Abstract
Left ventricular ejection fraction (LVEF) is an important prognostic indicator of cardiovascular outcomes. It is used clinically to determine the indication for several therapeutic interventions. LVEF is most commonly derived using in-line tools and some manual assessment by cardiologists from standardized echocardiographic views. LVEF is typically documented in free-text reports, and variation in LVEF documentation pose a challenge for the extraction and utilization of LVEF in computer-based clinical workflows. To address this problem, we developed a computerized algorithm to extract LVEF from echocardiography reports for the identification of patients having heart failure with reduced ejection fraction (HFrEF) for therapeutic intervention at a large healthcare system. We processed echocardiogram reports for 57,158 patients with coded diagnosis of Heart Failure that visited the healthcare system over a two-year period. Our algorithm identified a total of 3910 patients with reduced ejection fraction. Of the 46,634 echocardiography reports processed, 97% included a mention of LVEF. Of these reports, 85% contained numerical ejection fraction values, 9% contained ranges, and the remaining 6% contained qualitative descriptions. Overall, 18% of extracted numerical LVEFs were ≤ 40%. Furthermore, manual validation for a sample of 339 reports yielded an accuracy of 1.0. Our study demonstrates that a regular expression-based approach can accurately extract LVEF from echocardiograms, and is useful for delineating heart-failure patients with reduced ejection fraction.
Keywords: Ejection fraction, Natural language processing, Regular expression, Cardiology, Echocardiogram
Introduction
Left ventricular ejection fraction (LVEF) is an important prognostic indicator of cardiovascular outcomes [1, 2]. Clinically, it is used to guide the therapeutic pathways for patients having heart failure [3]. LVEF is estimated manually by cardiologists during echocardiographic examinations, and is typically documented in free-text reports [4]. Variation in LVEF documentation from the echocardiograms pose a challenge for the extraction and utilization of LVEF in computer-based clinical workflows.
Significance of left ventricular ejection fraction (LVEF)
During each heartbeat, when the heart contracts, it ejects blood from the two pumping chambers called ventricles, and the ventricles refill with blood when the heart relaxes. The term “ejection fraction” (EF) refers to the percentage of blood that is pumped out of a filled ventricle with each heartbeat. A LVEF of 55% or higher is considered normal under physiologic loading conditions, with an EF of 50% or lower being considered reduced. Notably, EFs between 50 and 55% are considered “borderline” [5, 6].
Several imaging modalities can be used to measure LVEF, including echocardiography, magnetic resonance imaging (MRI), computed tomography (CT), radionuclide angiography, and gated myocardial perfusion single-photon emission computed tomography (GSPECT) [4]. Currently, MRI is widely considered the gold standard for EF measurement, largely due to the utility of tomographic techniques in overcoming the need for radiation exposure and dealing with the geometric complexity of the cardiac chambers, particularly of the right ventricle [4, 7, 8].
EF on echocardiography is typically measured only in the left ventricle (LV), as the left ventricle is fully accessible using standard echocardiographic views and the geometry of the chamber allows robust estimates of EF as a parameter from orthogonal 2D images using the prolate ellipse as a model. While these estimates have been shown to be reliable, there remains a degree of operator dependence in most echocardiographic assessments of EF. Nevertheless, echocardiog-raphically estimated EF remains the most widely used approach, due to its low cost, absence of radiation and increasingly wide availability.
Therapeutic pathways for patients with reduced ejection fraction
The management of patients with reduced EF, whether they have the symptoms of heart failure or not, has been an area of major advance in the last two decades. Randomized control trials have identified multiple drug classes which reduce mortality and morbidity in those with HF and reduced EF (HFrEF). Several subsets of HFrEF are also known to benefit from the implantation of automated implantable cardiodefi-brillators. Similarly, trials combining EF with electrical information have identified subsets of patients who benefit from very specific forms of cardiac electrical resynchronization through implanted pacemakers. Together, these advances have firmly anchored the modern management of heart failure around echocardiographic data.
Challenge for LVEF extraction
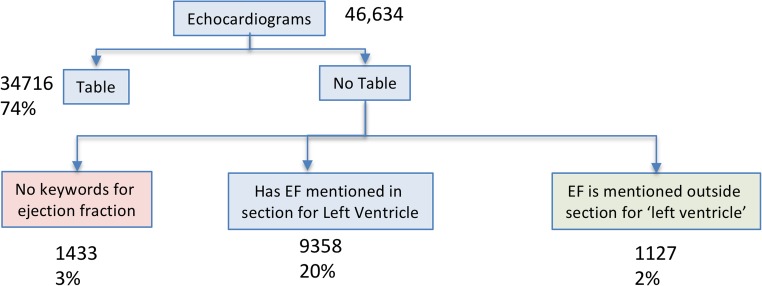
Ejection fraction (EF) is often reported in free-text format, and its extraction remains challenging due to considerable variation in the manner of documentation in echocardiography reports. EF is reported in different areas of reports, or in different formats, either as a number range or as qualitative descriptions (see Table 1).
| Type of Mention | Example Excerpts |
|---|---|
| Number | The left ventricular ejection fraction is 60% Ejection Fraction 20% (A) (Range: 50 - 75) lv ejection fraction 66% |
| Range | Estimated left ventricular ejection fraction is 45-50% The LVEF is visually estimated at 30-35% LVEF by visual estimation is around 35% |
| Qualitative | LVEF appears at the lower limits of normal Left ventricular systolic function is moderately impaired Left ventricular systolic function is moderately decreased |
Table 1.
Distribution of high level echocardiogram categorization of based on mentions of anchor terms for ejection fraction
| Group Description | Count | % |
|---|---|---|
| EF in tabular pattern | 34,716 | 74 |
| No section of left ventricle | 1127 | 2 |
| Left ventricle section | 9358 | 20 |
| No keywords for ejection fraction | 1433 | 3 |
| Total | 46,634 | 100 |
Previous research on extracting LVEF
The earliest effort to automatically extract information from echocardiograms was by Chung (2005), who developed an information extraction system to identify 10 medical concepts and their associated values from narrative echocardiogram reports [9]. The system used UMLS through a MetaMap API. Evaluation of this approach using 403 manually annotated reports determined that the system possessed 78% recall and 99% precision.
In 2011, Garvin developed an NLP system for quality measurement using rules to capture EF in a project entitled ‘Automated Data Acquisition for Heart Failure’, undertaken by the US Department of Veterans Affairs. The investigators utilized a random sample of 765 echocardiograms from seven VA medical centers [10]. This system was called ‘Capture with UIMA of Needed Data using Regular Expressions for EF’ (CUIMANDREef). The training and test document sets were annotated by two to three experts, which categorized documents with similar characteristics into five distinct formats by manual inspection, considering outline, headers, and the location EF data for developing the algorithm. The system used rules to assess combinations of concepts in a document, and gave positive weight toward a score to classify outputs as being consistent with an EF >40% or an EF <40%. Section headings and locations in documents were used to resolve instances when multiple mentions of LVEF were present, giving precedence to the LVEF recorded in the conclusion section. Their system had accuracy of 99.8%. In a subsequent study, Mystre compared the regular expression-based approach of CUIMANDREef with a sequence tanning approach to detect references to EF. The latter was found to perform better, with an F1-measure of 95.0% (versus 89.1%) [11]. Additionally, Kim investigated domain adaptation approaches for EF extraction and reported an improvement in EF extraction accuracy [12].
Furthermore, Gobbel reported that the use of machine learning to assist annotators halved the time necessary for annotating HF-related concepts [13]. Recently, several studies have focused on extraction of EF [10, 14–18]. Notably, Patterson developed a regular expression-based NLP system using Java UIMA architecture for extracting LVEF along with 26 other cardiology concepts from echocardiograms, radiology notes, and clinical notes. The system was able to extract LVEF with a precision of 0.96-1.0.
Xie implemented an algorithm at Kaiser Permanente Medical Center that first segments ECHO reports into sections and sentences, and then searches for phrases suggested by a domain expert to extract numerical values or qualitative descriptions of EF in the vicinity of the phrase. The final stage involved determining if the EF was historical or negated using simple phrases. For validation, the algorithm output was compared with the annotation of a cardiologist for a random sample of 200 patients, and concluded that the system exhibited high accuracy, with sensitivity and precision values of 0.95 and 0.97, respectively [17].
The above studies to extract information from the echocardiogram have been carried out as a part of a broader area of research referred to as EHR-based phenotyping, as there is an increasing realization that the EHR data is not readily amenable for analysis, and information processing techniques are required to utilize the data for epidemiological research as well as for clinical decision support [19–22].
We have developed a regular expression-based NLP system to extract LVEF from echocardiogram reports. The intended use of our algorithm was to identify patients presenting heart failure with reduced ejection fraction (HFrEF) for driving a population-based therapeutic intervention program. We implemented the algorithm using Python scripts using Apache Spark cluster [23]. (see Appendix). The Python scripts and sample echocardiograms are available as open source (link to Github repository: https://github.com/waghsk/lvef-paper).
Methods
This study was conducted at Partners Healthcare, Boston, and was approved by the institutional review board.
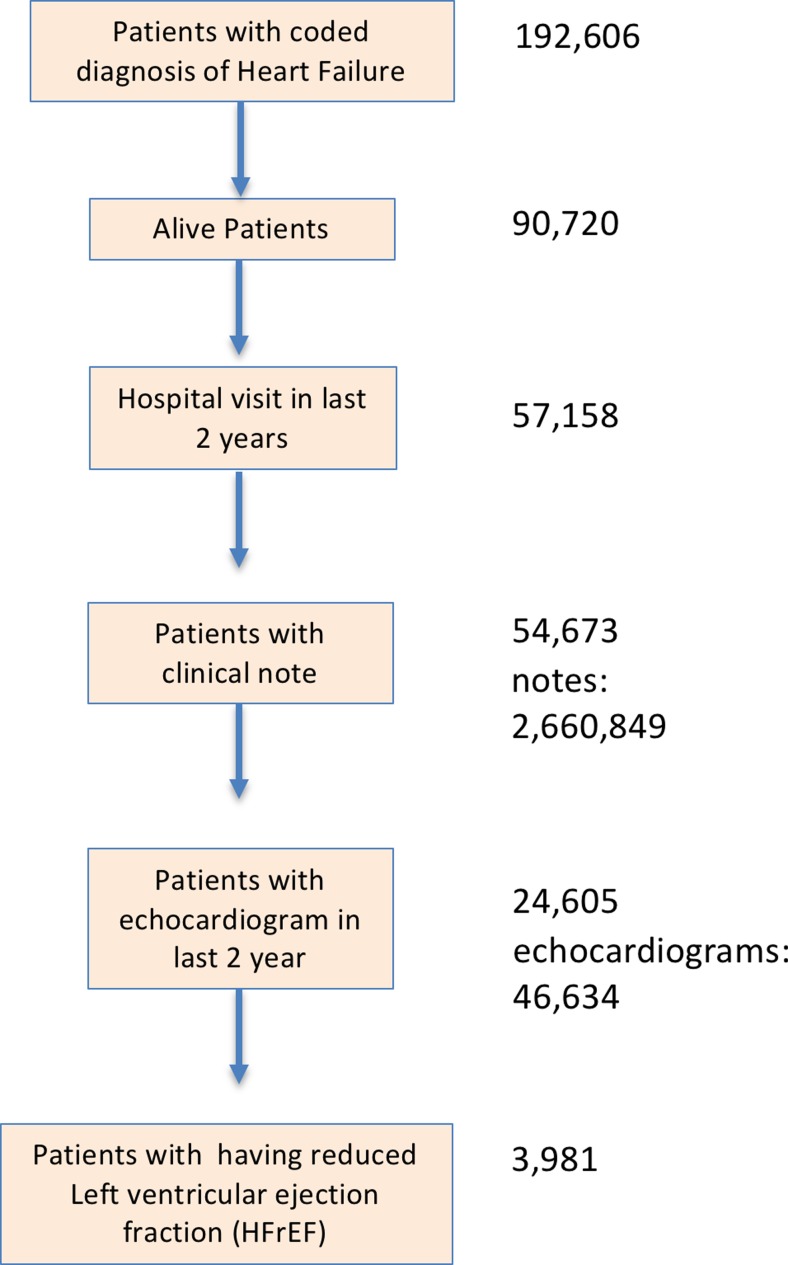
We queried the institutional research patient data registry (RPDR) to obtain a data set of 190,000 patients with coded heart failure, and further filtered this set to patients that were currently alive and had visited the health system in past two years [24]. Clinical notes for these patients were filtered to identify echocardiograms using codes for report-type that appear as meta-data fields. Then, we identified the codes for echocardiograms by manually examining random samples for each report type.
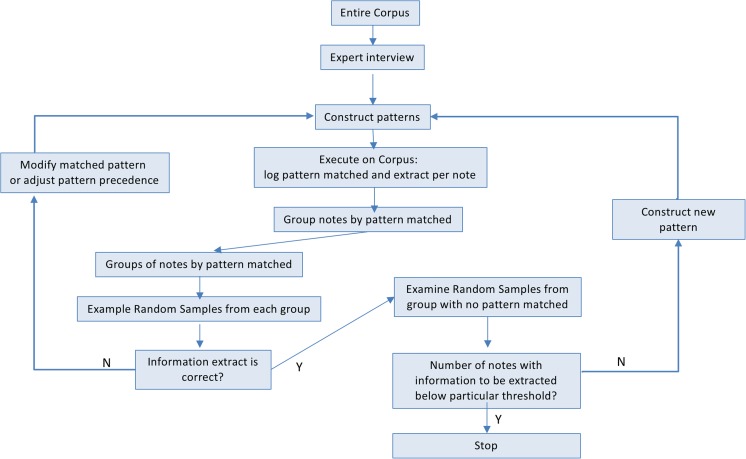
The study team included clinical experts with experience in manually extracting LVEF from the EHR chart. These experts also suggested that most notes had a table listing the ejection fraction or a paragraph with a heading for left ventricle that contained the EF. Accordingly, we designed a logic to extract LVEF by searching for 1) a tabular pattern, 2) a section for the left ventricle with numerical and range patterns, and 3) qualitative expressions in decreasing order of precedence. We took an iterative approach to implement the algorithm for LVEF extraction using regular expressions (see Fig. 2). The iterative approach is as follows: Implement code for a pattern, and execute on the entire corpus to generate a log, and then match results for each note. Based on the log output, the corpus was divided into a positive group that matched the pattern and a negative group that corresponded to ‘missing pattern’. A sample of the positive group was examined to ensure that the extracted value is accurate, and a random sample was then examined from the negative group to either extend the existing patterns or construct a new one. This process was continued until no inaccurate extractions existed in the positive group, and the accuracy of the negative group attained the desired threshold. A detailed distribution of the patterns is provided in Appendix 1.
Fig. 2.
Distribution of high level echocardiograms categorization based on mentions of anchor terms for ejection fraction
After computing the LVEF value output for each echocardiogram, we identified latest LVEF value for each patient by sorting the echocardiograms by date and ignoring those from which EF could not be extracted. The extracted LVEF values were then divided into two categories: reduced EF (rEF) and not-reduced EF (-rEF) (see Table 2). Patients were correspondingly classified as having ‘Heart Failure with reduced Ejection Fraction’ (HFrEF) or not-HFrEF. The latter group is composed of patients presenting heart failure with mid-range or preserved ejection fraction’ (HFpEF or HFpEF).
Table 2.
Distribution of pattern types for left ventricular ejection fraction mentions
| EF Pattern Data Type | Count | % |
|---|---|---|
| Numerical | 38,267 | 85 |
| Range | 4061 | 9 |
| Qualitative | 2768 | 6 |
| No pattern matched | 105 | <1 |
| Total | 45,201 | 100 |
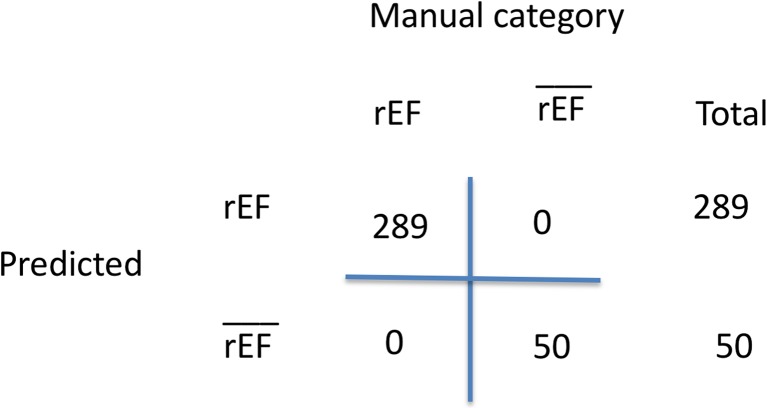
For validation, two sets of the most recent echocardiogram were manually annotated for the LVEF classes of LVEF<=40 and LVEF>40. The first set comprised 289 of the most recent echocardiogram reports classified as LVEF. The second set consisted of 50 of the most recent echocardiograms classified as LVEF>40. Then, manual annotations were compared with the system output to compute the confusion matrix and algorithm accuracy.
Results
Patients with coded heart failure diagnosis
We obtained all EHR data for patients that i) have ‘heart failure’ on their problem list, ii) have visited the hospital in the previous two years, and iii) are alive. Specifically, in ICD-10-CM, heart failure is coded as series i50, with 6 subcategories that further define the actual condition: combined systolic and diastolic heart failure, diastolic heart failure, left ventricular failure, systolic heart failure, other types of HF including (right heart failure), and unspecified heart failure. In addition to the ICD-10-CM codes for HF, we included any legacy ICD-9-CM, longitudinal medical record (LMR), and other institutional codes mapped into the ICD-10 codes.
‘Diagnoses \ Diseases of the circulatory system (i00-i99) \ Other forms of heart disease (i30-i52) \ Heart failure (i50)’.
Echocardiogram detection
To identify echocardiograms from notes in the dataset, we searched notes for the phrase ‘ejection fraction’, and grouped notes by their meta-data type field, sorting them in order of decreasing frequency. Then, we examined a random sample of codes with over 1% frequency to determine if the codes corresponded to echocardiograms. There were 629 different codes. We extracted 3 samples for each of the codes, and manually examined the samples to delineate 37 codes corresponding to echocardiograms. The codes for echocardiograms were generally found to contain the prefix “ECH”. By filtering the notes using the echocardiogram codes and restricting the time period to the previous 2 years, we obtained a total of 46,634 echocardiograms belonging to 24,605 patients.
Algorithm for extraction of LVEF from echocardiogram
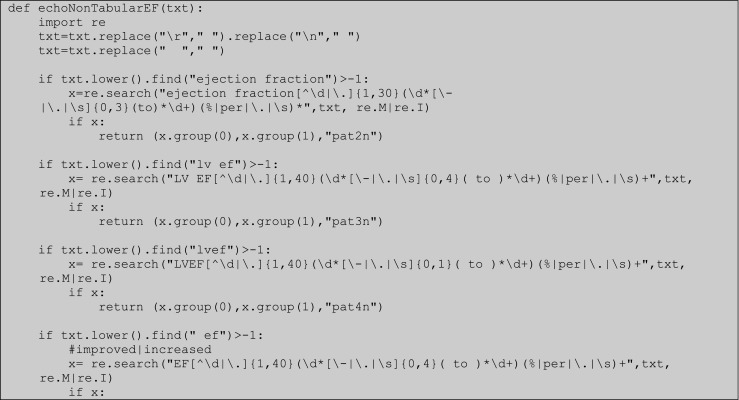
Section headings were identified in the notes by decomposing the corpus into lines and sorting out the most frequently occurring lines. These were then manually examined to construct a set of section headings. Using the section headings, we sliced each note in the corpus into sections and searched for the phrases ‘ejection fraction’, ‘lvef’, and ‘ef’ within sections with the heading ‘left ventricle’. We resorted to a regular expression-based approach, and analyzed the entire corpus using the approach summarized in Fig. 1.
Fig. 1.
Steps to develop regular expression patterns for extracting LVEF
The logic for parsing out EF from echocardiograms is described as follows. First, using meta-data, we ensured that the note to be parsed is an echocardiogram. Then, we searched for a tabular pattern with EF in the note. In the absence of a tabular pattern, we identified the section for left ventricle and searched for numerical and range patterns for EF. If none of these were located, we then searched for prose expressions. In the absence of such expressions, a log noted that no pattern was found (Table 1 and Fig. 2).
First, we developed regular expressions to extract EF from tables in echocardiograms, and then focused on the remaining echocardiograms to develop patterns for quantitative and qualitative descriptions of EF. Consequently, 4 major groups of patterns emerged: i) numerical, ii) range, iii) qualitative, and iv) no pattern (see Table 2).
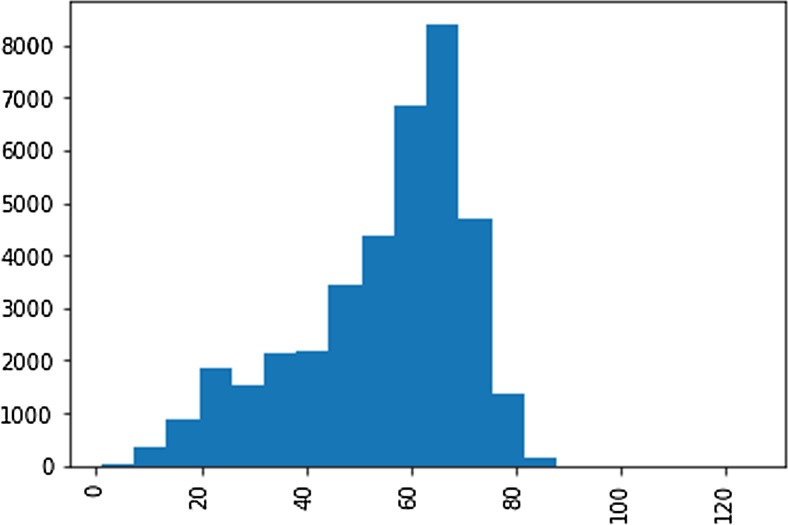
We classified the extracted EF ‘snippets’ to two classes, corresponding to EF ≤ 40 or EF > 40. First, the snippets with a single number were mapped. Next, the ranges (e.g. ‘40-50’ or ‘40 to 50’) were converted to an average. For qualitative expressions, we developed a lookup table to map the extracted text to the categories. The numerical values, ranges, and prose extracts had a distribution of 85, 9, and 6%, respectively. Less than 1% of notes with keywords for ejection fraction had no patterns matched for EF extraction. (Figs. 3, 4 and 5, Table 3)
Fig. 3.
Distribution of numerical ejection fractions extracted from echocardiograms
Fig. 4.
Consort diagram for the present study. Values to the right indicate the number of patients
Fig. 5.
Confusion matrix for system validation
Table 3.
Distribution of numerical LVEF values for HF patient cohorts
| Group Name | Range | Count | % |
|---|---|---|---|
| HFrEF | # < 40 | 6977 | 18 |
| HFmEF | #40-50 | 5443 | 14 |
| HFpEF | # > 50 | 25,847 | 68 |
| #total numerical | 38,267 |
Manual validation to computer performance of the LVEF extraction algorithm
We compared the manual annotations with the algorithm output to compute the confusion matrix and accuracy of the algorithm. All instances of echocardiograms were correctly classified by the LVEF extraction algorithm, providing an accuracy of 1.0 (Fig. 5).
Discussion
Our results demonstrate that an NLP algorithm using a regular expression-based approach was highly accurate in extracting LVEF from echocardiogram reports. This corroborates previous reports of high accuracy for LVEF extraction in other healthcare systems [16, 17, 25]. Use of regular expressions to extract information, though labor intensive, is known to yield high accuracy. Our approach described in Fig. 1 is helpful to systematically identify the patterns, while considering pattern prevalence to minimize the manual effort. An alternative approach is to automatically discover regular expressions from a training corpus, which is an active area of research [26–28].
We determined that 74% of extracted LVEF values were mentioned in tabular form, while this metric was 70% at Kaiser Permanente [17]. This difference is possibly due to the analysis of Xie being performed on a historical corpus, while the corpus of the present study was limited to the previous 2 years. We anticipate that improved integration of echocardiography software with EHR may have facilitated the granularity of EF reporting.
Many of previous approaches to LVEF extraction involved the creation of a gold standard, manually annotated by a domain expert that was then used to develop a text processing algorithm [25]. In contrast, we used an iterative approach, analyzing the entire corpus of echocardiograms to develop the EF extraction algorithm [17]. The developed algorithm was validated by manual inspection of a system output sample.
The disadvantage of developing a manually annotated corpus to guide system development is that it is labor intensive and can be expected to exhibit redundant instances of dominant patterns, as a random sample of instances will not be representative of the distribution of patterns to be extracted. For example, in the present study, a 70% random sample of instances for gold standard creation would have contained ejection fractions reported in the tabular form. However, these instances only corresponded to a small proportion of the regular expressions used in the system.
Knowing the prevalence of patterns in the text corpus is helpful for optimizing the effort invested in the implementation of processing logic for a certain pattern. Furthermore, such patterns can be escaped by logging an unknown pattern exception for the message. As shown in Fig. 1, we considered pattern prevalence distribution to guide the process of developing regular expressions for these patterns. The prevalence of the patterns for documenting EF is likely due to templates used during the data entry process. An important design feature of our algorithm was to give precedence to tabular mentions of EF, and only then consider section headers. This has been reported as being useful in previous studies [17, 25].
Limitations
We restricted the scope of our analysis to the extraction of LVEF from echocardiograms. LVEFs can be found in echocardiograms as well as visit notes, wherein physicians summarize the findings of echocardiograms. As the study and associated interventions required objective evidence of low EF, we restricted the source of LVEF mentions to echocardiograms. A second limitation of our study is that a significant proportion of patients are expected to have echocardiograms from institutions outside the health system, which triggered referral to our hospitals for intervention. However, we restricted the analysis to echocardiograms conducted within the healthcare system to obviate the complexities of dealing with reporting variations outside the healthcare system.
Conclusion
Our study demonstrates that a regular expression-based approach can accurately extract LVEF from echocardiograms. This algorithm can be utilized to delineate a cohort of HFrEF patients for implementing therapeutic interventions.
Appendix 1
Python script to split the echocardiogram into sections and to identify the section header.
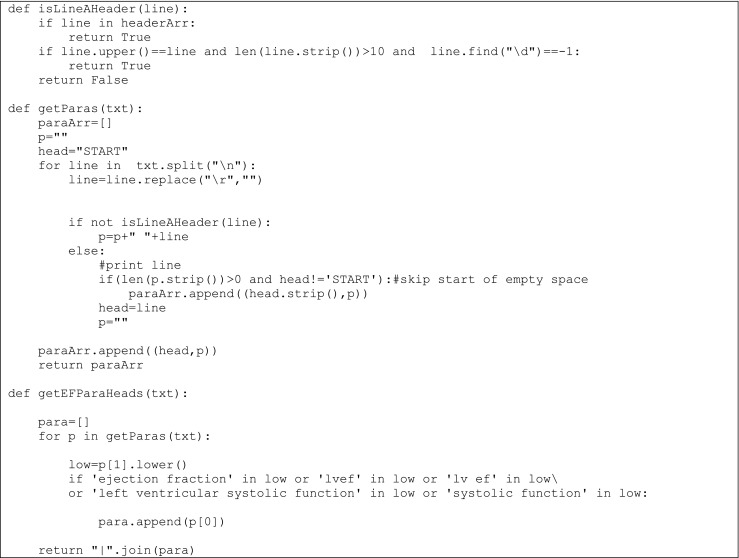
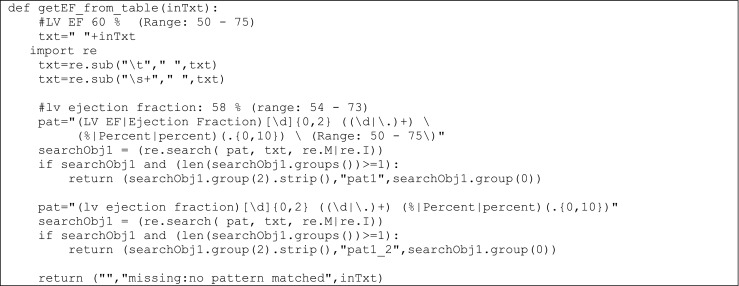
Python script to extract LVEF from echocardiogram from tabular format.
Distribution of patterns for extraction of LVEF from tabular form.
| Pattern | Count |
|---|---|
| pat1 | 34,509 |
| missing: no pattern matched | 11,918 |
| pat1_2 | 207 |
| Total | 46,634 |
Python script to extract LVEF from echocardiogram reports with a section for the left ventricle.

Distribution of patterns observed in echocardiogram reports with and without a section/paragraph for the left ventricle.
| Pattern | In Section for the Left Ventricle | Outside Section for the Left Ventricle |
|---|---|---|
| pat2n | 5482 | 489 |
| pat8c | 2450 | 212 |
| pat4n | 1159 | 258 |
| pat3n | 86 | 1 |
| pat7c | 77 | 16 |
| pattern not found | 52 | 53 |
| pat5n | 41 | 96 |
| pat4_1c | 11 | 2 |
| Total | 9358 | 1127 |
Appendix 2
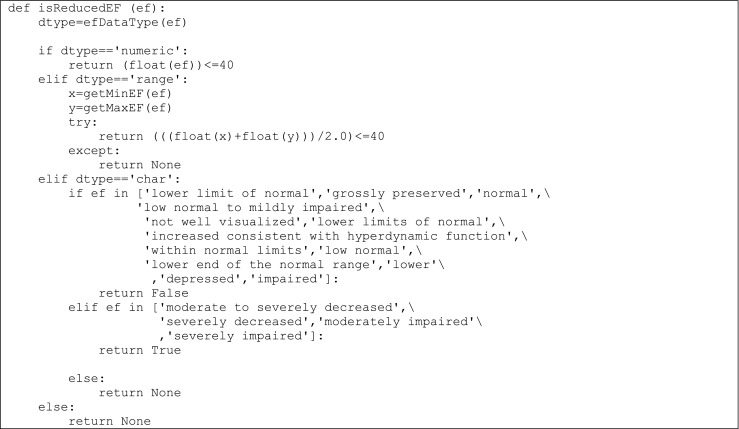
Python script to map numerical, range, and qualitative LVEF extracts to binary categories of reduced LVEF.
Funding
This study was supported by NIH grants R01-HG009174 and R00-LM011575.
Conflict of Interest
The authors declare that they have no conflict of interest.
Informed consent
The institutional Review Board had approved a waiver of informed consent for this study.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Footnotes
This article is part of the Topical Collection on Systems-Level Quality Improvement
References
- 1.Fogel MA. Use of ejection fraction (or lack thereof), morbidity/mortality and heart failure drug trials: a review. International Journal of Cardiology. 2002;84:119–132. doi: 10.1016/s0167-5273(02)00134-1. [DOI] [PubMed] [Google Scholar]
- 2.Sweitzer NK, Lopatin M, Yancy CW, Mills RM, Stevenson LW. Comparison of Clinical Features and Outcomes of Patients Hospitalized With Heart Failure and Normal Ejection Fraction (≥55%) Versus Those With Mildly Reduced (40% to 55%) and Moderately to Severely Reduced (<40%) Fractions. Am J Cardiol. 2008;101:1151–1156. doi: 10.1016/j.amjcard.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yancy, C. W. et al., 2017 ACC/AHA/HFSA Focused Update of the 2013 ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of America. Circulation136, 2017. 10.1161/cir.0000000000000509. [DOI] [PubMed]
- 4.Foley TA, et al. Measuring left ventricular ejection fraction-techniques and potential pitfalls. European Cardiology. 2012;8:108–114. doi: 10.15420/ecr.2012.8.2.108. [DOI] [Google Scholar]
- 5.Gaasch WH, Delorey DE, Kueffer FJ, Zile MR. Distribution of Left Ventricular Ejection Fraction in Patients With Ischemic and Hypertensive Heart Disease and Chronic Heart Failure. Am J Cardiol. 2009;104:1413–1415. doi: 10.1016/j.amjcard.2009.06.064. [DOI] [PubMed] [Google Scholar]
- 6.Dunlay SM, Roger VL, Weston SA, Jiang R, Redfield MM. Longitudinal Changes in Ejection Fraction in Heart Failure Patients With Preserved and Reduced Ejection Fraction. Circulation: Heart Failure. 2012;5:720–726. doi: 10.1161/circheartfailure.111.966366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang H, et al. Accuracy of left ventricular ejection fraction by contemporary multiple gated acquisition scanning in patients with cancer: comparison with cardiovascular magnetic resonance. Journal of Cardiovascular Magnetic Resonance. 2017;19:34. doi: 10.1186/s12968-017-0348-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood PW, Choy JB, Nanda NC, Becher H. Left Ventricular Ejection Fraction and Volumes: It Depends on the Imaging Method. Echocardiography. 2014;31:87–100. doi: 10.1111/echo.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung J, Murphy S. Concept-value pair extraction from semi-structured clinical narrative: a case study using echocardiogram reports. American Medical Informatics Association. 2005;2005:131–135. [PMC free article] [PubMed] [Google Scholar]
- 10.Garvin JH, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. Journal of the American Medical Informatics Association. 2012;19:859–866. doi: 10.1136/amiajnl-2011-000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mystre S (2012) Comparing Methods for left Ventricular Ejection Fraction Clinical Information Extraction. TBI_CRI
- 12.Kim Y, Garvin J, Heavirland J, Meystre SM (2013) Improving heart failure information extraction by domain adaptation. Stud Health Technol Inform 192:185–189 [PubMed]
- 13.Gobbel GT, Garvin J, Reeves R, Cronin RM, Heavirland J, Williams J, Weaver A, Jayaramaraja S, Giuse D, Speroff T, Brown SH, Xu H, Matheny ME. Assisted annotation of medical free text using RapTAT. J Am Med Inform Assoc. 2014;21(5):833–841. doi: 10.1136/amiajnl-2013-002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, et al. Extraction of left ventricular ejection fraction information from various types of clinical reports. Journal of biomedical informatics. 2017;67:42–48. doi: 10.1016/j.jbi.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meystre, S. M. et al., Congestive heart failure information extraction framework for automated treatment performance measures assessment. Journal of the American Medical Informatics Association24, 2017. 10.1093/jamia/ocw097. [DOI] [PMC free article] [PubMed]
- 16.Patterson OV, et al. Unlocking echocardiogram measurements for heart disease research through natural language processing. BMC Cardiovascular Disorders. 2017;17:151. doi: 10.1186/s12872-017-0580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie F, Zheng C, Shen A, Chen W. Extracting and analyzing ejection fraction values from electronic echocardiography reports in a large health maintenance organization. Health Informatics Journal. 2017;23:319–328. doi: 10.1177/1460458216651917. [DOI] [PubMed] [Google Scholar]
- 18.Nath Chinmoy, Albaghdadi Mazen S., Jonnalagadda Siddhartha R. A Natural Language Processing Tool for Large-Scale Data Extraction from Echocardiography Reports. PLOS ONE. 2016;11(4):e0153749. doi: 10.1371/journal.pone.0153749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson AE, et al. Electronic health record phenotyping improves detection and screening of type 2 diabetes in the general United States population: A cross-sectional, unselected, retrospective study. Journal of biomedical informatics. 2016;60:162–168. doi: 10.1016/j.jbi.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao Katherine P., Ananthakrishnan Ashwin N., Kumar Vishesh, Xia Zongqi, Cagan Andrew, Gainer Vivian S., Goryachev Sergey, Chen Pei, Savova Guergana K., Agniel Denis, Churchill Susanne, Lee Jaeyoung, Murphy Shawn N., Plenge Robert M., Szolovits Peter, Kohane Isaac, Shaw Stanley Y., Karlson Elizabeth W., Cai Tianxi. Methods to Develop an Electronic Medical Record Phenotype Algorithm to Compare the Risk of Coronary Artery Disease across 3 Chronic Disease Cohorts. PLOS ONE. 2015;10(8):e0136651. doi: 10.1371/journal.pone.0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pathak Jyotishman, Kho Abel N, Denny Joshua C. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. Journal of the American Medical Informatics Association. 2013;20(e2):e206–e211. doi: 10.1136/amiajnl-2013-002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. Journal of the American Medical Informatics Association. 2011;18:580–587. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Jimmy, Dyer Chris. Data-Intensive Text Processing with MapReduce. Synthesis Lectures on Human Language Technologies. 2010;3(1):1–177. doi: 10.2200/S00274ED1V01Y201006HLT007. [DOI] [Google Scholar]
- 24.Murphy SN, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) Journal of the American Medical Informatics Association: JAMIA. 2010;17:124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garvin JH, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. Journal of the American Medical Informatics Association: JAMIA. 2012;19:859–866. doi: 10.1136/amiajnl-2011-000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartoli A, Lorenzo A, Medvet E, Tarlao F. Inference of Regular Expressions for Text Extraction from Examples. IEEE Transactions on Knowledge and Data Engineering. 2015;28:1217–1230. doi: 10.1109/TKDE.2016.2515587. [DOI] [Google Scholar]
- 27.Bui D, Zeng-Treitler Q. Learning regular expressions for clinical text classification. Journal of the American Medical Informatics Association. 2014;21:850–857. doi: 10.1136/amiajnl-2013-002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartoli A, Lorenzo A, Medvet E, Tarlao F. Active Learning of Regular Expressions for Entity Extraction. IEEE Transactions on Cybernetics. 2017;48:1067–1080. doi: 10.1109/tcyb.2017.2680466. [DOI] [PubMed] [Google Scholar]