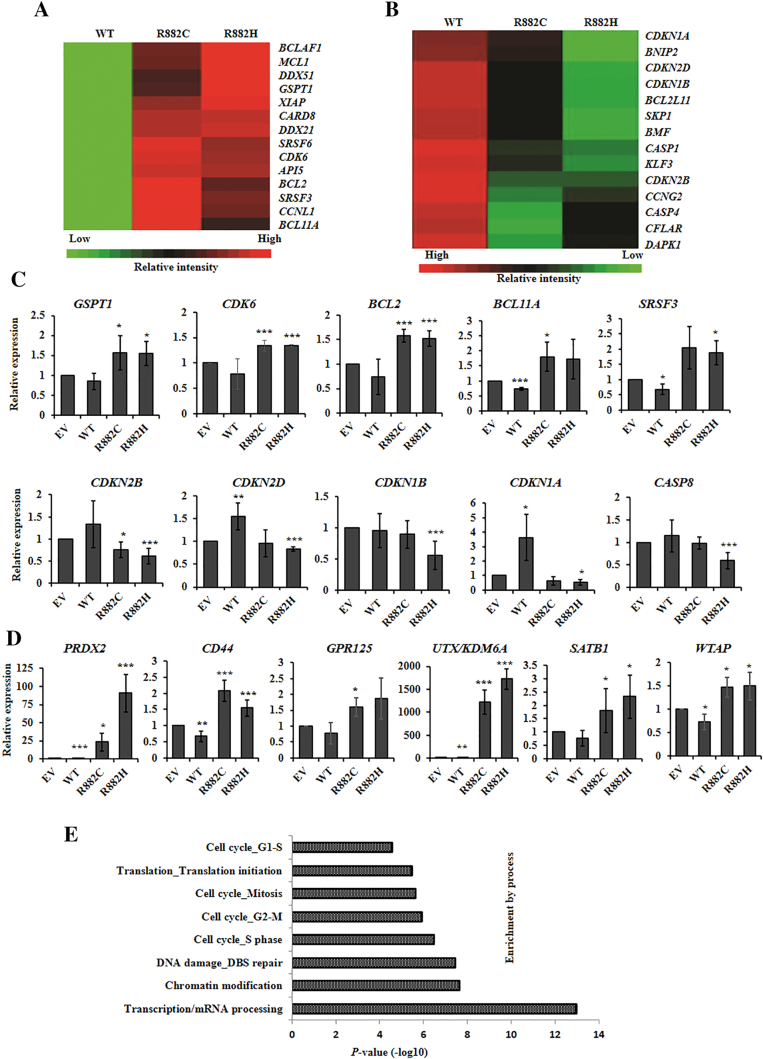
To understand the mechanism underlying DNMT3A-mutants phenotypic flaws, U937 cells transduced with EV, WT-, and mutants DNMT3A were subjected to the global gene expression microarray analysis. Gene expression microarray data revealed that 2262 and 2992 genes were upregulated (>1.5-fold) in either R882C- or R882H-expressing cells compared to EV and WT cells, respectively (Dataset S1). Similarly, 1862 and 2123 genes were downregulated (<0.6-fold) in either R882C- or R882H-expressing U937 cells compared to EV- and DNMT3A-WT–expressing cells, respectively (Dataset S2). The expression levels of 1171 genes in R882H cells were differentially expressed (upregulated 580 genes and downregulated 591 genes) compared to both EV and WT-expressing cells (Dataset S3). Among them, cell cycle facilitator and apoptosis inhibitor genes including GSPT1, BCL2, CDK6, and SRSF3 were upregulated (Figure 3A). Conversely, cell cycle inhibiting and apoptosis facilitator genes including CDKN1A, CDKN1B, CDKN2D, CASP1, CASP4, and CCNG2 were downregulated in DNMT3A-R882H/C–expressing cells (Figure 3B), suggesting that cells transduced with R882H/C were involved to regulate cell cycle and apoptosis. A portion of differential gene expression from microarray data was validated using qRT-PCR analyses of transduced U937 cells (Figure 3, C and D). The results of qRT-PCR for selected genes were compatible with the corresponding microarray data with a small number of exceptions. qRT-PCR is an analytically sensitive method of gene expression measurement platform, and there might be discrepancies between microarray and qRT-PCR data. This disagreement may be due to both biological and technical considerations including the cDNA synthesis, PCR primers, microarray probes, sensitivity, and the quality of the raw RNA between independent experiments. Upregulated genes in R882H/C-expressing cells compared to WT (Dataset S1) were used for Gene Ontology (GO) classification. Interestingly, many dysregulated genes (FUS, MLL, TPM3, EGR1, IKZF1, BCl2, CD44, WFDC1, VEGFA, and NUP98) involved in gene translocation or chromosome aberrations occurred frequently in AML and mixed-lineage leukemia. In addition, several upregulated genes were enriched in different cellular and molecular process including cell cycle, DNA damage repair, and transcription/chromatin modification (Figure 3E). Analysis based on the Hematopoietic Fingerprints Database [19] revealed 33 and 49 genes upregulated in both R882C and R882H expressing U937 cells compared to EV and WT, respectively. Among them, 12 genes were upregulated in R882H cells compared to both EV and DNMT3A-WT expressing cells (Dataset S4). Genes such as GPR125, ZBTB16, and UTX/KDM6A were reported to associate with hematopoietic malignancies [20], [21], [22].
Figure 3.
DNMT3A-R882 mutants deregulate cell cycle and apoptosis-associated genes in U937 cells. (A) Heatmap representation of cell-cycle facilitator and apoptosis-inhibiting genes identified as being differentially expressed in U937 cells transduced with DNMT3A-WT and mutants. Red indicates upregulated genes compared to WT (green). (B) Heatmap for gene expression in U937 cells transduced with DNMT3A-WT and mutants. Genes associated with cell-cycle inhibiting and apoptosis facilitators are clustered. Green indicates downregulated genes compared to WT- (red). (C and D) Quantitative RT-PCR results for 10 genes involved in cell cycle regulation and apoptosis (C) and representative genes associated with hematopoietic malignancies (D) showing the same patterns observed in gene-expression microarray analysis of WT- and mutants transduced U937 cells. (E) GO analyses of upregulated genes in R882C/H-expressing U937 cells showing a series of genes enriched in different cellular and molecular processes including cell cycle progression and DNA damage. Error bars represent ± S.D. of the mean of three to five experiments. *P ≤ .05, **P ≤ .01, ***P ≤ .005 compared to EV.
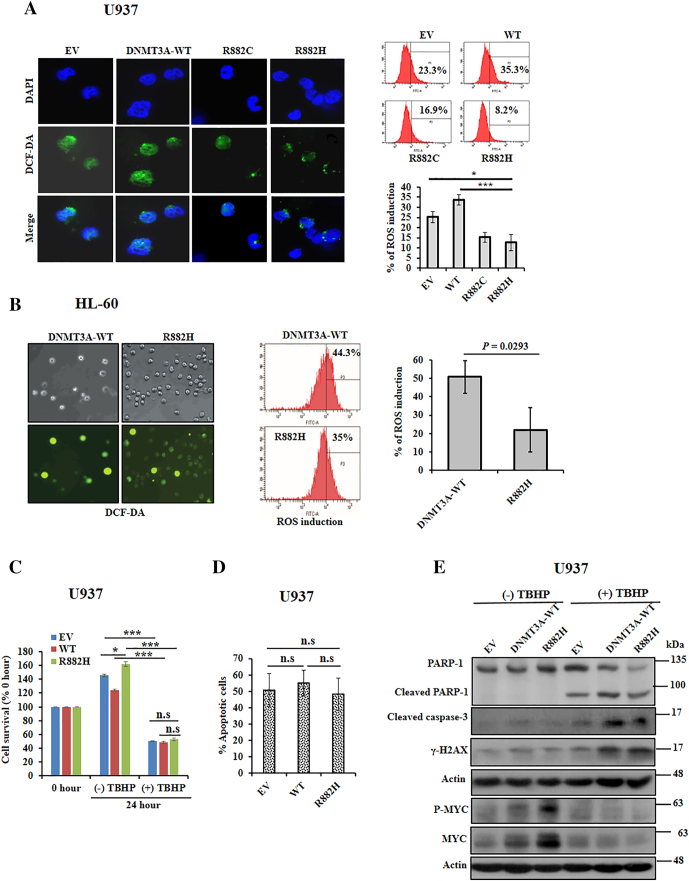
DNMT3A-R882 Mutants Augmented PRDX2, Reduce ROS Production, and Override the ROS-Mediated Apoptosis in the Presence of an Oxidizing Agent in Myeloid Cells
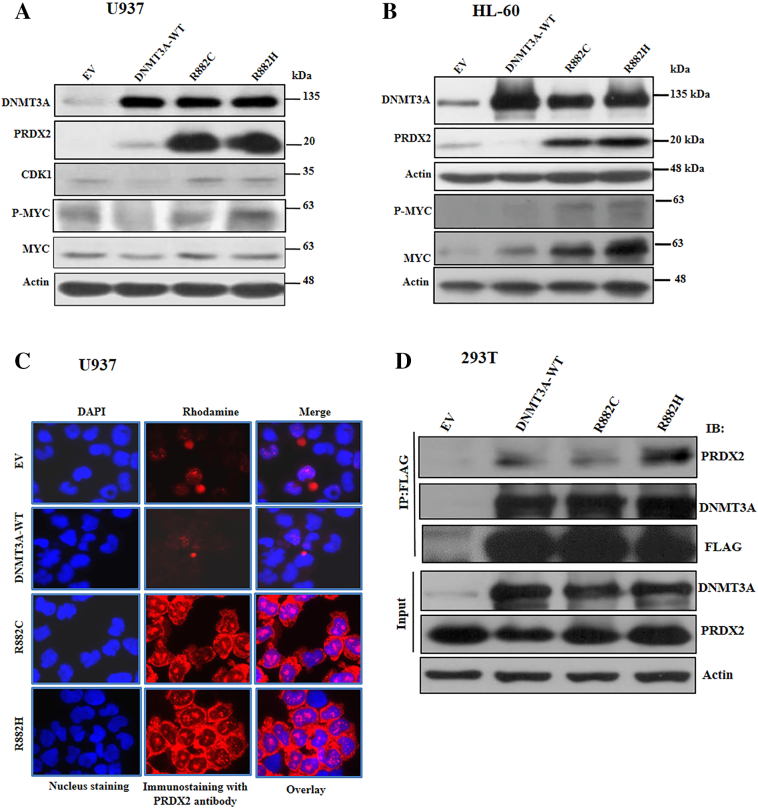
From gene expression microarray data analysis, we observed that the antioxidant protein PRDX2 was upregulated in DNMT3A-mutants–transduced U937 cells compared to EV and WT-expressing cells. The expression of PRDX2 was validated in U937 and HL-60 cells transduced with DNMT3A-WT, R882H/C, and control. The immunoblot data showed a marked increase of PRDX2 in R882H/C mutants expressing U937 and HL-60 cells compared to DNMT3A-WT or EV (Figure 4, A and B). Immunofluorescence data revealed that PRDX2 protein expression was augmented at cytoplasm of mutant cells compared to EV or DNMT3A-WT cells (Figure 4C). Moreover, several leukemia cell lines expressed both PRDX2-mRNA and protein in higher to moderate levels than expressed in U937 cells (Supplementary Figure S3, A and B). To explore the mechanism of PRDX2 upregulation in myeloid cells, we found that both WT- and mutant-DNMT3A proteins could form a complex with PRDX2 (Figure 4D and Supplementary Figure S1). PRDX2 is an antioxidant protein which regulates cellular ROS. To examine whether upregulation of PRDX2 in DNMT3A-mutated cells affects their intracellular ROS accumulation, we tested the internal generation of ROS using 2′,7′-dichlorofluorescein diacetate (DCF-DA) as a ROS scavenger. We found that either U937 or HL-60 cells transduced with mutants DNMT3A suppressed intracellular ROS accumulation compared with EV or WT-transduced cells (Figure 5, A and B). In consequence, we showed that the elevated PRDX2 expression was reduced with increased accumulation of ROS in the presence of ATRA in U937 cells transduced with WT- and mutant-DNMT3A (Supplementary Figure S4, A and B). To examine the connection between DNMT3A mutation and antioxidant defense by PRDX2 regulation, we exposed transduced U937 cells with TBHP, a stable form of H2O2, and checked cell proliferation, apoptosis, and apoptotic signaling. Cells treated with TBHP attenuated cell proliferation, enhanced apoptosis, and activated apoptosis-related signaling (Figure 5, C-E). Moreover, treatment of TBHP overrode the effects of mutant DNMT3A on cell proliferation, apoptosis, and signaling as there was not much difference between DNMT3A-mutant and WT-transduced cells. We also found that MYC and phosphorylation of MYC expression was decreased on the treatment of TBHP, suggesting the antioxidant effect of PRDX2 on MYC expression.
Figure 4.
DNMT3A-R882 mutants augmented PRDX2 expression in myeloid leukemia cells. (A and B) Immunoblot showing overexpression of DNMT3A-mutant into U937 (A) and HL-60 (B) cells augmented protein expression of PRDX2 with increasing MYC and P-MYC expression. β-Actin was used as a control for equal loading. (C) Cytoplasmic localization of PRDX2 in U937 cells transduced with EV-, WT-, and mutant-DNMT3A; immunostained with anti-PRDX2 (red) and DAPI (blue) (original magnification: ×1000). All the images were taken with same contrast and exposure time. (D) Co-immunoprecipitation data showing FLAG-tagged WT- and mutant DNMT3A both interacted with PRDX2 in 293T cells. Cell extracts were incubated with DNAse prior to immunoprecipitation (Supplementary Fig. S1 for effectiveness of DNAse treatment).
Figure 5.
DNMT3A-R882 mutants reduce ROS production and overrode the ROS-mediated apoptosis in the presence of an oxidizing agent. (A) Stably transduced U937 cells incubated with H2DCF-DA for 30 minutes at 37°C incubator; fluorescent oxidized DCF (green) and DAPI (blue) were photographed with fluorescence microscopy (left panel, original magnification: ×1000). All the images were taken with same contrast and exposure time. Flow cytometry detecting the generation of fluorescent oxidized DCF; representative flow cytometry (right upper panel) and percentage of ROS induction are shown (right lower panel). (B) Stably transduced HL-60 cells stained with H2DCF-DA; staining cells were photographed with phase-contrast microscopy (left upper panel) and fluorescence microscopy (left lower panel); original magnification: ×100. Flow cytometry detecting the generation of fluorescent oxidized DCF; representative flow cytometry and percentage of ROS induction (right panel) are shown. (C) Inhibition of cell survival following 200 μM TBHP treatment for 24 hours. (D) After treatment of 200 μM TBHP for 24 hours, cells were harvested and labeled with FITC-Annexin V as described in methods. Flow cytometry was used to detect Annexin V–positive cells. (E) Apoptosis triggering by TBHP treatment; transformed U937 cells were incubated with 1 mM TBHP for 2 hours followed by a 2-hour incubation without TBHP. Cell lysates were prepared, and apoptosis inducing proteins including MYC and P-MYC expressions were checked by immunoblot. All studies were repeated at least once. The mean ± S.D. of at least two replicates was plotted; *P < .05, ***P < .01. n.s., not significant.
Both DNMT3A-WT and mutants protein interacted with PRDX2; however, DNMT3A-R882C/H protein upregulated PRDX2 expression in two myeloid cell lines, U937 and HL-60. Simultaneously, MYC protein level was also increased in mutated cells. To check PRDX2 as a direct target of DNMT3A or MYC which regulated PRDX2 level, we knocked down MYC from U937-EV and U937-DNMT3A-R882C-expressing cells using two different shRNA and checked PRDX2 protein expression. We found that knock down of MYC from either EV or R882C-expressing U937 cells did not affect PRDX2 expression (Supplementary Figure S5), which indicated that PRDX2 was a direct target of DNMT3A rather than due to MYC level.
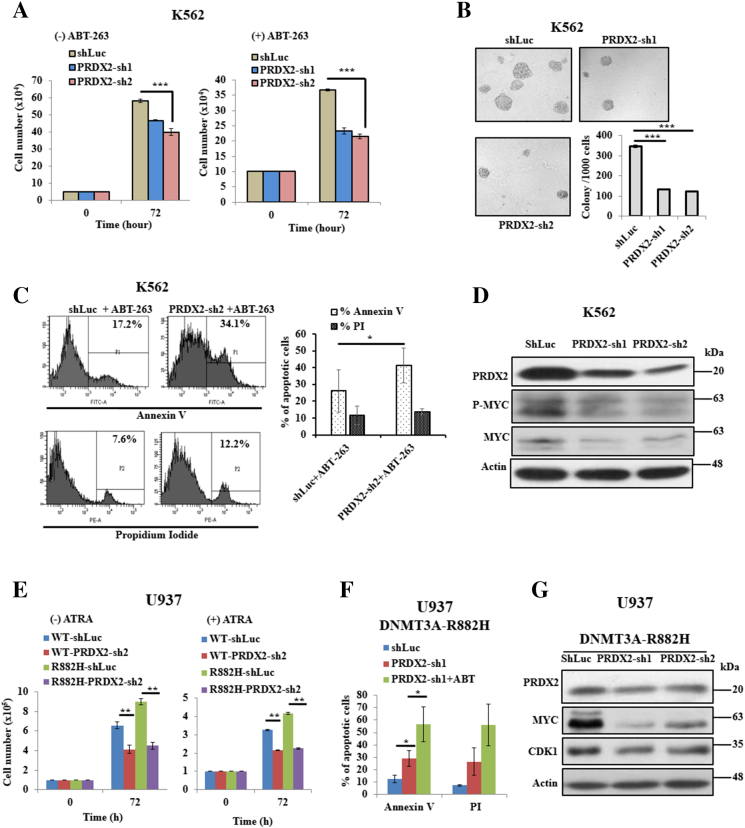
PRDX2 Blocks Apoptosis and Enhances Proliferation in Myeloid Leukemia Cells
We examined that both mRNA and protein expressions of PRDX2 in K562 cells were higher than those of other hematopoietic cell lines (Supplementary Figure S3, A and B). Knock down of PRDX2 suppressed cell proliferation, differentiation, and colony formation of K562 cells compared to scrambled cells (Figure 6, A and B; Supplementary Figure S6, A and B). Furthermore, knock down of PRDX2 enhanced ABT-263–induced apoptosis and decreased phosphorylation of MYC level of K562 cells (Figure 6, C and D). In addition, knock down of Prdx2 from murine myeloid 32D cells decreased cell proliferation, and increased apoptotic cells and alteration of apoptotic-related protein expression (Supplementary Figure S6, C-E). As U937 cells expressed reduced level of PRDX2 and transduction of DNMT3A-MT upregulated this protein, we knocked down PRDX2 from U937 cells transduced with WT- and mutant DNMT3A. In experiments using U937 cells transduced with R882H, PRDX2 knock down abolished the proliferative advantage and ATRA resistance of R882H-transduced U937 cells (Figure 6E). Moreover, knock down of PRDX2 terminated impairment of drug-induced apoptosis of R882H-transduced U937 cells and reduced the expression of MYC and CDK1 (Figure 6, F and G). Similar results were found using murine myeloid 32D cells (Supplementary Figure S7, A-C).
Figure 6.
PRDX2 blocks apoptosis of myeloid leukemia cells. (A) Silenced PRDX2 using two independent shRNA and scrambled (shLuc) K562 cells were treated with 300 nM ABT-263 or without drug for 72 hours, and cell proliferation was measured by trypan blue exclusion method. (B) Colony formation ability was assayed in Methocult medium after being stably silenced of PRDX2 in K562 cells. After 7 days, colonies were photographed and counted manually; original magnification: ×100. (C) PRDX2-silenced and scrambled K562 cells treated with 300 nM ABT-263 for 72 hours. Apoptosis cells were analyzed using Annexin V and PI staining by flow cytometric analysis. Representative flow cytometry analysis (left panel) and the % of Annexin V and PI-positive cells are shown (right panel). (D) Immunoblot data showing the PRDX2 silenced efficiency in K562 cells and protein expression of P-MYC and MYC after knock down of PRDX2 in K562 cells. β-Actin was used as a control for equal loading. (E) Proliferation of U937 cells transduced with WT- or R882H and knocked down of PRDX2 from transduced cells. (F) Apoptosis analysis of R882H-expressed U937 cells knocked down with shLuc or shPRDX2 and treatment of 300 nM ABT-263 for 72 hours. (G) Immunoblot data showing the PRDX2 silenced efficiency in U937 cells transduced with R882H and protein expression of MYC and CDK1. β-Actin was used as a control for equal loading. All studies were repeated at least once. The mean ± S.D. of at least two replicates was plotted. *P < .05; **P < .03; ***P < .01.
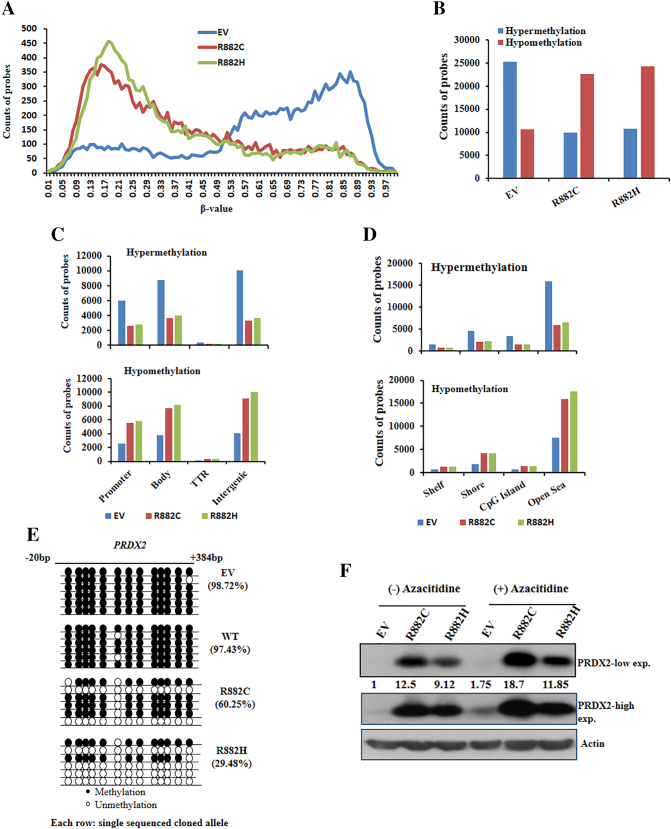
DNMT3A-R882 Mutants Induce Modifications of Genomic Methylation Patterns
We next analyzed DNA methylation status of transduced U937 cells to assess whether DNMT3A-R882H/C mutants altered gene expression profiles due to their changes of methyltransferase activity. Indeed, both DNA hypomethylation and hypermethylation features were observed in the specific region throughout the whole genome (Figure 7A). Overall, R882H/C mutations were more hypomethylated compared to control (Figure 7B). Also, the changes in hypo- and hypermethylation patterns were seen in the context of gene structure, namely, promoter, gene body, the transcriptional termination region, and the intergenic region. We found that R882H/C mutations were more hypomethylated in the intergenic and gene body regions, whereas control cells were hypermethylated in those regions (Figure 7C). We then examined the methylation patterns in four regions defined by the distance from CpG islands [23] such as CpG islands, Shore, Shelf, and Open Sea regions. Most of the hypo- and hypermethylation patterns were identified in the Open Sea region (Figure 7D). In the context of gene methylation patterns, we found that PRDX2 gene was differentially methylated in both gene body and promoter regions (Supplementary Figure S8) of DNMT3A-mutant U937 cells. However, there was no major change of methylation patterns of some differentially expressed genes (GSPT1, API5, CASP4, and CDKN1A) in EV, WT, and R882C/H mutants U937 cells (Supplementary Figure S9). Gene-targeted bisulfite sequencing experiment revealed that PRDX2 was hypomethylated in the gene body-associated CpG islands of R882H/C-transduced U937 cells relative to WT- and EV-transduced U937 cells (Figure 7E), suggesting upregulation of PRDX2 in mutant cells due to either promoter or gene body hypomethylation. Moreover, we found that treatment of demethylating agent azacitidine induced PRDX2 expression, confirming epigenetic upregulation of PRDX2 in DNMT3A-MTs cells (Figure 7F).
Figure 7.
DNMT3A-R882 mutants induce modifications of genomic methylation patterns and proposed mechanism of impairment of apoptosis in myeloid cells. (A) Distribution of genomic methylation patterns (β-value) in whole genome of U937 cells transduced with EV control, R882C, and R882H is shown. (B) Hypo- and hypermethylation probes counts obtained from U937 cells transduced with control and mutant DNMT3A shown in bar diagram. β-value <0.25 and >0.75 considered as hypomethylation and hypermethylation peaks, respectively. (C) The total hypermethylation and hypomethylation probes counted in each region defined by genomic structure shown in bar graph. (D) Methylation patterns in four regions defined by the distance from CpG islands, such as CpG islands, Shore, Shelf, and Open Sea region, are shown. (E) Validation of CpG methylation of PRDX2 gene by genomic bisulfite methylation sequencing showing PRDX2 gene was hypomethylated in DNMT3A mutant U937 cells. (F) Immunoblot data showing the PRDX2 expression induced by the treatment of 10 μM AZACITIDINE (Sigma) for 72 hours in U937 cells transduced with EV and DNMT3A-R882H/C. β-Actin was used as a control for equal loading. The number below the lanes indicates relative band intensity normalized to actin.
Discussion
DNA methylation is a basic epigenetics mechanism that is involved in gene expression, chromatin structure remodeling, and gene stability [24]. Among the leukemia-associated epigenetic regulators, recent findings suggested that DNMT3A mutations play a crucial role in clonal and malignant hematopoiesis [25], [26]. Despite tremendous progress in DNMT3A-related studies, understanding the molecular mechanism of DNMT3A mutation in AML still remains to be explored. In the present study, we demonstrated that DNMT3A-R882H/C mutations resulted in the impairment of apoptosis through aberrant expression of PRDX2 which was epigenetically upregulated in mutated cells.
Cell differentiation may be associated with apoptosis; induction of differentiation and apoptosis are potential approach in the treatment of AML. During apoptosis, H2A.X is phosphorylated on serine residue 139 in the nucleus and regulates caspase-3 activity which induces DNA fragmentation [27], [28]. We found that DNMT3A-R882H/C mutations deregulated the protein expression of γH2A.X in myeloid cells. In addition, with the treatment of ATRA or ABT-263 in transduced U937 cells, both caspase-3 activation and phosphorylation of H2A.X were enhanced. ABT-263 is a second-generation potent bioavailable apoptosis-inducing drug, which binds to the antiapoptotic family members BCL-2 and BCL-XL with high affinity [29]. ATRA-induced apoptosis in myeloid cells is a consequence of destabilization of antiapoptotic protein BCL-2 [30]. Previous study demonstrated that DNMT3A-R882H was more interacted with CDK1 compared to WT-protein and increased activity and stability of CDK1 which regulated cell cycle progression [13]. We observed that with the treatment of ATRA, both CDK1 and BCL-2 were downregulated, whereas WT-DNMT3A–transduced U937 cells expressed less amount of those proteins which facilitated apoptosis compared to mutant-DNMT3A cells. Furthermore, mutant-DNMT3A–expressed cells impaired the activation of cleaved poly (ADP-ribose) polymerase (PARP)-1 in the presence of ATRA or ABT-263 which indicated the chemoresistant nature of this mutant. Our finding is consistent with the recent report that DNMT3A-R882 mutation promoted chemoresistance and relapse through impaired DNA-damage sensing [31].
The impairment of ATRA-induced differentiation by DNMT3A-mutanted HL-60 human myeloid leukemia cells has been described [9]; however, how DNMT3A controls growth arrest and apoptosis during ATRA-induced differentiation in myeloid cells was not reported. Diallyl trisulfide–induced apoptosis induction of U937 cells through generation of ROS has been reported [32]. ATRA-induced differentiation and apoptosis of HL-60 cells were correlated to PRDX2 expression with ROS accumulation [33]. Previous study demonstrated that HL-60 cells are deficient of PRDX2 which inhibit apoptosis by locally scavenging ROS [33]. We found that both U937 and HL-60 cells expressed lower PRDX2 levels compared to other leukemia cells. We showed that overexpression of DNMT3A-mutants in U937 or HL-60 cells upregulated both mRNA and protein levels of PRDX2, which was correlated with accumulation of ROS. Upregulation of an antioxidant protein PRDX2 in DNMT3A mutant cells impaired apoptosis, which was augmented by pretreatment with TBHP, an oxidizing agent; consequently the attenuation of cell proliferation. In addition, treatment of TBHP overrode the effects of mutant DNMT3A on the activation of DNA damage and apoptosis signaling in DNMT3A mutant cells, suggesting the connection between DNMT3A mutant and antioxidant defense by PRDX2. Immunoprecipitation studies also revealed that PRDX2 protein was interacted with both WT- and mutants-DNMT3A. However, overexpression of R882C/H, but not WT-DNMT3A, induces higher level of PRDX2 in the U937 or HL-60 cells, suggesting the level of PRDX2 becomes dependent on the level of DNMT3A-R882C/H mutant. Moreover, we showed that knock down of PRDX2 decreased cell proliferation and stimulated apoptosis in DNMT3A mutant cells similar to WT- cells. We found that MYC expression was deregulated in DNMT3A mutant cells, and this was attenuated with either the treatment of TBHP or knockdown of PRDX2. Relation between MYC expression and regulation of PRDX2 was reported [34]. The transcription factor MYC directly binds to the promoter region of PRDX2 which regulates PRDX2 expression [34]. The metabolism of oxygen and production of ROS have been implicated in diverse physiological processes including cancer. Previously, it was known that lower ROS levels in cancer stem cells are associated with increased expression of free radical scavenging systems, which may contribute to tumor radioresistance [35]. It was also found that low ROS concentration was critical for self-renewal activity of hematopoietic stem cells [36]. For the first time, we found that knock down of PRDX2 enhanced PMA-induced megakaryocytic differentiation and augmented drug-induced apoptosis in K562 cells. It has been reported that the accumulation of intracellular ROS enhanced megakaryocytic differentiation induced by PMA in the K562cells [37]. Previous study demonstrated that downregulation of PRDX1 and PRDX2 inhibited cell growth and induced apoptosis of lymphoma cells [38]. PRDX2 regulated oxidative and metabolic stress, and oncogenic role of this protein was reported in several solid cancers [39], [40]. However, very limited data are known about this protein in leukemia and other myeloid neoplasms. A more recent study demonstrated that CDK2-PRDX2 axis blocked AML differentiation [41]. In contrast, others described PRDX2 as an epigenetically silenced-tumor suppressor gene in AML [42]. Our data demonstrated the specific role of DNMT3A-mutant in ROS-mediated apoptosis by PRDX2 pathway in myeloid cells of which the underlying mechanism warrants further investigation.
From gene expression microarray and DNA methylation studies, we observed that DNMT3A-R882 mutants had both DNA methylation-dependent and -independent effects and exhibited distinct mechanism for leukemogenesis. Also there was no direct correlation between methylation and differential gene expression in DNMT3A-R882 mutants U937 cells. Many cell cycle–promoting and apoptosis-inhibiting genes were upregulated in R882H-mutant cells, however; there was no difference in methylation status of those genes between WT- and mutant cells. The lack of correlation between methylation patterns and differential gene expression in Dnmt3A-null HSCs had been reported previously [11]. Previous epigenomic analysis of chronic lymphocytic leukemia cells showed that DNA hypomethylation occurred in the gene body and correlated with functional and clinical implications of leukemogenesis [43]. We demonstrated that exogenous expression of R882 mutants caused upregulation of antioxidant protein PRDX2 with accompanying global hypomethylation and specifically hypomethylation of promoter region or the gene body-associated CpG islands in U937 cells.
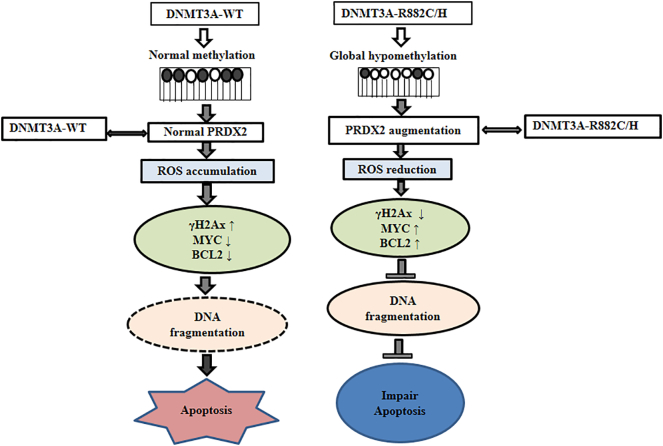
Taken together, our in vitro cell line model provided evidences that DNMT3A-R882 mutants not only impaired differentiation but also diminished apoptosis induction by disturbing the DNA methylation program and deregulation of cell-cycle and apoptosis-regulating genes. Importantly, DNMT3A-R882H/C mutation-induced hypomethylation augmented PRDX2 expression which reduced ROS accumulation in cells and might decrease apoptosis in part by increasing antioxidant capacity of cells (Figure 8). Pharmacological intervention targeting PRDX2 may be a useful approach to increase drug sensitivity and apoptosis as a consequence of reduced cell proliferation in myeloid cells harboring DNMT3A-R882 mutations.
Figure 8.
The proposed mechanism of impairment of apoptosis by DNMT3A-R882C/H mutant in myeloid cells. DNMT3A-R882 mutation-induced hypomethylation increased PRDX2 expression which reduced ROS accumulation in cells decreasing apoptosis by increasing antioxidant capacity of cells.
The following are the supplementary data related to this article.
List of upregulated genes (>1.5-fold) in either R882C or R882H compared to EV and DNMT3A-WT, respectively.
List of downregulated genes (<0.6-fold) in either R882C or R882H compared to EV and DNMT3A-WT, respectively.
List of upregulated (>1.5-fold) and downregulated (<0.6-fold) genes in R882H cells compared to both EV and DNMT3A-expressing U937 cells.
List of genes matched with Hematopoietic Fingerprint Database [19].
Supplementary material
Funding Sources
This work was supported by grant from the Ministry of Science and Technology, Taiwan (MOST 103-2321-B-182-015), grant from the Ministry of Health and Welfare, Taiwan (MOHW103-TD-B-111-09), grant from Mackay Memorial Hospital, Taipei, Taiwan (MMH-E-105-09), and grants from Chang Gung Memorial Hospital, Linkou, Taiwan (OMRPG380031 and OMRPG3C0021).
Conflict of Interest
The authors declare no interest conflict of this study.
Acknowledgements
The authors would like to thank APExBIO (an Apoptosis and Epigenetics Company) for providing us the ABT-263 drug for apoptosis analyses.
References
- 1.Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood. 2016;127:42–52. doi: 10.1182/blood-2015-07-604512. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA. DNA methylation and cancer. Oncogene. 2002;21:5358–5360. doi: 10.1038/sj.onc.1205597. [DOI] [PubMed] [Google Scholar]
- 3.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 6.Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- 7.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 8.Thol F, Damm F, Ludeking A, Winschel C, Wagner K, Morgan M, Yun H, Gohring G, Schlegelberger B, Hoelzer D. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 9.Koya J, Kataoka K, Sato T, Bando M, Kato Y, Tsuruta-Kishino T, Kobayashi H, Narukawa K, Miyoshi H, Shirahige K. DNMT3A R882 mutants interact with polycomb proteins to block haematopoietic stem and leukaemic cell differentiation. Nat Commun. 2016;7 doi: 10.1038/ncomms10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R, Wang P, Parton T, Zhou Y, Chrysovergis K, Rockowitz S, Chen WY, Abdel-Wahab O, Wade PA, Zheng D. Epigenetic perturbations by Arg882-mutated DNMT3A potentiate aberrant stem cell gene-expression program and acute leukemia development. Cancer Cell. 2016;30:92–107. doi: 10.1016/j.ccell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Wang YY, Dai YJ, Zhang W, Zhang WN, Xiong SM, Gu ZH, Wang KK, Zeng R, Chen Z. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc Natl Acad Sci U S A. 2014;111:2620–2625. doi: 10.1073/pnas.1400150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai SC, Shih LY, Liang ST, Huang YJ, Kuo MC, Huang CF, Shih YS, Lin TH, Chiu MC, Liang DC. Biological activities of RUNX1 mutants predict secondary acute leukemia transformation from chronic myelomonocytic leukemia and myelodysplastic syndromes. Clin Can Res. 2015;21:3541–3551. doi: 10.1158/1078-0432.CCR-14-2203. [DOI] [PubMed] [Google Scholar]
- 19.Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu JF, Yen TH, Chen Y, Huang YJ, Hsu CL, Liang DC, Shih LY. Involvement of Gpr125 in the myeloid sarcoma formation induced by cooperating MLL/AF10(OM-LZ) and oncogenic KRAS in a mouse bone marrow transplantation model. Int J Cancer. 2013;133:1792–1802. doi: 10.1002/ijc.28195. [DOI] [PubMed] [Google Scholar]
- 21.Wasim M, Carlet M, Mansha M, Greil R, Ploner C, Trockenbacher A, Rainer J, Kofler R. PLZF/ZBTB16, a glucocorticoid response gene in acute lymphoblastic leukemia, interferes with glucocorticoid-induced apoptosis. J Steroid Biochem Mol Biol. 2010;120:218–227. doi: 10.1016/j.jsbmb.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benyoucef A, Palii CG, Wang C, Porter CJ, Chu A, Dai F, Tremblay V, Rakopoulos P, Singh K, Huang S. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016;30:508–521. doi: 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 24.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 25.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem. 2000;275:9390–9395. doi: 10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]
- 28.Lu C, Zhu F, Cho YY, Tang F, Zykova T, Ma WY, Bode AM, Dong Z. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol Cell. 2006;23:121–132. doi: 10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Chen Y, Wan J, Liu X, Yu C, Li W. ABT-263 enhances sorafenib-induced apoptosis associated with Akt activity and the expression of Bax and p21((CIP1/WAF1)) in human cancer cells. Br J Pharmacol. 2014;171:3182–3195. doi: 10.1111/bph.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otake Y, Sengupta TK, Bandyopadhyay S, Spicer EK, Fernandes DJ. Retinoid-induced apoptosis in HL-60 cells is associated with nucleolin down-regulation and destabilization of Bcl-2 mRNA. Mol Pharmacol. 2005;67:319–326. doi: 10.1124/mol.104.006080. [DOI] [PubMed] [Google Scholar]
- 31.Guryanova OA, Shank K, Spitzer B, Luciani L, Koche RP, Garrett-Bakelman FE, Ganzel C, Durham BH, Mohanty A, Hoermann G. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22:1488–1495. doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi YH, Park HS. Apoptosis induction of U937 human leukemia cells by diallyl trisulfide induces through generation of reactive oxygen species. J Biomed Sci. 2012;19:50. doi: 10.1186/1423-0127-19-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chikamori K, Hill JE, Grabowski DR, Zarkhin E, Grozav AG, Vaziri SA, Wang J, Gudkov AV, Rybicki LR, Bukowski RM. Downregulation of topoisomerase IIbeta in myeloid leukemia cell lines leads to activation of apoptosis following all-trans retinoic acid-induced differentiation/growth arrest. Leukemia. 2006;20:1809–1818. doi: 10.1038/sj.leu.2404351. [DOI] [PubMed] [Google Scholar]
- 34.Wu F, Tian F, Zeng W, Liu X, Fan J, Lin Y, Zhang Y. Role of peroxiredoxin2 downregulation in recurrent miscarriage through regulation of trophoblast proliferation and apoptosis. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 37.Ojima Y, Duncan MT, Nurhayati RW, Taya M, Miller WM. Synergistic effect of hydrogen peroxide on polyploidization during the megakaryocytic differentiation of K562 leukemia cells by PMA. Exp Cell Res. 2013;319:2205–2215. doi: 10.1016/j.yexcr.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trzeciecka A, Klossowski S, Bajor M, Zagozdzon R, Gaj P, Muchowicz A, Malinowska A, Czerwoniec A, Barankiewicz J, Domagala A. Dimeric peroxiredoxins are druggable targets in human Burkitt lymphoma. Oncotarget. 2016;7:1717–1731. doi: 10.18632/oncotarget.6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stresing V, Baltziskueta E, Rubio N, Blanco J, Arriba MC, Valls J, Janier M, Clezardin P, Sanz-Pamplona R, Nieva C. Peroxiredoxin 2 specifically regulates the oxidative and metabolic stress response of human metastatic breast cancer cells in lungs. Oncogene. 2013;32:724–735. doi: 10.1038/onc.2012.93. [DOI] [PubMed] [Google Scholar]
- 40.Kim H, Lee TH, Park ES, Suh JM, Park SJ, Chung HK, Kwon OY, Kim YK, Ro HK, Shong M. Role of peroxiredoxins in regulating intracellular hydrogen peroxide and hydrogen peroxide-induced apoptosis in thyroid cells. J Biol Chem. 2000;275:18266–18270. doi: 10.1074/jbc.275.24.18266. [DOI] [PubMed] [Google Scholar]
- 41.Ying M, Shao X, Jing H, Liu Y, Qi X, Cao J, Chen Y, Xiang S, Song H, Hu R. Ubiquitin-dependent degradation of CDK2 drives the therapeutic differentiation of AML by targeting PRDX2. Blood. 2018;131:2698–2711. doi: 10.1182/blood-2017-10-813139. [DOI] [PubMed] [Google Scholar]
- 42.Agrawal-Singh S, Isken F, Agelopoulos K, Klein HU, Thoennissen NH, Koehler G, Hascher A, Baumer N, Berdel WE, Thiede C. Genome-wide analysis of histone H3 acetylation patterns in AML identifies PRDX2 as an epigenetically silenced tumor suppressor gene. Blood. 2012;119:2346–2357. doi: 10.1182/blood-2011-06-358705. [DOI] [PubMed] [Google Scholar]
- 43.Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of upregulated genes (>1.5-fold) in either R882C or R882H compared to EV and DNMT3A-WT, respectively.
List of downregulated genes (<0.6-fold) in either R882C or R882H compared to EV and DNMT3A-WT, respectively.
List of upregulated (>1.5-fold) and downregulated (<0.6-fold) genes in R882H cells compared to both EV and DNMT3A-expressing U937 cells.
List of genes matched with Hematopoietic Fingerprint Database [19].
Supplementary material