Bacteria have evolved numerous means of survival in adverse environments with dormancy, as represented by “persistence” and the “viable but nonculturable” (VBNC) state, now recognized to be common modes for such survival. VBNC cells have been defined as cells which, induced by some stress, become nonculturable on media that would normally support their growth but which can be demonstrated by various methods to be alive and capable of returning to a metabolically active and culturable state.
KEYWORDS: antibiotic tolerance, antibiotic resistance, dormancy, drug discovery, persistence, persister, VBNC, viable but nonculturable
ABSTRACT
Bacteria have evolved numerous means of survival in adverse environments with dormancy, as represented by “persistence” and the “viable but nonculturable” (VBNC) state, now recognized to be common modes for such survival. VBNC cells have been defined as cells which, induced by some stress, become nonculturable on media that would normally support their growth but which can be demonstrated by various methods to be alive and capable of returning to a metabolically active and culturable state. Persister cells have been described as a population of cells which, while not being antibiotic resistant, are antibiotic tolerant. This drug-tolerant phenotype is thought to be a result of stress-induced and stochastic physiological changes as opposed to mutational events leading to true resistance. In this review, we describe these two dormancy strategies, characterize the molecular underpinnings of each state, and highlight the similarities and differences between them. We believe these survival modes represent a continuum between actively growing and dead cells, with VBNC cells being in a deeper state of dormancy than persister cells.
INTRODUCTION
Bacteria exist in environments with constant fluctuations. Over billions of years, these perpetual selective pressures have molded one of the greatest characteristics of microorganisms, their ability to survive through rapid adaptation. As individuals, bacteria may seem fragile and sedate; however, as a population, they are expert survivalists and in countless ways support life on earth. It is not surprising then that this integral aspect of bacterial life has been a topic of great interest for microbiologists since their discovery over a century ago. The overarching lesson from years of research is our appreciation for the ability of bacterial populations to adapt through rapid, purposeful, and concerted changes in gene expression and with the help of their genomic plasticity. Although still not thoroughly understood, one of the most impressive and effective adaptive maneuvers a bacterium can perform is to enter into a state of dormancy. In fact, bacteria spend the majority of their lives in a state of low metabolic activity with little or no growth but remain ready to divide when their environment permits (1). Although classically, and in the context of sporulating bacteria, dormancy has been thought of as a robust and definitive response to the environment, recent insights have blurred the lines of dormant life in nonsporulating organisms. In many species, dormancy is not necessarily an on/off switch but rather a progressive, stepwise process that changes dynamically with time and can be stochastic or a result of environmental cues. Specifically, we refer to two dormancy phenomena, the viable but nonculturable (VBNC) state and antibiotic persisters (AP). They have been extensively described and characterized as two highly stress-tolerant physiological states. Although the VBNC state and AP were discovered decades ago, they were initially a source of controversy and have only recently become a more fundamental part of our understanding of microbial physiology. As we learn more about these alternate states of life, it is becoming increasingly clear that these modes of existence play a significant role in our ability to treat infections.
The VBNC state was first discovered in 1982 by Colwell and colleagues, when they demonstrated that bacteria under stressful conditions maintained a subpopulation that was unable to grow on media but was functionally viable (2). Since its discovery, the term “VBNC” was a nidus of controversy that largely stemmed from an outdated definition of viability, one that assumed that if cells lose their ability to grow on media, they are no longer alive. Of course, with the help of molecular techniques, we now understand that loss of culturability does not equal death. Nonetheless, this controversy of nomenclature has led to significant confusion on dormancy and the VBNC state. Our understanding of this physiological state is further complicated by the adoption of other terms to describe the same phenomenon, including “active but nonculturable” (ABNC), “nongrowing but metabolically active” (NGMA), “conditionally viable environmental cells” (CVEC), and even the generic term “dormant” has been used (3–6). In this review, we refer to this state with the original “VBNC” term, as this is the most descriptive, experimentally apparent, and widely used to describe this state. The VBNC state is now generally accepted as a bacterial mechanism of survival in the face of external stress, including antibiotics, and is discussed in further detail below.
Antibiotic persisters were first named by Joseph Bigger and initially discovered by Hobby and colleagues over 70 years ago when they described a small population of cells that were tolerant to antibiotics even though the majority of the population had succumbed to the treatment (7, 99). This drug-tolerant phenotype is thought to be a result of physiological changes that may be stochastic or environmentally induced, as opposed to antibiotic-resistant cells that carry a heritable factor allowing bacteria to resist a specific class of antibiotic (8). These persisters have attracted significant attention in the scientific community and in infectious disease medicine given their obvious potential to cause antibiotic failure in a less predictable manner.
Recent investigations into the molecular mechanisms that govern the dynamics of these physiological states have provided evidence that the VBNC state and AP may be physiologically related and part of a continuum of dormancy (9). Since our initial proposal of the “dormancy continuum hypothesis,” several independent investigations have reported findings in its support. In this review, we provide an up-to-date and succinct summary of the VBNC state and AP, analyze the most recent studies that relate the two, and provide a synthesis of their physiologic relatedness.
THE VBNC STATE
The VBNC state describes a subpopulation of bacteria that have transiently lost their ability to grow on routine laboratory media on which they were previously able to grow. These cells exhibit significantly lower, although present, metabolic activity than their actively growing counterparts, and they continue to maintain membrane integrity and produce proteins (10, 11). In Fig. 1, a prototypical VBNC experiment is depicted where cells lose culturability over time but a portion of the original population remains viable during a seemingly “lethal” stress. It is important to recognize that the VBNC state is not a laboratory phenomenon, as VBNC cells can be readily found in the environment and in infections (11–15). For example, increased cases of vibriosis are occurring in areas of the world due to temperature anomalies where vibriosis is typically nonexistent. This idea of environmental induction and resuscitation was also clearly shown when VBNC cells were monitored in situ in an estuarine environment, where cells were clearly shown to enter and resuscitate from the VBNC state based on changes in water temperature (25). To be an effective survival strategy, VBNC cells must be able to resuscitate when conditions are permissible to growth, which is the process by which these cells regain their culturability. Currently, over 100 species of bacteria have been shown to enter a nonculturable state, but only a subset of these species have been shown to resuscitate (i.e., confirmed to be VBNC). This is because (i) in some studies, nonculturable cells were detected but resuscitation was not attempted, (ii) the proper conditions to induce resuscitation are not yet known, or (iii) these cells are not in a true VBNC state in those particular examples. Indeed, some species have been shown to resuscitate only after a specific treatment (12, 16). Figure 2 shows a phylogenetically organized depiction of the most current list of organisms that have been detected in a nonculturable state. This depiction shows the breadth at which this phenomenon is found across domains of life and in most phyla of bacteria, suggesting that dormancy may be a universal mechanism for survival. Although we do not yet fully understand the molecular mechanisms governing the VBNC state, especially in this large organism set, a variety of molecular processes have been found to be important in the dynamics of the VBNC and AP and are further discussed below.
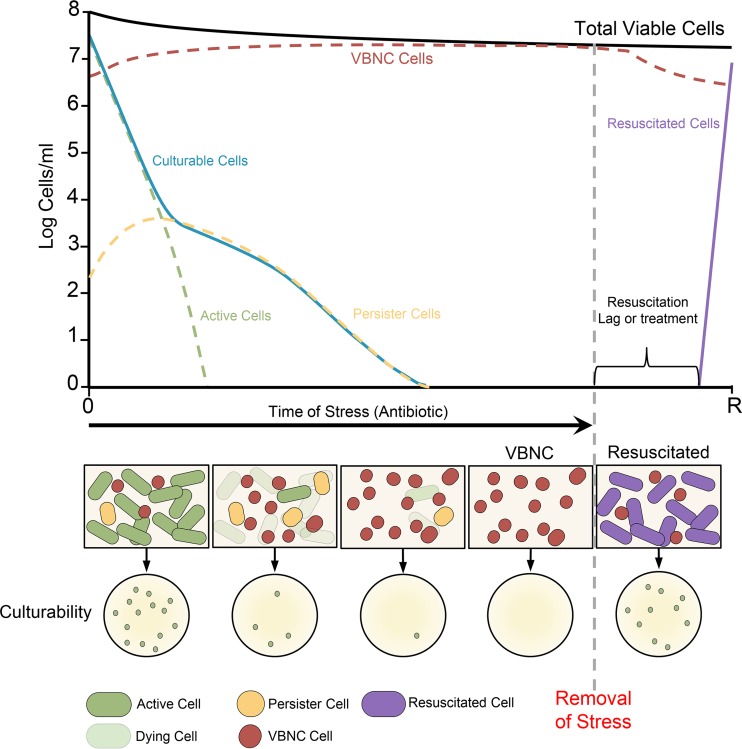
FIG 1.
Experimental dormancy dynamics of antibiotic persistence and the VBNC state. Persister cells are isolated by exposing a growing culture of cells to a lethal dose of antibiotics. The cells that remain culturable after treatment are called persisters. This typically produces the classic biphasic killing curve, with the slope of the initial phase (green dotted line) generated by the death of sensitive cells and the slope of the second phase (yellow dotted line) generated by the surviving persisters. Similarly, VBNC cells are isolated by applying a stress (e.g., cold temperature) or an antibiotic to a growing culture. Upon exposure to this stress, cells lose culturability in a varied amount of time (depending on the stress used) (blue line); however, a large portion of the population remains viable but nonculturable (red dotted line), as is determined by a variety of assays that test for gene expression, membrane stability, and metabolic activity. When the inducing stress is removed and adequate conditions are met (vertical dotted gray line), cells begin to alter their physiology toward resuscitation. After a lag period (dependent on the stress and bacterial species), cells regain the ability to grow on nutrient media.
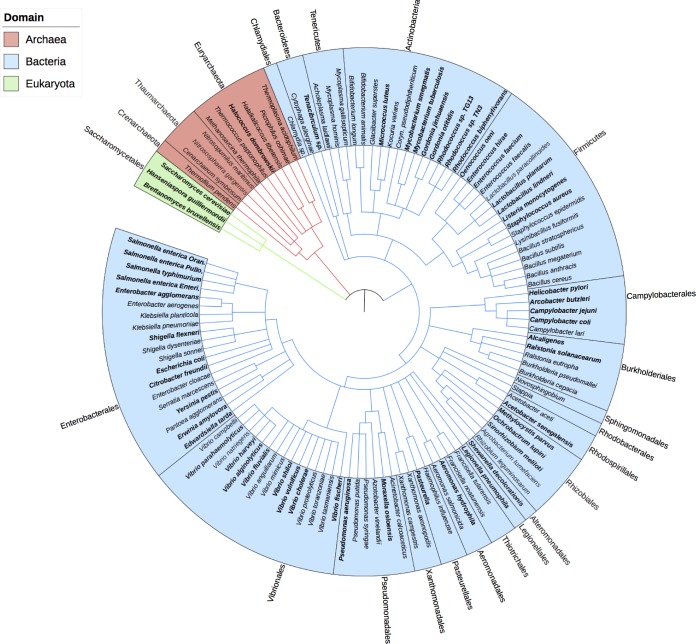
FIG 2.
The breadth of the VBNC state. This phylogenetic diagram was created using PhyloT (http://phylot.biobyte.de), using NCBI taxonomy. The VBNC state has been discovered in many bacteria but also in archaea and in fungi. Whether the described phenomenon is truly the same across these three domains of life has not yet been answered; however, this certainly upholds the idea that dormancy is a fundamental aspect of life on earth. Organisms written in bold have previously been shown to resuscitate from the VBNC state, while all others have either been detected in a nonculturable state or were induced but unable to resuscitate under the conditions provided. Coryn., Corynebacterium.
ANTIBIOTIC PERSISTENCE
Similar to the VBNC state, antibiotic persistence describes a subpopulation of cells that survive antibiotic treatment. The classic definition of persistence is derived from the biphasic killing curve that is produced when a logarithmic-phase culture is exposed to a lethal dose of antibiotics (Fig. 1). During the first phase of killing, there is a rapid decline in culturability due to death of the susceptible majority. In the second phase, there is a much slower and gradual decline in cell numbers. The cells in the second phase that remain viable and culturable are called the persisters and typically make up less than 1% of the original population. It is important to recognize that persisters are not a logarithmic-phase phenomenon. In fact, stationary-phase cultures harbor much higher numbers of persisters and may be a more realistic target for novel antimicrobials that are active against nongrowing cells. Significant work has been done in elucidating the molecular mechanisms governing persistence (8, 17–19), which are discussed and related to the VBNC state in further detail below. For excellent reviews on antibiotic persistence, please refer to references 8 and 19–21.
OVERLAPPING PHENOTYPIC CHARACTERISTICS
Although the VBNC state and AP were discovered independently and decades apart, these physiological states have many phenotypic similarities that warrant studying their relatedness. Here, we discuss the features that have led us to hypothesize that these two states of dormancy are closely related.
Formation of VBNC and AP cells.
It is thought that a VBNC population is formed during exposure to a stressful condition as a method of survival (11, 22–25). Alternatively, recent research has shown that there are large populations of VBNC cells in seemingly unstressed environments, such as logarithmic-phase cultures, suggesting a potential role for a stochastic transformation in physiology that then protects this subpopulation of cells from future unforeseen stressors (26–28). Based on current data, both means of VBNC formation, stochastic and stress induced, likely occur. VBNC cells have been shown to withstand a wide variety of stressors, including starvation, growth-inhibiting temperatures, decreased oxygen levels, suboptimal salinity, suboptimal pH, antibiotics, and internalization by macrophages (29–36). For several up-to-date reviews on the VBNC state, please refer to references 11, 33, and 37.
Conversely, it was traditionally thought that persisters arise stochastically within growing cultures. However, it is now known that cells can be induced to become persisters through exposure to stressful conditions, similar to the VBNC state (8). This is evidenced by the simple observation that stationary-phase cultures have much higher levels of persisters than cultures in the logarithmic phase. Besides predominating during stationary phase, persisters can be induced by starvation, carbon source transitions, macrophages, oxidative stress, DNA damage, suboptimal pH, and antibiotics (32, 39–43). Thus, there is very strong evidence that VBNC and AP cells are formed under similar stressful conditions, which strengthens the idea that they may be related.
Interestingly, not only are VBNC and AP cells induced by the same stresses, they are indeed found together. One study reported that there were ∼100-fold more VBNC cells than persisters in a logarithmic-phase culture of Escherichia coli (28). We have previously shown that VBNC cells are not only present with persisters at much higher numbers in logarithmic phase but also survive the antibiotic treatment alongside the persisters (26). A study by Gonçalves and de Carvalho observed the same phenomenon (27). These data certainly suggest that VBNC cells can form without the input of an obvious stress, similar to persisters. Given that VBNC cells are found at much higher numbers where there are persisters, it is possible that AP cells are in a state of transition from actively growing cells to a more dormant VBNC state.
Presence in biofilms.
Biofilms are one of the most predominant forms of bacterial life (44). These complex functional aggregates of bacteria have wide-ranging implications on biogeochemical cycles, biotechnology, and public health and play a role in active infection. For a thorough review on biofilms, please refer to reference 45. Importantly, bacteria within biofilms are much harder to eradicate with antibiotics than are planktonic cells (45). The complex architecture within biofilms produces stressful microenvironments leading to a spatiophysiologic heterogeneity of a population within a biofilm (45, 46). This is likely the reason why biofilms promote the presence of VBNC and persister cells. Indeed, central areas deep within biofilms tend to be hypoxic, resource limited, and acidic due to deposits of metabolic waste (46). Interestingly, these same stresses have independently been shown to induce both the VBNC state and AP (see “Formation of VBNC and AP cells,” above). Given that biofilm-related infections lead to significant morbidity and mortality, studying the role that these dormant populations play in biofilm biology is of significant interest.
Antibiotic tolerance.
As mentioned previously, the main phenotypic characteristic of persisters is their ability to tolerate high-dose antibiotics (20). In fact, this is how we know they exist. AP has gained significant attention in recent years because of the compelling evidence that persisters are significant contributors to relapsing infections and can lead to antibiotic failure (47–51). Moreover, studies have found that in relapsing urinary tract infections, which occur in 20 to 30% of patients within 4 months (52), the majority of cases are caused by the same isolate as the primary infections (53, 54). While surviving cells may hide intracellularly or within biofilms prior to clonal relapse, the idea that persisters play a significant role in human infection is difficult to contest.
Persisters are often multidrug tolerant but may vary in their susceptibilities to different drugs even in a comparison of the effect of two antibiotics on one culture (8). Since their initial discovery, Joseph Bigger hypothesized that persisters were nongrowing during the time of antibiotic treatment and therefore were able to survive and later grow when the antibiotic was no longer present (7). Nearly 60 years later, this hypothesis was supported through the use of microfluidics and single-cell imaging technology by Balaban and colleagues, who eloquently demonstrated that nongrowing cells were able to survive antibiotic treatment and resume growth after the antibiotic was removed (55). Following these findings, several other studies have attributed the lack of bacterial growth to their tolerability of antibiotics (20, 56). However, some studies have found that persistence may be much more complex than a simple lack of growth and provide evidence of active mechanisms of persistence, especially through the use of efflux pumps (57–59). Furthermore, a study by Collins and colleagues found that persisters, despite being dormant, are prepared for metabolite uptake, which could be harnessed in making these cells susceptible to antibiotic treatment (60).
The ability of VBNC cells to tolerate high-dose antibiotics has only recently been recognized. In fact, VBNC cells of Vibrio vulnificus have been shown to survive not only antibiotic treatment but also exposure to otherwise lethal levels of pH, heavy metals, heat, ethanol, and both osmotic and ionic challenges, establishing their propensity for stress resistance (59). Several studies have demonstrated the robust capability of VBNC cells of various species to survive antibiotics of multiple classes (15, 26, 36, 61–67). Similar to AP, several studies have demonstrated the presence of nonculturable cells at the clinical level (14, 68–72). Antibiotic failure due to VBNC cell survival and resuscitation is much harder to identify in the clinical laboratory, as VBNC cells are not detectable using standard culture-based methods. Therefore, it is difficult to recognize the specific clinical contribution of VBNC cells to instances of relapsing infection following antibiotic treatment. However, in a mouse urinary tract infection model, VBNC cells were detected after antibiotic treatment and were found to resuscitate within the host after antibiotic treatment ended, demonstrating the potential role of VBNC cells in relapsing infections (15). Similarly, studies on vibrios and in Campylobacter jejuni have found that VBNC cells can resuscitate within the host and retain their virulence (73–75). Furthermore, Liu and colleagues recently found that VBNC E. coli O157 strains continue to produce high levels of toxin (76). Taken together, the evidence is compelling that bacteria in the VBNC state are not simply a theoretical risk to public health but have an active role in relapsing infections and in antibiotic failure.
Similar to persisters, the antibiotic tolerance of VBNC cells most likely stems from their growth-arrested phenotype that leads to inactive drug targets. Interestingly, VBNC cells have also been shown to increase the expression of multidrug efflux pumps, which may be important in their drug tolerance (66, 77, 78). In fact, Lin and colleagues showed that VBNC cells had a higher expression of drug efflux pumps than culturable cells, and this led to a lower concentration of intracellular antibiotic in VBNC cells (66). Another study investigated the fatty acid composition of phospholipids in Staphylococcus aureus cells exposed to high-dose vancomycin and teicoplanin, which found that VBNC and persister cells exhibited similar changes in membrane lipid composition in response to these antibiotics (27). These findings indicate that antibiotic tolerance in the VBNC state may be an active process and one that is overlapping with the mechanisms of tolerance in persister cells.
OVERLAPPING MOLECULAR MECHANISMS
As suggested above, it is important to recognize that neither VBNC cells nor persisters are metabolically inept. Although there is a large decline in metabolic activity in these cells, they continue to energize their membranes and produce a more specialized set of proteins (10, 79). There are several mechanisms by which persister cells are thought to form, and there is growing evidence that these same mechanisms are involved in directing the VBNC state.
Stringent response.
The stringent response is a mechanism by which cells detect amino acid starvation and respond by slowing growth and increasing amino acid synthesis. This is done through the production of the alarmone guanosine pentaphosphate [(p)ppGpp], which inhibits cellular processes that consume significant resources, such as cell division, transcription, and translation. The alarmone alters gene expression by binding RNA polymerase and increasing the transcription of genes necessary for biosynthetic processes (80). Persister cells have been found to require the alarmone (p)ppGpp (81–83). In a proposed molecular model of persistence (Fig. 3), (p)ppGpp leads to the accumulation of polyphosphates, which in turn activate a Lon-mediated proteolytic process that disturbs the ratio of toxin-antitoxin systems (discussed further below) (84). Several studies have found that the stringent response also plays a role in the VBNC state. Mishra and colleagues found that relA, which is responsible for producing (p)ppGpp, is upregulated in the VBNC state (85). Similarly, mutants devoid of their ability to produce (p)ppGpp exhibited a significant reduction in their ability to enter the VBNC state, while strains that overproduced (p)ppGpp formed significantly higher numbers of VBNC cells (86). In a study on C. jejuni, mutants that were unable to produce polyphosphate exhibited decreased VBNC induction (87). Wan and colleagues measured intracellular (p)ppGpp and found that excess production of this alarmone was necessary for entry into the VBNC state (88). More work is required to elucidate the specific role of the stringent response in induction of VBNC; however, the above-mentioned evidence certainly points to an overlapping mechanism of induction involving the stringent response for both persisters and VBNC cells.
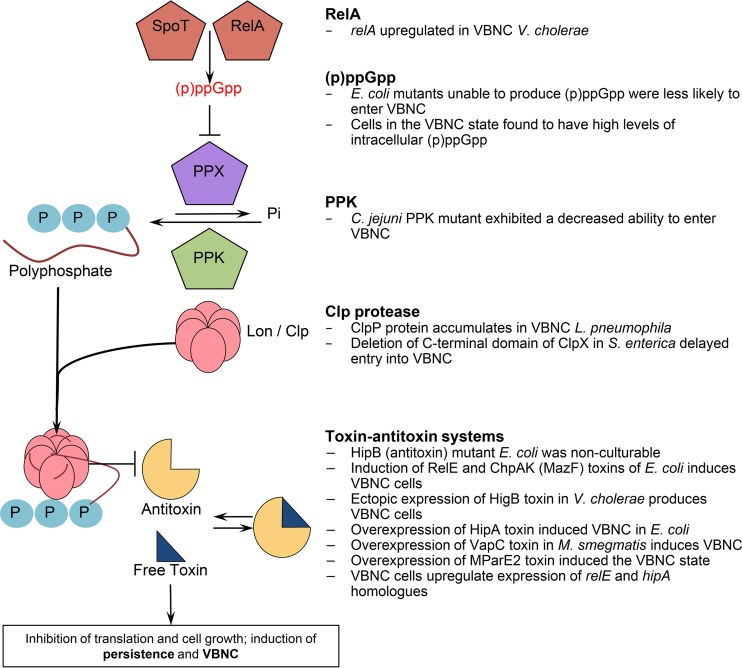
FIG 3.
Mechanism of persister formation with supporting evidence of its role in the VBNC state. In this model, the activation of RelA and SpoT due to amino acid starvation leads to the intracellular accumulation of the alarmone (p)ppGpp. This leads to the inhibition of exopolyphosphatase (PPX), an enzyme that degrades polyphosphates, causing their accumulation due to continued activity of polyphosphate kinase. Lon protease is activated by polyphosphate and begins to selectively degrade proteins, including antitoxins. This disturbs toxin-antitoxin ratios and allows free toxins to inhibit cell growth, leading to persistence. It is important to note that even if this exact mechanism is not at play, each component has independently been shown to have a significant role in AP and VBNC. Modified from reference 9. PPK, polyphosphate kinase.
Proteolysis.
Intracellular proteases have been found to play a significant role in persister cell formation. Specifically, Lon protease was shown to induce persistence through the degradation of type II antitoxins, thereby disrupting the ratio of toxin to antitoxin, allowing the free toxin to carry out its downstream effects (84). Some type II toxins inhibit translation through the cleavage of mRNA and tRNA, while others can inhibit other processes, such as that of DNA gyrase, all of which can lead to a reduction in growth and division (89). Lon protease has been shown to degrade antitoxins of E. coli (84). Interestingly, the role of Lon protease in the formation of VBNC cells has not yet been reported. However, another stress-induced protease, Clp, that may have a role in persistence (90) and has substrate recognition domains similar to those of Lon (91), has been shown to be important in VBNC induction. Clp protease was reported to degrade MazE, P1 Phd, and Kis antitoxins in E. coli and several antitoxins in S. aureus (92–94). Alleron and colleagues reported that Clp protease accumulated in VBNC cells (95), and another study found that mutants unable to produce Clp protease displayed a reduction in their ability to enter the VBNC state (96). Recently, Almagro-Moreno and colleagues found that proteolysis by a site 2 protease RspeP (YaeL) induced the VBNC state in Vibrio cholerae (97). These findings indicate that intracellular proteolysis is likely an important mechanism governing dormancy in both AP and the VBNC state.
Toxin-antitoxin systems.
Toxin-antitoxin (TA) systems have a significant role in governing persistence (17). For an excellent review on toxin-antitoxin systems, please refer to reference 89. It has been shown that the overproduction of toxin proteins results in a decrease in cell growth and subsequent increase in the number of persister cells (98). More definitive evidence produced by studying persisters at the single-cell level showed that these drug-tolerant cells exhibited increased expression of TA genes (100). Interestingly, TA systems are not the only mechanism shown to govern persister formation, and other mechanisms (discussed below) may govern persistence in Gram positives (17, 101). It is important to note that some of the work implicating TA systems in persister formation has been recently retracted, and there is new evidence confirming that TA systems do not play a major role in stochastic persister formation. However, there is independent evidence demonstrating the role of TA systems in forming persisters upon exposure to stress. Thus, we find that the role of TA systems in bacterial dormancy is strongly substantiated (102–104).
There is strong evidence that TA systems may also control the dynamics of the VBNC state. As with persisters, overexpression of toxin genes was shown to induce the formation of VBNC cells (11, 105–107). In a more concrete example, E. coli mutants of the HipB antitoxin but with functional HipA toxin were nonculturable altogether (100). Demidenok and colleagues showed that overexpression of the VapB antitoxin prevented cells from entering the VBNC state, while overexpression of the VapC toxin induced the VBNC state (108). Similarly, overexpression of the MParE2 toxin led to a decline in culturability due to entry into the VBNC state (109). Recently, we reported that VBNC cells of Vibrio vulnificus have significantly elevated expression of several homologues of the type II toxins of RelE and HipA compared to log-phase and stationary-phase cultures (26). In the same study, we found that both VBNC cells and AP were induced by incubation in human serum, which correlated with an upregulation of toxin gene expression (26). These findings demonstrate the importance of TA systems in both AP and the VBNC state, which significantly strengthens the idea that these two states are mechanistically related. Furthermore, it appears that TA genes are induced by host conditions indicating that in vitro studies on persistence may be underestimating the level of antibiotic tolerance that organisms display during infection.
ATP depletion and dormancy.
Recent studies have demonstrated that TA systems are less significant in controlling persistence in Gram-positive pathogens. It has been demonstrated that the depletion of ATP may be important in controlling persistence in these organisms (17, 101, 110). Although VBNC cells are found to have a reduction in intracellular ATP (111), to our knowledge, no studies have specifically investigated the role of ATP in orchestrating the VBNC state. A recent study by Pu and colleagues investigated the role of protein aggregation at the single-cell level to study their role in regulating bacterial dormancy and antibiotic tolerance (112). They found that cells displayed various “depths of dormancy” after antibiotic treatment, which correlated with the time necessary for the cell to regrow following treatment. Furthermore, the level of protein aggregation within cells correlated with the depth of dormancy. The more aggregates cells had, the longer it took to clear them and subsequently to resuscitate. Interestingly, they found that disaggregation occurs in an ATP-dependent manner, which is consistent with the idea that cells in a deeper state of dormancy and with lower intracellular ATP will take longer to resuscitate. These cells in deeper dormancy were indeed VBNC cells, while cells that were able to quickly clear their aggregates and recover were persisters (112). These results are quite interesting, as such distinctions between cell populations could be used as targets for therapeutic discovery (113).
DIFFERENCES BETWEEN VBNC AND AP
While we have highlighted a number of similarities in the physiology of VBNC and AP cells, it is important to note that while these two states are similar, they are not the same. There are few studies on the distinctions between these two states due to the difficulty in separating these two populations when they coexist; however, we provide further evidence below that supports the hypothesis that these two populations exist on a continuum, with VBNC cells existing in a deeper state of dormancy, In contrast, AP cells represent a more transient, early state of dormancy.
Resuscitation from the VBNC state and AP.
One of the major differences between VBNC and AP cells pertains to their resuscitation dynamics. While persisters are typically able to grow on nutrient media following a short lag period after the removal of antibiotics (55), VBNC cells are unable to grow on nutrient media even after removal of the inducing stress (26). They require a much longer resuscitation period away from nutrient media after removal of the inducing stress, while some species of VBNC bacteria require specific treatments (33). It is plausible that VBNC cells in deeper dormancy require more time or an external stimulus in order to restore toxin-antitoxin ratios, reestablish metabolic competence, and repair damaged proteins that are necessary for growth. In fact, one study found that the amount of free toxin within persister cells positively correlated with the time required to resume growth after removal of the stress (100), indicating that the amount of free toxin within a cell can determine the depth of dormancy. As noted above, Pu and colleagues determined that the amount of protein aggregates, a proxy for protein damage, correlated with depth of dormancy and time needed for resuscitation to occur after antibiotic removal (112). In a similar study, heat shock treatment led to the formation of two distinct populations that were distinguished using gradient centrifugation, a high-density fraction and a low-density fraction (114). The high-density population grew immediately after transfer to LB medium, as would be seen with persisters, while the low-density population required a significantly longer time to resuscitate prior to regaining the ability to grow on media, as expected of VBNC cells. They also found that once the cells resuscitated, they divided at the same rate, independent of the subpopulation from which they came, supporting the idea that the lag in regrowth is not due to slower-growing cells but rather the time necessary for cells to resuscitate. Interestingly, this study also showed that the low-density (VBNC) population was more tolerant to antibiotics than the high-density (persister) population. Similar to the findings from the study by Pu and colleagues, this study also quantified protein aggregates within cells but found no difference between VBNC and persister populations. They did, however, find that cells in the VBNC fraction had fewer ribosomes and more protein oxidation than did the persister fraction. Interestingly, another investigation found that cultures treated with antibiotics produced a heterogeneous population composed of cells that differed in their rates of resuscitation after antibiotic removal (115). Indeed, they found that the more ribosomes the cells had, the faster they were able to resuscitate. Together, these findings support the idea that VBNC cells are in a deeper form of dormancy than are persisters and therefore require more time, or a chemical or environmental signal, to resuscitate. Furthermore, resuscitation is likely dependent on oxidative damage and ribosome content within cells. These findings are quite interesting, and more work needs to be done to further determine the molecular mechanisms of resuscitation, as our understanding of this process may have a significant impact on our ability to treat chronic and relapsing infections.
Distinguishing VBNC and AP cells.
Based on the literature reviewed here, it is expected that VBNC cells are always present where there are persister subpopulations, and likely at a much higher number. Importantly, studies aimed at testing the ability of novel antimicrobials to eradicate persisters tend to use culturability as a measure of viability, which overlooks the presence of drug-tolerant VBNC populations and can be misleading. This is a critical oversight, since these cells make up a large portion of antibiotic-tolerant cells and likely contribute to antibiotic failure. When testing the ability of a drug to eradicate antibiotic-tolerant cells, both viability and culturability should be quantified, which would allow for the detection and enumeration of both persister and VBNC cells. Conversely, most studies on VBNC cells are done on pure VBNC cultures; therefore, the impact of persisters and their potential confusion with VBNC cells in these studies are eliminated. If sufficient stress is applied, it is possible to force the entire population into a completely nonculturable population, free of persisters, that can then be resuscitated when the stress is removed (26).
A recent study using single-cell imaging and microfluidics elegantly differentiated VBNC cells and persister cells under antibiotic exposure and identified differentially expressed promoters that could be used to distinguish VBNC and persister cells at the single-cell level (63). They provide strong evidence that VBNC cells are not dead and share molecular characteristics with persisters. Importantly, this new and effective approach by Bamford and colleagues allows for the identification and isolation of persisters and VBNC cells based on promoter activity and without the need to expose cells to an additional confounding stress, such as an antibiotic. This method can provide deeper insights into the molecular changes in cells as they transition from active growth to dormancy with much greater specificity. Furthermore, we can also foresee this methodology being used to study population dynamics during exposure to stress or antibiotics, which could be used to more rigorously test the dormancy continuum hypothesis. It may also prove to be invaluable in the setting of drug development and testing. We are hopeful that this and future developments in methodology, such as flow cytometry and fluorescence-activated cell sorting, will be used to further aid our understanding of these dormancy states and how they can be manipulated to eventually result in a reduction in relapsing infections and antibiotic failure.
Of note, the findings described by Bamford and colleagues (63) are in stark contrast to the findings of a study performed by Kim and colleagues, in which it was concluded that VBNC and persister cells describe the same dormant phenotype (116). In the study by Kim and colleagues, cells exposed to starvation produced a heterogeneous population of cells consisting of culturable and nonculturable (VBNC) cells. It was found that the culturable population was completely made up of persister cells after 14 days of starvation, as nearly all culturable cells survived antibiotic treatment. However, cells did not resuscitate on LB-agarose gel pads, and these same cells were then found to have an empty cytosol using TEM. For this reason, the investigators drew the conclusion that what most describe as VBNC cells are persister cells. However, a wealth of literature refutes this conclusion since, as mentioned previously, many studies have successfully obtained pure VBNC cultures without the presence of persisters, thereby eliminating any confusion that their presence would have on the interpretation of an experiment. In fact, these pure VBNC cultures are able to resuscitate when the stressful condition is removed (11, 12, 26, 62). Furthermore, it would not be expected that VBNC cells would resuscitate on agarose gel pads with LB, as this would be analogous to attempting to culture what are nonculturable cells. Additionally, resuscitation of starvation-induced VBNC cells of E. coli has classically been difficult, and only specific treatments have been successful at resuscitating them (34). An interesting finding in the Kim et al. study was that cells thought to be VBNC had cleared out cytoplasms as observed by electron microscopy, despite negative staining with propidium iodide (indicating that they had intact cell membranes). It has been previously reported that the nucleoid in dormant cells becomes very condensed (1), and the number of ribosomes present within the cytosol is drastically reduced. Therefore, the very thin sectioning required for electron microscopy could easily cut out focal aggregates of genomic material in cells and, along with the loss of electron-dense ribosomes, could give the appearance that the cellular cytosol is empty. This is a general limitation of microscopy and thus should be used and interpreted with caution as a proxy for determining cellular viability.
A note on resuscitation.
Resuscitation describes the molecular changes within cells as they exit dormancy and is not the same as regrowth. Regrowth on media, on the other hand, is the phenotypic determinant that a cell has resuscitated. In other words, a cell can exit from dormancy (resuscitate) without dividing, and it is only after a cell has resuscitated that it can then divide on nutrient media. In fact, transferring VBNC cells directly onto nutrient media without first allowing them to resuscitate will likely lead to their demise, given that dormant cells attempting to resuscitate poorly combat the oxidative stress imposed by nutrient media (11, 117).
The dormancy continuum hypothesis.
The findings discussed above provide further evidence for our previously proposed dormancy continuum hypothesis (9), which posits that a shared set of molecular mechanisms control both persistence and the VBNC state but that the two phenotypes are found in separate regions on the continuum. Figure 4 describes the relationship between active cells, persisters, and VBNC cells with respect to their culture status and general molecular characteristics. Figure 5 shows a theoretical model of population dynamics under stressful conditions and an estimation of population-level changes as cells transition from active growth to dormancy, and later resuscitation. We hope that these representations provide for a better understanding of the theorized population dynamics of bacterial dormancy in AP and VBNC cells. We are confident that novel methodological developments will soon allow us to test these population-level models of dormancy by allowing us to differentiate and quantify cells that are found along the entire dormancy continuum (63).
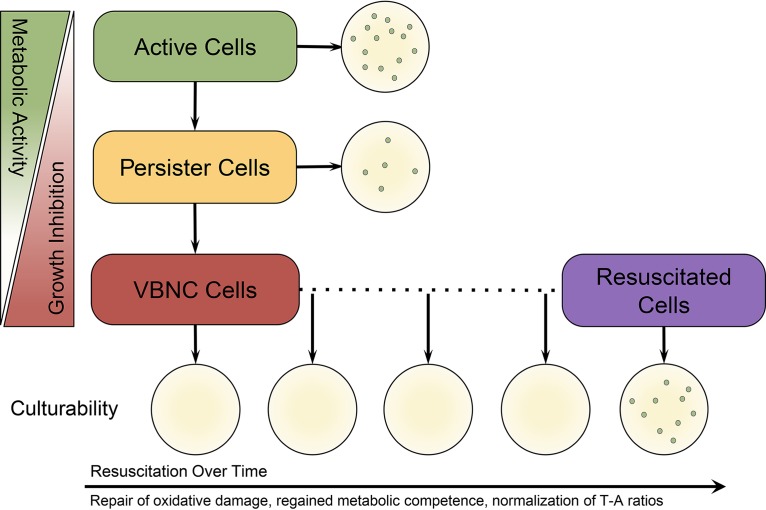
FIG 4.
The dormancy continuum hypothesis. Stochasticity and environmental cues lead to the degradation of proteins (including antitoxins), reduction in metabolic activity, and inhibition of growth. In the very beginning stages of dormancy, antibiotic-tolerant persister cells are produced. When these persister cells are transferred to laboratory media, their metabolic competence allows them to resuscitate and grow on media. However, if the stressful conditions continue, these persister cells go deeper into dormancy and lose their culturability (i.e., become VBNC) as their metabolism further declines, protein damage continues, and they become growth arrested. When VBNC cells are transferred to media, they do not produce colonies. However, if the inducing stress is removed and adequate time and conditions are provided for VBNC, cells will resuscitate and regain their ability to grow on media. Resuscitation is the molecular process by which cells repair oxidative damage, regain metabolic competence, and normalize their toxin-antitoxin ratios. After resuscitation has occurred, cells are once again able to produce colonies. Attempts at culturing these cells any time prior to the completion of the resuscitation process will result in no growth.
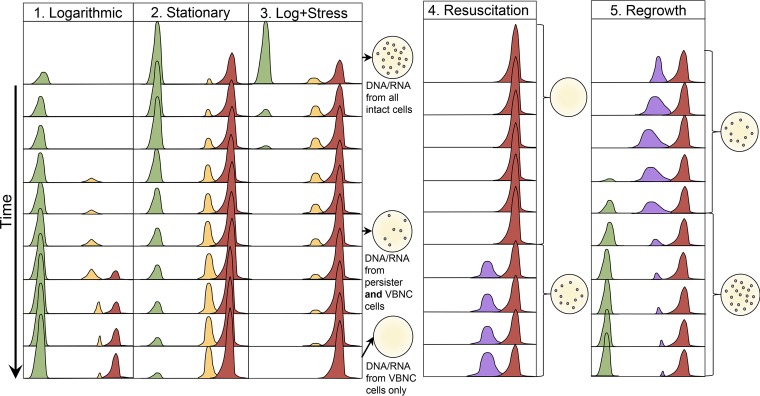
FIG 5.
Model of population dynamics as part of the dormancy continuum. Each column numbered 1 to 5 represents a different phase or treatment. Actively growing cells are represented by the green curve, persister cells are represented by the yellow curve, VBNC cells are represented by the red curve, and resuscitated cells are represented by the purple curve. Each set of curves represents a snapshot on an arbitrary time scale that proceeds downwards. 1) In logarithmic phase, persisters start to form and transition to the VBNC state. Persisters are in a transitory phase and exist at a low number, while VBNC cells represent deep dormancy and accumulate over time. 2) As stationary phase progresses, the number of transitioning persisters increase and reach a steady state with the VBNC population, while actively growing cells decline in number. 3) When logarithmic-phase cells encounter a stressful condition, some actively growing cells rapidly enter the persister state, while many susceptible cells die. Over time, with continued stress, these persisters will transition to the VBNC state. The drawings of petri plates to the right of the curves represent the culture status of the population. It is important to note that there are scenarios (pictured) when only persisters are culturable but DNA/RNA studies would be affected by genomic material from both persisters and VBNC cells. 4) Resuscitation is a variable process depending on the inducing stress and the species being studied but will eventually lead to regained culturability in a large portion of the remaining population. Even after resuscitation, some VBNC cells remain. Resuscitated cells may be in a unique physiological state and are likely antibiotic tolerant, as they are not actively dividing. 5) Regrowth occurs when resuscitated cells that have regained their ability to grow transition back to a rapidly growing phenotype when nutrients are supplied.
Some may feel that attempting to understand the relationship between these two phenomena is unnecessary. Our hope is that the findings described above shed light on the importance of this relationship. It is vital that we consider all of the viable cells that are in a culture or in an infection. If we are truly striving to develop methods to eradicate dormant cells from infections in an effort to reduce recurrence, we must focus on our ability to understand and target cells ranging the entire continuum.
REFERENCES
- 1.Bergkessel M, Basta DW, Newman DK. 2016. The physiology of growth arrest: uniting molecular and environmental microbiology. Nat Rev Microbiol 14:549–562. doi: 10.1038/nrmicro.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu HS, Roberts N, Singleton FL, Attwell RW, Grimes DJ, Colwell RR. 1982. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol 8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 3.Kamruzzaman M, Udden SM, Cameron DE, Calderwood SB, Nair GB, Mekalanos JJ, Faruque SM. 2010. Quorum-regulated biofilms enhance the development of conditionally viable, environmental Vibrio cholerae. Proc Natl Acad Sci U S A 107:1588–1593. doi: 10.1073/pnas.0913404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manina G, McKinney JD. 2013. A single-cell perspective on non-growing but metabolically active (NGMA) bacteria. Curr Top Microbiol Immunol 374:135–161. doi: 10.1007/82_2013_333. [DOI] [PubMed] [Google Scholar]
- 5.Nelson EJ, Chowdhury A, Flynn J, Schild S, Bourassa L, Shao Y, LaRocque RC, Calderwood SB, Qadri F, Camilli A. 2008. Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog 4:e1000187. doi: 10.1371/journal.ppat.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potgieter M, Bester J, Kell DB, Pretorius E. 2015. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol Rev 39:567–591. doi: 10.1093/femsre/fuv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 8.Fisher RA, Gollan B, Helaine S. 2017. Persistent bacterial infections and persister cells. Nat Rev Microbiol 15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 9.Ayrapetyan M, Williams TC, Oliver JD. 2015. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol 23:7–13. doi: 10.1016/j.tim.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Mali S, Mitchell M, Havis S, Bodunrin A, Rangel J, Olson G, Widger WR, Bark SJ. 2017. A proteomic signature of dormancy in the actinobacterium Micrococcus luteus. J Bacteriol 199:e00206-17. doi: 10.1128/JB.00206-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 12.Ayrapetyan M, Williams TC, Oliver JD. 2014. Interspecific quorum sensing mediates the resuscitation of viable but nonculturable vibrios. Appl Environ Microbiol 80:2478–2483. doi: 10.1128/AEM.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bari SM, Roky MK, Mohiuddin M, Kamruzzaman M, Mekalanos JJ, Faruque SM. 2013. Quorum-sensing autoinducers resuscitate dormant Vibrio cholerae in environmental water samples. Proc Natl Acad Sci U S A 110:9926–9931. doi: 10.1073/pnas.1307697110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lleo MM, Ghidini V, Tafi MC, Castellani F, Trento I, Boaretti M. 2014. Detecting the presence of bacterial DNA by PCR can be useful in diagnosing culture-negative cases of infection, especially in patients with suspected infection and antibiotic therapy. FEMS Microbiol Lett 354:153–160. doi: 10.1111/1574-6968.12422. [DOI] [PubMed] [Google Scholar]
- 15.Rivers B, Steck TR. 2001. Viable but nonculturable uropathogenic bacteria are present in the mouse urinary tract following urinary tract infection and antibiotic therapy. Urol Res 29:60–66. doi: 10.1007/s002400000151. [DOI] [PubMed] [Google Scholar]
- 16.Steinert M, Emody L, Amann R, Hacker J. 1997. Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl Environ Microbiol 63:2047–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes K, Maisonneuve E. 2012. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol 66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 19.Helaine S, Kugelberg E. 2014. Bacterial persisters: formation, eradication, and experimental systems. Trends Microbiol 22:417–424. doi: 10.1016/j.tim.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 21.Defraine V, Fauvart M, Michiels J. 2018. Fighting bacterial persistence: current and emerging anti-persister strategies and therapeutics. Drug Resist Updat 38:12–26. doi: 10.1016/j.drup.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Colwell RR. 2000. Viable but nonculturable bacteria: a survival strategy. J Infect Chemother 6:121–125. doi: 10.1007/PL00012151. [DOI] [PubMed] [Google Scholar]
- 23.Roszak DB, Colwell RR. 1987. Survival strategies of bacteria in the natural environment. Microbiol Rev 51:365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver JD. 2005. The viable but nonculturable state in bacteria. J Microbiol 43:93–100. [PubMed] [Google Scholar]
- 25.Oliver JD, Hite F, McDougald D, Andon NL, Simpson LM. 1995. Entry into, and resuscitation from, the viable but nonculturable state by Vibrio vulnificus in an estuarine environment. Appl Environ Microbiol 61:2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayrapetyan M, Williams TC, Baxter R, Oliver JD. 2015. Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infect Immun 83:4194–4203. doi: 10.1128/IAI.00404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonçalves FD, de Carvalho CC. 2016. Phenotypic modifications in Staphylococcus aureus cells exposed to high concentrations of vancomycin and teicoplanin. Front Microbiol 7:13. doi: 10.3389/fmicb.2016.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orman MA, Brynildsen MP. 2013. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob Agents Chemother 57:4398–4409. doi: 10.1128/AAC.00372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besnard V, Federighi M, Cappelier JM. 2000. Development of a direct viable count procedure for the investigation of VBNC state in Listeria monocytogenes. Lett Appl Microbiol 31:77–81. doi: 10.1046/j.1472-765x.2000.00771.x. [DOI] [PubMed] [Google Scholar]
- 30.Chaveerach P, ter Huurne AA, Lipman LJ, van Knapen F. 2003. Survival and resuscitation of ten strains of Campylobacter jejuni and Campylobacter coli under acid conditions. Appl Environ Microbiol 69:711–714. doi: 10.1128/AEM.69.1.711-714.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day AP, Oliver JD. 2004. Changes in membrane fatty acid composition during entry of Vibrio vulnificus into the viable but nonculturable state. J Microbiol 42:69–73. [PubMed] [Google Scholar]
- 32.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP.. 2014. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol 5:258. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinto D, Almeida V, Almeida Santos M, Chambel L. 2011. Resuscitation of Escherichia coli VBNC cells depends on a variety of environmental or chemical stimuli. J Appl Microbiol 110:1601–1611. doi: 10.1111/j.1365-2672.2011.05016.x. [DOI] [PubMed] [Google Scholar]
- 35.Xiao XL, Tian C, Yu YG, Wu H. 2013. Detection of viable but nonculturable Escherichia coli O157:H7 using propidium monoazide treatments and qPCR. Can J Microbiol 59:157–163. doi: 10.1139/cjm-2012-0577. [DOI] [PubMed] [Google Scholar]
- 36.Pasquaroli S, Zandri G, Vignaroli C, Vuotto C, Donelli G, Biavasco F. 2013. Antibiotic pressure can induce the viable but non-culturable state in Staphylococcus aureus growing in biofilms. J Antimicrob Chemother 68:1812–1817. doi: 10.1093/jac/dkt086. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X, Zhong J, Wei C, Lin CW, Ding T. 2017. Current perspectives on viable but non-culturable state in foodborne pathogens. Front Microbiol 8:580. doi: 10.3389/fmicb.2017.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Amato SM, Orman MA, Brynildsen MP. 2013. Metabolic control of persister formation in Escherichia coli. Mol Cell 50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppee JY, Ghigo JM, Beloin C. 2013. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 9:e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dörr T, Vulic M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vega NM, Allison KR, Khalil AS, Collins JJ. 2012. Signaling-mediated bacterial persister formation. Nat Chem Biol 8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y, Vulic M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 45.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. 2016. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 46.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 47.Grant SS, Hung DT. 2013. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 4:273–283. doi: 10.4161/viru.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conlon BP, Rowe SE, Lewis K. 2015. Persister cells in biofilm associated infections. Adv Exp Med Biol 831:1–9. doi: 10.1007/978-3-319-09782-4_1. [DOI] [PubMed] [Google Scholar]
- 49.Fauvart M, De Groote VN, Michiels J. 2011. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol 60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 50.Schumacher MA, Balani P, Min J, Chinnam NB, Hansen S, Vulic M, Lewis K, Brennan RG. 2015. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 524:59–64. doi: 10.1038/nature14662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van den Bergh B, Fauvart M, Michiels J. 2017. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol Rev 41:219–251. doi: 10.1093/femsre/fux001. [DOI] [PubMed] [Google Scholar]
- 52.Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med 113(Suppl 1A):5S–13S. [DOI] [PubMed] [Google Scholar]
- 53.Bingen E, Denamur E, Lambert-Zechovsky N, Braimi N, el Lakany M, Elion J. 1992. DNA restriction fragment length polymorphism differentiates recurrence from relapse in treatment failures of Streptococcus pyogenes pharyngitis. J Med Microbiol 37:162–164. doi: 10.1099/00222615-37-3-162. [DOI] [PubMed] [Google Scholar]
- 54.Russo TA, Stapleton A, Wenderoth S, Hooton TM, Stamm WE. 1995. Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women. J Infect Dis 172:440–445. doi: 10.1093/infdis/172.2.440. [DOI] [PubMed] [Google Scholar]
- 55.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 56.Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. 2012. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335:100–104. doi: 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. 2011. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pu Y, Ke Y, Bai F. 2017. Active efflux in dormant bacterial cells–new insights into antibiotic persistence. Drug Resist Updat 30:7–14. doi: 10.1016/j.drup.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Pu Y, Zhao Z, Li Y, Zou J, Ma Q, Zhao Y, Ke Y, Zhu Y, Chen H, Baker MA, Ge H, Sun Y, Xie XS, Bai F. 2016. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol Cell 62:284–294. doi: 10.1016/j.molcel.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu Y, Coates A. 2012. Nonmultiplying bacteria are profoundly tolerant to antibiotics, p 99–119. In Coates A. (ed), Handbook of experimental pharmacology, vol 211 Springer, Berlin, Germany. doi: 10.1007/978-3-642-28951-4_7. [DOI] [PubMed] [Google Scholar]
- 62.Nowakowska J, Oliver JD. 2013. Resistance to environmental stresses by Vibrio vulnificus in the viable but nonculturable state. FEMS Microbiol Ecol 84:213–222. doi: 10.1111/1574-6941.12052. [DOI] [PubMed] [Google Scholar]
- 63.Bamford RA, Smith A, Metz J, Glover G, Titball RW, Pagliara S. 2017. Investigating the physiology of viable but non-culturable bacteria by microfluidics and time-lapse microscopy. BMC Biol 15:121. doi: 10.1186/s12915-017-0465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chowdhury S, Hassan MM, Alam M, Sattar S, Bari MS, Saifuddin AK, Hoque MA. 2015. Antibiotic residues in milk and eggs of commercial and local farms at Chittagong, Bangladesh. Vet World 8:467–471. doi: 10.14202/vetworld.2015.467-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee S, Bae S. 2017. Molecular viability testing of viable but non-culturable bacteria induced by antibiotic exposure. Microb Biotechnol doi: 10.1111/1751-7915.13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin H, Ye C, Chen S, Zhang S, Yu X. 2017. Viable but non-culturable E. coli induced by low level chlorination have higher persistence to antibiotics than their culturable counterparts. Environ Pollut 230:242–249. doi: 10.1016/j.envpol.2017.06.047. [DOI] [PubMed] [Google Scholar]
- 67.Pasquaroli S, Citterio B, Cesare AD, Amiri M, Manti A, Vuotto C, Biavasco F. 2014. Role of daptomycin in the induction and persistence of the viable but non-culturable state of Staphylococcus aureus biofilms. Pathogens 3:759–768. doi: 10.3390/pathogens3030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boaretti M, Castellani F, Merli M, Lucidi C, Lleo MM. 2016. Presence of multiple bacterial markers in clinical samples might be useful for presumptive diagnosis of infection in cirrhotic patients with culture-negative reports. Eur J Clin Microbiol Infect Dis 35:433–441. doi: 10.1007/s10096-015-2556-x. [DOI] [PubMed] [Google Scholar]
- 69.Bullman S, O'Leary J, Corcoran D, Sleator RD, Lucey B. 2012. Molecular-based detection of non-culturable and emerging campylobacteria in patients presenting with gastroenteritis. Epidemiol Infect 140:684–688. doi: 10.1017/S0950268811000859. [DOI] [PubMed] [Google Scholar]
- 70.Cubero N, Esteban J, Palenque E, Rosell A, Garcia MJ. 2013. Evaluation of the detection of Mycobacterium tuberculosis with metabolic activity in culture-negative human clinical samples. Clin Microbiol Infect 19:273–278. doi: 10.1111/j.1469-0691.2012.03779.x. [DOI] [PubMed] [Google Scholar]
- 71.Zandri G, Pasquaroli S, Vignaroli C, Talevi S, Manso E, Donelli G, Biavasco F. 2012. Detection of viable but non-culturable staphylococci in biofilms from central venous catheters negative on standard microbiological assays. Clin Microbiol Infect 18:E259–E261. doi: 10.1111/j.1469-0691.2012.03893.x. [DOI] [PubMed] [Google Scholar]
- 72.Zoletti GO, Siqueira JF Jr, Santos KR. 2006. Identification of Enterococcus faecalis in root-filled teeth with or without periradicular lesions by culture-dependent and -independent approaches. J Endod 32:722–726. doi: 10.1016/j.joen.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 73.Baffone W, Casaroli A, Citterio B, Pierfelici L, Campana R, Vittoria E, Guaglianone E, Donelli G. 2006. Campylobacter jejuni loss of culturability in aqueous microcosms and ability to resuscitate in a mouse model. Int J Food Microbiol 107:83–91. doi: 10.1016/j.ijfoodmicro.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 74.Baffone W, Citterio B, Vittoria E, Casaroli A, Campana R, Falzano L, Donelli G. 2003. Retention of virulence in viable but non-culturable halophilic Vibrio spp. Int J Food Microbiol 89:31–39. doi: 10.1016/S0168-1605(03)00102-8. [DOI] [PubMed] [Google Scholar]
- 75.Oliver JD, Bockian R. 1995. In vivo resuscitation, and virulence towards mice, of viable but nonculturable cells of Vibrio vulnificus. Appl Environ Microbiol 61:2620–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu J, Zhou R, Li L, Peters BM, Li B, Lin CW, Chuang TL, Chen D, Zhao X, Xiong Z, Xu Z, Shirtliff ME. 2017. Viable but non-culturable state and toxin gene expression of enterohemorrhagic Escherichia coli O157 under cryopreservation. Res Microbiol 168:188–193. doi: 10.1016/j.resmic.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 77.Fida TT, Moreno-Forero SK, Heipieper HJ, Springael D. 2013. Physiology and transcriptome of the polycyclic aromatic hydrocarbon-degrading Sphingomonas sp. LH128 after long-term starvation. Microbiology 159:1807–1817. [DOI] [PubMed] [Google Scholar]
- 78.Postnikova OA, Shao J, Mock NM, Baker CJ, Nemchinov LG. 2015. Gene expression profiling in viable but nonculturable (VBNC) cells of Pseudomonas syringae pv. syringae. Front Microbiol 6:1419. doi: 10.3389/fmicb.2015.01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Babin BM, Atangcho L, van Eldijk MB, Sweredoski MJ, Moradian A, Hess S, Tolker-Nielsen T, Newman DK, Tirrell DA. 2017. Selective proteomic analysis of antibiotic-tolerant cellular subpopulations in Pseudomonas aeruginosa biofilms. mBio 8:e01593-17. doi: 10.1128/mBio.01593-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC, Marchal K, Beirlant J, Versees W, Hofkens J, Jansen M, Fauvart M, Michiels J. 2015. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 82.Amato SM, Brynildsen MP. 2014. Nutrient transitions are a source of persisters in Escherichia coli biofilms. PLoS One 9:e93110. doi: 10.1371/journal.pone.0093110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amato SM, Brynildsen MP. 2015. Persister heterogeneity arising from a single metabolic stress. Curr Biol 25:2090–2098. doi: 10.1016/j.cub.2015.06.034. [DOI] [PubMed] [Google Scholar]
- 84.Germain E, Roghanian M, Gerdes K, Maisonneuve E. 2015. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci U S A 112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Mishra A, Taneja N, Sharma M. 2012. Viability kinetics, induction, resuscitation and quantitative real-time polymerase chain reaction analyses of viable but nonculturable Vibrio cholerae O1 in freshwater microcosm. J Appl Microbiol 112:945–953. doi: 10.1111/j.1365-2672.2012.05255.x. [DOI] [PubMed] [Google Scholar]
- 86.Boaretti M, Lleo MM, Bonato B, Signoretto C, Canepari P. 2003. Involvement of rpoS in the survival of Escherichia coli in the viable but non-culturable state. Environ Microbiol 5:986–996. doi: 10.1046/j.1462-2920.2003.00497.x. [DOI] [PubMed] [Google Scholar]
- 87.Gangaiah D, Kassem II, Liu Z, Rajashekara G. 2009. Importance of polyphosphate kinase 1 for Campylobacter jejuni viable-but-nonculturable cell formation, natural transformation, and antimicrobial resistance. Appl Environ Microbiol 75:7838–7849. doi: 10.1128/AEM.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wan C, Zhang Q, Lee DJ, Wang Y, Li J. 2014. Long-term storage of aerobic granules in liquid media: viable but non-culturable status. Bioresour Technol 166:464–470. doi: 10.1016/j.biortech.2014.05.091. [DOI] [PubMed] [Google Scholar]
- 89.Harms A, Brodersen DE, Mitarai N, Gerdes K. 2018. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol Cell doi: 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 90.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 91.Smith CK, Baker TA, Sauer RT. 1999. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc Natl Acad Sci U S A 96:6678–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Diago-Navarro E, Hernandez-Arriaga AM, Kubik S, Konieczny I, Diaz-Orejas R. 2013. Cleavage of the antitoxin of the parD toxin-antitoxin system is determined by the ClpAP protease and is modulated by the relative ratio of the toxin and the antitoxin. Plasmid 70:78–85. doi: 10.1016/j.plasmid.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 93.Gottesman S. 1996. Proteases and their targets in Escherichia coli. Annu Rev Genet 30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 94.Donegan NP, Thompson ET, Fu Z, Cheung AL. 2010. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J Bacteriol 192:1416–1422. doi: 10.1128/JB.00233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alleron L, Khemiri A, Koubar M, Lacombe C, Coquet L, Cosette P, Jouenne T, Frere J. 2013. VBNC Legionella pneumophila cells are still able to produce virulence proteins. Water Res 47:6606–6617. doi: 10.1016/j.watres.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 96.Kusumoto A, Miyashita M, Kawamoto K. 2013. Deletion in the C-terminal domain of ClpX delayed entry of Salmonella enterica into a viable but non-culturable state. Res Microbiol 164:335–341. doi: 10.1016/j.resmic.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 97.Almagro-Moreno S, Kim TK, Skorupski K, Taylor RK. 2015. Proteolysis of virulence regulator ToxR is associated with entry of Vibrio cholerae into a dormant state. PLoS Genet 11:e1005145. doi: 10.1371/journal.pgen.1005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Christensen-Dalsgaard M, Jorgensen MG, Gerdes K. 2010. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol Microbiol 75:333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hobby GL, Meyer K, Chaffee E. 1942. Observations on the mechanism of action of penicillin. Exp Biol Med 50:281–285. [Google Scholar]
- 100.Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci U S A 107:12541–12546. doi: 10.1073/pnas.1004333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shan Y, Brown Gandt A, Rowe SE, Deisinger JP, Conlon BP, Lewis K. 2017. ATP-dependent persister formation in Escherichia coli. mBio 8:e02267-16. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alkasir R, Ma Y, Liu F, Li J, Lv N, Xue Y, Hu Y, Zhu B. 14 June 2018. Characterization and transcriptome analysis of Acinetobacter baumannii persister cells. Microb Drug Resist doi: 10.1089/mdr.2017.0341. [DOI] [PubMed] [Google Scholar]
- 103.Qian H, Yao Q, Tai C, Deng Z, Gan J, Ou HY. 2018. Identification and characterization of acetyltransferase-type toxin-antitoxin locus in Klebsiella pneumoniae. Mol Microbiol 108:336–349. doi: 10.1111/mmi.13934. [DOI] [PubMed] [Google Scholar]
- 104.Rycroft JA, Gollan B, Grabe GJ, Hall A, Cheverton AM, Larrouy-Maumus G, Hare SA, Helaine S. 2018. Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection. Nat Commun 9:1993. doi: 10.1038/s41467-018-04472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Christensen-Dalsgaard M, Gerdes K. 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol Microbiol 62:397–411. doi: 10.1111/j.1365-2958.2006.05385.x. [DOI] [PubMed] [Google Scholar]
- 106.Korch SB, Hill TM. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J Bacteriol 188:3826–3836. doi: 10.1128/JB.01740-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pedersen K, Christensen SK, Gerdes K. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol 45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 108.Demidenok OI, Kaprelyants AS, Goncharenko AV. 2014. Toxin-antitoxin vapBC locus participates in formation of the dormant state in Mycobacterium smegmatis. FEMS Microbiol Lett 352:69–77. doi: 10.1111/1574-6968.12380. [DOI] [PubMed] [Google Scholar]
- 109.Gupta M, Nayyar N, Chawla M, Sitaraman R, Bhatnagar R, Banerjee N. 2016. The chromosomal parDE2 toxin-antitoxin system of Mycobacterium tuberculosis H37Rv: genetic and functional characterization. Front Microbiol 7:886. doi: 10.3389/fmicb.2016.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Braetz S, Schwerk P, Thompson A, Tedin K, Fulde M. 2017. The role of ATP pools in persister cell formation in (fluoro)quinolone-susceptible and -resistant strains of Salmonella enterica ser. Typhimurium. Vet Microbiol 210:116–123. doi: 10.1016/j.vetmic.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 111.Zhao F, Wang Y, An H, Hao Y, Hu X, Liao X. 2016. New insights into the formation of viable but nonculturable Escherichia coli O157:H7 induced by high-pressure CO2. mBio 7:e00961-16. doi: 10.1128/mBio.00961-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pu Y, Li Y, Ma Q, Tian T, Zhao Z, McVey A, Bai F. 2017. Dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. bioRxiv doi: 10.1101/233890. [DOI] [PubMed]
- 113.Hurdle JG, O'Neill AJ, Chopra I, Lee RE. 2011. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat Rev Microbiol 9:62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bruhn-Olszewska B, Szczepaniak P, Matuszewska E, Kuczynska-Wisnik D, Stojowska-Swedrzynska K, Moruno Algara M, Laskowska E. 2018. Physiologically distinct subpopulations formed in Escherichia coli cultures in response to heat shock. Microbiol Res 209:33–42. doi: 10.1016/j.micres.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 115.Kim JS, Yamasaki R, Song S, Zhang W, Wood TK. 12 March 2018. Single cell observations show persister cells wake based on ribosome content. Environ Microbiol 20:2085–2098. doi: 10.1111/1462-2920.14093. [DOI] [PubMed] [Google Scholar]
- 116.Kim JS, Chowdhury N, Yamasaki R, Wood TK. 19 February 2018. Viable but non-culturable and persistence describe the same bacterial stress state. Environ Microbiol 20:2038–2048. doi: 10.1111/1462-2920.14075. [DOI] [PubMed] [Google Scholar]
- 117.Kong IS, Bates TC, Hulsmann A, Hassan H, Smith BE, Oliver JD. 2004. Role of catalase and oxyR in the viable but nonculturable state of Vibrio vulnificus. FEMS Microbiol Ecol 50:133–142. doi: 10.1016/j.femsec.2004.06.004. [DOI] [PubMed] [Google Scholar]