Abstract
Saccharomyces cerevisiae is a commonly used model organism for understanding eukaryotic gene function. However, the close proximity between yeast genes can complicate the interpretation of yeast genetic data, particularly high-throughput data. In this study, we examined the interplay between genes encoding components of the PAF1 complex and VPS36, the gene located next to CDC73 on chromosome XII. The PAF1 complex (Cdc73, Paf1, Ctr9, Leo1, and Rtf1, in yeast) affects RNA levels by affecting transcription, histone modifications, and post-transcriptional RNA processing. The human PAF1 complex is linked to cancer, and in yeast, it has been reported to play a role in telomere biology. Vps36, part of the ESCRT-II complex, is involved in sorting proteins for vacuolar/lysosomal degradation. We document a complex set of genetic interactions, which include an adjacent gene effect between CDC73 and VPS36 and synthetic sickness between vps36Δ and cdc73Δ, paf1Δ, or ctr9Δ. Importantly, paf1Δ and ctr9Δ are synthetically lethal with deletions of other components of the ESCRT-II (SNF8 and VPS25), ESCRT-I (STP22), or ESCRT-III (SNF7) complexes. We found that RNA levels of VPS36, but not other ESCRT components, are positively regulated by all components of the PAF1 complex. Finally, we show that deletion of ESCRT components decreases the telomere length in the S288C yeast genetic background, but not in the W303 background. Together, our results outline complex interactions, in cis and in trans, between genes encoding PAF1 and ESCRT-II complex components that affect telomere function and cell viability in yeast.
Electronic supplementary material
The online version of this article (10.1007/s00294-018-0828-6) contains supplementary material, which is available to authorized users.
Keywords: Saccharomyces cerevisiae, PAF1 complex, ESCRT machinery, Adjacent gene effect, Telomeres
Introduction
The budding yeast Saccharomyces cerevisiae is the best understood eukaryotic model organism. Its genome is compact and composed of over 6000 genes in 12 Mb of DNA, approximately 100-fold less dense than the human genome (Herrero et al. 2016). Such a high density of genes in yeast often leads to adjacent gene effects where deletion of one gene affects the function of the adjacent gene, for example by interfering with 5′ UTR (untranslated regions) or 3′ UTRs sequences (Ben-Shitrit et al. 2012; Atias et al. 2016). Adjacent gene effects can confound interpretation of genetic data, especially in large-scale studies. In yeast, CDC73, encoding a component of the PAF1 complex, is adjacent to VPS36, encoding a component of the ESCRT machinery and the two genes have been suggested to show an adjacent gene effect (Ben-Shitrit et al. 2012).
The PAF1 complex is a conserved transcriptional elongation factor that binds RNA polymerase II and is comprised of Cdc73, Paf1, Ctr9, Leo1, and Rtf1 (Betz et al. 2002; Jaehning 2010; Tomson and Arndt 2013). In budding yeast, Cdc73 and Rtf1 are the main interfaces for the physical interaction between the PAF1 complex and RNA pol II (Nordick et al. 2008; Xu et al. 2017). The PAF1 complex affects transcript abundance (Squazzo et al. 2002; Chen et al. 2009; Kim et al. 2010; Crisucci and Arndt 2011; Xu et al. 2017), histone modifications (Krogan et al. 2003a), and poly(A) site utilization (Penheiter et al. 2005).
In yeast, there is evidence that the PAF1 complex affects telomere function. Specifically, cdc73Δ, paf1Δ, and ctr9Δ cells show reduced levels of TLC1 RNA (the RNA template of telomerase) (Mozdy et al. 2008). In addition, Paf1 and Ctr9, but not Cdc73, are required to maintain low levels of TERRA (telomere repeat containing RNA) (Rodrigues and Lydall 2018). Finally, genetic screens have shown that Cdc73, Leo1, and Rtf1 are important for the fitness of telomere defective cells (carrying a cdc13-1 temperature-sensitive allele of the telomere capping gene CDC13) (Addinall et al. 2011).
It is interesting to note that although the five proteins of the PAF1 complex associate with RNA pol II, deletion of individual PAF1 complex components in yeast leads to distinct phenotypes (Betz et al. 2002; Rodrigues and Lydall 2018). Besides the already mentioned differences in TLC1 and TERRA levels in strains lacking different PAF1 complex components, there is also a strong difference in fitness amongst the different PAF1 deletion mutants. For example, ctr9Δ and paf1Δ cause severe growth defects, while cdc73Δ and rtf1Δ cause mild growth defects and leo1Δ causes no growth defects (Betz et al. 2002; Rodrigues and Lydall 2018). There are at least three plausible, not mutually exclusive, explanations for why deletion of different PAF1 complex components lead to distinct phenotypes: (1) the existence of subcomplexes; (2) complex components perform specific and independent functions; and/or (3) the existence of an adjacent gene effect.
Data from numerous experimental systems support the idea that the different PAF1 complex components perform different functions and the more severe defects of cells lacking Paf1 or Ctr9 have been attributed to the scaffolding properties of these components (Nordick et al. 2008; Tomson and Arndt 2013; Xu et al. 2017; Rodrigues and Lydall 2018). Indeed, structural studies in human and yeast cells have revealed Ctr9 to be the main scaffold protein for the PAF1 complex, and loss of Paf1 causes a significant decrease in Ctr9 protein levels (Nordick et al. 2008; Chu et al. 2013; Xu et al. 2017). However, it has also been suggested that a complete CDC73 deletion, often used in yeast studies, is affecting the function of VPS36, one of its adjacent genes (Ben-Shitrit et al. 2012). Large-scale surveys have shown that cdc73Δ and vps36Δ strains share many characteristics, for example reduced telomere length (Askree et al. 2004; Rog et al. 2005; Dieckmann et al. 2016), heat sensitivity (Betz et al. 2002; Sinha et al. 2008), and decreased resistance to hygromycin B and hydroxyurea (Betz et al. 2002; Dudley et al. 2005; Fell et al. 2011; Ejzykowicz et al. 2017).
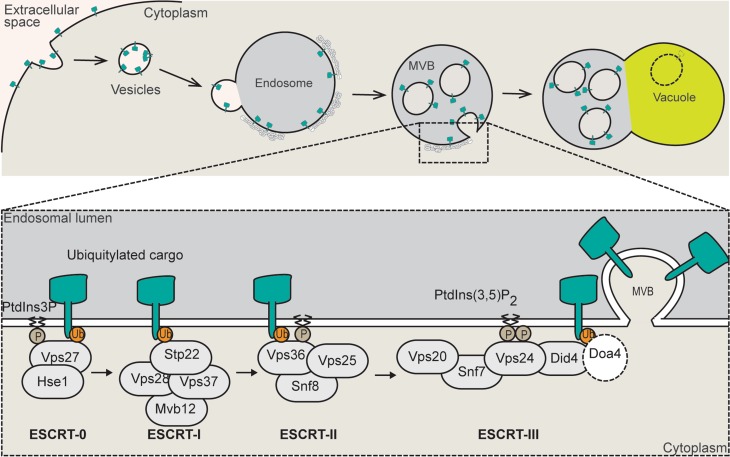
VPS36 is widely conserved among eukaryotes and encodes a protein that is part of the ESCRT (Endosomal Sorting Complexes Required for Transport)-II complex (Teo et al. 2004). The main function for the ESCRT machinery (ESCRT-0, ESCRT-I, ESCRT-II and ESCRT-III) is in the remodelling of membranes, which is important to its role in the multivesicular body (MVB) pathway of protein degradation (Schmidt and Teis 2012) (Fig. 1). The MVB pathway sorts ubiquitylated membrane proteins for degradation in vacuoles/lysosomes (Schmidt and Teis 2012). During this process, each of the ESCRT complexes assembles on endosomes in a sequential manner (Schmidt and Teis 2012). Together with MVB formation, the ESCRT machinery contributes to other membrane scission events like retroviral particle release and midbody abscission during cytokinesis (Caballe and Martin-Serrano 2011). Finally, the ESCRT machinery was also suggested to promote gene transcription due to its presence at the 3′ end of coding regions, and the increased sensitivity to 6-azauracil of ESCRT mutants (Song et al. 2014).
Fig. 1.
ESCRT machinery role in the multivesicular body pathway. Cartoon showing the function of the yeast ESCRT machinery in the sorting of ubiquitylated membrane proteins (cargo) for degradation through the multivesicular body (MVB) pathway. The ESCRT machinery is composed of ESCRT-0 (Vps27 and Hse1), ESCRT-I (Stp22, Mvb12, Vps37, and Vps28), ESCRT-II (Snf8, Vps25, and Vps36), and ESCRT-III (Snf7, Vps20, Vps24, and Did4). Doa4, an ubiquitin-specific protease, is not classified as part of the ESCRT machinery but is part of the MVB pathway. ESCRT-0 recognises Phosphatidyl Inositol 3 phosphate (PtdIns3P) and ubiquitylated cargo in the endosomal membrane. The remaining ESCRT complexes (-I, -II, and -III) sequentially interact with the cargo leading to the cargo deubiquitylation and formation of vesicles inside the endosome (MVB). The vesicles and their cargo are degraded when the MVB fuses with the vacuole/lysosome.
Adapted from (Williams and Urbe 2007; López-Reyes et al. 2011; Schmidt and Teis 2012; Schuh and Audhya 2014)
In this study, we demonstrate an adjacent gene effect between CDC73 and VPS36, and its effect in the fitness of telomere defective cells. We describe a number of complex interactions between the components of the PAF1 complex and VPS36 in budding yeast. We report, for the first time, synthetic lethal genetic interactions between VPS36 and two components of the PAF1 complex: PAF1 and CTR9. Importantly, this synthetic lethality extends to other components of the ESCRT machinery: SNF8 and VPS25 (ESCRT-II), STP22 (ESCRT-I) and SNF7 (ESCRT-III). Interestingly, in the yeast genetic background used in this study, loss of ESCRT components does not affect telomere length. Finally, we show that VPS36 RNA levels are controlled by the PAF1 complex.
Materials and methods
Yeast strains
Standard procedures for yeast culture, mating, and tetrad dissection were followed (Adams et al. 1997). All experiments were performed using Saccharomyces cerevisiae W303 strains as listed in Table S1. Gene disruptions were made in diploids (DDY227, S1 Table) using one-step PCR to insert a kanMX, natMX, or hphMX cassette into the genome (Goldstein and McCusker 1999). Gene disruptions were confirmed by PCR. Oligonucleotide sequences are described in Table S2.
Yeast growth assays
Colonies from germination plates were inoculated in liquid YEPD (supplemented with adenine) and grown until saturation overnight at 23 °C. Serial dilutions in water were spotted onto YEPD plates using a replica plating device. Plates were incubated for 2 (or 3) days at the appropriate temperatures before being photographed. Unless stated otherwise, a single plate per temperature was used (round plates typically fit between 8 and 16 strains, while rectangular plates fit between 16 and 32 strains).
Analysis of telomere structure
Southern blot analysis was used to assess telomere length and performed as previously described. Genomic DNA was extracted, digested with XhoI, and then run overnight on a 1% agarose gel at 1 V/cm. Southern transfer was performed using the Biorad Vacuum Blotter according to the manufacturer’s indications. Y′+TG probe labelling and Southern detection were made according to the DIG High Prime DNA Labelling and Detection Starter Kit II (Roche) manufacturer’s instructions. The probe has approximately 1 kb with ~ 880 bp of Y′ and 120 bp of TG repeats, and was cut from pDL1574 using XhoI and BamHI (Dewar and Lydall 2010).
Quantitative RT-PCR
RNA was isolated essentially as described (Collart and Oliviero 2001) followed by purification using the RNEasy Mini Kit (QIAGEN, 74,104) and DNase I digestion (Invitrogen, 18068-015). Quantitative RT-PCR was carried out using the Superscript III Platinum SYBR Green One-Step qRT-PCR kit (Invitrogen, 11736-059). RNA samples were normalized relative to the BUD6 mRNA levels as previously described (Holstein et al. 2014).
Results
Neighbouring gene effect between CDC73 and adjacent gene VPS36
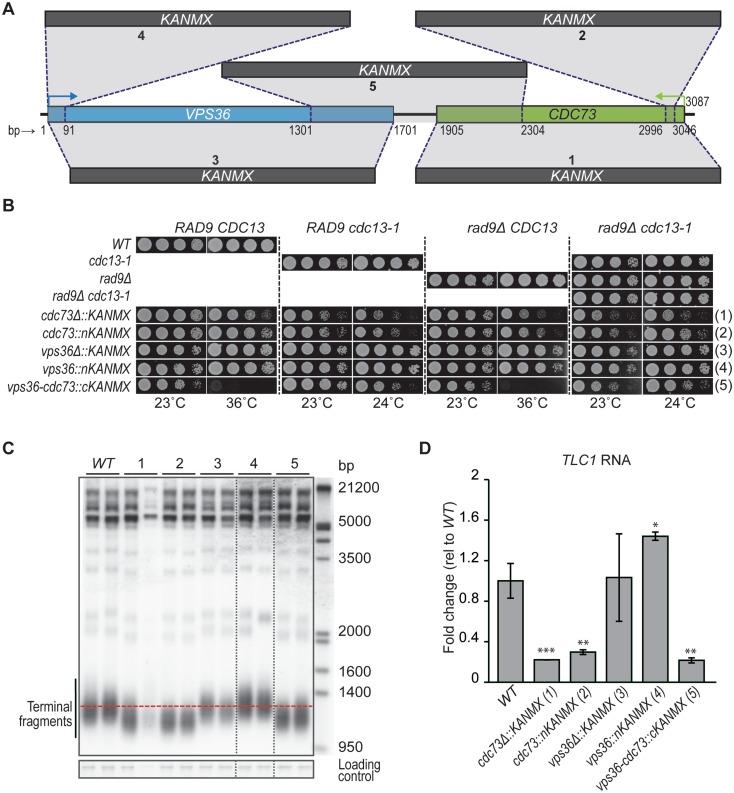
CDC73 and VPS36 point towards each other, with the stop codons separated by 204 bp (Fig. 2a), and vps36Δ and cdc73Δ have been suggested to demonstrate an adjacent gene effect (Ben-Shitrit et al. 2012). Importantly, both genes are reported to affect telomere length and the fitness of telomere defective cells (Mozdy et al. 2008; Addinall et al. 2011; Dieckmann et al. 2016). To clarify the contributions of CDC73 and VPS36 to the fitness of telomere defective, cdc13-1 cells, five related gene disruption constructs were created (Fig. 2a). Constructs 1 and 3 have CDC73 or VPS36 ORFs completely deleted (cdc73Δ or vps36Δ); constructs 2 and 4 have interruptions in the N-termini of CDC73 or VPS36 ORFs (farthest from each other, therefore, less likely to affect the adjacent gene) (cdc73Δn or vps36Δn). Construct 5 has a simultaneous deletion of the C-termini of CDC73 and VPS36 (predicted to disrupt the function of both genes) (cdc73Δc-vps36Δc). The effects of the constructs were analysed in WT (CDC13), cdc13-1 (causing a temperature-sensitive telomere defect), rad9Δ (causing a defect in the DNA damage checkpoint), and rad9Δ cdc13-1 backgrounds to determine their effects in cell fitness. Their effects on telomere length and TLC1 RNA levels were also measured.
Fig. 2.
CDC73 deletion affects VPS36 function. a Map of CDC73 and VPS36, together with five different constructs used for disruptions. b Haploid strains containing constructs in a combined with CDC13 RAD9, cdc13-1 RAD9, CDC13 rad9Δ, or cdc13-1 rad9Δ were grown overnight at 23 °C and serial dilutions spotted onto solid YEPD plates. Pictures were taken after 2 days of incubation at different temperatures. At each temperature, the strains were grown on the same YEPD plate. Four different strains for each genotype were analysed and a representative strain is shown (the full spot test can be seen in Fig. S1). c Telomeric Southern blot was performed using a Y’+TG probe to analyse the telomere structure of the strains with deletions described in a. Overnight cultures (grown at 23 °C) were used to isolate DNA. All lanes are from the same gel and vertical dashed lines represent the place where the gel was cut and pasted for presentation purposes. d RT-qPCR analysis of TLC1 RNA levels. RNA from two independent strains was measured except for WT and vps36Δ::KANMX strains, where the two independent strains were analysed twice. Each value was normalized to the levels of BUD6 RNA. The mean is indicated and the “error” bars indicate the two independent measurements, or average deviation. Statistical analyses used the two-tailed unpaired T test (*P < 0.05, **P < 0.01 and ***P < 0.001)
Importantly, and consistent with the existence of an adjacent gene effect, there is a small but consistent fitness difference between cdc73Δ and cdc73Δn strains in all combinations of cdc13-1 and rad9Δ backgrounds (Fig. 2b). Overall, cdc73Δn cells (most likely with VPS36 unaffected and, therefore, presumably the CDC73 null phenotype) are fitter than cdc73Δ cells. As reported before, vps36Δ cdc13-1 cells are slightly fitter than cdc13-1 cells at 27 °C (Fig. S1B) (Dieckmann et al. 2016) and we see no detectable fitness difference between vps36Δ and vps36Δn cells in cdc13-1 mutants (compare constructs 3 and 4, Fig. 2b). However, the strongest fitness phenotype observed was in cdc73Δc-vps36Δc cells at high temperatures (Fig. 2b). cdc73Δc-vps36Δc cells (CDC13 RAD9), missing both Cdc73 and Vps36, are considerably less fit than cdc73Δ or cdc73Δn cells (construct 5 versus 1 and 2) (Fig. 2b, 36 °C), suggesting that loss of the full CDC73 coding sequence only partially reduces Vps36 function (in cdc73Δ cells). We conclude that VPS36 and CDC73 most likely exhibit both an adjacent gene effect and a synthetic genetic interaction.
We next measured telomere length and TLC1 RNA levels to test if the CDC73-VPS36 adjacent gene effect contributed to the short telomeres of cdc73Δ and vps36Δ cells (Rog et al. 2005; Mozdy et al. 2008; Dieckmann et al. 2016). As expected, the three constructs that inactivate Cdc73 (constructs 1, 2, 5, Fig. 2a) each caused short telomeres and similarly decreased TLC1 RNA levels (Fig. 2c, d). Surprisingly, and different from previous reports, loss of Vps36 in vps36Δ and vps36Δn cells (constructs 3 and 4, Fig. 2a) did not decrease telomere length (Fig. 2c) (Askree et al. 2004; Rog et al. 2005; Dieckmann et al. 2016). If anything, vps36Δn, but not vps36Δ, slightly increased telomere length and TLC1 levels (Fig. 2c, d, constructs 3 and 4). These observations are consistent with an adjacent gene effect on telomere length and can be explained by opposing effects of Vps36 and Cdc73 on telomere length. If it is assumed that vps36Δn causes the null phenotype (increased TLC1 and telomere length), then full-length deletion, vps36Δ, reduces CDC73 function counteracting the increase in TLC1 (telomere length).
The effect of Vps36 on TLC1 RNA levels is not shared with other ESCRT components (Fig. S2), suggesting this is a specific function of Vps36. Overall, telomere length and TLC1 levels correlate with each other, but not with the fitness defects, across the different deletions (Fig. 2c, d versus Fig. 2b). For instance, cdc73Δc-vps36Δc cells show the strongest fitness defects, but, from the strains missing Cdc73 (cdc73Δc-vps36Δc, cdc73Δ or cdc73Δn), are perhaps the ones with the longest telomeres (Fig. 2b, c, constructs 1 and 2 versus construct 5). We conclude that the adjacent gene effect does not contribute to Cdc73 regulation of TLC1 RNA and telomere shortening is not the reason for cdc73-vps36Δc synthetic fitness defects.
Overall, our data suggest that the standard cdc73Δ mutation (construct 1) is partially interfering with VPS36 activity, most likely by affecting the VPS36 3′UTR. Furthermore, analysis of telomere lengths suggests that the standard VPS36 deletion affects CDC73 function. These observations confirm the existence of an adjacent gene effect between CDC73 and VPS36. We also report a synthetic fitness defect in cells simultaneously depleted of Vps36 and Cdc73, which is likely independent of the telomere length of these cells.
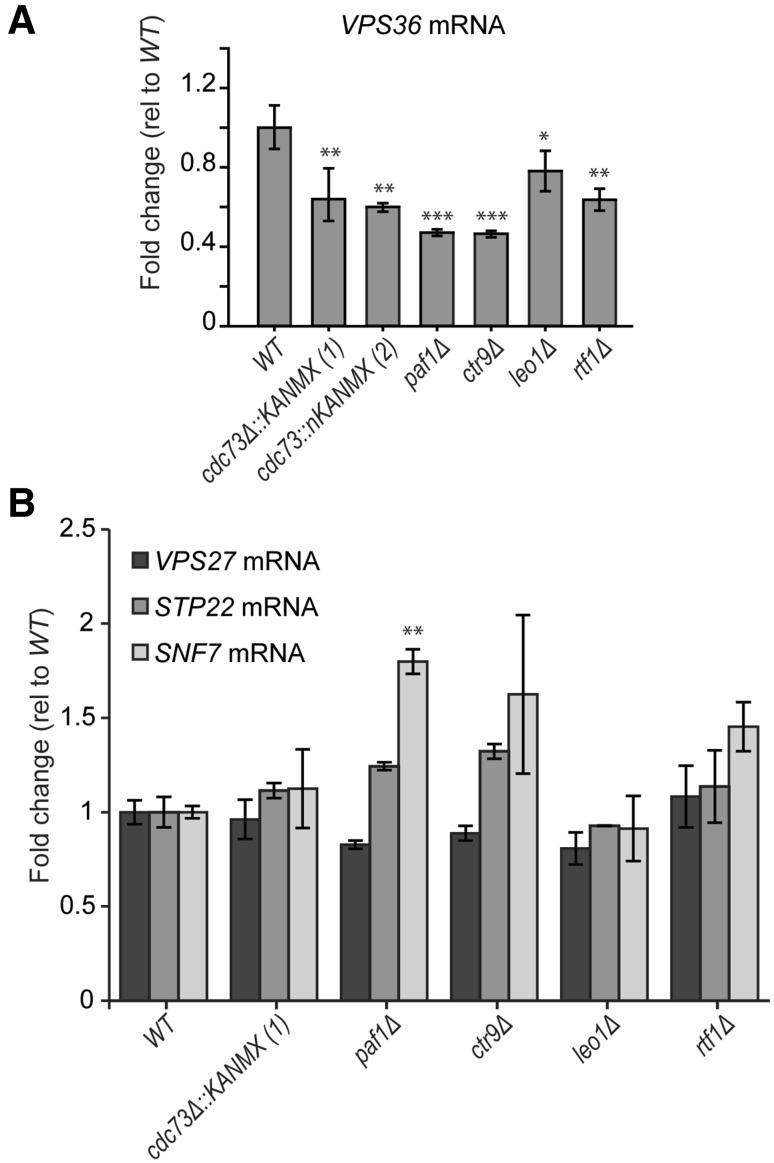
The PAF1 complex regulates VPS36 transcription
To test if the knockout of CDC73 was affecting VPS36 transcription and/or RNA stability, VPS36 RNA levels were measured in strains carrying the two disruptions of CDC73 (constructs 1 and 2, Fig. 2a). VPS36 RNA was also measured in strains with the other PAF1 components deleted, since the PAF1 complex is involved in transcriptional regulation and altered VPS36 RNA levels in cdc73Δ cells could be partially due to defects in the PAF1 complex activity. Interestingly, VPS36 RNA is decreased when each of the PAF1 complex components is deleted, with little difference between deletion or N-termini disruption of CDC73 (constructs 1 and 2, Fig. 3a). The lack of difference between cdc73Δ and cdc73Δn suggests that the adjacent gene effect does not affect VPS36 RNA levels, but, perhaps, an altered VPS36 3′-UTR could affect translation and/or mRNA localization. Finally, to test if all ESCRT machinery is under PAF1-dependent transcriptional regulation, the RNA levels of other ESCRT components were next analysed in PAF1 complex mutants. VPS27 (ESCRT-0), STP22 (ESCRT-I) and SNF7 (ESCRT-III) RNA levels were not reduced by deletion of any of the PAF1 complex components (Fig. 3b). In fact, SNF7 mRNA levels were significantly increased in paf1Δ mutants (and probably in ctr9Δ mutants). This is unusual, because paf1Δ decreases levels of most transcripts (Xu et al. 2017).
Fig. 3.
PAF1 complex controls VPS36 expression. a, b RT-qPCR analysis of VPS36 (a), STP22 (b), VPS27 (b) and SNF7 (b) RNA levels. RNA from two independent strains was measured as described in Fig. 2d. Statistical analyses used the two-tailed unpaired T test (*P < 0.05, **P < 0.01 and ***P < 0.001)
We conclude that the adjacent gene effect between CDC73 and VPS36 does not affect VPS36 RNA levels, but could, for instance, affect VPS36 mRNA localization and/or translation. In addition, we show that VPS36 RNA is regulated by the PAF1 complex.
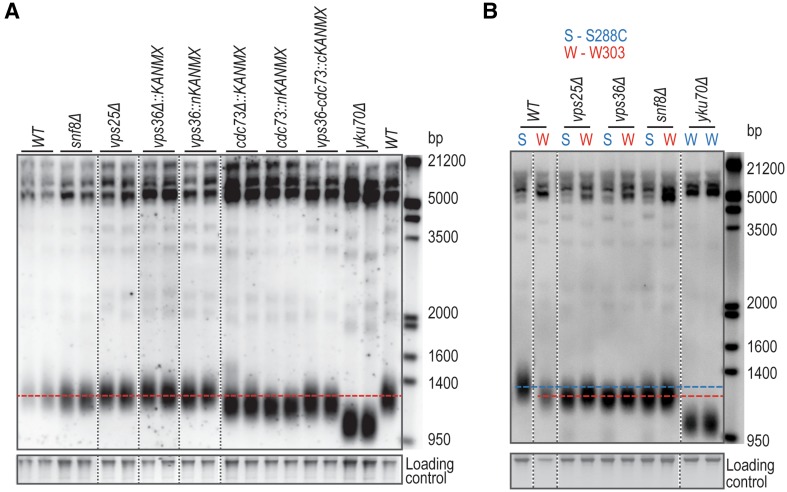
ESCRT complex does not affect telomere length of S. cerevisiae W303 cells
The absence of a telomere length phenotype in vps36Δ cells observed in Fig. 2c is not in agreement with the previously reported short telomeres observed in ESCRT mutants (Askree et al. 2004; Rog et al. 2005; Dieckmann et al. 2016). Therefore, we asked if normal telomere length was also observed in cells with mutations in other components of the ESCRT-II complex. To test this, the telomere length in snf8Δ or vps25Δ strains, defective in other components of the ESCRT-II complex, was measured. Interestingly, no strong effects on telomere length were observed upon deletion of any of the ESCRT-II complex components (Fig. 4a). On the other hand, as expected, CDC73 deletion or N-terminal disruption and YKU70 deletion led to shorter telomeres (Fig. 4a). Our experiments were performed in the W303 genetic background, and cells were cultured at 23 °C, whereas the published experiments were in the BY4742 (S288C) background, and cells were most likely cultured at 30 °C. To test if telomere length in ESCRT mutants is particularly sensitive to temperature, the telomere length of the same mutants analysed in Fig. 2 was measured after growth at 30 °C. There was still no observable telomere length decrease in any of the ESCRT complex mutants cultivated at 30 °C (Fig. S3). We next compared the telomere length of WT, vps25Δ, vps36Δ, and snf8Δ cells in the W303 and S288C genetic backgrounds (at 30 °C). As previously reported, WT cells of the S288C genetic background have longer telomeres than W303 cells (Fig. 4b) (Lebel et al. 2009). Interestingly, when compared to WT cells, the telomeres of the ESCRT mutants are relatively short in the S288C background but relatively normal in the W303 background (Fig. 4b). We conclude that the previously reported telomere length regulation by the ESCRT machinery is not conserved in S. cerevisiae W303 cells and is most likely strain background-dependent.
Fig. 4.
ESCRT-II mutants have normal telomeres in S. cerevisiae W303 cells. a Southern blot as described in Fig. 2c. b Telomere southern blot of WT, vps25Δ, vps36Δ and snf8Δ mutations in S288C and W303 genetic backgrounds. S288C strains are from the deletion database. Southern blot performed as described in Fig. 2c, except that the cells were cultured at 30 °C. In each panel, all lanes are from the same gel and vertical dashed lines represent the place where the gel was cut and pasted for presentation purposes
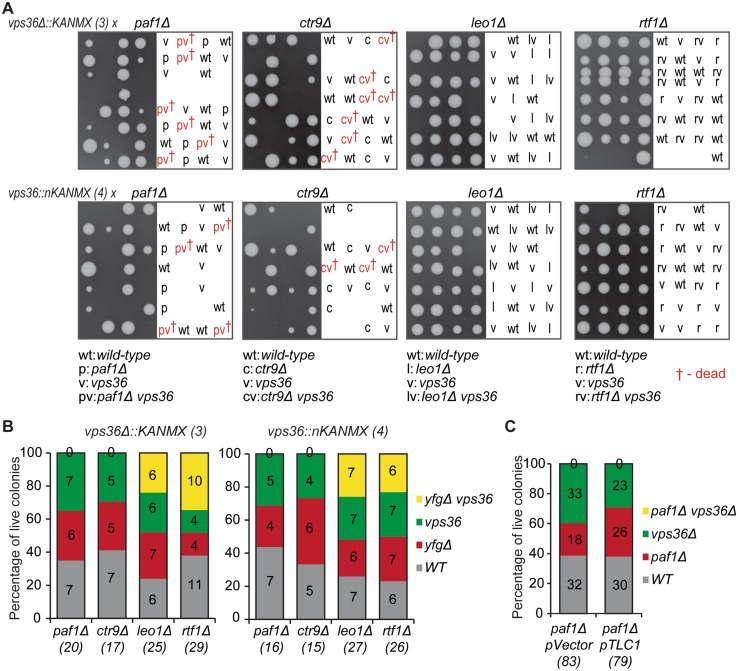
vps36Δ is synthetically lethal with paf1Δ or ctr9Δ but not with cdc73Δ, rtf1Δ or leo1Δ
Given that VPS36 and CDC73 contribute independently to maintain cell fitness, particularly at high temperature, we wanted to know what role, if any, VPS36 played in the fitness of other PAF1 complex deletion strains. To address this question, we crossed vps36Δ and vps36Δn (Fig. 2a, disruptions 3 and 4) to strains carrying paf1Δ, ctr9Δ, rtf1Δ, or leo1Δ mutations. Note that fitness of cdc73Δ vps36Δ double mutants was previously tested and found to be decreased when compared to single mutants (Fig. 2b). Interestingly, vps36Δ paf1Δ or vps36Δ ctr9Δ double mutants could not be identified, but all other double mutants were viable (Fig. 5a, quantification in Fig. 5b). Although this synthetic lethality between vps36Δ and paf1Δ or ctr9Δ has not been previously reported, both PAF1 and CTR9 deletions were reported to show synthetic growth defects with deletions affecting ubiquitylation pathways (UBP6) or multivesicular body/protein sorting pathways (VPS21, VPS71, and VPS72) (Krogan et al. 2003b; Laribee et al. 2005; Collins et al. 2007; Hang et al. 2011). Since Vps36 (as a component of ESCRT-II) is involved in sorting ubiquitylated proteins for degradation through the multivesicular body pathway, the findings here reported are in line with the previously described genetic interactions of PAF1/CTR9 and members of the multivesicular body degradation pathway (Krogan et al. 2003b; Laribee et al. 2005; Collins et al. 2007; Hang et al. 2011). Together, all these results show that Vps36 and the PAF1 complex, especially Paf1 and Ctr9, work in independent pathways to support cell viability.
Fig. 5.
Vps36 is needed for the viability of paf1Δ or ctr9Δ but not rtf1Δ or leo1Δ cells. a vps36Δ or vps36Δn (Fig. 2a, constructs 3 and 4) were crossed to paf1Δ, ctr9Δ, rtf1Δ, or leo1Δ strains and diploid cells were obtained. Diploids were sporulated and tetrads dissected onto YEPD plates. Germinated spores grew for 6 days at 23 °C before being photographed. Genotypes of each spore are indicated on the right when possible. Genotype of inviable spores was inferred using the expected 2:2 Mendelian segregation. b Quantification of A. Percentage of colonies of each genotype is shown (relative to the total number of viable colonies). The total number of colonies genotyped are shown in brackets. c A vps36Δ::KANMX paf1Δ heterozygous diploid was transformed with a centromeric plasmid carrying TLC1 (pDL751) or a vector plasmid (pDL1713). Random spore analysis was used to obtain haploid strains: the diploids carrying the plasmids were sporulated and plated onto –URA plates. After 7 days at 23 °C, colonies were genotyped. The percentage of each genotype was plotted as in b
CDC73, PAF1, and CTR9 deletions decrease TLC1 RNA levels, leading to short telomeres and growth defects (Mozdy et al. 2008). Therefore, we asked if the synthetic sickness/lethality between PAF1 and ESCRT mutants was caused by a TLC1-dependent mechanism. If so, then overexpression of TLC1 in vps36Δ paf1Δ mutants may render these cells more viable. To test this hypothesis, vps36Δ paf1Δ heterozygous diploids were transformed with a 2 µm TLC1 plasmid and the presence of viable vps36Δ paf1Δ (pTLC1) haploid progeny was assessed after sporulation. However, there was no effect of TLC1 plasmid (versus vector) on the viability of vps36Δ paf1Δ (from 79 viable spores carrying TLC1, none was vps36Δ paf1Δ) (Fig. 5c).
We conclude that Vps36 and Paf1/Ctr9 work in independent pathways to maintain cell viability, with simultaneous lack of Vps36 and Paf1 or Ctr9 leading to cell death. Furthermore, the synthetic lethality between vps36Δ and paf1Δ/ctr9Δ is unlikely to be related to the decreased levels of TLC1 in paf1Δ cells.
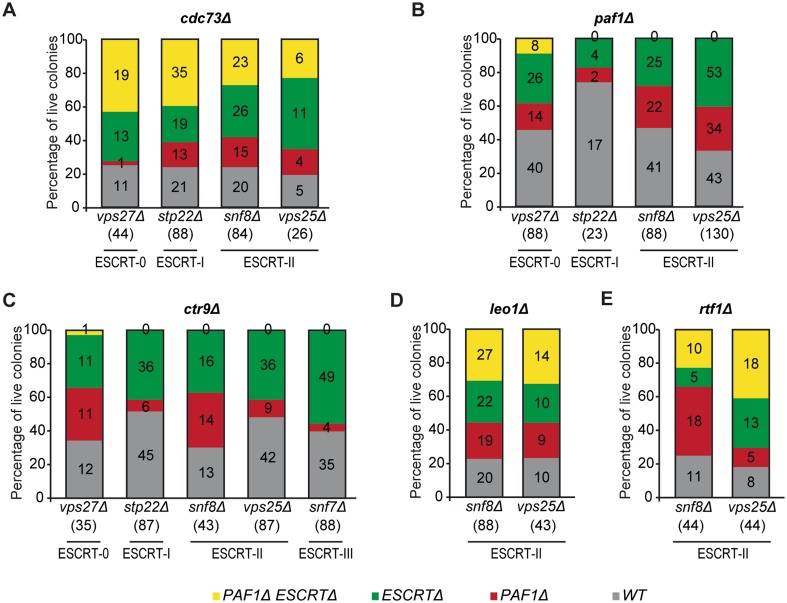
paf1Δ and ctr9Δ are synthetically lethal with deletions of ESCRT-I, ESCRT-II and ESCRT-III components
To test if the synthetic lethality between paf1Δ/ctr9Δ and vps36Δ is conserved with other ESCRT components, we analysed the progeny of diploids heterozygous for deletions of PAF1 complex components and VPS27 (ESCRT-0), STP22 (ESCRT-I), SNF8 (ESCRT-II), VPS25 (ESCRT-II), and SNF7 (ESCRT-III) (Fig. 1). Interestingly, and in agreement with the interactions observed between PAF1/CTR9 and VPS36, PAF1 and CTR9 deletion in cells carrying deletions of any of the tested ESCRT-I to -III components led to synthetic lethality (Fig. 6). In most cases, the double mutants divided 3–5 times before stopping divisions, as measured by micro-colony analysis (Fig. S4). The fact that these cells can divide a number of times before death suggests that they accumulate defects that eventually lead to cell death. Interestingly, 8 (of 88) paf1Δ vps27Δ and 1 (of 35) ctr9Δ vps27Δ cells were viable, suggesting that Vps27 (ESCRT-0) is not as important as downstream ESCRT components for the viability of paf1Δ or ctr9Δ cells. We conclude that PAF1 complex (through Paf1 and Ctr9) works with the ESCRT machinery, in independent pathways, to maintain cell viability.
Fig. 6.
paf1Δ and ctr9Δ are synthetically lethal with deletions affecting ESCRT-I, ESCRT-II and ESCRT-III components. a–e Strains carrying deletions of different components of the ESCRT machinery complex were crossed with cdc73Δ (a), paf1Δ (b), ctr9Δ (c), leo1Δ (d), or rtf1Δ (e) strains. Data were analysed as described in Fig. 5b
Discussion
Budding yeast is a powerful model organism to systematically study eukaryotic gene function and interactions, with many gene functions being conserved in human cells. For instance, yeast genome-wide screens have been used extensively for analysis and discovery of new gene interactions in various fields: transcription elongation and chromatin modification (Krogan et al. 2003b), DNA replication (Dubarry et al. 2015), and telomere length maintenance (TLM) (Ungar et al. 2009). However, there are caveats associated with the use of systematic yeast genetic approaches. For instance, the deletion of one gene ORF might affect its adjacent gene (adjacent gene effect). Indeed, it has been suggested that 10% of all budding yeast genes demonstrate an adjacent gene effect (Ben-Shitrit et al. 2012; Atias et al. 2016). Another issue is related to the genetic backgrounds used. Since the principal yeast deletion library was created in the S288C genetic background, most of the genome-wide data available is in this background (Giaever et al. 2002). However, other commonly used yeast genetic backgrounds like W303, SK1, Y55, among others, show important differences, such as telomere length (Lebel et al. 2009). Thus, it is important to test for the role of adjacent gene effects and of genetic backgrounds.
CDC73 encodes a component of the PAF1 complex, involved in transcription regulation, and is adjacent to VPS36, encoding a component of the ESCRT machinery, involved in membrane remodelling (mainly to sort ubiquitylated proteins to the vacuole). Bioinformatic analysis identified CDC73 and VPS36 as potentially demonstrating an adjacent gene effect, since deletion of either causes short telomeres and the ORFs are only separated by 204 bp (Ben-Shitrit et al. 2012). On the other hand, proteins that work with Cdc73 in the PAF1 complex, and with Vps36 in the ESCRT machinery also affect telomere length, suggesting that there may not be an adjacent gene effect on telomere length.
To clarify whether there is an adjacent gene effect between CDC73 and VPS36 we compared the fitness and telomere lengths of cells with different CDC73 and VPS36 ORF disruptions. We established that complete CDC73 deletion partially reduces VPS36 function to affect fitness. Since we did not see any change in VPS36 mRNA levels in strains with CDC73 ORF deletion or an N-terminal disruption, it is likely that CDC73 ORF deletion affects VPS36 mRNA localization and/or translation. Complete VPS36 deletion partially reduces CDC73 function to affect telomere length (without noticeable effects on fitness). We conclude that CDC73 and VPS36 show somewhat complex adjacent gene effects.
While investigating the adjacent gene effect between CDC73 and VPS36, we noted that in the W303 yeast genetic background, ESCRT mutants do not affect telomere length, in contrast to what has been reported for the S288C yeast genetic background where ESCRT mutations caused short telomeres (Rog et al. 2005; Ungar et al. 2009; Dieckmann et al. 2016). This difference shows that the relatively well studied role of ESCRT mutants in telomere length is not conserved within different S. cerevisiae genetic backgrounds; therefore, it might not be preserved in other species (like mammals) (Rog et al. 2005; Dieckmann et al. 2016). However, it is possible that ESCRT genes affect telomere function in both S288C and W303, but that it is only possible to see an effect on telomere length when telomeres are long (S288C), rather than short (W303). Thus, more work is needed to understand the true role of the ESCRT machinery in telomere biology.
In addition to the adjacent gene effect, we describe other interactions between the PAF1 complex (composed of Cdc73, Paf1, Ctr9, Leo1, and Rtf1) and VPS36 (ESCRT machinery). First, we report that all components of the PAF1 complex regulate VPS36 RNA levels. In addition, we found that vps36Δ exacerbates the temperature sensitivity of cdc73Δ mutants. Furthermore, there is a synthetic lethal interaction in cells with defective ESCRT-I (stp22Δ), ESCRT-II (vps36Δ, snf8Δ and vps25Δ), and ESCRT-III (snf7Δ) machineries and ctr9Δ or paf1Δ mutations. LEO1 and RFT1 do not show synthetic genetic interactions with genes encoding ESCRT components, in agreement with their minor role in the PAF1 complex integrity (Jaehning 2010; Tomson and Arndt 2013; Rodrigues and Lydall 2018). These show that the ESCRT machinery functions redundantly with Ctr9/Paf1 to maintain yeast cell viability.
How the PAF1 complex and the ESCRT machinery function together to maintain yeast cell fitness is not yet clear. In yeast, ESCRT-0 is as important as the other ESCRT complexes (-I, -II, and -III) in the MVB formation (to degrade ubiquitylated cargo) (Schmidt and Teis 2012). Therefore, the fact that paf1Δ/ctr9Δ are not synthetically lethal with vps27Δ (ESCRT-0) suggests that defective MVB pathway is not the major cause for the synthetic lethality of paf1Δ/ctr9Δ with ESCRT-I, ESCRT-II, and ESCRT-III mutants. PAF1 and CTR9 deletions lead to severe alterations in the transcriptome, which is believed to be the cause of the strong fitness defects of these mutants (Xu et al. 2017; Rodrigues and Lydall 2018). The higher sensitivity of ESCRT deletion mutants to 6-azauracil and the fact that ESCRT proteins associate with 3′ regions of actively transcribed genes suggested that the ESCRT components could also play a role in transcription elongation (Song et al. 2014). Thus, paf1Δ vpsΔ and ctr9Δ vpsΔ might have worse transcriptional defects leading to cell death. Supporting the idea that the ESCRT machinery is important in cells with transcription defects, histone deacetylases (HDA1, HDA2, and HDA3), and mRNA surveillance genes (PAP2) were found to be essential in snf8Δ, stp22Δ, vps25Δ, and vps36Δ cells (Pan et al. 2006; Lin et al. 2008).
In conclusion, we report that the PAF1 complex and the ESCRT machinery work together to maintain the fitness of W303 yeast cells. We confirm that deletion of CDC73 affects VPS36 and vice versa. Therefore, these particular adjacent genes, encoding PAF1 and ESCRT machinery components, interact both in cis and in trans. Thus, these studies indicate how complex it can be to interpret the effects of single gene deletions in budding yeast, and have implications for systematic studies in all cell types. Finally, we show that ESCRT mutations do not affect the telomere length of W303 yeast cells, while confirming that they decrease the telomere length of S288C yeast cells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Marie Curie Initial Training Network CodeAge project [GA316354 (CodeAge) FP7-PEOPLE-2012-ITN] and the Biotechnology and Biological Sciences Research Council [BB/M002314/1].
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in yeast genetics. New York: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Addinall SG, Holstein EM, Lawless C, Yu M, Chapman K, Banks AP, Ngo HP, Maringele L, Taschuk M, Young A, et al. Quantitative fitness analysis shows that NMD proteins and many other protein complexes suppress or enhance distinct telomere cap defects. PLoS Genetics. 2011;7:e1001362. doi: 10.1371/journal.pgen.1001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atias N, Kupiec M, Sharan R. Systematic identification and correction of annotation errors in the genetic interaction map of Saccharomyces cerevisiae. Nucleic Acids Res. 2016;44:e50. doi: 10.1093/nar/gkv1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shitrit T, Yosef N, Shemesh K, Sharan R, Ruppin E, Kupiec M. Systematic identification of gene annotation errors in the widely used yeast mutation collections. Nat Methods. 2012;9:373–378. doi: 10.1038/nmeth.1890. [DOI] [PubMed] [Google Scholar]
- Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol Genet Genom MGG. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- Caballe A, Martin-Serrano J. ESCRT machinery and cytokinesis: the road to daughter cell separation. Traffic. 2011;12:1318–1326. doi: 10.1111/j.1600-0854.2011.01244.x. [DOI] [PubMed] [Google Scholar]
- Chen YX, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Gene Dev. 2009;23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X, Qin X, Xu H, Li L, Wang Z, Li F, Xie X, Zhou H, Shen Y, Long J. Structural insights into Paf1 complex assembly and histone binding. Nucleic Acids Res. 2013;41:10619–10629. doi: 10.1093/nar/gkt819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Oliviero S (2001) Preparation of yeast RNA. In: Ausubel FM et al (eds) Current protocols in molecular biology, Chap 13: Unit 13, p 12 [DOI] [PubMed]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Crisucci EM, Arndt KM. The roles of the Paf1 complex and associated histone modifications in regulating gene expression. Genetics Res Int. 2011;2011:707641. doi: 10.4061/2011/707641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar JM, Lydall D. Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping. EMBO J. 2010;29:4020–4034. doi: 10.1038/emboj.2010.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann AK, Babin V, Harari Y, Eils R, Konig R, Luke B, Kupiec M (2016) Role of the ESCRT complexes in telomere biology. MBio [DOI] [PMC free article] [PubMed]
- Dubarry M, Lawless C, Banks AP, Cockell S, Lydall D (2015) Genetic networks required to coordinate chromosome replication by DNA polymerases alpha, delta, and epsilon in Saccharomyces cerevisiae. G3 5:2187–2197 [DOI] [PMC free article] [PubMed]
- Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol. 2005;1:0001. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejzykowicz DE, Locken KM, Ruiz FJ, Manandhar SP, Olson DK, Gharakhanian E. Hygromycin B hypersensitive (hhy) mutants implicate an intact trans-Golgi and late endosome interface in efficient Tor1 vacuolar localization and TORC1 function. Curr Genet. 2017;63:531–551. doi: 10.1007/s00294-016-0660-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell GL, Munson AM, Croston MA, Rosenwald AG (2011) Identification of yeast genes involved in k homeostasis: loss of membrane traffic genes affects k uptake. G3 1:43–56 [DOI] [PMC free article] [PubMed]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Hang LE, Liu X, Cheung I, Yang Y, Zhao X. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol. 2011;18:920–926. doi: 10.1038/nsmb.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SM, Amode R, Brent S, et al. Ensembl comparative genomics resources. Database J Biol Databases Curation. 2016;2016:bav096. doi: 10.1093/database/bav096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein EM, Clark KR, Lydall D. Interplay between nonsense-mediated mRNA decay and DNA damage response pathways reveals that Stn1 and Ten1 are the key CST telomere-cap components. Cell Rep. 2014;7:1259–1269. doi: 10.1016/j.celrep.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/S1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Molecular cell. 2003;12:1565–1576. doi: 10.1016/S1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Lebel C, Rosonina E, Sealey DC, Pryde F, Lydall D, Maringele L, Harrington LA. Telomere maintenance and survival in saccharomyces cerevisiae in the absence of telomerase and RAD52. Genetics. 2009;182:671–684. doi: 10.1534/genetics.109.102939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 2008;22:2062–2074. doi: 10.1101/gad.1679508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Reyes I, Bañuelos C, Betanzos A, Orozco E (2011) A bioinformatical approach to study the endosomal sorting complex required for transport (ESCRT) machinery in protozoan parasites: the entamoeba histolytica case. Bioinformatics—trends and methodologies, pp 290–292
- Mozdy AD, Podell ER, Cech TR. Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Mol Cell Biol. 2008;28:4152–4161. doi: 10.1128/MCB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordick K, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryotic Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Rodrigues J, Lydall D. Paf1 and Ctr9, core components of the PAF1 complex, maintain low levels of telomeric repeat containing RNA. Nucleic Acids Res. 2018;46:621–634. doi: 10.1093/nar/gkx1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Smolikov S, Krauskopf A, Kupiec M. The yeast VPS genes affect telomere length regulation. Curr Genet. 2005;47:18–28. doi: 10.1007/s00294-004-0548-y. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Teis D. The ESCRT machinery. Curr Biol. 2012;22:R116–R120. doi: 10.1016/j.cub.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh AL, Audhya A. The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit Rev Biochem Mol Biol. 2014;49:242–261. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha H, David L, Pascon RC, Clauder-Munster S, Krishnakumar S, Nguyen M, Shi G, Dean J, Davis RW, Oefner PJ, et al. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics. 2008;180:1661–1670. doi: 10.1534/genetics.108.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Duan R, Ryu HY, Ahn SH. The yeast ESCRT complexes are involved in the regulation of transcription elongation. Genes Genom. 2014;36:335–343. doi: 10.1007/s13258-013-0171-8. [DOI] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. The EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim Biophys Acta. 2013;1829:116–126. doi: 10.1016/j.bbagrm.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar L, Yosef N, Sela Y, Sharan R, Ruppin E, Kupiec M. A genome-wide screen for essential yeast genes that affect telomere length maintenance. Nucleic Acids Res. 2009;37:3840–3849. doi: 10.1093/nar/gkp259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Xu Y, Bernecky C, Lee CT, Maier KC, Schwalb B, Tegunov D, Plitzko JM, Urlaub H, Cramer P. Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat Commun. 2017;8:15741. doi: 10.1038/ncomms15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.