Abstract
MicroRNAs (miRs), which regulate target gene expression at the post-transcriptional level, play a crucial role in inducing biological effects upon high-dose ionizing radiation. Yet, the miR expression profiles in response to repeated low-dose radiation (LDR) in vivo have not been elucidated. This study investigated the response profiles of 11 miRs with functions involved in metabolism, DNA damage and repair, inflammation, and fibrosis in mouse liver, heart, and testis upon repeated LDR exposure for 4 months. The expression profiles were evaluated using stem-loop quantitative reverse transcription polymerase chain reaction immediately and at 2 months after LDR exposure. The expression profiles varied significantly at both time points. At the organ level, the heart was the most affected, followed by the liver and testis, in which significant miR upregulation related to DNA damage response was found. Metabolism-related miRs decreased in the liver and increased in the testis. The current results showed immediate and long-lasting alterations in the miR expression profiles in response to repeated LDR in different organs.
Keywords: microRNA, ionizing radiation, low dose
Introduction
MicroRNAs (miRs) are conserved, endogenous, small noncoding RNAs comprised of 16 to 22 nucleotides.1 At the post-transcriptional level, miRs bind to the 3′-untranslated regions of target mRNAs to negatively regulate their expression; in fact, 30% to 60% of protein-coding genes are regulated in this manner. The miRs are involved in most physiological and pathological processes, including development, differentiation, autophagy, apoptosis, DNA damage response, metabolism, and inflammation.2 They also initiate and are involved in an organism’s response to ionizing radiation.3
Low-dose radiation (LDR) triggers specific biological responses that are distinct from those caused by high-dose radiation (HDR). The mechanisms of biological responses to HDR have been studied extensively.4–8 Yet, the responses to LDR at the molecular, cellular, tissue, or organism levels are not fully understood. To date, increasing evidence indicates significant changes of various miRs upon irradiation.3,9,10 However, the LDR-induced biological responses of miRs in different organs of the same animal model remain unidentified.
In this study, the expression levels of 11 miRs were systematically and comprehensively examined using stem-loop quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays with hydrolysis probes immediately and at 2 months after repeated LDR exposure for 4 months in mouse liver, heart, and testis tissues.
Materials and Methods
Animals
Thirty-two male FVB/NJ mice, aged 12 weeks old, were purchased from Jackson Laboratory and were used for this study. All mice were housed at the University of Louisville Research Resources Center at 22°C with a 12-hour light/dark cycle. A diet of standard rodent chow and tap water were provided. The mice were divided into 2 groups: sham (n = 16) and LDR (n = 16) groups. In the LDR group, the mice were administered 28 mGy of γ-rays (source: cesium-137) every 3 days for 4 months. Half of the irradiated mice (n = 8) were sacrificed for renal function, histological, and biochemical measurements, whereas the remaining mice (n = 8) were kept for an additional 2 months without further irradiation. Identical examinations were performed at 2 months after LDR exposure. All experimental procedures were carried out in accordance with the guidelines provided by the US National Institutes of Health Care and Use of Laboratory Animals (National Academy Press, Washington, DC, 1996). Approval by the Institutional Animal Care and Use Committee of the University of Louisville (no.: 12067) was received.
Experimental Design and Setup
To approximate the dose delivered in medical examinations, 28 mGy, which is equivalent to a whole-body computed tomography examination, was selected as the LDR dose.11 A Gammacell 40 Exactor (Nordion Intl Inc, Canada) was used to deliver the whole-body LDR at a dose rate of 0.84 Gy/min.
The types of tissue were selected based on their functions: (1) liver for metabolism with cell proliferation, (2) heart for metabolism without active cell proliferation, and (3) testis for active cell proliferation.
MiR Selection
Most miRs play multiple roles and act on various downstream targets. The functions of miRs can significantly overlap. In this study, 11 miRs with functions predominantly involved in the regulation of metabolism, DNA damage and repair, cell-cycle control, as well as inflammation were analyzed (Table 1): (1) miR-34a and miR-185 regulate lipid metabolism, and miR-375 is involved in glucose-regulated insulin secretion and is required for normal pancreatic genesis; (2) miR-421, miR-193a, miR-21, and miR-199a are involved in active DNA damage response pathways; and (3) miR-199a, miR-146a, miR-155, and miR-221/222 participate in inflammation progression. In addition, miR-21 and miR-199a are related to fibrosis.12,13
Table 1.
Selected MicroRNAs and Their Key Target Genes Involved in Metabolism, DNA Damage Response, and Inflammation.
| MicroRNA | Target | Biological Effects |
|---|---|---|
| miR-34a | SIRT-1, HNF4α, MYC, CCND1, CCNE2, MAPK, ACSL1, CD44, PPARα | Regulate lipid and lipoprotein metabolism, promote cell division, senescence, and apoptosis |
| miR-375 | PDK1, GSK3, IGFR, JAK2, CCND2, ATG7 | Regulate glucose-stimulated insulin secretion, promote pancreatic development, contribute to cell growth, autophagy, and apoptosis |
| miR-185 | SREBP-1c, SREBP-2, Six1, DNMT1, SOCS3, STIM1 | Regulate lipid metabolism, improve insulin sensitivity, alter cell-cycle progression, and sensitizes cells to apoptosis |
| miR-193a | Mcl-1, CDK2, WT-1 | Promote apoptosis, active DNA damage response pathway |
| miR-421 | ATM, PINK1, FXR, SMAD4, FOXO4 | Involved in DNA despair response, cell proliferation, and apoptosis and regulate radiosensitivity |
| miR-21 | PTEN, cdc25a, TGF-β, EGFR, ERK, PPARα | Have influence on cell-cycle control, DNA damage repair, apoptosis, autophagy; enhance radiosensitivity in some cells; and contribute to fibrotic process |
| miR-199a | HIF-1α, VEGFA, GLUT-4, COX-2, FZD7 | Regulate angiogenesis, contribute to fibrotic process, and involved in cell proliferation and survival |
| miR-146a | NF-κB, TNF-α, IL-1β, SMAD4, TRAF6 | Involved in inflammation, tumor progression, promote autophagy, and regulate apoptosis |
| miR-155 | SHIP1, SOCS1, NF-κB | Regulate autoimmune inflammation and leukemogenesis |
| miR-221/222 | PTEN, ADIDOR1, p27, p57, TIMP2, PUMA | Regulate cell cycle inhibition, cell proliferation, apoptosis, radiosensitivity; regulate endothelial inflammatory response |
RNA Extraction
Prepared tissues were homogenized in 1 mL of Trizol reagent (Invitrogen, Carlsbad, CA) and mixed vigorously with 0.2 mL of chloroform for 15 seconds to isolate the RNAs. The aqueous phase was isolated by centrifugation at 4°C and 12 000 rpm for 15 minutes. RNA was precipitated with 0.5 mL of isopropyl alcohol, washed with 75% ethanol by centrifugation at 4°C and 7500 rpm for 5 minutes, and dissolved in 20 to 40 μL of nuclease-free water. A Nanodrop spectrophotometer (Thermo) at 260/280 nm was used to determine the concentration and purity of the RNA.
qRT-PCR Analysis of MiRs
A TaqMan miR assay kit (Applied Biosystems, Foster City, California) was used to validate the miR expression analysis. Briefly, each RT reaction contained 5 ng of total purified RNA, 1× RT buffer, 5× stem-loop RT primer, 0.25 mM of each dNTP, 50 U of MultiScribe reverse transcriptase, and 3.8 U of RNase inhibitor. The reactions were incubated for 30 minutes at 16°C, 30 minutes at 42°C, and 5 minutes at 85°C and were kept at 4°C. An ABI 7500 real-time PCR system and TaqMan miR assays were used to amplify the complementary DNA for quantitation of miR-34a, miR-375, miR-185, miR-21, miR-421, miR-193a, miR-199a, miR-146a, miR-155, miR-221, and miR-222. The expression of U6 was used as an endogenous control. All assays were performed in triplicate. All procedures were performed according to the manufacturer’s instructions. The 2−▵▵CT method was employed to determine the relative quantitative level and was expressed as a fold-difference to the relevant control (recognized as 1 [0.00]).
Data Analysis
All statistical tests were performed using Origin 7.5 data analysis and graphing software (OriginLab, Northampton, Massachusetts). One-way analysis of variance was used for comparisons among groups, followed by post hoc pairwise comparisons using the Student t test. Data were expressed as the mean (standard deviation). P < .05 was considered statistically significant.
Results
The expression levels of each miR analyzed immediately and at 2 months after LDR exposure were normalized relative to that in the control group. Repeated LDR exposure for 4 months in normal mice induced different changes of these miRs immediately and at 2 months after LDR exposure among the different organs, respectively. The results are described in terms of the organs and time points.
Alteration of MiR Expression in Liver Tissue Upon LDR
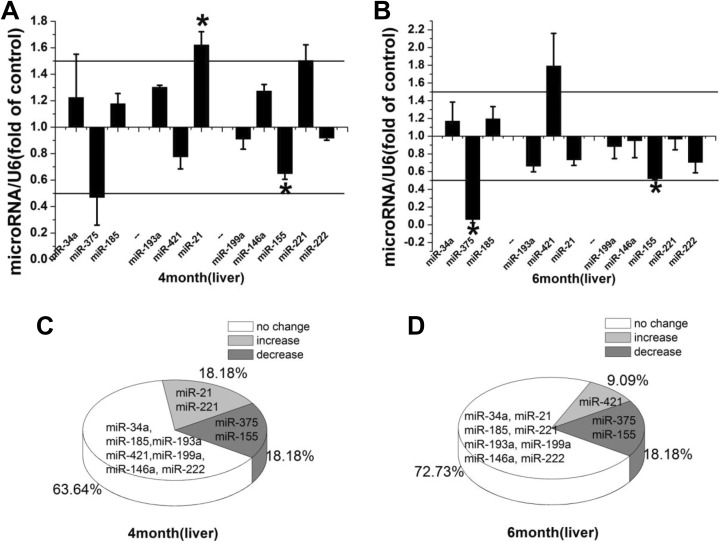
MicroRNAs play important roles in normal liver development, physiology, and pathophysiology.14,15 In response to LDR, miR-21 and miR-221 were upregulated (18.18%; 2/11) and miR-375 and miR-155 were downregulated (18.18%; 2/11), while the remaining miRs did not show significant changes in expression (63.64%; 7/11; Figure 1A and C). At 2 months after LDR exposure, the expression of miR-421 was increased, whereas the expression of both miR-375 and miR-155 was significantly decreased (Figure 1B). These composed a new profile of 9.09% upregulated miRs, 18.18% downregulated miRs, and 72.73% unchanged miRs (Figure 1D).
Figure 1.
A, B, Relative miR expression levels in the mouse liver immediately and at 2 months after repeated LDR exposure. Relative miR expression is represented by fold-change compared to the control. Student t test: *P < .05. C, D, Profile of alteration in miR expression immediately and at 2 months after LDR exposure. miR indicates microRNA; LDR, low-dose radiation.
Alteration of MiR Expression in Heart Tissue Upon LDR
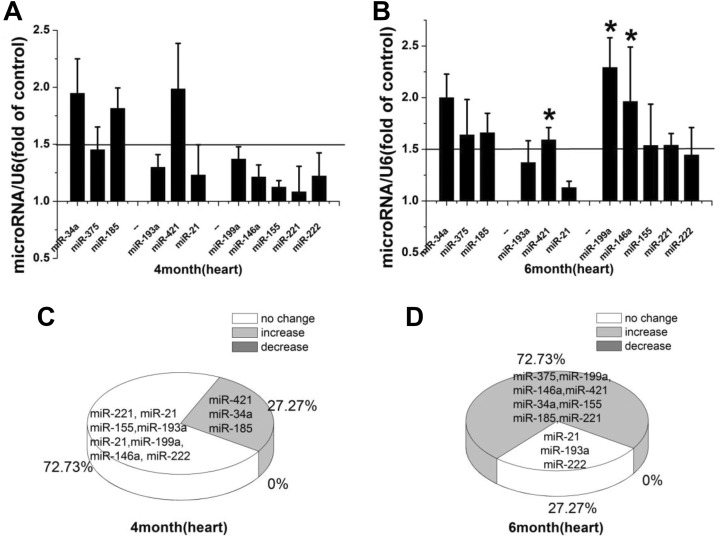
MiR-421, miR-34a, and miR-185 were upregulated (27.27%, 3/11), while no changes in expression were found for the remaining miRs analyzed immediately after LDR exposure (Figure 2A and C). In total, 8 miRs were upregulated (72.73%, 8/11) at 2 months after LDR exposure (Figure 2B and D). Significant upregulation was found for miR-199a, miR-146a, miR-421, and miR-34a, whereas the expression levels of miR-375, miR-185, miR-155, and miR-221 were upregulated without significance.
Figure 2.
A, B, Relative miR expression in the mouse heart immediately and at 2 months after repeated LDR exposure. Relative miR expression is represented by fold-change compared to the control. Student t test: *P < .05. C, D, Profile of alteration in miR expression immediately and at 2 months after LDR exposure. miR indicates microRNA; LDR, low-dose radiation.
Alteration of MiR Expression in Testis Tissue Upon LDR
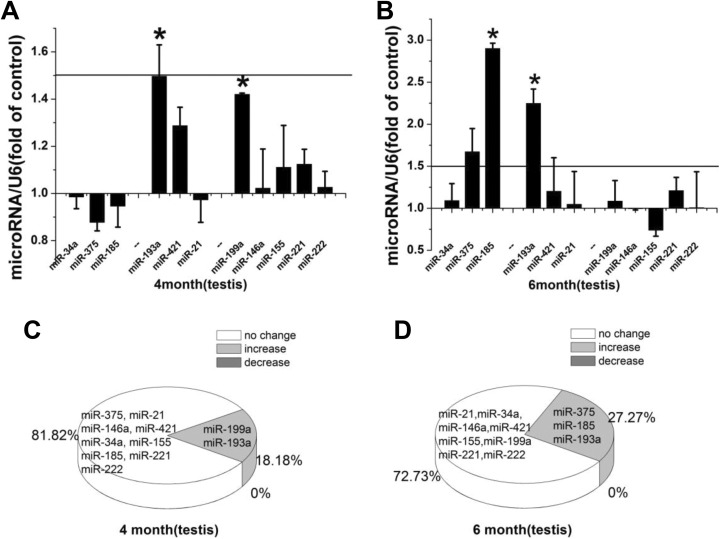
During spermatogenesis, miRs are crucial for translational regulation.16 Immediately after LDR exposure, miR-193a and miR-199a were significantly upregulated (Figure 3A). At 2 months after LDR, miR-185 and miR-193a were significantly upregulated, and miR-375 also showed apparent upregulation (Figure 3B). The percentages of upregulated miRs immediately and at 2 months after LDR were 18.18% and 27.27%, respectively (Figure 3C and D).
Figure 3.
A, B, Relative miR expression in the mouse testis immediately and at 2 months after repeated LDR exposure. Relative miR expression is represented by fold-change compared to the control. Student t test: *P < .05. C, D, Profile of alteration in miR expression immediately and at 2 months after LDR exposure. miR indicates microRNA; LDR, low-dose radiation.
Comparison of MiR Expression in Various Tissues and the Time Response to LDR
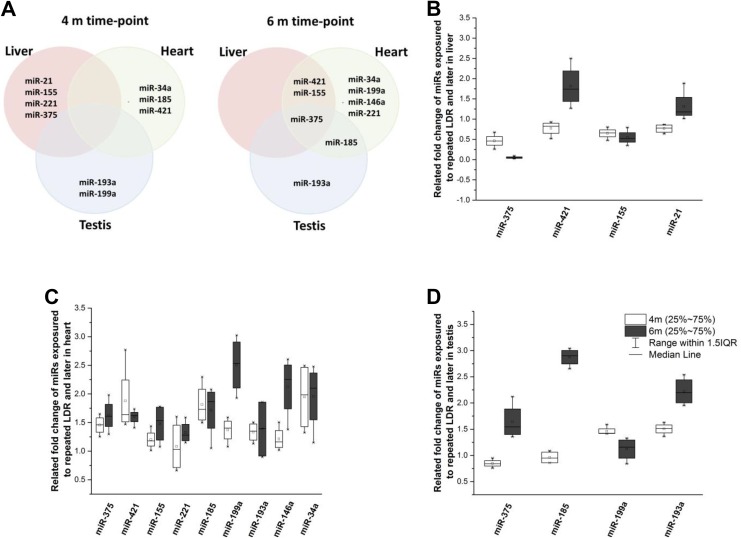
The expression of miRs that was 0.5-fold lower or 1.5-fold higher than that of the control was compared among organs and across time points, as shown in Figure 4. Four (miR-21, miR-155, miR-221, and miR-375), 3 (miR-34a, miR-185, and miR-421), and 2 (miR-193a and miR-199a) miRs were altered in the liver, heart, and testis, respectively, immediately after LDR exposure, showing that there was no common alteration in miR expression found among the tissue types. However, at 2 months after LDR, common late-responding miRs were observed. MiR-375 had a late response in all 3 organs. In addition, miR-421 and miR-155 showed changes in the liver and heart, whereas miR-185 was altered in the heart and testis.
Figure 4.
A, Profile of alteration in miR expression in the liver, heart, and testis tissues. B-D, Changes in miR expression from immediately after repeated LDR exposure (4-month time point) to 2 months after LDR exposure (6-month time point) in the liver, heart, and testis. miR indicates microRNA; LDR, low-dose radiation.
Throughout the experimental period, 2 miRs (miR-155 and miR-375) remained downregulated in the liver. Moreover, miR-21 and miR-421 had drastic changes in expression throughout the period. MiR-421 was first downregulated and then upregulated, whereas miR-21 was first upregulated and then downregulated (Figure 4B). In heart tissue (Figure 4C), 3 miRs (miR-34a, miR-421, and miR185) were upregulated and remained upregulated at 2 months after LDR. Of note, 5 different miRs (miR-146a, miR-155, miR-199a, miR-221, and miR-375) showed no immediate response to LDR but were upregulated at 2 months after LDR. In the testis (Figure 4D), only 2 miRs (miR-193a and miR-199a) were upregulated throughout the experiment, in which miR-193a expression increased further. On the other hand, miR-185 and miR-375 were upregulated significantly only at 2 months after LDR.
Discussion
Repeated whole-body LDR exposure to mice apparently altered the miR expression in the liver, heart, and testis tissues. This study examined the immediate and late responses of miR expression for various tissue types upon LDR.
Immediate Effects of LDR on MiR Expression in the Liver
Upregulation of miR-21 and miR-221 expression in the liver was found after LDR. MiR-21 is a well-defined survival factor during hepatocellular carcinoma development and liver injury.17,18 The overexpression of miR-21 during human liver regeneration suggests that miR-21 is associated with hepatic cell survival and regeneration.19 Reportedly, miR-21 is oncogenic and is involved in the cellular response to ionizing radiation.20 Consistent with this study, here we also showed upregulated expression of miR-21 by repeated LDR, suggesting the possible role of miR-21 in stimulating cell proliferation.
Evidence indicates that miR-221 is negatively associated with the endothelial nitric oxide synthase (eNOS) level in endothelial cells21 and is involved in the mitogen-activated protein kinase kinase/extracellular signal-regulated kinase (ERK) pathway to inhibit the proliferation of endothelial progenitor cells through p21-activating kinase 1.22 Therefore, miR-221 might regulate nitric oxide production by reducing eNOS expression or phosphorylation. Moreover, miR-221 promotes the proliferation and migration of endothelial tip cells during vascular development.23 Another study has shown that miR-221 inhibits nitric oxide synthesis and activates nuclear factor (NF)-κB signaling.24 These results suggest that the positive feedback of miR-221 and NF-κB is possible to enhance the inflammatory response. Besides, miR-221 has been shown to participate in glucose/fat metabolism. In humans, overexpression of miR-221 upregulated several proteins involved in fat metabolism, similar to peroxisome proliferator-activated receptor activation.25 miR-221 is reported to affect radiosensitivity by targeting phosphatase and tensin homolog/protein kinase B pathway.26 The liver is heavily involved in metabolism and cell proliferation, which are relatively insensitive to ionizing radiation. Upregulation of miR-221 might suggest an alteration in glucose/fat metabolism and the involvement of inflammation after whole-body LDR.
The downregulation of miR-375 and miR-155 has many effects. For instance, miR-375 regulates insulin secretion, pancreatic islet development, and alveolar epithelial cell transdifferentiation27,28 or adipocyte differentiation probably related to regulating phosphoinositide-dependent protein kinase 1, myotrophin, and ERK1/2 expression and function.29,30 However, limited, if any, studies have investigated the expression of miR-375 upon whole-body LDR, and the present data showed downregulation of miR-375 in the liver and upregulation in the heart and testis. These results suggest that miR-375 is a target after LDR; however, the exact mechanism of the different regulation among tissues remains to be elucidated.
MiR-155 showed distinct expression profiles in hematopoietic lineage differentiation, immunity, inflammation, cancer, and cardiovascular diseases.31 Although miR-155 has been shown to be intensively involved in regulating inflammation-associated processes,32,33 anti-inflammatory proteins have been found to be the target genes of miR-155, for example, the suppressor of cytokine signaling 1.34 In line with previous reports, the downregulation of miR-155 suggested inhibition of the inflammatory response upon LDR.35,36
Immediate Effects of LDR on MiR Expression in the Heart
Previously, miR-421 expression was mainly found in multiple cancer cell types.37–39 Moreover, Wang et al have reported that mitochondrial fragmentation and apoptosis are regulated by miR-421 in vitro and in vivo through targeting Pink1.40 When miR-421 interference using antagomirs or an miR-insensitive expression vector for ATM was used, radiation sensitivity was dramatically reduced.41 Thus, miR-421 could be a potential marker for radiosensitivity and heart that contains cells rich in mitochondria, and our findings may indicate that the heart was sensitive to LDR-related mitochondrial alterations.
MiR-34a has been shown to be hypermethylated in cancers, whereas ectopic miR-34a induces G1 cell-cycle arrest, senescence, and apoptosis, suggesting its tumor-suppressive nature.42 SIRT1, which regulates the activity of adenosine monophosphate kinase and is a known regulator of energy metabolism, has been shown to be suppressed by miR-34a.43 Furthermore, it has been demonstrated that downregulation of miR-34a alters lipid metabolism in the liver of nonalcoholic fatty liver disease.44 Moreover, downregulation of miR-34a has been shown to reduce cell death and fibrosis in acute myocardial infarction, thus improving the recovery of myocardial function.45 Therefore, upregulation of miR-34a could be a sensitive biomarker of heart injury.
Selective high-density lipoprotein cholesterol uptake is repressed by miR-185 through the direct targeting of high-density lipoprotein scavenger receptor class B type I in human hepatic cells.46 In prostate cancer cells47 and in hepatocytes,48 fatty acid metabolism, lipogenesis, and cholesterogenesis have been shown to be regulated by miR-185 through targeting SREBP1 and SREBP2. Another study has demonstrated that miR-185 can enhance radiation-induced apoptosis and proliferation inhibition.49 Our study is the first to report the upregulation of miR-185 in the heart in response to LDR.
Immediate Effects of LDR on MiR Expression in the Testis
In a study, ionizing radiation induced miR-193a, which downregulates Mcl-1 and eventually leads to apoptosis by increasing reactive oxygen species and DNA damage.50 Other studies indicated significant downregulation of miR-193a in non-small cell lung cancer, which was associated with proliferation, metastasis, and apoptosis.51,52 The testis is known as one of the organs with predominantly active cell proliferation. Thus, miR-193a might be a potential biomarker for harmful effects of LDR.
Previous studies have indicated that miR-199a expression is modulated during development, growth, and regeneration in cancers.53–55 Specifically, miR-199a-5p is involved in cell proliferation, migration, and apoptosis in a hypoxic tumor microenvironment.54 MiR-199a-5p is also heavily involved in the process of fibrosis. Furthermore, it has been linked to multiple processes associated with fibrogenesis, including cell proliferation, migration, invasion, and differentiation into myofibroblasts. Therefore, the upregulation of miR-199a observed in the mouse testis might suggest fibrosis upon LDR.
Late Effects of LDR on MiR Expression in the Liver, Heart, and Testis
The effect of LDR at 2 months after exposure was investigated. Our results indicated that some of the immediate responses of miRs returned to a normal level, while others remained dysregulated. Interestingly, the expression of a few miRs showed drastic changes at 2 months after LDR exposure (Figure 4B–D).
Among the altered miRs, 8 miRs were changed, independent of the postirradiation time point, including miR-21 and miR-221 in the liver; miR-375, miR-199a, miR-146a, miR-221, and miR-155 in the heart; and miR-199a, miR-185, and miR-375 in the testis. Meanwhile, the expression of miR-21 and miR-221 in the liver and miR-199a in the testis returned to control levels at 2 months after LDR exposure.
These miRs with drastic alteration throughout the experiment are involved in the DNA repair response, cell proliferation, apoptosis, as well as regulation of radiosensitivity. These signaling pathways were activated with the upregulation of miR-221 in the liver after LDR exposure.41 Sustained upregulation was found in 5 miRs (miR-375, miR-199a, miR-155, miR-146a, and miR-221) in the heart tissue, and these miRs are reported to correlate with heart development and disease. Studies have shown that miR-375 is significantly upregulated in maternal serum at 18 to 22 weeks of gestation in a fetus with congenital heart defects (CHD);56 in addition, it is associated with heart development and the occurrence of CHD through the Notch signaling pathway.57 MiR-155 can regulate endothelial inflammation,58 whereas the expression of miR-155 is upregulated in atherosclerotic plaques and is altered in ischemic stroke.59 On the other hand, miR-199a has been shown to be frequently upregulated in pressure-overloaded hypertrophic hearts.60 In addition, dysregulation of miR-146a has been found in a few vascular diseases, including abdominal aortic aneurysm, atherosclerosis, and coronary artery disease.61–63 Lastly, regulated miRs in the testis (miR-375 and miR-185) are involved in metabolism regulation. The detailed roles, functions, and biological effects of altered expression of these novel miRs in response to LDR in mice should be further studied for a better understanding of the effects of LDR.
LDR Affects MiR Expression and Their Functions
The miRs that had altered expression upon LDR are mainly involved in metabolism, DNA damage response, cell-cycle regulation, and inflammation. Previous studies have reported metabolic changes in cells after irradiation, such as downregulation of the citric acid cycle, pyruvate, and fatty acid metabolism pathways in the mammary gland.64 Precancerous and cancer cells depend on glycolysis, which increases fatty acid synthesis and increases the rate of glutamine metabolism. These properties are often employed in cancer therapeutics.65 Prediction of the radiotherapeutic response is based on a progressive decrease in glucose metabolism in cancer cells.66 MiR-34a, miR-185, and miR-375 are known to regulate these metabolic processes.48,67,68 In this study, LDR led to significant modifications in gene expression at the transcriptional level. MiR-375 was regulated in all 3 tissue types, miR-185 was upregulated in both the heart and the testis, while an apparent increase in miR-34a expression was found in the heart. Our findings agreed with previous reports and have important implications in understanding and assessing the health risks of radiation exposure.69,70
The DNA damage response is activated upon DNA damage to prevent the accumulation of gene mutations and chromosomal rearrangements, which are related to carcinogenesis.71 Many efforts have been made to analyze alterations of miR expression profiles upon ionizing radiation. Recent studies have detected many miRs that respond to ionizing radiation in various biological contexts.72–74 A number of miRs that modulate a cell’s sensitivity to radiotherapy have been identified.75 Validated targets of misregulated miRs fall into the cell cycle and apoptosis categories. The increased expression of select miRs also has been linked to cell-cycle arrest, apoptosis, and the DNA damage response.
Inflammation maintains tissue integrity through a homeostatic mechanism. The underlying immunological mechanisms and the relationship between ionizing radiation and inflammation are complex and multifactorial on both the cellular and biochemical levels. Inflammatory events vary in benignancy and malignancy. Published data indicate that LDR therapy influences murine osteoclast differentiation and may reduce bone destruction.76 Without affecting basic functionalities, such as phagocytosis, the phenotype of inflammatory cells can be switched to an anti-inflammatory status by LDR therapy. Recently, metabolic risk factors, such as decreased ApoE lipoproteins, were found to enhance proinflammatory and prothrombotic late responses in locally irradiated hearts.77–79 Another study also has suggested that LDR has the potential to prevent diabetic cardiovascular complications.80 Our study showed that the expression of inflammation-related miRs, including miR-155, miR146, and miR221, were changed by LDR. However, various aspects of the response upon LDR were not investigated. Further studies are needed to clarify this issue in vitro and in vivo.
Conclusion
The current study revealed alteration in miR expression in the liver, heart, and testis tissues upon whole-body repeated LDR of mice. All 3 of these tissue types showed different miR expression profiles, with the heart showing the greatest number of responding miRs. Several miRs that may be associated with the regulation of metabolism, DNA damage response, and inflammation were identified. These data improved the understanding of cellular and genetic changes induced by LDR.
Acknowledgments
The authors are grateful to Dr Jing Chen for her excellent assistance with the experimental procedures and animal maintenance.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by grants from the National Natural Science Foundation of China (81302379, X.L.; 81302380, D.Y.), the Key Project of Science and Technology Research of the Ministry of Education (311015, J.C.), and the National Institutes of Health (1R01DK 091338-01A1, L.C.).
ORCID iD: Lu Cai  http://orcid.org/0000-0003-3048-1135
http://orcid.org/0000-0003-3048-1135
References
- 1. Himoto T, Nomura T, Tani J, et al. Exacerbation of insulin resistance and hepatic steatosis deriving from zinc deficiency in patients with HCV-related chronic liver disease. Biol Trace Elem Res. 2015;163(1-2):81–88. [DOI] [PubMed] [Google Scholar]
- 2. Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget, 2015;6(11):8474–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr Opin Genet Dev. 2013;23(1):12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonner WM. Phenomena leading to cell survival values which deviate from linear-quadratic models. Mutat Res. 2004;568(1):33–39. [DOI] [PubMed] [Google Scholar]
- 5. Matsumoto H, Hamada N, Takahashi A, Kobayashi Y, Ohnishi T. Vanguards of paradigm shift in radiation biology: radiation-induced adaptive and bystander responses. J Radiat Res. 2007;48(2):97–106. [DOI] [PubMed] [Google Scholar]
- 6. Marples B, Collis SJ. Low-dose hyper-radiosensitivity: past, present, and future. Int J Radiat Oncol Biol Phys. 2008;70(5):1310–1318. [DOI] [PubMed] [Google Scholar]
- 7. Dauer LT, Brooks AL, Hoel DG, Morgan WF, Stram D, Tran P. Review and evaluation of updated research on the health effects associated with low-dose ionising radiation. Radiat Prot Dosim. 2010;140(2):103–136. [DOI] [PubMed] [Google Scholar]
- 8. Morgan WF, Bair WJ. Issues in low dose radiation biology: the controversy continues. A perspective. Radiat Res. 2013;179(5):501–510. [DOI] [PubMed] [Google Scholar]
- 9. Czochor JR, Glazer PM. MicroRNAs in cancer cell response to ionizing radiation. Antioxid Redox Signal. 2014;21(2):293–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marta GN, Garicochea B, Carvalho AL, Real JM, Kowalski LP. MicroRNAs, cancer and ionizing radiation: where are we? Rev Assoc Med Bras. 2015;61(3):275–281. [DOI] [PubMed] [Google Scholar]
- 11. Lell MM, Wildberger JE, Alkadhi H, Damilakis J, Kachelriess M. Evolution in computed tomography: the battle for speed and dose. Invest Radiol. 2015;50(9):629–644. [DOI] [PubMed] [Google Scholar]
- 12. Zhao J, Tang N, Wu K, et al. MiR-21 simultaneously regulates ERK1 signaling in HSC activation and hepatocyte EMT in hepatic fibrosis. PLos One. 2014;9(10):e108005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Henaoui IS, Cauffiez C, Aubert S, et al. [miR-199a-5p in idiopathic pulmonary fibrosis]. Med Sci (Paris). 2013;29(5):461–463. [DOI] [PubMed] [Google Scholar]
- 14. Schueller F, Roy S, Vucur M, Trautwein C, Luedde T, Roderburg C. The role of miRNAs in the pathophysiology of liver diseases and toxicity. Int J Mol Sci. 2018;19(1):pii: E261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Y, Verfaillie CM. MicroRNAs: the fine modulators of liver development and function. Liver Int. 2014;34(7):976–990. [DOI] [PubMed] [Google Scholar]
- 16. Chen X, Li X, Guo J, Zhang P, Zeng W. The roles of microRNAs in regulation of mammalian spermatogenesis. J Anim Sci Biotechnol. 2017;8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu C, Yu J, Yu S, et al. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J Hepatol. 2010;53(1):98–107. [DOI] [PubMed] [Google Scholar]
- 18. Francis H, McDaniel K, Han Y, et al. Regulation of the extrinsic apoptotic pathway by microRNA-21 in alcoholic liver injury. J Biol Chem. 2014;289(40):27526–27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song G, Sharma AD, Roll GR, et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology. 2010;51(5):1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Halimi M, Parsian H, Asghari SM, et al. Clinical translation of human microRNA 21 as a potential biomarker for exposure to ionizing radiation. Transl Res. 2014;163(6):578–584. [DOI] [PubMed] [Google Scholar]
- 21. Rippe C, Blimline M, Magerko KA, et al. MicroRNA changes in human arterial endothelial cells with senescence: relation to apoptosis, eNOS and inflammation. Exp Gerontol. 2012;47(1):45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang X, Mao H, Chen JY, et al. Increased expression of microRNA-221 inhibits PAK1 in endothelial progenitor cells and impairs its function via c-Raf/MEK/ERK pathway. Biochem Biophys Res Commun. 2013;431(3):404–408. [DOI] [PubMed] [Google Scholar]
- 23. Nicoli S, Knyphausen CP, Zhu LJ, Lakshmanan A, Lawson ND. . miR-221 is required for endothelial tip cell behaviors during vascular development. Dev Cell. 2012;22(2):418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen CF, Huang J, Li H, et al. MicroRNA-221 regulates endothelial nitric oxide production and inflammatory response by targeting adiponectin receptor 1. Gene. 2015;565(2):246–251. [DOI] [PubMed] [Google Scholar]
- 25. Meerson A, Traurig M, Ossowski V, Fleming JM, Mullins M, Baier LJ. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-alpha. Diabetologia. 2013;56(9):1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang J, Han L, Ge Y, et al. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol. 2010;36(4):913–920. [DOI] [PubMed] [Google Scholar]
- 27. Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4(1):9–12. [DOI] [PubMed] [Google Scholar]
- 28. Abdelmohsen K, Hutchison ER, Lee EK, et al. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol Cell Biol. 2010;30(17):4197–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fernandez-Valverde SL, Taft RJ, Mattick JS. MicroRNAs in beta-cell biology, insulin resistance, diabetes and its complications. Diabetes. 2011;60(7):1825–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dehwah MA, Xu A, Huang Q. MicroRNAs and type 2 diabetes/obesity. J Genet Genomics. 2012;39(1):11–18. [DOI] [PubMed] [Google Scholar]
- 31. Faraoni I, Antonetti FR, Cardone J, Bonmassar E. . MiR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792(6):497–505. [DOI] [PubMed] [Google Scholar]
- 32. Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179(8):5082–5089. [DOI] [PubMed] [Google Scholar]
- 33. Marques-Rocha JL, Milagro FI, Mansego ML, Zulet MA, Bressan J, Martínez JA. Expression of inflammation-related miRNAs in white blood cells from subjects with metabolic syndrome after 8 wk of following a Mediterranean diet-based weight loss program. Nutrition. 2016;32(1):48–55. [DOI] [PubMed] [Google Scholar]
- 34. Cardoso AL, Guedes JR, Pereira de Almeida L, Pedroso de Lima MC. MiR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology. 2012;135(1):73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frischholz B, Wunderlich R, Ruhle PF, et al. Reduced secretion of the inflammatory cytokine IL-1beta by stimulated peritoneal macrophages of radiosensitive Balb/c mice after exposure to 0.5 or 0.7 Gy of ionizing radiation. Autoimmunity. 2013;46(5):323–328. [DOI] [PubMed] [Google Scholar]
- 36. Hong EH, Song JY, Lee SJ, et al. Low-dose gamma-radiation inhibits IL-1beta-induced dedifferentiation and inflammation of articular chondrocytes via blockage of catenin signaling. IUBMB Life. 2014;66(2):128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lerebours F, Cizeron-Clairac G, Susini A, et al. MiRNA expression profiling of inflammatory breast cancer identifies a 5-miRNA signature predictive of breast tumor aggressiveness. Int J Cancer. 2013;133(7):1614–1623. [DOI] [PubMed] [Google Scholar]
- 38. Lin L, Lin Y, Jin Y, Zheng C. Microarray analysis of microRNA expression in liver cancer tissues and normal control. Gene. 2013;523(2):158–160. [DOI] [PubMed] [Google Scholar]
- 39. Wu JH, Yao YL, Gu T, et al. MiR-421 regulates apoptosis of BGC-823 gastric cancer cells by targeting caspase-3. Asian Pac J Cancer Prev. 2014;15(13):5463–5468. [DOI] [PubMed] [Google Scholar]
- 40. Wang K, Zhou LY, Wang JX, et al. E2F1-dependent miR-421 regulates mitochondrial fragmentation and myocardial infarction by targeting Pink1. Nat Commun. 2015;6:7619. [DOI] [PubMed] [Google Scholar]
- 41. Mansour WY, Bogdanova NV, Kasten-Pisula U, et al. Aberrant overexpression of miR-421 downregulates ATM and leads to a pronounced DSB repair defect and clinical hypersensitivity in SKX squamous cell carcinoma. Radiother Oncol. 2013;106(1):147–154. [DOI] [PubMed] [Google Scholar]
- 42. Lodygin D, Tarasov V, Epanchintsev A, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7(16):2591–2600. [DOI] [PubMed] [Google Scholar]
- 43. McCubbrey AL, Nelson JD, Stolberg VR, et al. MicroRNA-34a negatively regulates efferocytosis by tissue macrophages in part via SIRT1. J Immunol. 2016;196(3):1366–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ding J, Li M, Wan X, et al. Effect of miR-34a in regulating steatosis by targeting PPARalpha expression in nonalcoholic fatty liver disease. Sci Rep. 2015;5:13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boon RA, Iekushi K, Lechner S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495(7439):107–110. [DOI] [PubMed] [Google Scholar]
- 46. Wang L, Jia XJ, Jiang HJ, et al. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol. 2013;33(10):1956–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li X, Chen YT, Josson S, et al. MicroRNA-185 and 342 inhibit tumorigenicity and induce apoptosis through blockade of the SREBP metabolic pathway in prostate cancer cells. PLos One. 2013;8(8):e70987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang XC, Zhan XR, Li XY, Yu JJ, Liu XM. MicroRNA-185 regulates expression of lipid metabolism genes and improves insulin sensitivity in mice with non-alcoholic fatty liver disease. World J Gastroenterol. 2014;20(47):17914–17923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang J, He J, Su F, et al. Repression of ATR pathway by miR-185 enhances radiation-induced apoptosis and proliferation inhibition. Cell Death Dis. 2013;4:e699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kwon JE, Kim BY, Kwak SY, Bae IH, Han YH. Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting Mcl-1. Apoptosis. 2013;18(7):896–909. [DOI] [PubMed] [Google Scholar]
- 51. Wang J, Yang B, Han L, et al. Demethylation of miR-9-3 and miR-193a genes suppresses proliferation and promotes apoptosis in non-small cell lung cancer cell lines. Cell Physiol Biochem. 2013;32(6):1707–1719. [DOI] [PubMed] [Google Scholar]
- 52. Yu T, Li J, Yan M, et al. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34(4):413–423. [DOI] [PubMed] [Google Scholar]
- 53. Xu N, Zhang J, Shen C, et al. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423(4):826–831. [DOI] [PubMed] [Google Scholar]
- 54. Raimondi L, Amodio N, Di Martino MT, et al. Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: in vitro and in vivo anti-tumor activity. Oncotarget. 2014;5(10):3039–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mata R, Palladino C, Nicolosi ML, et al. IGF-I induces upregulation of DDR1 collagen receptor in breast cancer cells by suppressing MIR-199a-5p through the PI3K/AKT pathway. Oncotarget. 2016;7(7):7683–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu S, Cao L, Zhu J, et al. Identification of maternal serum microRNAs as novel non-invasive biomarkers for prenatal detection of fetal congenital heart defects. Clin Chim Acta. 2013;424:66–72. [DOI] [PubMed] [Google Scholar]
- 57. Wang L, Song G, Liu M, et al. MicroRNA-375 overexpression influences P19 cell proliferation, apoptosis and differentiation through the Notch signaling pathway. Int J Mol Med. 2016;37(1):47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhu N, Zhang D, Chen S, et al. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis. 2011;215(2):286–293. [DOI] [PubMed] [Google Scholar]
- 59. Hunsberger JG, Fessler EB, Wang Z, Elkahloun AG, Chuang DM. Post-insult valproic acid-regulated microRNAs: potential targets for cerebral ischemia. Am J Transl Res. 2012;4(3):316–332. [PMC free article] [PubMed] [Google Scholar]
- 60. el Azzouzi H, Leptidis S, Dirkx E, et al. The hypoxia-inducible microRNA cluster miR-199a approximately 214 targets myocardial PPARdelta and impairs mitochondrial fatty acid oxidation. Cell Metab. 2013;18(3):341–354. [DOI] [PubMed] [Google Scholar]
- 61. Schroen B, Heymans S. Small but smart—microRNAs in the centre of inflammatory processes during cardiovascular diseases, the metabolic syndrome, and ageing. Cardiovasc Res. 2012;93(4):605–613. [DOI] [PubMed] [Google Scholar]
- 62. Chen J, Yuan L, Fan Q, Su F, Chen Y, Hu S. Adjuvant effect of docetaxel on the immune responses to influenza A H1N1 vaccine in mice. BMC Immunol. 2012;13:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kin K, Miyagawa S, Fukushima S, et al. Tissue- and plasma-specific microRNA signatures for atherosclerotic abdominal aortic aneurysm. J Am Heart Assoc. 2012;1(5):e000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luzhna L, Kovalchuk O. Low dose irradiation profoundly affects transcriptome and microRNAme in rat mammary gland tissues. Oncoscience. 2014;1(11):751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Giovacchini G, Picchio M, Schipani S, et al. Changes in glucose metabolism during and after radiotherapy in non-small cell lung cancer. Tumori. 2009;95(2):177–184. [DOI] [PubMed] [Google Scholar]
- 67. Fernandez-Hernando C. Emerging role of microRNAs in the regulation of lipid metabolism. Hepatology. 2013;57(2):432–434. [DOI] [PubMed] [Google Scholar]
- 68. Yang Z, Cappello T, Wang L. Emerging role of microRNAs in lipid metabolism. Acta Pharm Sin B. 2015;5(2):145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lall R, Ganapathy S, Yang M, et al. Low-dose radiation exposure induces a HIF-1-mediated adaptive and protective metabolic response. Cell Death Differ. 2014;21(5):836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bakshi MV, Azimzadeh O, Barjaktarovic Z, et al. Total body exposure to low-dose ionizing radiation induces long-term alterations to the liver proteome of neonatally exposed mice. J Proteome Res. 2015;14(1):366–373. [DOI] [PubMed] [Google Scholar]
- 71. van Jaarsveld MT, Wouters MD, Boersma AW, et al. DNA damage responsive microRNAs misexpressed in human cancer modulate therapy sensitivity. Mol Oncol. 2014;8(3):458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Simone NL, Soule BP, Ly D, et al. Ionizing radiation-induced oxidative stress alters miRNA expression. PLos One. 2009;4(7):e6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, Huber PE. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat Oncol. 2010;5:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Niemoeller OM, Niyazi M, Corradini S, et al. MicroRNA expression profiles in human cancer cells after ionizing radiation. Radiat Oncol. 2011;6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chistiakov DA, Chekhonin VP. Contribution of microRNAs to radio- and chemoresistance of brain tumors and their therapeutic potential. Eur J Pharmacol. 2012;684(1-3):8–18. [DOI] [PubMed] [Google Scholar]
- 76. Kulzer L, Rubner Y, Deloch L, et al. Norm- and hypo-fractionated radiotherapy is capable of activating human dendritic cells. J Immunotoxicol. 2014;11(4):328–336. [DOI] [PubMed] [Google Scholar]
- 77. Monceau V, Meziani L, Strup-Perrot C, et al. Enhanced sensitivity to low dose irradiation of ApoE−/− mice mediated by early pro-inflammatory profile and delayed activation of the TGFbeta1 cascade involved in fibrogenesis. PLos One. 2013;8(2):e57052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mathias D, Mitchel RE, Barclay M, et al. Low-dose irradiation affects expression of inflammatory markers in the heart of ApoE−/− mice. PLos One. 2015;10(3):e0119661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Patties I, Haagen J, Dorr W, Hildebrandt G, Glasow A. Late inflammatory and thrombotic changes in irradiated hearts of C57BL/6 wild-type and atherosclerosis-prone ApoE-deficient mice. Strahlenther Onkol. 2015;191(2):172–179. [DOI] [PubMed] [Google Scholar]
- 80. Zhang C, Jin S, Guo W, et al. Attenuation of diabetes-induced cardiac inflammation and pathological remodeling by low-dose radiation. Radiat Res. 2011;175(3):307–321. [DOI] [PubMed] [Google Scholar]