Abstract
Given the constantly improving cost and speed of genome sequencing, it is reasonable to expect that personal genomes will soon be known for many millions of humans. This stands in stark contrast with our limited ability to interpret the sequence variants which we find. Although it is, perhaps, easiest to interpret variants in coding regions, knowledge of functional impact is unknown for the vast majority of missense variants. While many computational approaches can predict the impact of coding variants, they are given a little weight in the current guidelines for interpreting clinical variants. Laboratory assays produce comparatively more trustworthy results, but until recently did not scale to the space of all possible mutations. The development of deep mutational scanning and other multiplexed assays of variant effect has now brought feasibility of this endeavour within view. Here, we review progress in this field over the last decade, break down the different approaches into their components, and compare methodological differences.
Keywords: Deep mutational scanning, MAVE, Variant effect, VUS, Variants of uncertain significance
Introduction
Linking genotype to phenotype is a very difficult problem. The parts of the human genome which we understand best are protein-coding genes, yet they only constitute a small fraction of the whole. Impacts of mutations in other functional elements such as splice sites, promoters, or regulatory sequences are more difficult to assay, not to mention the vast stretches of intergenic space. While one might expect a priori that any given intergenic variant is unlikely to bear functional significance, a large number of loci identified as correlated with diseases in genome-wide association studies (GWAS) are found within these regions (Maurano et al. 2012; Edwards et al. 2013). While many of these cases may stem from linkage disequilibrium to coding or splice-altering variants (Taşan et al. 2015), more functions yet unknown may lie hidden within this vast space. Even for protein-coding sequences, the problem is far from simple. Alleles with simple Mendelian behaviour are the exception rather than the rule. Most phenotypes are complex, i.e., they emerge through the interplay of many different genetic or environmental factors. Conversely, many genes are also pleiotropic, i.e., they are involved in more than one mechanism (Chesmore et al. 2018). As a result of this complexity, mutations found in different people may have different quantitative or qualitative effects—phenomena that are correspondingly termed variable expressivity and incomplete penetrance. Similarly, two different mutations within the same coding sequence will often differ by effect. Depending on how the translated protein is affected (e.g., catastrophic folding failure, alteration of a molecular interaction interface or active site, or a subtle change on an unused surface), the effects may differ in severity or in rare cases may even result in the emergence of qualitatively different behaviours.
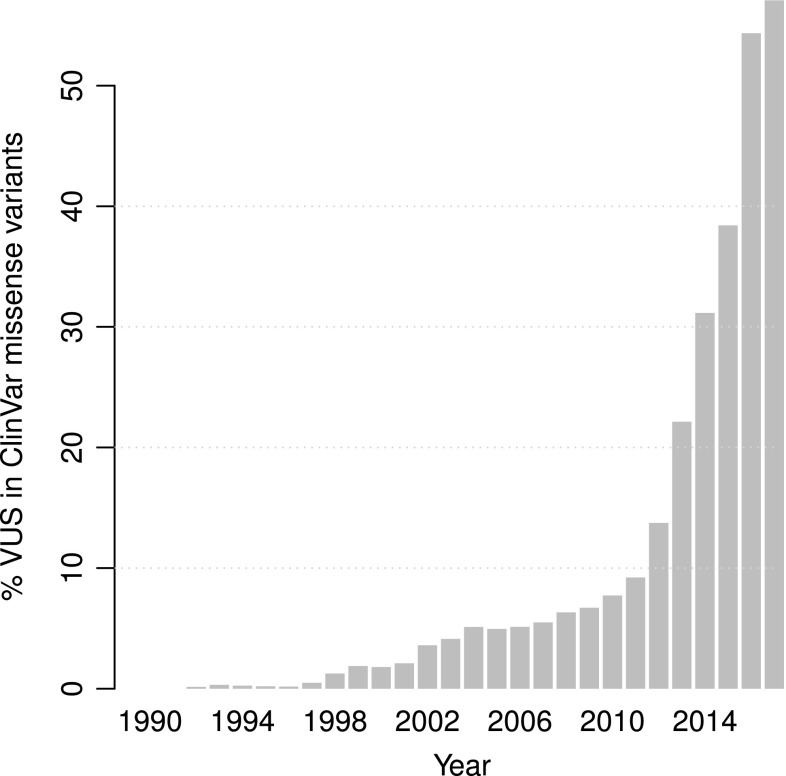
Given the much greater difficulty of interpreting non-coding regions, clinical applications have so far largely concentrated on protein-coding genes. Sequencing panels for known disease-associated genes and even whole-exome sequencing (WES) are widely commercially available. A number of different standards for classifying mutations with respect to their potential health impacts have been proposed; most prominently, the American College of Medical Genetics and Genomics (ACMG) standard (Richards et al. 2015). It defines categories stretching from “pathogenic” to “benign”, including the ‘gray zone’ category of “variant of uncertain significance” (VUS). Even though the mutational landscape for a handful of genes, such as BRCA1 are explored better than others due to their established relevance and potential for taking clinical action (Cheon et al. 2014), the majority of clinical variants across all genes are currently classified as VUS. In ClinVar alone, VUS make up over 50% of entries for missense variants (Fig. 1), despite ClinVar guidelines that actively discourage submission of unclassified variants. In a recent study using gene panels assessing germline cancer risk loci (Maxwell et al. 2016), over 98% of missense variants were classified as VUS. Not only can these uncertainties burden patients with unnecessary anxiety (Cheon et al. 2014), they also call into question the value of sequencing in the clinic if the majority of findings are not actionable. With increasing use of WES and WGS as opposed to targeted gene panels, this problem is only going to get worse. According to the 1000 Genomes Project data, every person carries 100–400 missense variants that are so rare that they have likely never been seen before in the clinic (The 1000 Genomes Project Consortium 2015). In the absence of the previous observations, they would automatically be added to the long list of VUSs.
Fig. 1.
Percentage of variants of uncertain significance (VUS) among missense allele Clinvar records over time from 1990 until 2017
Like its sister standards, the ACMG guidelines also recognize different methods of gathering evidence towards a variant’s classification. These can be broadly summarized as (1) frequency of observation in affected or unaffected individuals; (2) laboratory assays; and (3) in silico prediction. Out of these three categories, in silico prediction used to be the only option that easily scaled to cover all possible variants and could be applied proactively. However, it is also considered one of the weakest forms of evidence. Over the last decade, however, a new type of high-throughput laboratory assay has emerged: Multiplexed Assays of Variant Effect (MAVEs) (Starita et al. 2017), which promise to massively increase the scalability of those methods that the ACMG considers in the highest tiers of evidence. In the following, we will first recapitulate some of the more popular in silico approaches and then discuss MAVEs, breaking down the methodological variety in the existing studies, describing some of the newest developments and their implications for the future.
In silico approaches to variant function assessment
A number of algorithms exist that offer predictions as to the deleteriousness of mutations, with prominent examples including PolyPhen-2 (Adzhubei et al. 2013), SIFT (Ng and Henikoff 2001), and PROVEAN (Choi et al. 2012). PolyPhen-2 employs a simple (naive Bayes) machine learning method based on evolutionary conservation and protein structural features. It uses a set of previously reported pathogenic alleles as a positive training set and differences between human genes and their mammalian homologues as a negative training set. By contrast, SIFT (Sorting Intolerant From Tolerant) only uses evolutionary conservation. The tool uses multiple sequence alignments to calculate position-specific score matrices for each gene which are then normalized and transformed into probability values. PROVEAN (PROtein Variation Effect ANalyzer) similarly only takes into account sequence alignments. However, rather than just computing a position-specific score, PROVEAN calculates the difference in alignment quality between using the wild-type or variant sequence against clusters of homologous sequences. The average distance is then interpreted as indicative of the deleteriousness of the variant.
While the three tools succeed in making good predictions, their reliability is unfortunately still not high enough to serve as a basis of clinical decision making. We and others recently performed an independent comparison of these tools on a set of well-established disease-causing variants as well as rare polymorphisms with no known disease association (Sun et al. 2016). The study examined the trade-off between precision (the fraction of pathogenic variant predictions that were correct) and sensitivity (the fraction of pathogenic variants that were predicted to be pathogenic). A high precision can be considered especially important when considering taking clinical action based on a prediction. When compared at a minimum precision level of 90%, PolyPhen-2 and PROVEAN only reach a sensitivity of 19 and 21%, respectively, while SIFT did not achieve 90% precision at any score threshold. Consistent with these limitations, the ACMG currently considers only cases in which multiple methodologically orthogonal prediction algorithms agree as weak evidence in a supporting role for VUS re-classification (Richards et al. 2015).
Multiplex assays of variant effect (MAVE)
An alternative to the in silico methods above are functional assays in the laboratory. Such assays are, indeed, useful tools for the classification of variants of uncertain significance. Assessment of the effects of variants observed in the clinic has led to many high-impact discoveries such as drug resistance variants in cancer genes (Solit et al. 2006; Azam et al. 2003; Shah et al. 2002; Kohsaka et al. 2017). However, experimental assays of variant function have generally been ’reactive’, in the sense that measurements are carried out only after (often long after) the first clinical presentation of a variant, owing to the resource- and time-intensive nature of this testing. However, as more variants are discovered, it may be more useful to take a proactive experimental approach: Building an atlas of the functional effects of all possible variants, including those that have never before been observed in a patient. One may object that it would not be economical to screen variants that may never actually be observed in a patient. However, a simple back-of-envelope calculation given the size of the human population and the frequency of de novo mutation (Acuna-Hidalgo et al. 2016) shows that every missense variant that can possibly exist (i.e., it is not fundamentally incompatible with life) can be expected to occur on average in
Yet, assaying all possible variants in known disease genes would require massive parallelization. Such efforts have recently gained much traction, having their foundations laid in the winter of 2010/11 with three papers by Fowler et al. (2010), Ernst et al. (2010), and Hietpas et al. (2011) that collectively pioneered a technology initially termed Deep Mutational Scanning (DMS). These seminal papers have since inspired a growing number of similar efforts by other groups. While the earliest studies of this kind focused on coding regions, multiple groups have since begun interrogating the effects of non-coding variants, e.g., on promoter activity (Kwasnieski et al. 2012; Maricque et al. 2017), autonomously replicating sequences (Liachko et al. 2013; Hoggard et al. 2016), splicing (Julien et al. 2016; Ke et al. 2018), or the behaviour of RNAs (Li et al. 2016; Puchta et al. 2016). The term “Multiplex Assays of Variant Effect” (MAVE) was coined by Starita et al. (2017) to encompass these high-throughput functional assays for a wider range of variant types.
Table 1 lists a selection of MAVE studies and their respective scales, the growth of which is shown in Fig. 2. MAVE screens can be broken down into a number of experimental and computational components: (1) mutagenesis and library creation; (2) selection of functional variants; (3) sequencing of the selected and control populations; (4) scoring and computational analysis (see Fig. 3). In the following sections, we will review the different previous implementations of these components in detail.
Table 1.
List of MAVE studies, their respective target spaces, and achieved levels of coverage
| Reference | Target | Search space | Coverage (%) |
|---|---|---|---|
| Fowler et al. (2010) | YAP65 | WW domain | |
| Ernst et al. (2010) | Synthetic PDZ domain | 10 AAs | |
| Hietpas et al. (2011) | Hsp90 | 9 AAs | |
| Fujino et al. (2012) | Fab antibody fragment | Fragment | 79 |
| Adkar et al. (2012) | Ccdb | whole protein | |
| McLaughlin et al. (2012) | PSD95 | PDZ domain | |
| Schlinkmann et al. (2012) | GPCR | Whole protein | |
| Whitehead et al. (2012) | Synthetic protein | 51AA (whole protein) | 99 |
| Traxlmayr et al. (2012) | IgG1 | CH2/CH3 domains | |
| Araya et al. (2012) | YAP65 | WW domain | |
| Deng et al. (2012) | TEM1 | Whole protein | |
| Kwasnieski et al. (2012) | Rhodopsin promoter | 52 bp cis-regulatory element | |
| Wu et al. (2013) | Neuraminidase | SNP accessible | |
| Roscoe et al. (2013) | Ubiquitin | Whole protein | |
| Starita et al. (2013) | Ub.E3 E4B | Whole protein | |
| Procko et al. (2013) | Synthetic protein | 60 AA | |
| Tinberg et al. (2013) | Synthetic protein | 40AA | 90 |
| Jiang et al. (2013) | Hsp90 | Substrate binding loop | |
| Kim et al. (2013) | Mat alpha | Degron region | |
| Melamed et al. (2013) | Pab1 | RRM domain | |
| Forsyth et al. (2013) | Antibody for EGFR | Whole protein | |
| Jacquier et al. (2013) | TEM1 | whole protein | 64 |
| Hietpas et al. (2013) | Hsp90 | pos. 528–590 | |
| Liachko et al. (2013) | ARS1 | 100 bp | |
| Wagenaar et al. (2014) | BRAF | 77 AAs | 99.65 |
| Firnberg et al. (2014) | TEM1 -lactamase | Whole protein | |
| Olson et al. (2014) | G-protein (GB1) | IgG-binding domain | |
| Melnikov et al. (2014) | APH(3’)II (kinase) | Whole protein | |
| Bloom (2014) | Influenza nucleoprotein | Whole protein | |
| Thyagarajan and Bloom (2014) | Influenza hemagglutinin | Whole protein | |
| Qi et al. (2014) | NS5A | IA domain | |
| Roscoe and Bolon (2014) | Ubiquitin | Whole protein | |
| Reich et al. (2015) | Bcl- ligands | Peptide library | N/A |
| Stiffler et al. (2015) | TEM1 -Lactamase | Whole protein | |
| Doud et al. (2015) | Influenza nucleoprotein | Whole protein | |
| Kitzman et al. (2015) | Gal4 | DB domain | |
| Starita et al. (2015) | BRCA1 | RING domain | |
| Rockah-Shmuel et al. (2015) | M.HaeIII | Whole protein | 38 |
| Wu et al. (2015) | PA | Whole protein | 94 |
| Mishra et al. (2016) | Hsp90 | ATPase domain | |
| Doud and Bloom (2016) | Hemagglutinin | Whole protein | |
| Mavor et al. (2016) | Ubiquitin | Whole protein | |
| Majithia et al. (2016) | PPAR | Whole protein | |
| Julien et al. (2016) | FAS/CD95 | Exon 6 | 95 |
| Li et al. (2016) | tRNA Arg-CCU | whole gene | |
| Sarkisyan et al. (2016) | GFP | whole protein | |
| Tripathi et al. (2016) | CcdB | whole protein | 87 |
| Puchta et al. (2016) | snoRNA U3 | pos. 7-333 | |
| Brenan et al. (2016) | Mapk1/Erk2 | Whole protein | 99 |
| Steinberg and Ostermeier (2016) | TEM1 | Whole protein | 39–50 |
| Hoggard et al. (2016) | miniARS317/301 | bp | |
| Ma et al. (2017) | BCR-ABL | 8AAs | |
| Matreyek et al. (2017) | GFP | whole protein | |
| Klesmith et al. (2017) | TEM1,LGK | whole proteins | |
| Chan et al. (2017) | IGPS | 8 -strands | |
| Bandaru et al. (2017) | H-Ras | pos. 2–166 | |
| Weile et al. (2017) | UBE2I,SUMO1,TPK1,CALM1/2/3 | whole proteins | 100 |
| Mighell et al. (2018) | PTEN | Whole protein | |
| Plesa et al. (2018) | PPAT | Whole protein | |
| Matreyek et al. (2018) | PTEN,TPMT | Whole protein | |
| Ke et al. (2018) | DHFR | exon 5 | |
| Starita et al. (2018) | BRCA1 | 302 AAs | |
| Kotler et al. (2018) | TP53 | DNA-binding domain |
Fig. 2.
Variant effects covered in MAVE studies. Top: the total number of variant effects covered in MAVE studies up to a given year. For 2018, the solid bar indicates the current state, while the dashed outline represents an extrapolation for the rest of the year. Bottom: the number of variant effects reported in individual studies, where colour indicates the study’s saturation of its respective target space
Fig. 3.

Generalized workflow of a typical MAVE experiment. Steps marked in gray are downstream computational procedures not found in every study, but contribute to the quality of the data. The proportions of studies using different mutagenesis, selection, and sequencing methods are broken down in pie charts. Colors serve to visually differentiate different categories but do not bear meaning
Mutagenesis approaches
A variety of saturation mutagenesis methods have previously been applied in MAVE studies; some more technically challenging than others. The simplest method is error-prone PCR amplification (Cadwell and Joyce 1994; Mohan et al. 2011). While this has the advantage of being an inexpensive and facile procedure, it will almost exclusively result in the generation of point mutations and as such will not generate all possible amino acid replacements. One may argue that the evaluation of VUS does not require insight into amino acid substitutions that cannot be achieved by a single-nucleotide change, as they are unlikely to occur in the clinic. However, the preference for transitions over transversions in many error-prone PCR protocols can lead to uneven representations of variants, so that codon-level mutagenesis can lead to more even representation amongst those missense variants that are achievable by single nucleotide change. In addition, multiple nucleotide changes do occur within single codons, a non-negligible 2% of the time (Kaplanis et al. 2018). Moreover, exploring all possible amino acid changes offers the potential for valuable insights into what biochemical properties may be most important for each amino acid at each position.
Another set of methods often employed are scaled-up versions of site-directed mutagenesis approaches (Hutchison et al. 1978; Seyfang and Jin 2004; Firnberg and Ostermeier 2012), with one popular example being Kunkel mutagenesis (Kunkel 1985). The Kunkel approach uses a strain of E. coli that has been modified to produce high levels of uridine and lacks the ability to excise these bases from DNA. A phage vector carrying the desired template sequence is transfected into the cells resulting in its replication with a high uracil incorporation rate. The thus-uracilated plasmid can then be used as a template for primer extension, with primers containing the mutations of interest, and subsequently introduced into wild-type E. coli which will degrade the uracilated template, thus enriching the mutant copies. A number of derivatives of Kunkel mutagenesis have since been developed to bring its output to scale supporting saturated libraries, most notably Pfunkel (Firnberg and Ostermeier 2012). To address the full spectrum of amino acids at a given position, oligonucleotides carrying degeneracy codons (Pal and Fellouse 2005) are often used. Particularly popular is the use of NNK and NNS degeneracies, which have long been used in biochemistry (Scott and Smith 1990; Barbas et al. 1992). Here, S denotes either Guanine or Cytosine, and K denotes either Guanine or Thymine in the third position of the degenerate codon. Either of these options only enables 32 out of all 64 possible codons, covering all 20 possible amino acids while avoiding two of the three possible stop codons (TGA and TAA). An alternative to degeneracy codes is the use of custom oligonucleotide arrays covering all possible (or desired) options of codon changes explicitly (Kitzman et al. 2015). While this option allows for the precise control of desired mutations, it is currently too expensive to be applicable for more than a handful of genes at a time.
Another saturation mutagenesis method often applied in Deep Mutational Scanning is EMPIRIC (“Extremely Methodical and Parallel Investigation of Randomized Individual Codons”) (Hietpas et al. 2011). In this method, rather than using PCR amplification, oligonucleotide cassettes carrying the variants of interest are directly ligated at the appropriate positions. This is achieved by designing the underlying vector, such that it omits the cassette sequence. Instead, it carries a restriction site at the equivalent position, which can be cut to create sticky ends. Pairs of oligos carrying the variants of interest can be synthesized, such that they can assemble into a fitting cassette that integrates with the vector. EMPIRIC is one example of a mutagenesis method that was explicitly developed to be used in Deep Mutational Scanning. Another example is PALS (“Programmed ALlelic Series”) (Kitzman et al. 2015), which aims to limit the number of amino acid changes per library clone to only one. Oligos carrying the variants of interest are annealed to uracilated templates and linearly amplified with strand-displacing polymerase. In a second step, the template is degraded using uracil-DNA glycosylase and an antisense strand is generated in a second linear amplification step. The product is denatured and yet again hybridized with uracilated template allowing it to be extended towards the other end of the template. Finally, the template is degraded again and the now full-length mutagenized strands are amplified.
Yet another approach, which recently gained popularity, is dubbed “inverse PCR” (Jain and Varadarajan 2014). This method uses circular templates and pairs of oligos, one of which carries the mutagenic degenerate sequence, while the other points directly away from it. This primer setup appears like a directional inversion of that used in a regular PCR, thus lending the method its name. A first amplification step produces a set of linear products which serve as templates for the second, exponential amplification step, after which the final product is circularized. The authors compared the method to similar approaches using overlapping primers and found the inverse PCR method to display superior efficiency. This approach has become popular recently, and multiple MAVE maps have relied on it (Puchta et al. 2016; Matreyek et al. 2018).
A more recent development is “POPCode” (Weile et al. 2017), which expands upon the site-directed approach described by Seyfang and Jin (2004). Here, a set of oligos carrying all possible codon replacements are designed, such that their melting temperatures are uniform. They are hybridized to a uracilated template, in similar fashion to the PALS approach; however, they are allowed to directly compete with each other, enabling either single or multiple variants per molecule, depending on oligonucleotide concentrations. Non-strand-displacing polymerase is used to fill the gaps and seal the remaining nicks, followed by the degradation of the template using uracil-DNA glycosylase. A useful feature of this approach is the availability of a webtool that automates POPCode oligo design, such that each oligo arm surrounding the degeneracy has a similar melting temperature (Weile et al. 2017).
In addition to the various mutagenesis methods discussed here, it may be noted that complete variant libraries are also recently becoming commercially available via gene synthesis (Kosuri et al. 2010). A current limitation is the rate of point mutations and indels (Plesa et al. 2018), which makes it inappropriate for achieving frameshift-free coding regions with 1 aa change per clone for longer proteins. However, it is possible that, with increased interest in gene synthesis applications, these options may become more accurate and economical in the future.
Finally, with the rise of CRISPR/Cas9 and other gene editing tools, new methods are emerging that are able to introduce variants directly into endogenous gene loci, at efficiencies which are beginning to allow saturation mutagenesis. Findlay et al. (2014) first demonstrated this idea on a small scale, mutagenizing a small range of codons in BRCA1. A large-scale implementation of this idea is soon to be published (Findlay et al. 2018). Although this is currently more resource-intensive than introduction of an mutagenized library generated ex vivo, the advantages of studying variation in the context of the native gene locus are potentially great. For example, the function of variants that depend on the action of distal enhancers would be missed using mutagenized constructs introduced at a safe harbor site that is far from the endogenous locus.
Selection approaches
The most central component of a MAVE study is the selection process. The selection schemes used in previous studies can be sorted into four broad categories: (1) in vitro display methods (such as phage display or ribodisplay); (2) competition-based methods that couple a protein property under investigation (such as molecular interactions, toxicity, or overall functionality) to host cell fitness; (3) cell sorting based on fluorescence-labeled reporters; (4) transcript-abundance-based methods.
Phage display (Smith 1985) and ribodisplay (Mattheakis et al. 1994) couple the genetic information of each given variant to the physical protein itself and select according to the protein’s ability to bind to a fixed interactor. In phage display, this is achieved by the protein being displayed on the surface of a phage that contains the corresponding gene, while ribodisplay stalls a cluster of ribosomes on the variant mRNA with the corresponding protein still attached. Variants that are unable to bind to the interactor-coated surface are washed away and thus depleted. This can be done in multiple rounds, as the associated genetic information can be replicated again after selection (via viral propagation in bacteria for phage display or via PCR in ribodisplay). Fowler et al. (2010) employed phage display in their seminal Deep Mutational Scanning study of the binding of the YAP65-WW domain to its cognate peptide target. However, since display methods are only feasible for small proteins or fragments thereof, more recent studies have instead employed more scalable methods.
The most frequently applied selection mechanisms are fitness-based. In these cases, a particular property of the variant protein is coupled to its host cell’s ability to thrive in competitive growth. Of these methods, functional complementation (Lee and Nurse 1987; Osborn and Miller 2007) and Yeast-2-Hybrid (Y2H) (Fields and Song 1989) are among the most frequently applied. While complementation couples fitness to a protein’s overall ability to perform its biological role in a model organism, Y2H couples fitness to the ability of the protein to maintain a specific protein–protein interaction.
The largest share of growth-based selection methods in MAVE studies employs functional complementation, and most use the yeast Saccharomyces cerevisiae as their model system (see Fig. 3). The assay is based on the premise that some human genes can be used to rescue the deletion of their orthologues in yeast. That is, a fitness defect resulting from the inactivation of the yeast gene is alleviated by the artificial expression of the human gene. Therefore, any relative changes in fitness upon expressing a variant of the human gene can be interpreted as the variant’s effect on the protein’s overall ability to function. We and others recently examined the applicability of functional complementation in yeast to the assessment of disease variants (Sun et al. 2016). This study found that functional complementation assays in yeast offered sensitive and accurate predictions despite yeast and humans being diverged by 1 billion years. Indeed, yeast complementation outperformed in silico methods like PolyPhen-2 and PROVEAN in terms of disease variant prediction by a wide margin. At a threshold of 90% precision (as discussed in “In silico approaches to variant function assessment”), the complementation assay achieved a sensitivity of over 60%, as compared to 19 and 21% for the two in silico methods, respectively. It is consistent with these findings that the ACMG considers functional assays among the strongest sources of evidence for variant classification (Richards et al. 2015).
A limitation of functional complementation in yeast is that currently only human disease-implicated genes have been found to be amenable to the assay (Sun et al. 2016). However, the existence of synthetic lethal genetic interactions for many yeast genes may allow for the design of strains with sensitized backgrounds providing new complementation assays. In addition, CRISPR screens have in recent years revealed many genes for which growth phenotypes exist directly in human cell lines (Hart et al. 2015; Blomen et al. 2015; Wang et al. 2014); opening the possibility of performing functional complementation directly in these cell lines. A number of studies have since exploited the possibility of complemention assays directly in human cells (e.g., Wagenaar et al. 2014; Qi et al. 2014; Brenan et al. 2016). However, while future variant analysis is likely to trend towards complementation in mammalian cell models as opposed to yeast, the latter assays are not obsolete. Sun et al. (2016) found that a selectable phenotype that is potentially compatible with MAVE using human cell-based complementation has been identified for less than half of all human disease genes. Thus, for many disease genes, yeast-based complementation or Y2H may be the only viable option for a MAVE.
A popular condition-dependent extension to complementation is selection for drug resistance (Wu et al. 2013; Wagenaar et al. 2014), but other fitness-based selection methods have been used in MAVEs as well. For example, Adkar et al. (2012) used the toxicity of CCDB in E. coli, while Kim et al. (2013) select according to degron activity by fusing the degron to an auxotrophic marker. Finally, a number of MAVE studies have been performed on viral genes, by selecting for virus propagation efficiency (Bloom 2014; Thyagarajan and Bloom 2014).
Another popular growth-based assay is Yeast-2-Hybrid (Y2H). It is a binary protein interaction assay also performed within the yeast S. cerevisiae. Y2H is based on the reconstitution of two fragments of the transcription factor Gal4 fused to two proteins of interest. A successful interaction of the two proteins allows Gal4 to induce the expression of a reporter; usually, an auxotrophy marker. When comparing different variants of the same protein interacting with the same partner, reporter expression has even been shown to be proportional to binding affinity (Yang et al. 1995). This proportional relationship allows for quantitative interpretation of Y2H results under these specific circumstances.
One objection to the use of Y2H as an assay for variant function assessment is that it does not measure all aspects of a protein’s functionality, but rather only its ability to physically associate with a given interaction partner. However, in addition to detecting variants that specifically affect binding, e.g., via changes to the binding interface, this approach should also detect many other variants that broadly impact protein function, e.g., those that cause protein mis-folding or instability. Indeed, in a recent examination of the Y2H performance of common disease-associated variants, we found that approximately two out of three disease variants in proteins with multiple interaction partners lose some or all of their protein interactions (Sahni et al. 2015).
Another selection mechanism is the use of fluorescence-activated cell sorting (FACS) (Julius et al. 1972). Here, surface markers for which abundance is proportional to the activity of the studied protein are targeted with fluorescently labeled antibodies, such that cells can be sorted accordingly, as has been performed by Schlinkmann et al. (2012) and Majithia et al. (2016). FACS-based selection has also been used to gain read-outs of protein stability and abundance (Matreyek et al. 2018).
In addition to assays that measure general properties of proteins, more specialized methods also exist, which assess specific molecular functions. For example, Starita et al. (2015) developed an assay that quantifies the ubiquitination activity of BRCA1’s RING domain. Most recently, they also developed an assay to test the same protein’s DNA-repair activity (Starita et al. 2018).
In terms of selection for the properties of non-coding regions, a number of technologies have been developed. Massively Parallel Reporter Assays (MPRAs) (Kwasnieski et al. 2012; Maricque et al. 2017) for example place libraries of mutagenized promoter sequences upstream of barcoded regions, the expression of which is measured using RNA-Seq, while the initial abundance of corresponding cells is measured by DNA sequencing of the same loci. The ratio of RNA-Seq to DNA-Seq reads can then be used to calculate the effect of promoter variant on expression. Similarly, splicing assays such as employed by Julien et al. (2016) or Ke et al. (2018) also use RNA-Seq to measure the fraction of transcripts in which the exon of interest is spliced in.
Sequencing strategies
The experimental step immediately following selection in a MAVE experiment is sequencing. Next-generation sequencing technology can be considered the key technological advance that made Deep Mutational Scanning possible. Many studies use a fairly simple approach by performing deep shotgun sequencing of the library (Ernst et al. 2010; Hietpas et al. 2011; Fujino et al. 2012). However, a major problem with this approach is that, without knowing which reads originate from which DNA molecule, each read can only be considered by itself, making it difficult to distinguish real mutations from sequencing error. To address this problem, different solutions have emerged. In cases where the amplicon is short enough, paired-end ‘duplex’ sequencing can be exploited to use information from both strands for variant calling. In the simplest case, this is achieved by requiring both reads to agree on the base call in question, as in the case of Whitehead et al. (2012) and Weile et al. (2017). A less stringent, but potentially more sensitive alternative as used by Fowler et al. (2010) is to perform Bayesian inference on the quality scores associated with the base calls in each read pair. This way, a variant may still be identified if one of the two reads reported a wild-type base call with low confidence.
Where the length of the nucleotide sequence in question exceeds the read-length capabilities of short-read sequencing technologies, other strategies are required. A notable borderline case can be found in Olson et al. (2014) where only a partial overlap between read pairs was achieved and variant calls outside of the overlap region were of lower quality. Other studies have used more involved approaches. A popular paradigm is the association of molecular barcodes with each clone within the MAVE library. While this simplifies the readout of the experiment (as only the barcodes need to be sequenced and counted), it adds the requirement of identifying which barcode belongs to which genotype. In most cases, this is addressed using “subassembly” (Hiatt et al. 2010), a high-throughput amplicon sequencing approach based on attaching random tags to amplicons. The DNA is then amplified, sheared, and ligated to adapters, so that paired-end sequencing can be used to identify the random tag together with each read. This allows reads to be sorted according to which original tagged molecule they belong, which, in turn, enables separate assemblies for each input molecule to be computed. The resulting high-quality virtual reads are long enough to cover both ORF and barcode locus. Another subassembly approach is “KiloSeq” (Weile et al. 2017) which works using an array-based format where well-specific tags are attached to the amplicons, followed by Tn5 tagmentation and re-amplification of tag-bearing fragments. Another barcode-based method, called EMPIRIC-BC was described by Mavor et al. (2016), where the amplicon in question was short enough not to require subassembly. Here, a long read can cover the entire ORF, while a second, short read can identify the barcode.
An alternative approach to covering longer stretches of DNA is to subdivide them into smaller regions that can be sequenced separately from each other. For example, Doud and Bloom (2016) amplify each region with primers carrying random tags. This way, if multiple reads contain the same tag, they are highly likely to originate from PCR copies of the same original molecule and can be used to make more accurate variant calls. This approach is often dubbed “tag-clustering”. While this approach has the advantage of being less labour-intensive than barcoding each individual clone in the MAVE library, it can only detect variants co-occurring within the same region of the sequence. Thus the library must be designed in such a way that either only a single mutation occurs within each clone or that it is large enough that effects of many background variants are averaged out. This approach can also be used in combination with duplex sequencing, as performed in Weile et al. (2017), where it is called “DMS-TileSeq”.
In a benchmark study, Zhang et al. (2016) evaluated some of these approaches. They found that duplex sequencing decreases the rate of transition and transversion base-calling errors tenfold while decreasing indel errors by 100-fold. By contrast, tag clustering lowered transition and transversion errors 20 fold, but had a little impact on indel errors. A combination of both approaches in which tagged reads are first compared against their paired partners and then clustered is found to perform best. The authors also examined the effect of quality score filtering. While this method had moderate impact when applied to raw reads, read pairing and tag clustering benefited little from it.
Computational analysis
Most MAVE studies use custom scripts to process the sequencing readout and calculate the selection advantage for each variant. Nonetheless, a few published software packages exist. The EMPIRIC mutagenesis and screening method provides its own software package for data processing (Hietpas et al. 2011), though it is not generally applicable to other MAVE methods. The dms_tools package (Bloom 2015) offers the same services, but is tailored more towards methods using regionally focused sequencing. Finally, Enrich (Fowler et al. 2011) offers a generalized solution applicable to most DMS frameworks. A second version that adds a more sophisticated statistical analysis including the assessment of measurement confidence levels (Rubin et al. 2016).
The DMS-BarSeq and DMS-TileSeq methods used in Weile et al. (2017) also come with publicly available analysis pipelines. Most importantly, they offer imputation of missing values using machine learning. A Random Forest model (Breiman 2001) was created using physicochemical and structural features of the affected amino acids as well as position-specific biases of the existing map, yielding surprisingly accurate predictions that surpassed those of Polyphen-2 (Adzhubei et al. 2013) and PROVEAN (Choi et al. 2012). This most recent innovation has since been adapted for use with different predictive features by Mighell et al. (2018).
Going beyond the scope of predicting the effects of variants omitted within a mapped gene, Gray et al. (2018) applied machine learning to extrapolating maps for new genes. Using a gradient-boost model (Friedman 2001) trained on similar features as the imputation method from Weile et al. (2017), they implemented a new cross-validation scheme that swaps out whole proteins to be more sensitive towards the detection of overfitting. While predictions were generally more reliable than established computational predictors, accuracy was highly variable across proteins, with some performing better than others. This behaviour may be alleviated as more DMS data sets become available for training.
Beyond the potential utility of the variant effect maps in the clinic, they also lend themselves to extract new insights from computational biology. Bloom (2017) recently developed a method to detect signatures of evolutionary selection within these maps far exceeding the sensitivity of comparing orthologous sequences alone. Meanwhile, Wu et al. (2016) developed a method to calculate direct estimates for the folding energy effects of variants by examining their intragenic genetic interactions within variant effect maps.
Conclusion
Since their inception in 2010, MAVEs have produced a steadily increasing wealth of variant effect maps. Recent years have seen an increasing trend of targeting clinically relevant genes. The utility of these maps towards the eventual goal of clinical variant assessment has been demonstrated in multiple studies (Starita et al. 2015; Majithia et al. 2016; Weile et al. 2017). Since then, an arsenal of different methodologies have been developed to capture a wider spectrum of sequence types and functions. In addition, new computational methods continue to improve the quality and reliability of the data produced.
However, a number of issues are still apparent. Many of the studies listed in Table 1 do not make their data easily available. While some provide full access to final results, some only provide raw data in the NCBI short-read archive (SRA), the majority require interested parties to contact the authors personally. There is a clear need for an open data repository that makes MAVE data available to the public and allows for downstream probabilistic integration and analysis. Similarly, the issue of reagent availability remains as a challenge. In most cases, the saturation mutagenesis libraries generated are not made available via common repositories. Furthermore, due to the large diversity of methodologies employed, libraries cannot generally be expected to be compatible across platforms.
Another complicating factor is the fact that the assays underlying different MAVE studies are quite diverse and measure different aspects of a protein’s behaviour. As a consequence, they cannot be easily compared with each other. In addition, the achieved coverage of possible amino acid changes varies from map to map. Finally, many maps do not control the quality of measurements. Therefore, the confidence levels underlying different parts of these maps are often unknown. While generalized frameworks have been proposed that would increase the potential comparability and interpretability across maps (Rubin et al. 2016; Weile et al. 2017), they are not implemented by most studies. Here, a centralized repository could also be of help, as it could serve as a basis for re-analysis of data with the latest tools.
Acknowledgements
The authors would like to thank Doug Fowler, Alan Rubin, Jesse Bloom, Amy Caudy, Igor Stagljar, Lincoln Stein, and Dan Bolon for their input and collaboration. The authors gratefully acknowledge funding by the One Brave Idea Initiative, the National Human Genome Research Institute of the National Institutes of Health (NIH/NHGRI) Center of Excellence in Genomic Science (CEGS) Initiative (HG004233), the Canadian Excellence Research Chairs (CERC) Program, and the Ontario Ministry of Research and Innovation (MRI).
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Contributor Information
Jochen Weile, Phone: +1-416-586-4800, Email: jochen.weile@mail.utoronto.ca.
Frederick P. Roth, Phone: +1-416-946-5130, Email: fritz.roth@utoronto.ca
References
- Acuna-Hidalgo R, Veltman JA, Hoischen A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 2016;17:241. doi: 10.1186/s13059-016-1110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkar BV, Tripathi A, Sahoo A, Bajaj K, Goswami D, Chakrabarti P, Swarnkar MK, Gokhale RS, Varadarajan R. Protein model discrimination using mutational sensitivity derived from deep sequencing. Structure. 2012;20(2):371–381. doi: 10.1016/j.str.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Adzhubei Ivan, Jordan Daniel M., Sunyaev Shamil R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Current Protocols in Human Genetics. 2013;76(1):7.20.1-7.20.41. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya CL, Fowler DM, Chen W, Muniez I, Kelly JW, Fields S. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. PNAS. 2012;109(42):16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112(6):831–843. doi: 10.1016/S0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- Bandaru P, Shah NH, Bhattacharyya M, Barton JP, Kondo Y, Cofsky JC, Gee CL, Chakraborty AK, Kortemme T, Ranganathan R, Kuriyan J. Deconstruction of the Ras switching cycle through saturation mutagenesis. eLife. 2017;6:e27810. doi: 10.7554/eLife.27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas CF, Bain JD, Hoekstra DM, Lerner RA. Semisynthetic combinatorial antibody libraries: a chemical solution to the diversity problem. PNAS. 1992;89(10):4457–4461. doi: 10.1073/pnas.89.10.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen V. A., Majek P., Jae L. T., Bigenzahn J. W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F. R., Olk N., Stukalov A., Marceau C., Janssen H., Carette J. E., Bennett K. L., Colinge J., Superti-Furga G., Brummelkamp T. R. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350(6264):1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- Bloom JD. An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol Biol Evol. 2014;31(8):1956–1978. doi: 10.1093/molbev/msu173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD. Software for the analysis and visualization of deep mutational scanning data. BMC Bioinf. 2015;16:168. doi: 10.1186/s12859-015-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD. Identification of positive selection in genes is greatly improved by using experimentally informed site-specific models. Biol Direct. 2017;12:1. doi: 10.1186/s13062-016-0172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Brenan L, Andreev A, Cohen O, Pantel S, Kamburov A, Cacchiarelli D, Persky NS, Zhu C, Bagul M, Goetz EM, Burgin AB, Garraway LA, Getz G, Mikkelsen TS, Piccioni F, Root DE, Johannessen CM. Phenotypic characterization of a comprehensive set of MAPK1/ERK2 missense mutants. Cell Rep. 2016;17(4):1171–1183. doi: 10.1016/j.celrep.2016.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell RC, Joyce GF. Mutagenic PCR. Genome Res. 1994;3(6):S136–S140. doi: 10.1101/gr.3.6.S136. [DOI] [PubMed] [Google Scholar]
- Chan YH, Venev SV, Zeldovich KB, Matthews CR. Correlation of fitness landscapes from three orthologous TIM barrels originates from sequence and structure constraints. Nat Commun. 2017;8:14614. doi: 10.1038/ncomms14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon JY, Mozersky J, Cook-Deegan R. Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med. 2014;6:121. doi: 10.1186/s13073-014-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesmore K, Bartlett J, Williams SM. The ubiquity of pleiotropy in human disease. Hum Genet. 2018;137(1):39–44. doi: 10.1007/s00439-017-1854-z. [DOI] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLos One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Huang W, Bakkalbasi E, Brown NG, Adamski CJ, Rice K, Muzny D, Gibbs RA, Palzkill T. Deep sequencing of systematic combinatorial libraries reveals -lactamase sequence constraints at high resolution. J Mol Biol. 2012;424(3):150–167. doi: 10.1016/j.jmb.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud MB, Bloom JD. Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses. 2016;8(6):155. doi: 10.3390/v8060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doud MB, Ashenberg O, Bloom JD. Site-specific amino acid preferences are mostly conserved in two closely related protein homologs. Mol Biol Evol. 2015;32(11):2944–2960. doi: 10.1093/molbev/msv167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93(5):779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Gfeller D, Kan Z, Seshagiri S, Kim PM, Bader GD, Sidhu SS. Coevolution of PDZ domain-ligand interactions analyzed by high-throughput phage display and deep sequencing. Mol BioSyst. 2010;6(10):1782–1790. doi: 10.1039/C0MB00061B. [DOI] [PubMed] [Google Scholar]
- Fields S, Song OK. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513(7516):120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, Janizek JD, Huang X, Starita LM, Shendure J (2018) Accurate functional classification of thousands of BRCA1 variants with saturation genome editing. bioRxiv 294520. 10.1101/294520 [DOI] [PMC free article] [PubMed]
- Firnberg E, Ostermeier M. PFunkel: efficient, expansive, user-defined mutagenesis. PLoS One. 2012;7(12):e52031. doi: 10.1371/journal.pone.0052031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol Biol Evol. 2014;31(6):1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth CM, Juan V, Akamatsu Y, DuBridge RB, Doan M, Ivanov AV, Ma Z, Polakoff D, Razo J, Wilson K, Powers DB. Deep mutational scanning of an antibody against epidermal growth factor receptor using mammalian cell display and massively parallel pyrosequencing. mAbs. 2013;5(4):523–532. doi: 10.4161/mabs.24979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, Fields S. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7(9):741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Araya CL, Gerard W, Fields S. Enrich: software for analysis of protein function by enrichment and depletion of variants. Bioinformatics. 2011;27(24):3430–3431. doi: 10.1093/bioinformatics/btr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman Jerome H. machine. The Annals of Statistics. 2001;29(5):1189–1232. doi: 10.1214/aos/1013203451. [DOI] [Google Scholar]
- Fujino Y, Fujita R, Wada K, Fujishige K, Kanamori T, Hunt L, Shimizu Y, Ueda T. Robust in vitro affinity maturation strategy based on interface-focused high-throughput mutational scanning. Biochem Biophys Res Commun. 2012;428(3):395–400. doi: 10.1016/j.bbrc.2012.10.066. [DOI] [PubMed] [Google Scholar]
- Gray VE, Hause RJ, Luebeck J, Shendure J, Fowler DM. Quantitative missense variant effect prediction using large-scale mutagenesis data. Cell Syst. 2018;6(1):116–124.e3. doi: 10.1016/j.cels.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163(6):1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Hiatt JB, Patwardhan RP, Turner EH, Lee C, Shendure J. Parallel, tag-directed assembly of locally derived short sequence reads. Nat Methods. 2010;7(2):119–122. doi: 10.1038/nmeth.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas RT, Jensen JD, Bolon DNA. Experimental illumination of a fitness landscape. PNAS. 2011;108(19):7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas RT, Bank C, Jensen JD, Bolon DNA. Shifting fitness landscapes in response to altered environments. Evolution. 2013;67(12):3512–3522. doi: 10.1111/evo.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard T, Liachko I, Burt C, Meikle T, Jiang K, Craciun G, Dunham MJ, Fox CA. High throughput analyses of budding yeast ARSs reveal new DNA elements capable of conferring centromere-independent plasmid propagation. G3 Genes Genomes Genet. 2016;6(4):993–1012. doi: 10.1534/g3.116.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA, Phillips S, Edgell MH, Gillam S, Jahnke P, Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978;253(18):6551–6560. [PubMed] [Google Scholar]
- Jacquier H, Birgy A, Nagard HL, Mechulam Y, Schmitt E, Glodt J, Bercot B, Petit E, Poulain J, Barnaud G, Gros PA, Tenaillon O. Capturing the mutational landscape of the beta-lactamase TEM-1. PNAS. 2013;110(32):13067–13072. doi: 10.1073/pnas.1215206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain PC, Varadarajan R. A rapid, efficient, and economical inverse polymerase chain reaction-based method for generating a site saturation mutant library. Anal Biochem. 2014;449:90–98. doi: 10.1016/j.ab.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Jiang L, Mishra P, Hietpas RT, Zeldovich KB, Bolon DNA. Latent effects of Hsp90 mutants revealed at reduced expression levels. PLos Genet. 2013;9(6):e1003600. doi: 10.1371/journal.pgen.1003600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, Miñana B, Baeza-Centurion P, Valcárcel J, Lehner B. The complete local genotype-phenotype landscape for the alternative splicing of a human exon. Nat Commun. 2016;7:11558. doi: 10.1038/ncomms11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius MH, Masuda T, Herzenberg LA. Demonstration that antigen-binding cells are precursors of antibody-producing cells after purification with a fluorescence-activated cell sorter. PNAS. 1972;69(7):1934–1938. doi: 10.1073/pnas.69.7.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplanis J, Akawi N, Gallone G, McRae JF, Prigmore E, Wright CF, Fitzpatrick DR, Firth HV, Barrett JC, Hurles ME (2018) Exome-wide assessment of the functional impact and pathogenicity of multi-nucleotide mutations. bioRxiv. 10.1101/258723 [DOI] [PMC free article] [PubMed]
- Ke S, Anquetil V, Zamalloa JR, Maity A, Yang A, Arias MA, Kalachikov S, Russo JJ, Ju J, Chasin LA. Saturation mutagenesis reveals manifold determinants of exon definition. Genome Res. 2018;28(1):11–24. doi: 10.1101/gr.219683.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Miller CR, Young DL, Fields S. High-throughput analysis of in vivo protein stability. Mol Cell Proteom. 2013;12(11):3370–3378. doi: 10.1074/mcp.O113.031708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzman JO, Starita LM, Lo RS, Fields S, Shendure J. Massively parallel single-amino-acid mutagenesis. Nat Meth. 2015;12(3):203–206. doi: 10.1038/nmeth.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klesmith JR, Bacik JP, Wrenbeck EE, Michalczyk R, Whitehead TA. Trade-offs between enzyme fitness and solubility illuminated by deep mutational scanning. PNAS. 2017;114(9):2265–2270. doi: 10.1073/pnas.1614437114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka S, Nagano M, Ueno T, Suehara Y, Hayashi T, Shimada N, Takahashi K, Suzuki K, Takamochi K, Takahashi F, Mano H. A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci Transl Med. 2017;9(416):eaan6566. doi: 10.1126/scitranslmed.aan6566. [DOI] [PubMed] [Google Scholar]
- Kosuri S, Eroshenko N, LeProust EM, Super M, Way J, Li JB, Church GM. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat Biotechnol. 2010;28(12):1295–1299. doi: 10.1038/nbt.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotler E, Shani O, Goldfeld G, Lotan-Pompan M, Tarcic O, Gershoni A, Hopf TA, Marks DS, Oren M, Segal E. A systematic p53 mutation library links differential functional impact to cancer mutation pattern and evolutionary conservation. Mol Cell. 2018;71(1):178–190.e8. doi: 10.1016/j.molcel.2018.06.012. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. PNAS. 1985;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci. 2012;109(47):19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327(6117):31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- Li C, Qian W, Maclean CJ, Zhang J. The fitness landscape of a tRNA gene. Science. 2016;352(6287):837–840. doi: 10.1126/science.aae0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Youngblood RA, Keich U, Dunham MJ. High-resolution mapping, characterization, and optimization of autonomously replicating sequences in yeast. Genome Res. 2013;23(4):698–704. doi: 10.1101/gr.144659.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Boucher JI, Paulsen J, Matuszewski S, Eide CA, Ou J, Eickelberg G, Press RD, Zhu LJ, Druker BJ, Branford S, Wolfe SA, Jensen JD, Schiffer CA, Green MR, Bolon DN. CRISPR-Cas9-mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy. PNAS. 2017;114(44):11751–11756. doi: 10.1073/pnas.1708268114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majithia Amit R, Tsuda Ben, Agostini Maura, Gnanapradeepan Keerthana, Rice Robert, Peloso Gina, Patel Kashyap A, Zhang Xiaolan, Broekema Marjoleine F, Patterson Nick, Duby Marc, Sharpe Ted, Kalkhoven Eric, Rosen Evan D, Barroso Inês, Ellard Sian, Kathiresan Sekar, O'Rahilly Stephen, Chatterjee Krishna, Florez Jose C, Mikkelsen Tarjei, Savage David B, Altshuler David. Prospective functional classification of all possible missense variants in PPARG. Nature Genetics. 2016;48(12):1570–1575. doi: 10.1038/ng.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricque BB, Dougherty JD, Cohen BA. A genome-integrated massively parallel reporter assay reveals dna sequence determinants of cis-regulatory activity in neural cells. Nucleic Acids Res. 2017;45(4):e16. doi: 10.1093/nar/gkw942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek KA, Stephany JJ, Fowler DM. A platform for functional assessment of large variant libraries in mammalian cells. Nucleic Acids Res. 2017;45(11):e102–e102. doi: 10.1093/nar/gkx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek KA, Starita LM, Stephany JJ, Martin B, Chiasson MA, Gray VE, Kircher M, Khechaduri A, Dines JN, Hause RJ, Bhatia S, Evans WE, Relling MV, Yang W, Shendure J, Fowler DM (2018) Multiplex assessment of protein variant abundance by massively parallel sequencing. bioRxiv 211011. 10.1101/211011 [DOI] [PMC free article] [PubMed]
- Mattheakis LC, Bhatt RR, Dower WJ. An in vitro polysome display system for identifying ligands from very large peptide libraries. PNAS. 1994;91(19):9022–9026. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., Wang H., Reynolds A. P., Sandstrom R., Qu H., Brody J., Shafer A., Neri F., Lee K., Kutyavin T., Stehling-Sun S., Johnson A. K., Canfield T. K., Giste E., Diegel M., Bates D., Hansen R. S., Neph S., Sabo P. J., Heimfeld S., Raubitschek A., Ziegler S., Cotsapas C., Sotoodehnia N., Glass I., Sunyaev S. R., Kaul R., Stamatoyannopoulos J. A. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavor D, Barlow K, Thompson S, Barad BA, Bonny AR, Cario CL, Gaskins G, Liu Z, Deming L, Axen SD, Caceres E, Chen W, Cuesta A, Gate RE, Green EM, Hulce KR, Ji W, Kenner LR, Mensa B, Morinishi LS, Moss SM, Mravic M, Muir RK, Niekamp S, Nnadi CI, Palovcak E, Poss EM, Ross TD, Salcedo EC, See SK, Subramaniam M, Wong AW, Li J, Thorn KS, Conchúir SO, Roscoe BP, Chow ED, DeRisi JL, Kortemme T, Bolon DN, Fraser JS. Determination of ubiquitin fitness landscapes under different chemical stresses in a classroom setting. eLife. 2016;5:e15802. doi: 10.7554/eLife.15802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KN, Hart SN, Vijai J, Schrader KA, Slavin TP, Thomas T, Wubbenhorst B, Ravichandran V, Moore RM, Hu C, Guidugli L, Wenz B, Domchek SM, Robson ME, Szabo C, Neuhausen SL, Weitzel JN, Offit K, Couch FJ, Nathanson KL. Evaluation of ACMG-guideline-based variant classification of cancer susceptibility and non-cancer-associated genes in families affected by breast cancer. Am J Hum Genet. 2016;98(5):801–817. doi: 10.1016/j.ajhg.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491(7422):138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D, Young DL, Gamble CE, Miller CR, Fields S. Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)-binding protein. RNA. 2013;19(11):1537–1551. doi: 10.1261/rna.040709.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Rogov P, Wang L, Gnirke A, Mikkelsen TS. Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. Nucl Acids Res. 2014;42(14):e112–e112. doi: 10.1093/nar/gku511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mighell TL, Evans-Dutson S, O’Roak BJ (2018) A saturation mutagenesis approach to understanding PTEN lipid phosphatase activity and genotype-phenotypes relationships. bioRxiv 255265. 10.1101/255265 [DOI] [PMC free article] [PubMed]
- Mishra P, Flynn JM, Starr TN, Bolon DNA. Systematic mutant analyses elucidate general and client-specific aspects of Hsp90 function. Cell Rep. 2016;15(3):588–598. doi: 10.1016/j.celrep.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan U, Kaushik S, Banerjee UC. PCR based random mutagenesis approach for a defined DNA sequence using the mutagenic potential of oxidized nucleotide products. Open Biotechnol J. 2011;5(1):21–27. doi: 10.2174/1874070701105010021. [DOI] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CA, Wu NC, Sun R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr Biol. 2014;24(22):2643–2651. doi: 10.1016/j.cub.2014.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Miller JR. Rescuing yeast mutants with human genes. Br Funct Genom. 2007;6(2):104–111. doi: 10.1093/bfgp/elm017. [DOI] [PubMed] [Google Scholar]
- Pal Gabor, Fellouse Frederic. Drug Discovery Series. 2005. Methods for the Construction of Phage-Displayed Libraries; pp. 111–142. [Google Scholar]
- Plesa C, Sidore AM, Lubock NB, Zhang D, Kosuri S. Multiplexed gene synthesis in emulsions for exploring protein functional landscapes. Science. 2018;359(6373):343–347. doi: 10.1126/science.aao5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko E, Hedman R, Hamilton K, Seetharaman J, Fleishman SJ, Su M, Aramini J, Kornhaber G, Hunt JF, Tong L, Montelione GT, Baker D. Computational design of a protein-based enzyme inhibitor. J Mol Biol. 2013;425(18):3563–3575. doi: 10.1016/j.jmb.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta O, Cseke B, Czaja H, Tollervey D, Sanguinetti G, Kudla G. Network of epistatic interactions within a yeast snoRNA. Science. 2016;352(6287):840–844. doi: 10.1126/science.aaf0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Olson CA, Wu NC, Ke R, Loverdo C, Chu V, Truong S, Remenyi R, Chen Z, Du Y, Su SY, Al-Mawsawi LQ, Wu TT, Chen SH, Lin CY, Zhong W, Lloyd-Smith JO, Sun R. A quantitative high-resolution genetic profile rapidly identifies sequence determinants of hepatitis C viral fitness and drug sensitivity. PLOS Pathogens. 2014;10(4):e1004064. doi: 10.1371/journal.ppat.1004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich L, Dutta S, Keating AE. Sortcery—a high-throughput method to affinity rank peptide ligands. J Mol Biol. 2015;427(11):2135–2150. doi: 10.1016/j.jmb.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards Sue, Aziz Nazneen, Bale Sherri, Bick David, Das Soma, Gastier-Foster Julie, Grody Wayne W., Hegde Madhuri, Lyon Elaine, Spector Elaine, Voelkerding Karl, Rehm Heidi L. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine. 2015;17(5):405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockah-Shmuel L, Tóth-Petróczy A, Tawfik DS. Systematic mapping of protein mutational space by prolonged drift reveals the deleterious effects of seemingly neutral mutations. PLos Comput Biol. 2015;11(8):e1004421. doi: 10.1371/journal.pcbi.1004421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe BP, Bolon DNA. Systematic exploration of ubiquitin sequence, E1 activation efficiency, and experimental fitness in yeast. J Mol Biol. 2014;426(15):2854–2870. doi: 10.1016/j.jmb.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DN. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J Mol Biol. 2013;425(8):1363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AF, Lucas N, Bajjalieh SM, Papenfuss AT, Speed TP, Fowler DM (2016) Enrich2: a statistical framework for analyzing deep mutational scanning data. bioRxiv 075150, 10.1101/075150 [DOI] [PMC free article] [PubMed]
- Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, Peng J, Weile J, Karras GI, Wang Y, Kovács IA, Kamburov A, Krykbaeva I, Lam MH, Tucker G, Khurana V, Sharma A, Liu YY, Yachie N, Zhong Q, Shen Y, Palagi A, San-Miguel A, Fan C, Balcha D, Dricot A, Jordan DM, Walsh JM, Shah AA, Yang X, Stoyanova AK, Leighton A, Calderwood MA, Jacob Y, Cusick ME, Salehi-Ashtiani K, Whitesell LJ, Sunyaev S, Berger B, Barabási AL, Charloteaux B, Hill DE, Hao T, Roth FP, Xia Y, Walhout AJM, Lindquist S, Vidal M. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161(3):647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisyan KS, Bolotin DA, Meer MV, Usmanova DR, Mishin AS, Sharonov GV, Ivankov DN, Bozhanova NG, Baranov MS, Soylemez O, Bogatyreva NS, Vlasov PK, Egorov ES, Logacheva MD, Kondrashov AS, Chudakov DM, Putintseva EV, Mamedov IZ, Tawfik DS, Lukyanov KA, Kondrashov FA. Local fitness landscape of the green fluorescent protein. Nature. 2016;533(7603):397–401. doi: 10.1038/nature17995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlinkmann KM, Honegger A, Tureci E, Robison KE, Lipovsek D, Pluckthun A. Critical features for biosynthesis, stability, and functionality of a G protein-coupled receptor uncovered by all-versus-all mutations. Proc Natl Acad Sci. 2012;109(25):9810–9815. doi: 10.1073/pnas.1202107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249(4967):386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Seyfang A, Jin JH. Multiple site-directed mutagenesis of more than 10 sites simultaneously and in a single round. Anal Biochem. 2004;324(2):285–291. doi: 10.1016/j.ab.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2(2):117–125. doi: 10.1016/S1535-6108(02)00096-X. [DOI] [PubMed] [Google Scholar]
- Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, Golub TR, Sebolt-Leopold J, Sellers WR, Rosen N. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439(7074):358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita LM, Pruneda JN, Lo RS, Fowler DM, Kim HJ, Hiatt JB, Shendure J, Brzovic PS, Fields S, Klevit RE. Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis. Proc Natl Acad Sci. 2013;110(14):E1263–E1272. doi: 10.1073/pnas.1303309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita Lea M., Young David L., Islam Muhtadi, Kitzman Jacob O., Gullingsrud Justin, Hause Ronald J., Fowler Douglas M., Parvin Jeffrey D., Shendure Jay, Fields Stanley. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics. 2015;200(2):413–422. doi: 10.1534/genetics.115.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita LM, Ahituv N, Dunham MJ, Kitzman JO, Roth FP, Seelig G, Shendure J, Fowler DM. Variant interpretation: functional assays to the rescue. Am J Hum Genet. 2017;101(3):315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita LM, Islam MM, Banerjee T, Adamovich AI, Gullingsrud J, Fields S, Shendure J, Parvin JD (2018) A multiplexed homology-directed DNA repair assay reveals the impact of 1,700 BRCA1 variants on protein function. bioRxiv 295279. 10.1101/295279 [DOI] [PMC free article] [PubMed]
- Steinberg B, Ostermeier M. Shifting fitness and epistatic landscapes reflect trade-offs along an evolutionary pathway. J Mol Biol. 2016;428(13):2730–2743. doi: 10.1016/j.jmb.2016.04.033. [DOI] [PubMed] [Google Scholar]
- Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM-1 -lactamase. Cell. 2015;160(5):882–892. doi: 10.1016/j.cell.2015.01.035. [DOI] [PubMed] [Google Scholar]
- Sun S, Yang F, Tan G, Costanzo M, Oughtred R, Hirschman J, Theesfeld CL, Bansal P, Sahni N, Yi S, Yu A, Tyagi T, Tie C, Hill DE, Vidal M, Andrews BJ, Boone C, Dolinski K, Roth FP. An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res. 2016;26(5):670–680. doi: 10.1101/gr.192526.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taşan M, Musso G, Hao T, Vidal M, MacRae CA, Roth FP. Selecting causal genes from genome-wide association studies via functionally coherent subnetworks. Nat Methods. 2015;12(2):154–159. doi: 10.1038/nmeth.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium (2015) A global reference for human genetic variation. Nature 526(7571):68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed]
- Thyagarajan B, Bloom JD. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. eLife. 2014;3:e03300. doi: 10.7554/eLife.03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, Baker D. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501(7466):212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxlmayr MW, Hasenhindl C, Hackl M, Stadlmayr G, Rybka JD, Borth N, Grillari J, Rüker F, Obinger C. Construction of a stability landscape of the CH3 domain of human IgG1 by combining directed evolution with high throughput sequencing. J Mol Biol. 2012;423(3):397–412. doi: 10.1016/j.jmb.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Gupta K, Khare S, Jain PC, Patel S, Kumar P, Pulianmackal AJ, Aghera N, Varadarajan R. Molecular determinants of mutant phenotypes, inferred from saturation mutagenesis data. Mol Biol Evol. 2016;33(11):2960–2975. doi: 10.1093/molbev/msw182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar TR, Ma L, Roscoe B, Park SM, Bolon DN, Green MR. Resistance to vemurafenib resulting from a novel mutation in the BRAFV600e kinase domain. Pigment Cell Melanoma Res. 2014;27(1):124–133. doi: 10.1111/pcmr.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weile J, Sun S, Cote AG, Knapp J, Verby M, Mellor JC, Wu Y, Pons C, Wong C, Lieshout Nv, Yang F, Tasan M, Tan G, Yang S, Fowler DM, Nussbaum R, Bloom JD, Vidal M, Hill DE, Aloy P, Roth FP. A framework for exhaustively mapping functional missense variants. Mol Syst Biol. 2017;13(12):957. doi: 10.15252/msb.20177908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead TA, Chevalier A, Song Y, Dreyfus C, Fleishman SJ, De Mattos C, Myers CA, Kamisetty H, Blair P, Wilson IA, Baker D. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30(6):543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Young AP, Dandekar S, Wijersuriya H, Al-Mawsawi LQ, Wu TT, Sun R. Systematic identification of H274y compensatory mutations in influenza a virus neuraminidase by high-throughput screening. J Virol. 2013;87(2):1193–1199. doi: 10.1128/JVI.01658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Olson CA, Du Y, Le S, Tran K, Remenyi R, Gong D, Al-Mawsawi LQ, Qi H, Wu TT, Sun R. Functional constraint profiling of a viral protein reveals discordance of evolutionary conservation and functionality. PLoS Genet. 2015;11(7):e1005310. doi: 10.1371/journal.pgen.1005310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Anders OC, Sun R. High-throughput identification of protein mutant stability computed from a double mutant fitness landscape. Protein Sci. 2016;25(2):530–539. doi: 10.1002/pro.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Wu Z, Fields S. Protein-peptide interactions analyzed with the yeast two-hybrid system. Nucl Acids Res. 1995;23(7):1152–1156. doi: 10.1093/nar/23.7.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TH, Wu NC, Sun R. A benchmark study on error-correction by read-pairing and tag-clustering in amplicon-based deep sequencing. BMC Genom. 2016;17:108. doi: 10.1186/s12864-016-2388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]