Abstract
Background and Aims
Silicon has been proven to exert beneficial effects on plant growth and stress tolerance, and silicon accumulation varies among different plant species. Cucumber (Cucumis sativus) is a widely used dicot model for silicon accumulation, but little is known about the molecular mechanism of its silicon uptake. Previously, we isolated and characterized CsLsi1, a silicon influx transporter gene from cucumber. In this study, we cloned a putative silicon efflux transporter gene, CsLsi2, from cucumber and investigated its role in silicon uptake.
Methods
The expression pattern, transport activity, and subcellular and cellular localizations of CsLsi2 were investigated. The transport activity of CsLsi2 was determined in Xenopus laevis oocytes. The subcelluar and cellular localizations were conducted by transient expression of fused 35S::CsLsi2-eGFP in onion epidermal cells and expression of ProCsLsi2::CsLsi2-mGFP in cucumber, respectively.
Key Results
CsLsi2 was mainly expressed in the roots. Expression of CsLsi2-eGFP fusion sequence in onion epidermis cells showed that CsLsi2 was localized at the plasma membrane. Transient expression in Xenopus laevis oocytes showed that CsLsi2 demonstrated efflux but no influx transport activity for silicon, and the transport was energy-dependent. Expression of CsLsi2-mGFP under its own promoter revealed that CsLsi2 was mainly expressed on endodermal cells, showing no polar distribution. In combination with our previous work on CsLsi1, a model for silicon uptake in cucumber roots is proposed.
Conclusion
The results suggest that CsLsi2 is a silicon efflux transporter gene in cucumber. The coordination of CsLsi1 and CsLsi2 mediates silicon uptake in cucumber roots. This study may help us understand the molecular mechanism for silicon uptake in cucumber, one of the few dicots with a relatively high capacity for silicon accumulation.
Keywords: Cucumber (Cucumis sativus), silicon uptake, efflux transporter
INTRODUCTION
Silicon is the second most prevalent element in the Earth’s crust (Epstein, 1999). Because silicon is ubiquitous, it is difficult to verify its essentiality for higher plants. Therefore, silicon has not been considered a generally essential element for higher plants. Despite this, many studies have demonstrated that silicon is beneficial for plant growth and development, especially under adverse environmental conditions (Liang et al., 2015; Debona et al., 2017).
All plants grown in soil contain silicon in their tissues, but silicon accumulation in different plants varies considerably, with the concentration ranging from 0.1 % to 10 % in dry weight (Epstein, 1999). Lycophytes and early-diverging ferns accumulate more silicon than the later-evolving gymnosperms (Trembath-Reichert et al., 2015). In Angiospermae, monocots usually accumulate more silicon than dicots (Hodson et al., 2005). The distinct accumulation of silicon in different plants has been attributed to the difference in silicon uptake ability of roots (Takahashi et al., 1990; Mitani and Ma, 2005).
Silicon is generally absorbed in the form of monosilicic acid with a range from 0.1 to 0.6 mm in soil solution (Gunnarsson and Arnórsson, 2000; Mitani et al., 2005). The silicon uptake system in higher plants was first clarified in rice (Oryza sativus), a high silicon accumulator (Ma et al., 2006, 2007). In rice, OsLsi1 and OsLsi2 are the silicon influx transporter and efflux transporter, respectively, and they mediate silicon transport into and out of the cell across the plasma membrane (Ma et al., 2006, 2007). OsLsi1 and OsLsi2 coordinate together to mediate the radial transport of silicon into the xylem in the root (Ma and Yamaji, 2015). Homologous genes of OsLsi1 and/or OsLsi2 have also been functionally characterized in other plants, such as barley (Hordeum vulgare) (Chiba et al., 2009; Mitani et al., 2009a), maize (Zea mays) Mitani et al., 2009a, b), pumpkin (Cucurbita moschata) (Mitani et al., 2011; Mitani-Ueno et al., 2011), wheat (Triticum aestivum) (Montpetit et al., 2012), soybean (Glycine max) (Deshmukh et al., 2013) and cucumber (Cucumis sativus) (Sun et al., 2017).
Interestingly, Lsi1s and Lsi2s from different plant species have been localized at different cells of root tissues, despite exhibiting similar functions in mediating silicon transport across species. OsLsi1 is localized at the distal side in the exodermal and endodermal cells in rice roots (Ma et al., 2006, 2007). In maize, ZmLsi1 is localized on the distal side of root epidermal and hypodermal cells in seminal and crown roots, and it is also localized in cortical cells in lateral roots (Mitani et al., 2009b). In barley, HvLsi1 is localized on the distal side of epidermal and cortical cells in seminal roots, and the distal side of hypodermal cells in lateral roots (Chiba et al., 2009). Lsi2s in these plants also have different localizations. In rice, OsLsi2 is localized on the proximal side in both exodermal and endodermal cells in the root (Ma et al., 2006, 2007), whereas in maize and barley, the Lsi2s were only localized in endodermal cells, showing no polar distribution (Mitani et al., 2009a). The difference in cellular localizations of silicon transporters in different plants may be associated with their silicon uptake ability. Using a mathematical simulation model, Sakurai et al. (2015, 2017) concluded that the cellular localization of OsLsi1 and OsLsi2 to the exodermis and endodermis with polar distribution is the most efficient with regard to silicon uptake, making rice a typical silicon accumulator. Therefore, the silicon transporters in different plants need to be experimentally identified and characterized.
Cucumber is an intermediate silicon-accumulating plant and one of the high silicon-accumulating vegetable crops (Mitani and Ma, 2005). Its silicon uptake is lower than that of rice, but higher than that of tomato (Solanum lycopersicum) and faba bean (Vicia faba) (Liang et al., 2005; Mitani and Ma, 2005; Nikolic et al., 2007). Although previous physiological experiments have suggested that silicon uptake in cucumber roots is an active process and mediated by transporter(s) (Liang et al., 2005; Nikolic et al., 2007), little is known about the molecular mechanism involved. In our previous work, we have isolated and characterized CsLsi1, a silicon influx transporter gene in cucumber (Sun et al., 2017). In this study, we identified and characterized a silicon efflux transporter gene, CsLsi2. The model for silicon uptake in cucumber roots is also proposed and discussed.
MATERIALS AND METHODS
Plant materials
Cucumber (‘Mch-4’) seeds were provided by the cucumber research team at Northwest A&F University. Seed sterilization and seedling preparation were conducted as described previously (Sun et al., 2017). The plants were grown in a growth chamber (Southeast-RDN, Ningbo, China) with a 14-h photoperiod (26 °C) and 10-h darkness (18 °C) cycle. Seedlings at the two-leaf stage were transferred to 12-litre plastic boxes filled with diluted Hoagland’s solution (Hoagland and Arnon, 1950) containing 1.25 mm KNO3, 1.25 mm Ca(NO3)2, 0.5 mm MgSO4, 0.25 mm KH2PO4, 46.1 μm H3BO3, 0.24 μm (NH4)6Mo7O24, 9.1 μm MnCl2, 0.76 μm ZnSO4, 0.32 μm CuSO4 and 71 μm FeNa-EDTA. The plants were cultured in a glasshouse at the campus under natural sunlight. The temperature was set to 26 °C/18 °C (day/night). The nutrient solution pH was adjusted to 6.0 daily with diluted HCl or NaOH, and the solution was renewed every 5 d.
Isolation of CsLsi2
Seven days after being transplanted into nutrient solution, the cucumber roots were sampled, immediately frozen in liquid N2 and stored at −80 °C until use. Total RNA was extracted from root samples with a plant RNA extraction kit (Omega, Madison, WI, USA). The first strand of cDNA was synthesized with 1 µg of total RNA using reverse transcriptase (PrimeScript II 1st Strand cDNA Synthesis Kit, Takara, Kusatsu, Japan) with supplied oligo dT primers. The genomic DNA in cucumber roots was extracted using the sodium dodecyl sulphate (SDS) method described by Dellaporta et al. (1983). To identify the cucumber Lsi2 homologue from the cucumber genome, we used the coding sequence (CDS) of OsLsi2 (GenBank accession number AB222273.1) in a BLAST search against the cucumber genome and identified its homologous gene (GenBank accession number XM_004140721.2). We named this putative Lsi2 in cucumber as CsLsi2 in this paper. The CDS and promoter region (2000 bp) of CsLsi2 were isolated using TKs Gflex polymerase (Takara) and primers listed in Supplementary Data Table S2, with cDNA and genomic DNA as the template, respectively. We have submitted the promoter sequence to GenBank under accession number MG888662.
The nucleotide and deduced amino acid sequences were analysed using DNAMAN v.6 (Lynnon Biosoft, San Ramon, CA, USA), and the phylogenetic tree was constructed using the neighbor-joining algorithm with MEGA v.6 (Tamura et al., 2013).
Expression analysis of CsLsi2
To investigate the tissue expression characteristics of CsLsi2 and the expression response to exogenous silicon, cucumber seedlings grown in diluted Hoagland nutrient solution for 7 d were treated with or without 1 mm sodium silicate (pH 6.0). After 6 h of treatment, the lamina and petiole of newly expanded leaf, root and stem were sampled. Total RNA and first-strand cDNAs were prepared as described above for the isolation of CsLsi2. Quantitative real-time PCR (qPCR) for measuring CsLsi2 expression was performed on an ABI StepOne plus qPCR system (Applied Biosystems, Carlsbad, CA, USA) with SYBR Premix EX TaqTM (Takara). Ubiquitin extension protein was used as an internal control (Wan et al., 2010). The primers used are listed in Table S2.
Subcellular localization of CsLsi2
Both bioinformatic analysis and heterologous expression were used to investigate the subcellular localization of CsLsi2. The subcellular localization of CsLsi2 was first determined based on in silico analysis with the Plant-Ploc online service (Chou and Shen, 2010), and confirmed by introducing a fused CaMV 35S::CsLsi2-eGFP (enhanced green fluorescent protein) into onion epidermal cells. The CDS of CsLsi2 was amplified with primers containing SalI and BamHI restriction sites, and inserted into vector pTF486 with tagged eGFP sequence at the 3′ terminal end. The primers used are listed in Table S2.
Transient expression in onion epidermal cells was determined as described by Scott et al. (1999). Transient expression of empty vector (pTF486) was used as a positive control. Plasmolysis was conducted with 0.3 g mL−1 sugar solution before being observed. The fluorescent signal was examined with a confocal laser scanning microscope (IX83-FV1200, Olympus, Tokyo, Japan) at an excitation wavelength of 488 nm for GFP.
Transport activity analysis
We expressed CsLsi2 in Xenopus laevis oocytes to determine the transport activity of CsLsi2 for silicon. The CDS of CsLsi2 was inserted into vector pXßG-ev1 at the BglII site with the primers listed in Table S2. The construct was linearized with XbaI, and the complementary RNA (cRNA) with cap analogue was synthesized with Riboprobe System - T3 and Ribo m7G Cap Analog (both from Promega, Madison, WI, USA).
The isolation and culture of Xenopus laevis oocytes were performed as described previously (Sun et al., 2017). Germanium (Ge), an analogue of silicon, is used as a substrate for determination of silicon transport activity, as has been used in other studies (Nikolic et al., 2007; Mitani-Ueno et al., 2013). The influx transport activity of CsLsi2 was determined with a method described previously (Sun et al., 2017). The concentration of Ge was determined with inductively coupled plasma mass spectrometry (ICAP-Qc Thermofisher Scientific, Waltham, MA, USA).
To investigate the efflux transport activity of CsLsi2, the oocytes were first injected with 50 nL of 1 ng nL−1 of CsLsi2 cRNA or H2O (as a negative control) and cultured in modified Birth’s saline solution (MBS) at 18 °C for 24 h. Then 50 nL of 1.0 mm germanium oxide was injected into the same oocytes. These oocytes containing CsLsi2 cRNA and germanium oxide or containing H2O and germanium oxide were washed with MBS quickly to remove germanium oxide on the surface and cultured in fresh MBS at 18 °C for 30 min. The MBS culture medium was carefully collected for determination of Ge content. To determine whether the transport was energy-dependent, the oocytes injected with CsLsi2 cRNA were injected with germanium oxide as described above. These oocytes were then cultured in precooled MBS at 4 °C for 30 min, after which the medium was collected and determination of Ge content was conducted as above.
Cellular localization of CsLsi2 in cucumber roots
An expression vector containing ProCsLsi2::CsLsi2-mGFP (monomer GFP) cassette was transformed into cucumber (‘Mch-4’) to examine the cellular localization of CsLsi2 in roots. The transformation of cucumber was performed as described previously (Sun et al., 2017). Transgenic cucumber plants were genotyped by PCR with two pairs of primers (Table S2) overlapping by one intron (509 bp) of CsLsi2 with a Taqmix kit (Vazyme, Nanjing, China). The PCR products from T0 seedlings were sequenced by Shanghai Sangon (Shanghai, China) for further confirmation.
For analysis of CsLsi2 localization, the T1 generation of cucumber seedlings were prepared as described in Plant materials. To examine the cellular localization of CsLsi2-mGFP, the roots were embedded in the stem pith of rice-paper plant (Tetrapanax papyriferus) and cross-sectioned with a scalpel. Sectioned root samples were visualized under a fluorescence microscope (Olympus-BX51).
Statistics
The data were subjected to one-way ANOVA or t-test using SPSS software (SPSS Inc. 19.0). Differences were considered significant at P < 0.05.
RESULTS
A homologous Lsi2 exists in the cucumber genome
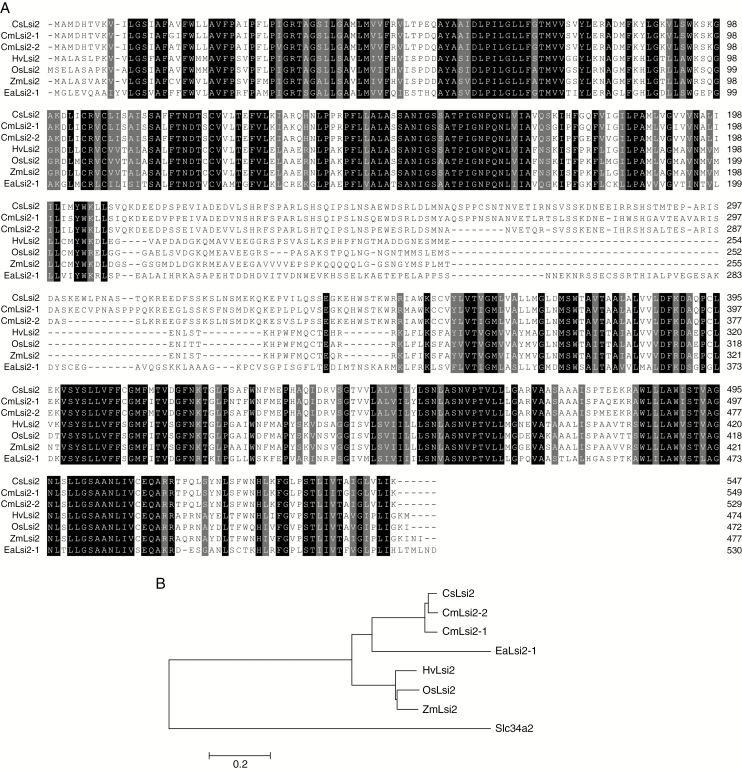
To examine whether a homologue of OsLsi2 exists in the cucumber genome, we performed a BLAST search with the OsLsi2 sequence. The results revealed that the cucumber genome also contains a copy of Lsi2 that exhibits high sequence identity to OsLsi2. This putative CsLsi2 is localized at chromosome 3, consisting of three exons and two introns. We next isolated CsLsi2 from the cucumber genotype ‘Mch-4’. Its complete CDS is 1644 bp long and shows 100 % identity with the predicted CDS of Lsi2 from cucumber ‘9930’ in GenBank (accession number XM_004140721.2). The deduced protein sequence of CsLsi2 consists of 547 amino acids, exhibiting 53.27 % and 89.95 % identity with rice OsLsi2 and pumpkin CmLsi2-2, respectively (Fig. 1A). Overall, there is a high sequence similarity among Lsi2s from different plants (Fig. 1A), whereas there is no obvious homology in Lsi2 sequences between rat and plants (Fig. S1). However, the transmembrane domains are similar (Fig. S2), which suggests that the Lsi2s from different plants and rat have conserved structure. Functional domain analysis showed that CsLsi2 shares 97.22 % and 97.78 % similarity in the predicted transmembrane regions with those of pumpkin CmLsi2-1 and CmLsi2-2, respectively, which suggests that CsLsi2 may have a similar function to that of CmLsi2s.
Fig. 1.
Protein sequence alignment and phylogenetic tree of CsLsi2 along with other identified silicon transporters. (A) Protein sequence alignment of Lsi2s from cucumber (Cucumis sativus) (Cs), pumpkin (Cucurbita moschata) (Cm), barley (Hordeum vulgare) (Hv), rice (Oryza sativa) (Os), maize (Zea mays) (Zm) and horsetail (Equisetum arvense) (Ea). (B) Phylogenetic tree of Lsi2s from different plant species and rat. Protein sequence alignment was performed with DNAMAN v.6; the phylogenetic tree was constructed using the neighbor-joining algorithm with MEGA v.6. The scale bar represents a substitution distance of 0.2. Gene IDs for each of the silicon transporter proteins are listed in Supplementary Data Table S1.
Phylogenetic analysis showed that CsLsi2 clustered with CmLsi2s from pumpkin, with the closest genetic relationship to CmLsi2-2 (Fig. 1B). In higher plants, Lsi2s from monocots including rice, maize and barely are closer to each other than those from dicot species (Fig. 1B), suggesting that the distinction of Lsi2s between dicots and monocots may be caused by evolutionary speciation.
CsLsi2 is mainly expressed in roots and is responsive to exogenous silicon
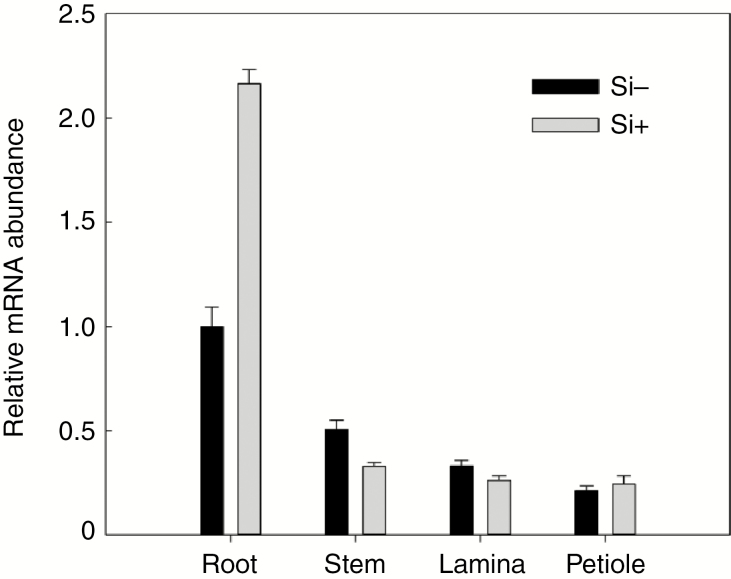
To examine the tissue expression pattern, we measured CsLsi2 transcript levels in cucumber seedlings in the absence of exogenous silicon. CsLsi2 transcripts were detected in the roots, stems, laminae and petioles of three-leaf cucumber seedlings, but with the highest level in roots (Fig. 2). Additionally, a CsLsi2 expression response to exogenous silicon was only observed in roots, where the transcript level doubled in the presence of applied silicon. This up-regulation of CsLsi2 in response to silicon was not observed in stems, laminae and petioles of the same seedlings (Fig. 2), suggesting a specific role of CsLsi2 in mediating silicon transport in cucumber roots.
Fig. 2.
Tissue expression pattern of CsLsi2 and its response to applied silicon. Roots, stems, laminae and petioles of new fully expanded leaves were sampled from cucumber seedlings with or without 1 mm exogenous silicon application for 6 h. Values are means ± s.d. of six replicates. The ubiquitin extension protein gene was used as an internal control.
CsLsi2 is a plasma membrane protein
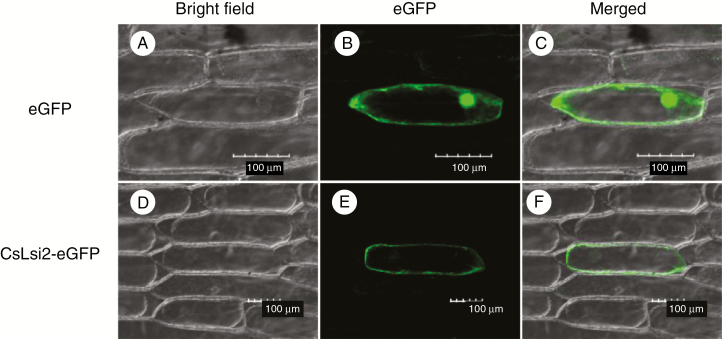
Bioinformatics analysis predicted that CsLsi2 was a plasma membrane protein like OsLsi2 (Ma et al., 2007). To confirm this prediction, we transiently expressed CsLsi2 fused with eGFP under the 35S promoter into onion epidermal cells. As a control, the empty vector containing only eGFP was also transformed into onion epidermal cells. A clear fluorescence signal of fused CsLsi2-eGFP protein was observed from the plasma membrane, while the green fluorescence from the control eGFP was detected at both the plasma membrane and nucleus (Fig. 3). These results suggested that CsLsi2 is a plasma membrane protein, increasing the likelihood that it is involved in mediating silicon transport.
Fig. 3.
Subcellular localization of CsLsi2 in onion epidermal cells. The constructs carrying 35S::CsLsi1-eGFP or 35S::eGFP were transferred into onion epidermal cells. (A–C) Cellular localizations of CsLsi2; eGFP transformation in D–F was used as a positive control. A and D show bright-field images; B and E show eGFP fluorescence; and C and F show merged images. Scale bars = 100 μm.
CsLsi2 is an energy-dependent silicon efflux transporter
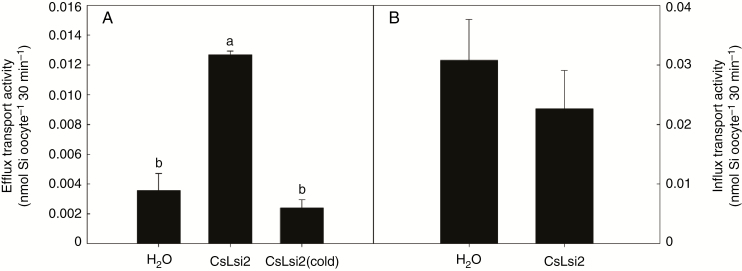
To determine whether CsLsi2 has transport activity for silicon, we transiently expressed CsLsi2 in Xenopus laevis oocytes. The oocytes were first injected with CsLsi2 cRNA or H2O (mock control), then injected with 1.0 mm Ge (an analogue of silicon). The culture medium of oocytes was used for determination of Ge concentration. If CsLsi2 has efflux activity for Ge, it will transport Ge out of oocytes, resulting in a higher Ge concentration in culture medium for oocytes injected with both CsLsi2 cRNA and Ge than those injected with H2O and Ge. As shown in Fig. 4A this was indeed the case, indicating that CsLsi2 has efflux transport activity for silicon. This result was generated from oocytes cultured at 18 °C, a temperature at which oocytes have high metabolic activity that could possibly provide energy for efflux transport. To further understand whether the efflux transport activity of CsLsi2 is energy-dependent, we performed similar tests with oocyte cultures at 4 °C, at which viability of oocytes would be maintained but metabolic activity was effectively inhibited. The result in Fig. 4A (cold) showed that there was no difference in Ge concentration in the culture medium of oocytes injected with CsLsi2 cRNA and Ge or oocytes injected with H2O and Ge, suggesting that silicon efflux mediated by CsLsi2 is an active process and requires energy.
Fig. 4.
Transport activity of CsLsi2 in Xenopus laevis oocytes. (A) Silicon efflux transport activity of CsLsi2. CsLsi2 cRNA and 1.0 mm Ge or just 1.0 mm Ge was injected into Xenopus oocytes. The oocytes were cultured in MBS, and the released Ge was measured to determine the transport activity. Cold (4 °C) treatment was applied to determine whether the efflux transport was energy-dependent. (B) Silicon influx transport activity of CsLsi2. Xenopus oocytes injected with CsLsi2 cRNA or water were cultured in MBS containing 1 mm Ge for 30 min, after which the absorbed Ge in Xenopus oocytes was measured. Values are means ± s.d. (n = 3). Different lower-case letters indicate a significant difference at P < 0.05.
We also examined whether CsLsi2 has influx activity for silicon as for CsLsi1 (Sun et al., 2017). Xenopus oocytes injected with CsLsi2 cRNA or water were cultured in MBS containing 1 mm Ge. If CsLsi2 has influx activity for silicon, the Ge concentration will be higher in oocytes injected with CsLsi2 cRNA than those injected with H2O only. We did not observe a significant difference in Ge concentration between the two groups of oocytes, indicating that CsLsi2 has no influx activity for silicon (Fig. 4B).
Tissue localization of CsLsi2 in cucumber roots
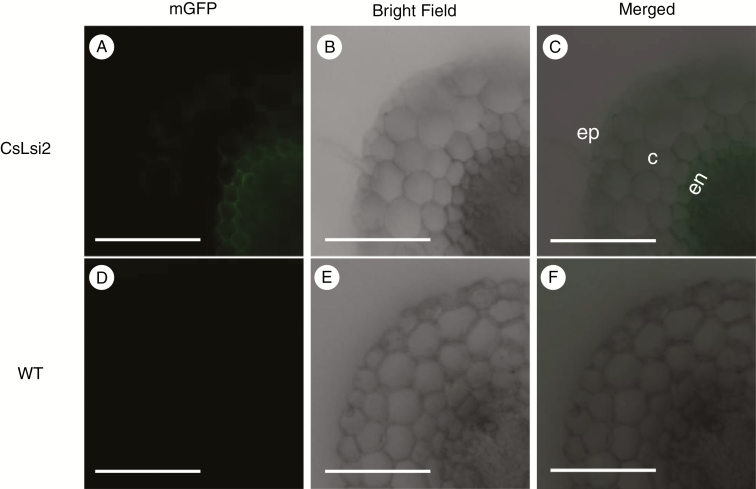
To better understand the role of CsLsi2 in radial silicon transport in the root, we generated transgenic cucumber plants carrying the ProCsLsi2::CsLsi2-mGFP cassette. When the roots of transgenic plants were cross-sectioned, fluorescence signal was observed on both sides of the endodermal cells without polarity (Fig. 5A–C). In contrast, no fluorescence signal was detected in the wild-type cucumber plants (Fig. 5D–F). These results suggest that CsLsi2 may function in silicon transport across the endodermis.
Fig. 5.
Cellular localization of CsLsi2 in cross-sections of transgenic cucumber root. (A–C) Localization of CsLsi2 in sectioned roots of transgenic cucumber expressing ProCsLsi2::CsLsi2-mGFP. Wild type (WT) was introduced as a control (D–F). c, cortex; ep, epidermis; en, endodermis; vc, vascular cylinder. Scale bars = 500 µm.
DISCUSSION
Silicon uptake systems have been explored in different plants, and silicon transporters Lsi1 and Lsi2 have been demonstrated to mediate radial transport of silicon in roots of plants such as rice (Ma et al., 2006, 2007), barely (Chiba et al., 2009; Mitani et al., 2009a), maize (Mitani et al., 2009a, b), wheat (Montpetit et al., 2012), pumpkin (Mitani et al., 2011; Mitani-Ueno et al., 2011) and soybean (Deshmukh et al., 2013). Silicon transporters have been intensively reported for monocots, probably because grass family members, like rice, can accumulate higher levels of silicon than dicots. Interestingly, although the silicon transporters from different species have conserved functional roles, different localizations of silicon transporters have been observed across different plant species. This distinct localization across plant species may be associated with their difference in silicon uptake capabilities. Although cucumber is a widely used model for silicon-accumulating dicots, studies on the molecular mechanism underlying silicon uptake have lagged behind those of monocot counterparts. Previously, we have identified a silicon influx transporter gene, CsLsi1 (Sun et al., 2017). Here we isolated and characterized a silicon efflux transporter gene, CsLsi2, and propose a model for radial transport of silicon in cucumber roots.
Sequence analysis showed that CsLsi2 and CmLsi2s from pumpkin, another cucurbitaceous plant, share very high similarity (97.22 % and 97.78 % with CmLsi2-1 and CmLsi2-2, respectively) in the predicted transmembrane regions (Fig. 1A; Fig. S2). Phylogenetic analysis also showed that CsLsi2 is closest to CmLsi2s among the analysed Lsi2s (Fig. 1B). These results suggest that CsLsi2 may have a similar function to CmLsi2s as a silicon efflux transporter. Phylogenetic analysis of Lsi2s in this study (Fig. 1B) and Lsi1s in our previous study (Sun et al., 2017) from different plants also showed that the group pattern of these two types of transporters coincides with plant taxonomy (Fig. 1B), which suggests that the orthologous silicon transporters such as Lsi1s and Lsi2s may evolve as plants evolve.
In this study, CsLsi2 was clearly observed at the plasma membrane via transient expression in onion epidermal cells (Fig. 3). This subcellular localization is consistent with those of characterized silicon transporters in other plant species (Ma and Yamaji, 2015). The result indicates that CsLsi2 may be involved in silicon transport across the plasma membrane. Transient expression of CsLsi2 in Xenopus laevis oocytes showed efflux transport activity for silicon, while no influx transport activity was observed. This clearly demonstrates that CsLsi2 functions as a silicon efflux transporter, which can transport silicon out of the cell. In this study, the efflux silicon transport activity of CsLsi2 was significantly decreased under cold conditions (Fig. 4). This result is consistent with that for OsLsi2 in rice, where efflux transport is an active process driven by the proton gradient across the plasma membrane (Ma et al., 2007). Under cold conditions, the active transport of nutrients would be reduced because of the decreased H+-ATPase activity of the plasma membrane (Lee et al., 2004). Therefore, our result suggests that CsLsi2-mediated silicon transport requires energy.
Experimentally identified Lsi2s remain very limited (Ma et al., 2007; Mitani et al., 2009a; Mitani-Ueno et al., 2011) and tissue expression patterns show some differences. For example, CmLsi2 in pumpkin is highly expressed in both roots and shoots (Mitani-Ueno et al., 2011), but Lsi2s in rice, barley and maize are mainly expressed in the roots (Ma et al., 2007; Mitani et al., 2009a). In this study, highest expression of CsLsi2 was in the root (Fig. 2), which is consistent with the observations in rice, barley and maize. The high expression of CsLsi2 in the roots implies that CsLsi2 may play a role in silicon uptake in cucumber.
Previous studies in rice, barley, maize and pumpkin have all demonstrated a down-regulation of Lsi2 expression by exogenous silicon (Ma et al., 2007; Mitani et al., 2009a; Yamaji and Ma, 2011). In this study, expression of CsLsi2 was increased by exogenous silicon in the roots, but it was not obviously changed in the stem, lamina or petiole (Fig. 2). These results imply that the effect of silicon on Lsi2 expression is dependent on plant species and tissues. Mitani-Ueno et al. (2016) found that expression of Lsi1 to exogenous silicon was closely related to a conserved sequence in the promoter region, but whether the response of Lsi2 is also associated with a certain promoter region remains to be investigated.
Previous studies have shown that Lsi2s in different plants demonstrate distinct cellular localizations in the roots (Ma and Yamaji, 2015). OsLsi2 in rice is localized at the exodermis and endodermis on the proximal side (Ma et al., 2007), and HvLsi2 in barely and ZmLsi2 in maize are localized only at the endodermis without polarity (Mitani et al., 2009a). In the present study, we found that CsLsi2 was localized at the plasma membrane of endodermal cells and showing no polarity, which is the same as for HvLsi2 and ZmLsi2. Given its role in efflux silicon transport (Fig. 4), CsLsi2 may function at the endodermis in the transport of silicon across endodermal cells into the stele for xylem loading.
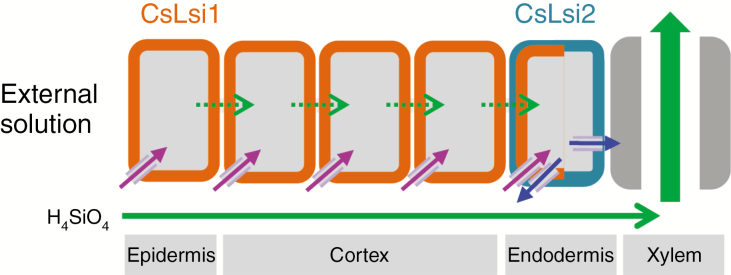
In higher plants, the silicon uptake system in roots has been well clarified in the monocotyledonous plants rice, barley and maize (Ma and Yamaji, 2015). For example, in rice roots, the silicon influx transporter OsLsi1 and efflux transporter OsLsi2 on the distal side and proximal side of the exodermis, respectively, mediate the movement of silicon into and out of these cells. Silicon then diffuses to near the endodermis, where OsLsi1 on the distal side transports silicon into these cells, and OsLsi2 on the proximal side mediates silicon transport into the stele (Ma et al., 2006, 2007). The molecular mechanism for silicon uptake in cucumber is still not well understood. In our previous work, we have identified CsLsi1 (a silicon influx transporter gene) in cucumber roots and found that it is expressed on the distal side of the endodermis and the cortical cells without polarity (Sun et al., 2017). Given the cellular localization and function of CsLsi2 in this study, we propose a silicon uptake model in cucumber roots (Fig. 6). In this model, there are two silicon transport pathways: an apoplasmic pathway and a cell-to-cell pathway. In the apoplasmic pathway, silicon moves from the external solution into the xylem through the apoplasmic free space by diffusion. Although the movement of water and nutrients in the apoplasmic space is usually blocked by Casparian bands at the endodermis and/or exodermis, this blockage is not complete. For example, Garcia et al. (1997) found that Na+ uptake is largely the consequence of Na+ leakage along the apoplasmic pathway to the xylem. Ranathunge et al. (2005) observed that the endodermis is permeable to ions in maize and rice roots. Therefore, it is possible that silicon can move from the external solution into the xylem through the apoplasmic pathway. In the cell-to-cell pathway, CsLsi1 mediates the transport of silicon into the epidermal or cortical cells from the external solution or the apoplasmic space. The absorbed silicon is then transported inward through the symplasmic pathway (via plasmodesmata) to the endodermal cells. CsLsi2 localized on the proximal side of the plasma membrane then transports silicon out of the endodermal cells into the stele for xylem loading.
Fig. 6.
Hypothetical model of silicon uptake in cucumber. The schematic diagram shows the putative silicon uptake process in cucumber roots. Purple and blue arrows represent the transporter-mediated silicon transport in root cells. The orange boxes represent the plasma membrane of epidermal and cortical cells where CsLsi1 functions as a silicon influx transporter, and the blue box represents the plasma membrane of endodermal cells where CsLsi2 functions as a silicon efflux transporter. The dotted and solid green arrows show the symplasmic (via plasmadesma) and apoplasmic flows, respectively, in the roots.
Similar to Lsi2s in barely and maize (Mitani et al., 2009a), CsLsi2 is localized on both sides of endodermal cells, without polarity (Fig. 5). Therefore, some silicon in endodermal cells may be transported back into the apoplasmic space among cortical cells (Fig. 6). In rice (a high silicon accumulator), due to the polar distribution of OsLsi2 (on the proximal side of endodermis), silicon in endodermal cells can be efficiently released into the stele (Ma et al., 2006, 2007). Using an in silico simulation model, Sakurai et al. (2015) found that the cellular localization of OsLsi1 and OsLsi2 at the exodermis and endodermis with a polar distribution is the most efficient with regard to silicon uptake. Therefore, the differences in polar localizations of Lsi2 in the roots between rice and cucumber, barely and maize may contribute to their differences in silicon accumulation: rice has a greater capability for silicon accumulation than cucumber, barley and maize, which are medium silicon accumulators.
In summary, our results suggest that CsLsi2 is a silicon efflux transporter in cucumber. Based on the functional characteristics of CsLsi1 and CsLsi2, we propose a silicon uptake model in cucumber roots, which may help us to understand the molecular mechanism for silicon uptake in this widely used dicot model for silicon accumulation.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Fig. S1. Protein sequence alignment of CsLsi2 along with other identified silicon transporters from plants and rat. Fig. S2. Predicted transmembrane domains of Lsi2s in plants and rat. Table S1. Gene ID and accessions for the silicon transporter proteins used in phylogenetic analysis. Table S2. Primers used in this study.
ACKNOWLEDGEMENTS
We thank Assoc. Prof. Huanwen Meng for providing the cucumber seeds and Dr Maki Katsuhara from Okayama University for providing the expression vector pXβG-ev1. We also thank Dr Yan Zhang from Northwest A&F University for advice on cucumber transformation. This work was supported by the National Natural Science Foundation of China (grant numbers 31471866, 31772290).
Conflict of Interest: none declared.
LITERATURE CITED
- Chiba Y, Mitani N, Yamaji N, Ma JF. 2009. HvLsi1 is a silicon influx transporter in barley. Plant Journal 57: 810–818. [DOI] [PubMed] [Google Scholar]
- Chou KC, Shen HB. 2010. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PloS ONE 5: e11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debona D, Rodrigues FA, Datnoff LE. 2017. Silicon’s role in abiotic and biotic plant stress. Annual Review of Phytopathology 55: 85–107. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. 1983. A plant DNAminipreparation: Version II. Plant Molecular Biology Reporter 1: 19–21. [Google Scholar]
- Deshmukh RK, Vivancos J, Guerin V, et al. 2013. Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Molecular Biology 83: 303–315. [DOI] [PubMed] [Google Scholar]
- Epstein E. 1999. Silicon. Annual Review of Plant Physiology and Plant Molecular Biology 50: 641–664. [DOI] [PubMed] [Google Scholar]
- Garcia A, Rizzo CA, Ud Din J, Bartos SL, Senadhira D, Flowers TJ, Yeo AR. 1997. Sodium and potassium transport to the xylem are inherited independently in rice, and the mechanism of sodium: potassium selectivity differs between rice and wheat. Plant, Cell and Environment 20: 1167–1174. [Google Scholar]
- Gunnarsson I, Arnórsson S. 2000. Amorphous silica solubility and the thermodynamic properties of H4SiO4 in the range of 0 °C to 350 °C at Psat.Geochimica et Cosmochimica Acta 64: 2295–2307. [Google Scholar]
- Hoagland DR, Arnon DI. 1950. The water-culture method for growing plants without soil. California Agricultural Experiment Station Circular 347: 1–32. [Google Scholar]
- Hodson MJ, White PJ, Mead A, Broadley MR. 2005. Phylogenetic variation in the silicon composition of plants. Annals of Botany 96: 1027–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Singh AP, Chung GC, Ahn SJ, Noh EK, Steudle E. 2004. Exposure of roots of cucumber (Cucumis sativus) to low temperature severely reduces root pressure, hydraulic conductivity and active transport of nutrients. Physiologia Plantarum 120: 413–420. [DOI] [PubMed] [Google Scholar]
- Liang Y, Si J, Romheld V. 2005. Silicon uptake and transport is an active process in Cucumis sativus. New Phytologist 167: 797–804. [DOI] [PubMed] [Google Scholar]
- Liang Y, Nikolic M, Bélanger R, Gong H, Song A. 2015. Chapter 5. Silicon-mediated tolerance to metal toxicity, pp. 83–122; Chapter 6. Silicon-mediated tolerance to salt stress, pp. 123–142; Chapter 7. Silicon-mediated tolerance to drought and low-temperature stress, pp. 143–160; Chapter 8. Silicon-mediated tolerance to other abiotic stresses, pp. 161–179. In: Silicon in agriculture. Springer: Dordrecht. [Google Scholar]
- Ma JF, Yamaji N. 2015. A cooperative system of silicon transport in plants. Trends in Plant Science 20: 435–442. [DOI] [PubMed] [Google Scholar]
- Ma JF, Tamai K, Yamaji N, et al. 2006. A silicon transporter in rice. Nature 440: 688–691. [DOI] [PubMed] [Google Scholar]
- Ma JF, Yamaji N, Mitani N, et al. 2007. An efflux transporter of silicon in rice. Nature 448: 209–212. [DOI] [PubMed] [Google Scholar]
- Mitani N, Ma JF. 2005. Uptake system of silicon in different plant species. Journal of Experimental Botany 56: 1255–1261. [DOI] [PubMed] [Google Scholar]
- Mitani N, Ma JF, Iwashita T. 2005. Identification of the silicon form in xylem sap of rice (Oryza sativa L.). Plant and Cell Physiology 46: 279–283. [DOI] [PubMed] [Google Scholar]
- Mitani N, Chiba Y, Yamaji N, Ma JF. 2009a Identification and characterization of maize and barley Lsi2-like silicon efflux transporters reveals a distinct silicon uptake system from that in rice. The Plant Cell 21: 2133–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani N, Yamaji N, Ma JF. 2009b Identification of maize silicon influx transporters. Plant and Cell Physiology 50: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani N, Yamaji N, Ago Y, Iwasaki K, Ma JF. 2011. Isolation and functional characterization of an influx silicon transporter in two pumpkin cultivars contrasting in silicon accumulation. Plant Journal 66: 231–240. [DOI] [PubMed] [Google Scholar]
- Mitani-Ueno N, Yamaji N, Ma JF. 2011. Silicon efflux transporters isolated from two pumpkin cultivars contrasting in Si uptake. Plant Signaling & Behavior 6: 991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani-Ueno N, Ogai H, Yamaji N, Ma JF. 2013. Physiological and molecular characterization of Si uptake in wild rice species. Physiologia Plantarum 151: 200–207. [DOI] [PubMed] [Google Scholar]
- Mitani-Ueno N, Yamaji N, Ma JF. 2016. High silicon accumulation in the shoot is required for down–regulating the expression of Si transporter genes in rice. Plant and Cell Physiology 57: 2510–2518. [DOI] [PubMed] [Google Scholar]
- Montpetit J, Vivancos J, Mitaniueno N, et al. 2012. Cloning, functional characterization and heterologous expression of TaLsi1, a wheat silicon transporter gene. Plant Molecular Biology 79: 35–46. [DOI] [PubMed] [Google Scholar]
- Nikolic M, Nikolic N, Liang Y, Kirkby EA, Romheld V. 2007. Germanium-68 as an adequate tracer for silicon transport in plants. characterization of silicon uptake in different crop species. Plant Physiology 143: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranathunge K, Steudle E, Lafitte R. 2005. Blockage of apoplastic bypass‐flow of water in rice roots by insoluble salt precipitates analogous to a Pfeffer cell. Plant, Cell and Environment 28: 121–133. [Google Scholar]
- Sakurai G, Satake A, Yamaji N, et al. 2015. In silico simulation modeling reveals the importance of the casparian strip for efficient silicon uptake in rice roots. Plant and Cell Physiology 56: 631–639. [DOI] [PubMed] [Google Scholar]
- Sakurai G, Yamaji N, Mitani-Ueno N, Yokozawa M, Ono K, Ma JF. 2017. A model of silicon dynamics in rice: an analysis of the investment efficiency of Si transporters. Frontiers in Plant Science 8: 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AC, Wyatt SE, Tsou P, Robertson D, Allen NS. 1999. Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125–1132. [DOI] [PubMed] [Google Scholar]
- Sun H, Guo J, Duan Y, Zhang T, Huo H, Gong H. 2017. Isolation and functional characterization of CsLsi1, a silicon transporter gene in Cucumis sativus. Physiologia Plantarum 159: 201–214. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Ma JF, Miyake Y. 1990. The possibility of silicon as an essential element for higher plants. Comments on Agricultural and Food Chemistry 2: 99–122. [Google Scholar]
- Trembath-Reichert E, Wilson JP, Mcglynn SE, Fischer WW. 2015. Four hundred million years of silica biomineralization in land plants. Proceedings of the National Academy of Sciences of the United States of America 112: 5449–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan HJ, Zhao ZG, Qian CT, Sui YH, Malik AA, Chen JF. 2010. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Analytical Biochemistry 399: 257–261. [DOI] [PubMed] [Google Scholar]
- Yamaji N, Ma JF. 2011. Further characterization of a rice silicon efflux transporter, Lsi2. Soil Science and Plant Nutrition 57: 259–264. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.