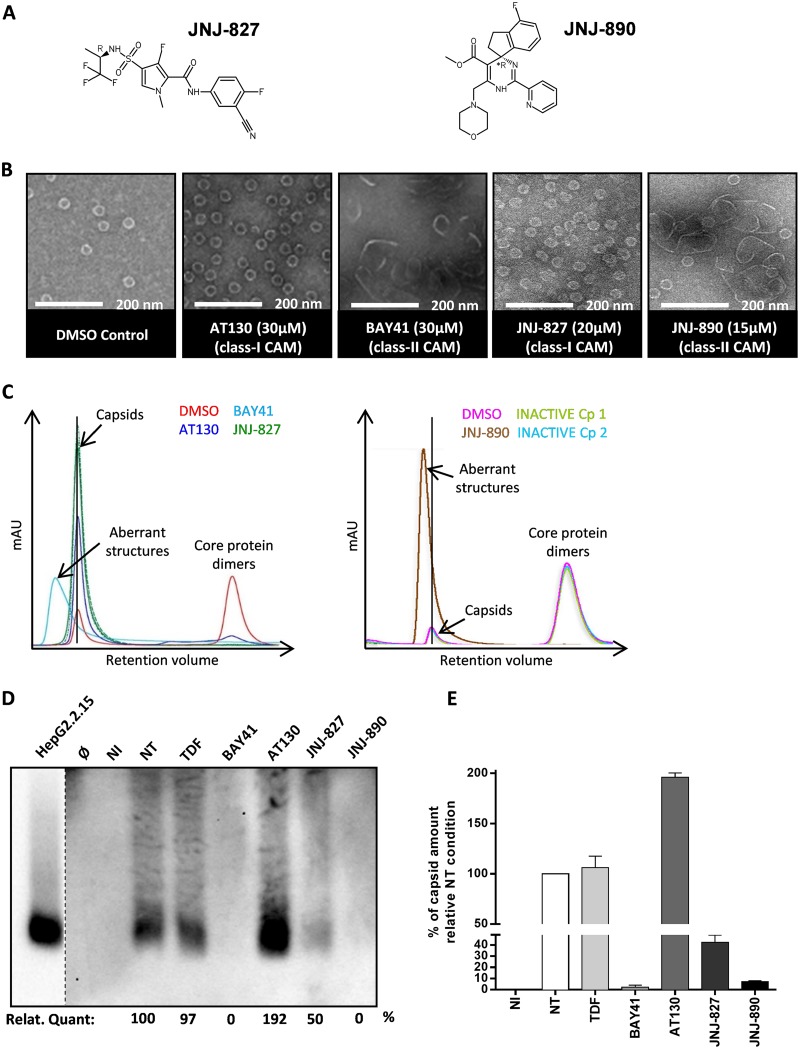
The assembly of hepatitis B virus (HBV) core protein (HBc) into capsids represents a critical step of viral replication. HBc has multiple functions during the HBV life cycle, which makes it an attractive target for antiviral therapies.
KEYWORDS: hepatitis B virus, core/capsid/HBc protein, nucleocapsid assembly, capsid assembly modulators, mechanism of action, HBV, antiviral, capsid inhibitors
ABSTRACT
The assembly of hepatitis B virus (HBV) core protein (HBc) into capsids represents a critical step of viral replication. HBc has multiple functions during the HBV life cycle, which makes it an attractive target for antiviral therapies. Capsid assembly modulators (CAMs) induce the formation of empty capsid or aberrant capsid devoid of pregenomic RNA (pgRNA) and finally block relaxed circular DNA neosynthesis and virion progeny. In this study, the novel CAMs JNJ-827 and JNJ-890 were found to be potent inhibitors of HBV replication with respective half-maximal effective concentrations of 4.7 and 66 nM, respectively, in HepG2.117 cells. Antiviral profiling in differentiated HepaRG (dHepaRG) cells and primary human hepatocytes revealed that these compounds efficiently inhibited HBV replication, as well as de novo establishment of covalently closed circular DNA (cccDNA). In addition to these two known effects of CAMs, we observed for the first time that a CAM, here JNJ-827, when added postinfection for a short-term period, significantly reduced hepatitis B e antigen (HBeAg) secretion without affecting the levels of cccDNA amount, transcription, and hepatitis B surface antigen (HBsAg) secretion. This inhibitory activity resulted from a direct effect of JNJ-827 on HBeAg biogenesis. In a long-term treatment condition using persistently infected dHepaRG cells, JNJ-827 and JNJ-890 reduced HBsAg concomitantly with a decrease in viral total RNA and pgRNA levels. Altogether, these data demonstrate that some CAMs could interfere with multiple functions of HBc in the viral life cycle.
INTRODUCTION
Chronic hepatitis B (CHB) infections caused by the hepatitis B virus (HBV), a member of the Hepadnaviridae family (1), remain a major public health problem worldwide. According to the World Health Organization, over 250 million people are chronic carriers of the virus (Fact Sheet 204). These patients have a higher risk of developing severe liver diseases, such as decompensated cirrhosis and hepatocellular carcinoma (HCC); the latter being the third leading cause of mortality worldwide (2, 3).
HBV is a noncytopathic DNA virus that specifically enters hepatocytes through the sodium taurocholate cotransporting polypeptide (NTCP) receptor to replicate and produce virion progeny (4). After delivery to the cytosol, the viral nucleocapsid is translocated to the nuclear pores for disassembly and release of the relaxed circular DNA (rcDNA) (5). Within the nucleus, the rcDNA is converted to a covalently closed circular DNA (cccDNA), which is “chromatinized” to form a long-lived life viral minichromosome that serves as the main template for all viral RNA transcript (pre-C, pregenomic [pg], pre-S, S, and X) synthesis. From these transcripts, seven proteins are translated: HBeAg (hepatitis B e antigen; secreted dimer protein); HBV Pol (viral polymerase); HBc/core (capsid protein); the large (which contains PreS1, PreS2, and S domains), medium (which contains PreS2 and S domains), and small surface envelope glycoproteins; and HBx (transcriptional transactivator) (1, 6). The pgRNA is encapsidated in a nucleocapsid and converted into rcDNA by the reverse transcriptase activity of the HBV polymerase, which represents the main replication step. The polymerase-bound pgRNA (Pol-pgRNA) serves as a specific intermediate for the association with several dimers of HBc (nucleation step) from which the fully matured nucleocapsid will arise containing rcDNA (7–9). Mature nucleocapsids containing rcDNA are then either used to produce progeny virions, following envelopment in HBV-envelope-protein-containing cellular membranes, or are redirected to the nucleus to amplify or maintain the cccDNA pool (this process is called “recycling”) (1, 10). The secretion of HBV RNA-containing virion-like particles was also recently reported both in vitro and in patient serum. These types of particles, which were shown to be produced in even higher quantity during nucleos(t)ide analogue (NA) treatment, have been very recently described as replication deficient (11). Importantly, the levels of HBV RNA virion-like particles could fluctuate during the natural history of HBV infections and be used as a biomarker of response to CHB treatments (12, 13).
Currently approved CHB treatments are limited to pegylated interferon alpha (Peg-IFN-α) and NAs. A 48-week Peg-IFN-α treatment leads to complete viral suppression (i.e., undetectable HBV DNA in blood) in approximately 25% of patients but is associated with significant side effects (14, 15). NA administration (e.g., tenofovir or entecavir) is well tolerated and induces a strong viral suppression in the majority of patients, but NAs usually have to be taken lifelong to prevent relapses (14, 15). Despite viral suppression, a functional cure (i.e., the loss of serum HBsAg [hepatitis B surface antigen] with or without seroconversion) is achieved in only 10% of treated patients after a 5-year follow up (14, 15). Therefore, the identification of new targets and development of new antiviral strategies are needed.
The core/HBc protein of HBV has recently reemerged as a promising antiviral target due to its multiple functions in the HBV life cycle (16). Indeed, besides its roles in capsid assembly and viral replication in the cytoplasm, nuclear HBc has been reported to modulate cccDNA transcription or posttranscriptional events, as well as host gene expression (17–23). It is also worth noting that HBc contains the major epitopes recognized by T cells during an HBV-targeted host immune response, thus further emphasizing the importance of this viral protein (1). Molecules targeting HBc, now generically named capsid assembly modulators (CAMs) or core protein allosteric modulators, are being developed as novel direct-acting antivirals. CAMs either induce the formation of morphologically intact empty capsids, referred to as class I mechanism of action (MoA) compounds (e.g., phenylpropenamide derivates such as AT130 [24, 25] and sulfamoylbenzamides derivates [26]) or the formation of aberrant empty structures, referred to as class II MoA compounds (e.g., heteroarypyrimidines [HAPs] such as Bayer41-4109 [here abbreviated BAY41] [27, 28]). Altogether, they mainly act by preventing and/or blocking the encapsidation of the Pol-pgRNA complex and thus its reverse transcription into rcDNA (29, 30). HAPs were the first CAMs demonstrating potent antiviral activity in vitro (31) and in vivo in both HBV transgenic mice (32) and HBV-infected chimeric mice (33). Importantly, CAMs efficiently inhibit replication of HBV mutants resistant to NAs (30, 34) and are active against multiple HBV genotypes (35). Moreover, CAMs inhibit the production of extracellular RNA-containing particles (36) and prevent de novo formation of cccDNA by acting at a postentry step (37). The inhibition of the establishment of HBV infection could be due to an accelerated breakdown of capsid in cytoplasm before rcDNA could be delivered to nucleus and converted into cccDNA or to a stabilization of the capsid structure, which would prevent uncoating at the nuclear pore (38, 39). In addition, it was suggested that CAMs could interfere with the nuclear functions of HBc, including the regulation of cccDNA transcription (40) and host gene expression (19). To date, several CAMs are in preclinical evaluations or have entered clinical trials, including GLS-4 (trial with China-CFDA; morphothiadine mesilate, HEC Pharm), NVR 3-778 (NCT02112799; Novira Therapeutics, now part of Janssen), JNJ-56136379 (NCT02662712; Janssen), ABI-H0731 (NCT02908191; Assembly Biosciences), and RO7049389 (NCT02952924; Hoffmann-La Roche).
The aim of this study was to determine whether CAM antiviral effects can extend beyond their two well-described MoA compounds in vitro, which are the inhibition of HBV replication and de novo cccDNA formation. To answer this question, two novel CAMs, JNJ-827 and JNJ-890, were profiled in vitro. Altogether, our data demonstrate that some CAMs could interfere with multiple functions of HBc during the viral life cycle.
RESULTS
In tubo and in cellulo effects of JNJ-827 and JNJ-890 on capsid assembly.
CAMs were reported to induce either the formation of morphologically intact HBV capsids (class I MoA compounds) (41) or the formation of pleiomorphic/aberrant structures (class II MoA compounds) (28). In this study, two novel CAMs, JNJ-61030827 and JNJ-54138890 (Fig. 1A), abbreviated here as JNJ-827 and JNJ-890, were tested for their anti-HBV activity and cytotoxicity. Before testing them for their antiviral effect in an infectious model, electron microscopy (EM) and size exclusion chromatography (SEC) analyses were performed to categorize JNJ-827 and JNJ-890 as having a class I or II MoA compounds. The compounds were incubated together with the recombinant HBV core assembly domain (amino acids 1 to 149) for 24 h in the presence of 150 mM NaCl before visualization by EM. Images were compared to those obtained in our previous study (37) with the phenylpropenamide AT130 (25) and the heteroaryldihydropyrimidine Bayer-41-4109 (here abbreviated as BAY41) (31) compounds with class I and class II MoA compounds, respectively. EM images showed the formation of morphologically intact capsids in the case of JNJ-827 or aberrant structures after treatment with JNJ-890 (Fig. 1B). SEC analysis confirmed these observations (Fig. 1C). The control sample without compound (dimethyl sulfoxide [DMSO]) showed, as expected, two absorbance peaks, one corresponding to capsid and the other to HBc dimers. Control class I MoA compound AT130 increased the formation of capsid as expected, whereas control class II MoA compound BAY41 altered the retention time, indicating the formation of aberrant structures. With JNJ-827, a unique peak was observed at the elution volume corresponding to morphologically intact capsids, whereas a peak was observed at the elution volume corresponding to aberrant structures for the sample treated with JNJ-890. Two inactive compounds (inactive Cp#1 and Cp#2) did not change the SEC profile. Altogether, these studies indicate that JNJ-827 is a class I CAM and that JNJ-890 is a class II CAM.
FIG 1.
Effect of JNJ-827 and JNJ-890 on capsid assembly in tubo and in cellulo. (A) Structures of JNJ-827 and JNJ-890. (B) EM images of in tubo capsid formation obtained using the indicated amounts of molecules as described in Materials and Methods. (C) Graphs of SEC analyses performed with the same amounts of compounds as in panel B. Compound 1 (Cp 1) and Cp 2 correspond to inactive controls of JNJ-827 and JNJ-890. (D) Differentiated HepaRG were infected with HBV (500 virus genome equivalent [vge]/ml) and then treated from day 7 postinfection for 7 days every 2 or 3 days (three treatments) with 10 μM concentrations of the indicated compounds. Noninfected (NI) and infected not treated (NT) controls were also added. Protein lysates were loaded and run in native conditions on an agarose gel, the proteins were transferred onto an enhanced chemiluminescence membrane, and HBc was detected by chemiluminescent immunoblotting with an anti-HBc antibody. The blot is representative of three experiments. (E) Quantification (means plus the standard errors of the mean [SEM]; n = 3) of the capsid signal by chemiluminescence determined using a ChemiDoc XRS+ System (Bio-Rad).
To study the effect of both CAMs on capsid assembly in cellulo, differentiated HepaRG (dHepaRG) cells were infected with HBV and then treated from day 7 postinfection for a total of 7 days, during which the compound was refreshed three times. Intracellular capsid formation was visualized by a native agarose gel migration assay. A cell lysate from HepG2.2.15 cells was used as a positive control. As expected, capsid formation was not affected with tenofovir (TDF), whereas there was no band migrating at that same height with BAY41 and JNJ-890 (Fig. 1D). Capsid formation was increased by 2-fold upon AT130 treatment, whereas JNJ-827, which was classified as class I, induced a 2-fold decrease in capsid formation (Fig. 1E).
In vivo studies investigating the subcellular distribution of HBc have reported both cytoplasmic and nuclear localization (42, 43). Here, we wanted to know whether CAM treatment could affect HBc subnuclear localization. To address this question, the cellular distribution of HBc was investigated by immunostaining using an antibody, which solely recognizes HBc as part of a capsid and not its monomeric or dimeric form (44). Assembled HBc was mainly found in the nucleus of nontreated HBV-infected dHepaRG (see Fig. S1 in the supplemental material). In untreated (NT) and in TDF- and AT130-treated cells, HBc staining was diffuse in the nucleus, whereas the BAY41 and JNJ-890 treatments induced HBc clustering (see Fig. S1 in the supplemental material). Interestingly, these HBc clusters colocalized with large nuclear bodies containing the promyelocytic leukemia (PML) protein and the proteasomal 20S subunit, two proteins associated with sites of protein modification/degradation (45). An intermediate phenotype was obtained with JNJ-827. Altogether, the data showed that CAM treatment can affect HBc nuclear localization and capsid formation in infected hepatocytes.
Anti-HBV activity and cytotoxicity of JNJ-827 and JNJ-890.
The half-maximal effective concentration (EC50) was determined in HepG2.117 cells (intracellular EC50 [iEC50]) and in HepG2.2.15 cells (extracellular EC50 [eEC50]) after quantification of total intracellular or extracellular HBV DNA by quantitative PCR (qPCR). The mean iEC50 values for JNJ-827, JNJ-890, BAY41, and AT130 were 4.7, 66, 67, and 1,020 nM, respectively, whereas the eEC50 values were 5.7, <391, 101, and 127 nM, respectively (Table 1). Moreover, we observed in infected dHepaRG cells a dose-dependent reduction in the amount of intracellular HBV DNA, without cytotoxicity and cell dedifferentiation, as measured by the cellular secretion of the apolipoprotein B (ApoB) (see Fig. S2 in the supplemental material). JNJ-827, JNJ-890, and AT130 did not show cytotoxicity in HepG2 cells at any of the concentrations tested (>25, >100, and >50 μM, respectively), whereas the 50% cytotoxic concentration (CC50) of BAY41 was 33.9 μM (Table 1). The selectivity index (SI) values (i.e., the CC50/iEC50 value) for JNJ-827 and JNJ-890 in HepG2 cells were >5,319 and >1,515, respectively (Table 1). For the remainder of the mechanistic studies, CAMs were used at a single, noncytotoxic concentration of 10 μM.
TABLE 1.
Anti-HBV activity and cytotoxicity in HepG2.117 and HepG2.2.15 cellsa
| Compound | Type | Mean (range) EC50 (nM) in HepG2.117 or HepG2.2.15 cells | Mean (range) CC50 (nM) |
Selectivity index |
||
|---|---|---|---|---|---|---|
| HepG2.117/2.15 | HepG2 | HepG2.117/2.15 | HepG2 | |||
| AT130 | Intra | 1,020 (120–4,620), n = 24 | ND | >50,000, n = 11 | ND | >49 |
| Extra | 127 (<98–207), n = 4 | ND | >393 | |||
| JNJ-827 | Intra | 4.7 (2.1–11), n = 55 | >25,000 (>5,000–>25,000), n = 3 | >25,000, n = 11 | >5,319 | >5,319 |
| Extra | 5.7 (<3.9–9.8), n = 11 | >4,386 | >4,386 | |||
| BAY41 | Intra | 67 (21–216), n = 467 | 33,900 (21,400–>50,000), n = 3 | 35,400 (10,600–>120,000), n = 56 | 506 | 528 |
| Extra | 101 (25–216), n = 99 | 336 | 350 | |||
| JNJ-890 | Intra | 66 (63–70), n = 2 | ND | >100,000, n = 4 | ND | >1,515 |
| Extra | <39, n = 2 | ND | >256 | |||
The anti-HBV activity of AT130, JNJ-827, BAY41, and JNJ-890 in stable HBV-replicating cells was tested in a dose-response assay. Intracellular (HepG2.117 cells) and extracellular (HepG2.2.15 cells) HBV DNA was extracted, and DNA levels were assessed using qPCR. In each experiment, the EC50s were determined based on the mean inhibition from two wells per compound concentration. The cytotoxicity for HepG2 cells was assessed using a resazurin readout. EC50, 50% effective concentration; CC50, 50% cytotoxic concentration; Intra, intracellular; Extra, extracellular; n, number of experiments; ND, not done.
Ex vivo effect of JNJ-827 and JNJ-890 in infected PHHs.
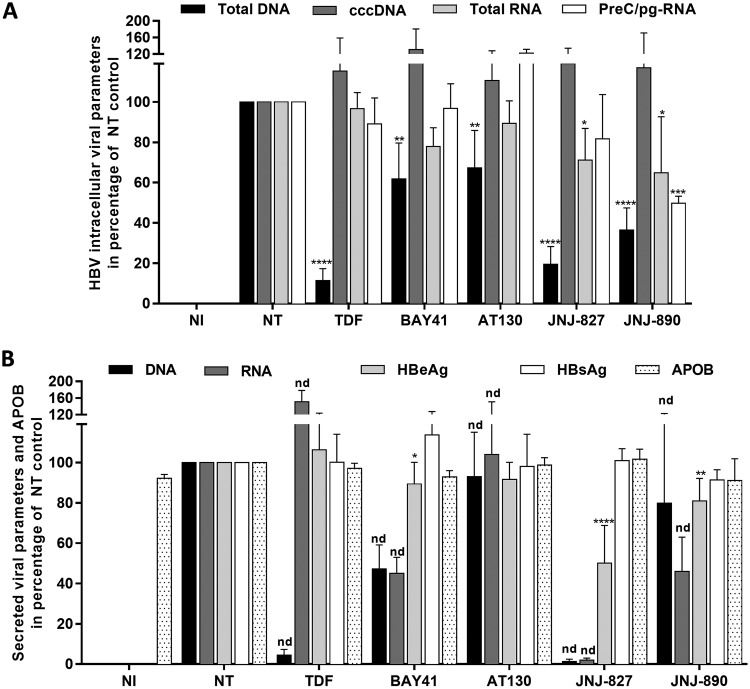
Primary human hepatocytes (PHHs) represent the golden-standard in terms of evaluating drug metabolism and hepatoxicity. Moreover, the susceptibility of the cell culture model to HBV infection makes it ideal for studying viral replication ex vivo (46). Three different batches of freshly isolated PHHs were infected with HBV and treated three times with JNJ-827, JNJ-890, control CAMs, and TDF from day 5 postinfection for a total of 7 days. JNJ-827 and JNJ-890 reduced total intracellular HBV DNA by 80 and 60%, respectively, whereas only weak decreases were observed with BAY41 and AT130, and 90% intracellular HBV DNA reduction was obtained with TDF (Fig. 2A). Interestingly, JNJ-827 and JNJ-890 induced also a weak but significant reduction in intracellular total HBV RNA levels, whereas JNJ-890 was the sole compound able to significantly reduce PreC/pg-RNA by 2-fold (Fig. 2A). All treatments had no effect on the established pool of cccDNA in this short treatment duration setting.
FIG 2.
Antiviral properties of JNJ-827 and JNJ-890 in PHHs. PHHs were mock infected (NI condition) or infected with HBV (200 vge/ml), cultivated during 5 days postinfection, and then not treated (NT condition) or treated three times for 7 days (treatment every 2 or 3 days) with 10 μM concentrations of the indicated compound. (A) Intracellular HBV total DNA, cccDNA, total RNA, and PreC/pgRNA levels were quantified by qPCR, qPCR FRET (fluorescence resonance energy transfer), and quantitative reverse transcription-PCR (RT-qPCR), respectively. (B) The HBeAg, HBsAg, and ApoB levels in cell culture supernatant were monitored by ELISA. Extracellular HBV DNA and RNA levels were quantified by qPCR or RT-qPCR after nucleic acid extraction from supernatant. The results are means plus the SEM (n = 3) performed with three different PHH donors. Statistics were determined using a two-tailed Mann-Whitney test, with the following calculated probabilities: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; and nd, not determined.
At the extracellular level, TDF, as already reported for lamuvidine (36), induced a >90% reduction of secreted HBV DNA, along with an accumulation of secreted HBV RNA (Fig. 2B). Among all the CAMs tested, JNJ-827 was the only compound able to strongly decrease both secreted HBV RNA and DNA levels. Moreover, JNJ-827 was the sole CAM observed to affect HBeAg secretion, whereas none of the molecules affected HBsAg secretion (Fig. 2B). The EC50 of JNJ-827 regarding HBeAg secretion inhibition was around 5 μM (see Fig. S3 in the supplemental material). None of the molecules tested were toxic at the concentrations tested, as indicated by the unchanged levels of HBsAg and ApoB in supernatants (Fig. 2B). Note that results obtained with the three individual batches of PHHs, for cccDNA as well as for secreted viral and ApoB parameters, are also presented as absolute quantifications in Fig. S4 in the supplemental material.
Altogether, these results identified JNJ-827 as having the most pronounced effects among the CAMs tested. Moreover, JNJ-827 and JNJ-890 featured interesting novel properties that were further investigated in the rest of the study.
Effect of JNJ-827 and JNJ-890 on the formation of cccDNA in a de novo infection setting.
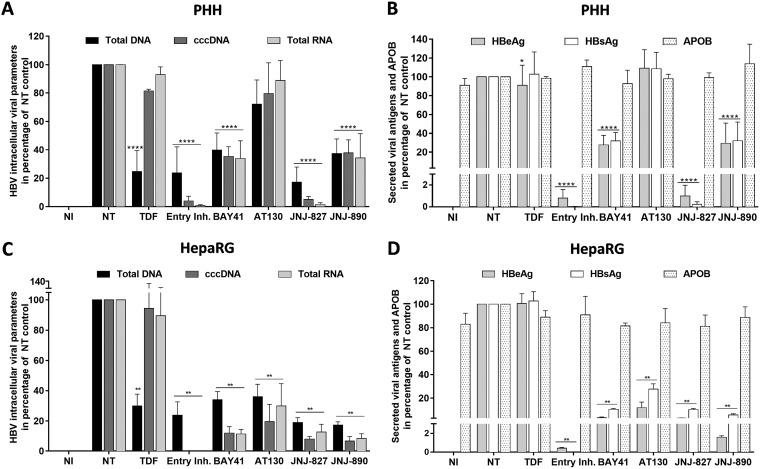
It was recently described that CAMs could also prevent cccDNA formation, thus defining a secondary MoA compound of these molecules based on a postentry mechanism (37). To evaluate JNJ-827 and JNJ-890 with respect to this MoA compound and compare with control CAMs or the PreS1 peptide (i.e., an entry inhibitor), which is known to inhibit HBV entry by interfering with hNTCP binding (47), compounds were added 1 day before and during viral inoculation of PHHs or dHepaRG cells. This is in contrast to what was done in previous experiments (Fig. 2) that aimed at evaluating the postinfection action of these molecules. TDF was added 24 h postinfection and used here as a technical control for cccDNA quantification, since NAs do not alter the cccDNA level. cccDNA quantification was performed at day 7 postinfection. None of the drugs were toxic in this setting at the concentration tested, as shown by an absence of variation in ApoB secretion (Fig. 3B and D). As expected, the PreS1 peptide prevented viral entry thus preventing cccDNA establishment and subsequent synthesis of viral intermediates (Fig. 3A). Interestingly, JNJ-827 treatment also reduced cccDNA formation by >90%, resulting in a concomitant decrease of intracellular HBV DNA and RNA levels (Fig. 3A), as well as HBeAg and HBsAg secretion (Fig. 3B). JNJ-890 and BAY41 were found to be far less potent than JNJ-827, whereas AT130 was similarly inefficient as TDF at preventing cccDNA establishment. The EC50s of JNJ-827 for cccDNA establishment in PHH were between 19 and 61 nM and around 100 nM for HBV total DNA, HBeAg, and HBsAg (see Fig. S5 in the supplemental material). Phenotypically, JNJ-827 and the PreS1 peptide behaved the same, indicating that a bona fide entry or a postentry inhibition could be equal in terms of antiviral effect. In the dHepaRG model, all CAMs were found to be efficient to inhibit cccDNA establishment (Fig. 3C), replication intermediates (Fig. 3C), and antigen production (Fig. 3D), suggesting that the compounds might be differentially metabolized in both cell types. Note that results obtained with the three individual batches of PHHs and three independent HepaRG differentiations, regarding cccDNA as well as secreted HBe/HBsAg and ApoB parameters, are also presented in absolute quantification in Fig. S6 and S7 in the supplemental material.
FIG 3.
Effect of JNJ-827 and JNJ-890 on HBV infection establishment in dHepaRG cells and PHHs. PHH (A and B) or dHepaRG (C and D) cells were pretreated for 24 h with 10 μM concentrations of the indicated CAMs or 100 nM preS1 peptide entry inhibitor and then inoculated with HBV (200 vge/ml) in the presence of drug for 24 h. One day after infection, the medium was changed, and the cells were cultured for 1 week. For the TDF control, cells were treated twice after infection. A noninfected (NI) control was included. (A and C) Intracellular HBV total DNA, cccDNA, and total RNA levels were quantified by qPCR, qPCR FRET, and RT-qPCR, respectively. (B and D) HBeAg, HBsAg, and ApoB levels in cell culture supernatant were monitored by ELISA. The results are means plus the SEM (n = 3) performed with three different PHH donors or three different differentiations of HepaRG. Statistics were determined using a two-tailed Mann-Whitney test, with the following calculated probabilities: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Long-term treatment with JNJ-827 and JNJ-890 affects viral RNA accumulation.
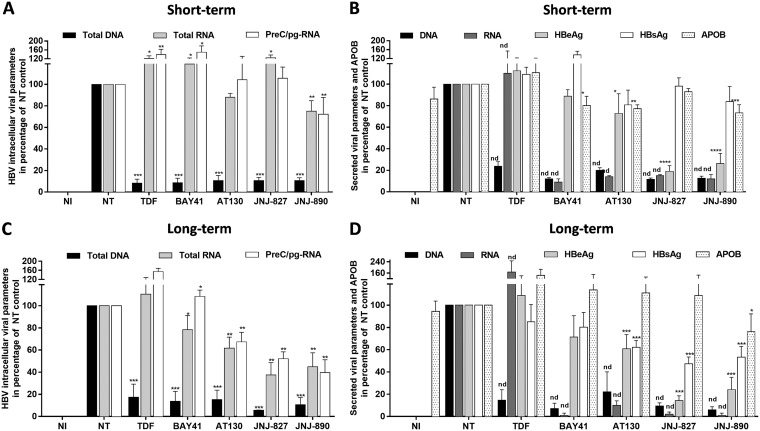
Short-term postinfection treatment with JNJ-827 and JNJ-890 in PHHs suggested a modest effect on viral RNA accumulation (Fig. 2A). It was previously speculated that CAM treatment could impact cccDNA transcription and/or posttranscriptional events (40). To further analyze this potential MoA of JNJ-827 and JNJ-890 compared to other CAMs, we made use of dHepaRG that can be maintained for weeks or months in culture in contrast to PHHs, which quickly dedifferentiate when cultured ex vivo. dHepaRG cells were infected with HBV and then treated from day 7 postinfection either for a total of 7 days (total of 3 administrations) or 1 month (total of 15 administrations). Under short-term conditions using dHepaRG, all CAMs induced a strong reduction of intracellular HBV DNA accumulation (Fig. 4A), which is in line with the observed, though less pronounced, effects in PHHs, most likely due to different metabolization properties of each cell type (compare Fig. 4A with Fig. 2A). This inhibition of nucleocapsid assembly was translated into a strong inhibition of secreted HBV DNA and RNA, yet without any effect on HBsAg and ApoB, secretions (Fig. 4B). Regarding the effect on intracellular RNA accumulation, only JNJ-890 led to a weak reduction in dHepaRG (Fig. 4A). It is also worth noting that a significant reduction in HBeAg secretion was observed with both JNJ-827 and JNJ-890, in the absence of any correlative inhibition of HBsAg (Fig. 4B). Such an inhibition was also observed in the PHH model for JNJ-827 (Fig. 2B), suggesting a peculiar feature of JNJ-827 and JNJ-890 with respect to HBeAg biogenesis and secretion.
FIG 4.
Antiviral properties of JNJ-827 and JNJ-890 in dHepaRG cells: short-term versus long-term treatment. dHepaRG were infected with HBV (200 vge/ml), cultured for 1 week, and then treated for 7 days (treatment each 2 or 3 days; 3 treatments in total) (A and B) or 1 month (treatment each 2 or 3 days; 15 treatments in total) (C and D) with a 10 μM concentration of the indicated compound. The antiviral effects of the compounds were monitored at endpoint. (A and C) Intracellular HBV total DNA, total RNA, and PreC/pgRNA levels were quantified by qPCR and RT-qPCR. (B and D) HBeAg, HBsAg, and ApoB levels in cell culture supernatant were monitored by ELISA. Extracellular HBV DNA and RNA levels were quantified by qPCR or RT-qPCR after nucleic acid extraction from the supernatant. The results are means plus the SEM (n = 3) performed with three different differentiations of HepaRG. Statistics were determined using a two-tailed Mann-Whitney test, with the following calculated probabilities: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; and nd, not determined.
In the long-term setting, JNJ-827 and JNJ-890 were able to reduce both intracellular total HBV RNA and PreC/pgRNA levels (Fig. 4C), as well as antigens and HBV DNA/RNA secretions (Fig. 4D). Intracellular total HBV RNA and PreC/pgRNA levels were also impacted by BAY41 and AT130, but to a lesser extent. All CAMs induced a strong inhibition of secreted HBV DNA and RNA, whereas TDF gave rise to an expected profile, i.e., decreasing DNA and increasing RNA secretion. JNJ-890 induced a weak but significant reduction of ApoB secretion (Fig. 4D). Altogether, these data suggest that JNJ-827 and JNJ-890 could reduce HBV RNA synthesis/stability in a long-term treatment context and that this reduction can be associated with a decrease in HBsAg production. Unfortunately, in dHepaRG the amount of cccDNA is very low (<0.05 copy/cell), making a reliable quantification of this parameter not possible. Note that results obtained with the three independent HepaRG differentiations in short- and long-term treatment conditions, regarding secreted HBe/HBsAg and ApoB parameters, are also presented in absolute quantification in Fig. S8 and S9 in the supplemental material.
Effect of JNJ-827 and JNJ-890 on HBeAg biogenesis and/or secretion.
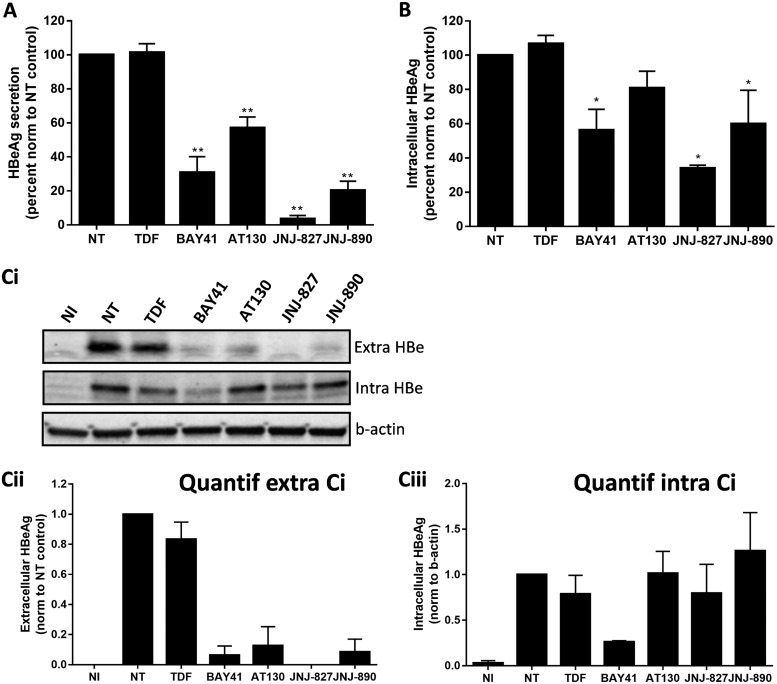
In both PHH and dHepaRG cells and a short-term treatment setting, we observed a reduction in HBeAg secretion with JNJ-827 without intracellular total HBV RNA decrease, leading us to speculate that this CAM has putative effects on HBeAg biogenesis/secretion. To specifically address this question, a HepaRG cell line expressing HBeAg alone in a tetracycline-dependent manner was used (HepaRG-TR-HBe). First, we confirmed that HBeAg secretion increased with the tetracycline amount (see Fig. S10A in the supplemental material). We choose to use 1 μg/ml of tetracycline, for which the HBeAg secretion level (about 150 NCU/ml) was comparable to what we obtained after an infection of dHepaRG cells. By native agarose gel migration assay using also a HepaRG-TR-HBc cell line as a positive control, we confirmed that the HepaRG-TR-HBe cells do not express HBc and as such do not produce capsid (see Fig. S10B in the supplemental material). HepaRG-TR-HBe cells were induced to secrete HBeAg for 1 day and then treated for 5 days (total of two treatments). We observed a reduction in HBeAg secretion of >90% with JNJ-827 (Fig. 5A). The same observation was made with other CAMs tested, but to a lower extent. As expected, TDF had no effect on HBeAg secretion. Note that intracellular quantification of p22/pre-HBeAg by enzyme-linked immunosorbent assay (ELISA) led to the same ranking of compounds, although with a lower level of inhibition (Fig. 5B).
FIG 5.
Effect of JNJ-827 and JNJ-890 on HBeAg biogenesis and/or secretion. HepaRG-TR-HBe cells were treated with tetracycline (1 μg/ml) for 48 h to allow expression of HBeAg from tetracycline-regulated promoter and then treated for 5 days (two treatments) with 10 μM concentrations of the indicated compounds. (A) The HBeAg secretion level in cell culture supernatant was monitored by ELISA. (B) Intracellular HBeAg amount was also monitored by ELISA. For panels A and B, the results are means plus the SEM (n = 3), and the statistics were determined using a two-tailed Mann-Whitney test (**, P < 0.01). (Ci) Evaluation of intracellular and extracellular HBeAg levels by immunoblotting (results from a representative experiment [n = 2]). (Cii and iii) Quantification of extracellular and intracellular HBeAg signal by chemiluminescence done with a ChemiDoc XRS+ System (Bio-Rad). The results are means plus the SEM (n = 2).
To further extend the analysis to the primary level of p22/pre-HBe/HBeAg expression, supernatants were also submitted to denaturing Western blotting after trichloroacetic acid precipitation to confirm that the observed effect was genuinely due to an inhibition of HBeAg secretion and not to a problem of detection of the protein by ELISA. At the intracellular level, p22/pre-HBeAg amounts were only affected by BAY41 treatment, whereas other CAMs only reduced extracellular accumulation (Fig. 5Ci, ii, and iii). Altogether, these data suggest that all CAMs tested, except for BAY41, do not tend to prevent the primary expression of p22/pre-HBeAg, while some of them can also potentially interfere with HBeAg biogenesis/secretion. In this respect, JNJ-827 was the most potent in this assay (i.e., use of HepaRG-TR-HBe), and this potency can translate into an inhibition of HBeAg secretion in HBV-infected PHHs (Fig. 2), suggesting that this molecule has a particular feature.
DISCUSSION
Treatment of CHB is limited to the use of NAs and Peg-IFN-α. Unfortunately, these therapeutic options lead to a functional cure (i.e., HBsAg loss with or without HBsAg to HBsAb seroconversion) in only 10% of cases after a 5-year follow-up (14, 15). Therefore, there is a need for new therapeutic options.
The core/HBc protein plays multiple roles during the HBV life cycle (e.g., forms nucleocapsid, interacts with cccDNA and human host genes, and may be involved in the regulation of their transcription) (1, 17, 19, 20). Interfering with the multiple functions of HBc using a direct antiviral agent represents a promising strategy to disrupt HBV replication at several steps in the viral life cycle. Currently, several CAMs are being evaluated in clinical trials, and many others are in preclinical development (additional information is available [http://www.hepb.org/treatment-and-management/drug-watch/]).
JNJ-827 and JNJ-890 are two novel CAMs. SEC and EM studies revealed that JNJ-827 possesses a class I MoA compound, whereas JNJ-890 possesses a class II MoA compound. However, and in contrast to the hallmark of a class I MoA compound, JNJ-827 was capable of reducing the amount of intracellular capsid by ∼2-fold, as visualized on a native agarose gel. This discrepancy might be explained by the much higher potency of JNJ-827 compared to the other class I CAM in the panel, AT130 (217-fold less active than JNJ-827 in HepG2.117). For the two other CAMs tested, i.e., JNJ-890 and BAY41, the iEC50s were similar.
In the present study, CAMs were mostly used at a high concentration (10 μM) to highlight novel antiviral effects and MoA. In HBV-infected PHHs we confirmed that JNJ-827 and JNJ-890 possess the two described antiviral effects of CAMs: inhibition of HBV replication in already established infections (primary mechanism) and inhibition of cccDNA de novo formation (secondary mechanism) (37, 38). JNJ-827 was the most potent CAM tested in stably HBV-transformed HepG2 cell lines, with an EC50 in the single-digit nanomolar range. Like the PreS1 peptide entry inhibitor, JNJ-827 inhibited cccDNA formation with an EC50 around 50 nM. BAY41 and JNJ-890 were similarly efficient in this model. The improved efficacy of JNJ-827 at inhibiting HBV infection by a postentry mechanism was likely due the higher potency of JNJ-827 (iEC50 at 4.7 nM), as reflected by the lack of inhibition with AT130 (iEC50 at 1,020 nM). It is worth noting that all CAMs, including AT130, were similarly efficient at preventing HBV infection in the dHepaRG model. This can either point to the difference in metabolic properties of these cells compared to PHH or the lower infection rate in dHepaRG cells compared to PHHs at the same multiplicity of infection (as an indication, HBsAg production ranges between 50 and 100 IU/ml in dHepaRG versus 1,000 to 5,000 IU/ml in PHHs when a multiplicity of infection of 100 genome equivalent of HBV/cell is used). The concentration necessary to achieve a 50% reduction of the formation of cccDNA (EC50 for cccDNA formation inhibition) was 10 times higher than that necessary to achieve the main inhibitory action (i.e., inhibition of HBV DNA neosynthesis). It remains to be determined whether this inhibition of the establishment of HBV infection is due to an accelerated breakdown of capsid in cytoplasm before rcDNA could be delivered to nucleus and converted into cccDNA (38, 39) or to a prevention of disassembly at the nuclear pore. In the first case, it would be interesting to determine whether rcDNA could serve as a pathogen-associated molecular pattern for innate sensing by the dsDNA sensing machinery.
In postinfection and short-term treatment conditions, among all CAMs tested, JNJ-827 was the most potent at inhibiting intracellular HBV replication (i.e., neosynthesized rcDNA) in PHHs. Very importantly, as reported both for NVR-3-1983 (36) in vitro and NVR-3-778 (48) in vivo, we found that, in PHHs, JNJ-827 strongly reduced both extracellular HBV DNA and RNA levels, whereas TDF only decreased extracellular HBV DNA levels (and increased the HBV RNA level), and other CAMs tested were inefficient. This is due to the fact that JNJ-827, in contrast to NAs, blocks the encapsidation of pgRNA, and therefore this results in a reduction of encapsidated DNA and RNA in the cell culture supernatant (11–13, 49). The function of secreted HBV-RNA virion-like particles in HBV life cycle is unclear. HBV RNA virion-like particles were recently reported to be mainly replication-deficient (11), but they could have immunological functions. Therefore, the inhibition of their release into the blood of CHB patients could be beneficial.
Nuclear HBc was suggested to regulate HBV transcription by interacting with cccDNA (17, 22). Interestingly, we found, like a very recently published study (50), that CAM treatment induces the relocalization of nuclear HBc to subnuclear sites staining positive for the 20S proteasomal subunit and PML bodies, which are the sites of protein posttranslational modification, activation, sequestration, or degradation (45). Here, we found that BAY41 treatment led to the formation of large HBc aggregates, whereas AT130 treatment had no effect or only a slight effect on nuclear HBc distribution. These observations correlate with the MoA of the compounds and suggest that CAMs could differentially disrupt nuclear functions of HBc. In this respect, in both dHepaRG and PHH cells, JNJ-890 was the sole CAM capable of decreasing intracellular total HBV RNA and PreC/pgRNA levels after 1 week of treatment. However, after 1 month of treatment of dHepaRG cells, all CAMs tended to decrease total HBV RNA levels, as well as HBeAg and HBsAg secretion, with JNJ-827 and JNJ-890 being again the most potent compounds tested. Even if we cannot exclude an effect on cccDNA amount after a long-term treatment (we failed to reliably detect cccDNA in this experimental context by qPCR-based methods), our observations suggest that HBc could be a positive regulator of cccDNA transcription or/and posttranscriptional events and suggest also that (i) CAMs interfere with the nuclear function of core in vitro and that (ii) compound class is not necessarily an indicator of the effect of CAM on HBc nuclear functions; compound potency could also be involved.
HBeAg is a secreted antigen sharing primary structure with HBc. The intracellular precursor of HBe (called p22 or pre-HBe) features 10 additional amino acids at the N-terminal extremity compared to HBc and needs to be processed by both N- and C-terminal maturation to give rise to secreted HBeAg (51, 52). Secreted HBeAg is in fact a dimer of p17 polypeptide. One possible function of secreted HBeAg could be to transiently suppress the immune response against HBV. This could be done by negatively regulating the TLR2 signaling pathway (51, 52) and/or by modulating the immune response against HBc (53, 54). HBeAg seroconversion is an immunological process that occurs during the natural history of HBV infections due to the occurrence of stop or PreC/core promoter mutations in the cccDNA integration events and can be increased upon treatment with Peg-IFN and NAs. It is associated with higher immune activity and is considered an important clinical endpoint during therapy (53, 54). In this study, we observed a strong and fast inhibition of HBeAg biogenesis/secretion by CAMs in the HepaRG-TR-HBe model, more particularly with JNJ-827, which was the only compound to recapitulate this phenotype in PHHs with an EC50 of around 5 μM. The inhibition is unlikely due to an inhibition of the precursor (p22/pre-HBe) synthesis, since Western blot analyses under denaturing conditions did not reveal any differences, but is likely mediated by a direct interaction of these compounds with the intracellular precursor of HBeAg, leading to an impaired processing and dimerization (i.e., biogenesis of the antigen), as well as to an impaired secretion. Although HBc and HBe monomer share very similar primary structures, the quaternary dimeric form of HBeAg is drastically different from HBc (55). This is a consequence of an intramolecular disulfide bridge between the cysteine 61 (common to HBeAg and HBc) and cysteine 7 (specific to HBeAg) that is specifically added in the lumen of the endoplasmic reticulum, in which HBeAg is translocated during translation (56). HBeAg dimers with the C7–C61 disulfide bridge do not assemble in capsid-like structures, while the reduced form does in the cytoplasm (56, 57). CAMs target HBc by binding to a hydrophobic pocket at the dimer-dimer interface (58–60). To explain how some CAMs target HBeAg, we can hypothesize that they could force HBeAg dimer-dimer assembly, leading to HBeAg aggregation, as observed for HBc, and could promote a fast degradation.
To summarize, CAMs possess two well-characterized mechanisms of action. They accelerate capsid assembly, which results in an inhibition of HBV replication (primary MoA) and prevent de novo formation of cccDNA by acting at a postentry step (secondary MoA). Here, we demonstrated that treatment with some CAMs can extend beyond these two mechanisms. Indeed, some CAMs can directly reduce secreted HBeAg and act also either at transcriptional or posttranscriptional levels, leading to a reduction in both total HBV RNA and PreC/pgRNA accumulation, as well as HBsAg production. These additional effects with some CAMs require further investigation and may lead to the development of second-generation compounds. Due to their multiple and additional effects compared to NAs, one could expect that some CAMs could lead to higher rates of functional cure in CHB patients, and clinical evaluation of this type of multifunctional CAMs is eagerly awaited.
MATERIALS AND METHODS
Compounds.
BAY41, AT130, JNJ-827, and JNJ-890 were provided by Janssen. JNJ-827 and JNJ-890 (see structure in Fig. 1A) were prepared according to methods described in the patent applications WO2014184350 and WO2013102655, respectively. The PreS1 peptide (myristoylated amino acids 2 to 48 from the N-terminal part of PreS1) was synthesized by GenScript, and TDF was obtained from Gilead Sciences (Foster City, CA). All compounds had an high-pressure liquid chromatography-certified purity of >95%.
Antiviral assay using HepG2.2.15 cells.
Determinations of the JNJ-827 and JNJ-890 EC50 and EC90 values were performed as previously described (35).
Cytotoxicity assay using HepG2 cells.
Determinations of the JNJ-827 and JNJ-890 CC50 and CC90 values were performed as previously described (35).
EM and SEC studies.
Determination of JNJ-827 and JNJ-890 effects on capsid formation in tubo were performed as previously described (35).
Culture of HepaRG cells and primary human hepatocytes and HBV infection.
Human liver progenitor HepaRG cells (61) and an engineered HepaRG-TR-HBe cell line expressing only HBeAg in a tetracycline-dependent manner (obtained by double transduction with lentiviral vectors [from Invitrogen; T-Rex System] containing tetracycline repressor and HBe expression transgenes [a detailed method is available on request]) were cultured as previously described (61). Primary human hepatocytes (PHHs) were isolated from surgical liver resections, after informed consent of patients (kindly provided by M. Rivoire [CLB, Lyon]; agreements DC-2008-99 and DC-2008-101) as previously described (62) and cultured in complete William's medium supplemented with 1.8% of DMSO. Differentiated HepaRG (dHepaRG) cells and PHHs were infected as previously described (63), with HBV genotype D inoculum prepared either from HepG2.2.15 or HepAD38 cells. The multiplicities of infection (expressed as virus genome equivalent/cell) are indicated in the figure legends.
Antiviral treatment of PHHs and dHepaRG cells.
JNJ-827, JNJ-890, and TDF (used at 10 μM), as well as PreS1 peptide (used at 100 nM), were evaluated for their effect on the establishment of HBV infections or on established HBV infections. Under the preinfection condition, CAMs and PreS1 peptide were added 1 day before and during infection. TDF was added twice postinfection. Cells and supernatants were harvested at day 6 postinfection. Under the postinfection condition, PHHs or dHepaRG cells were infected by HBV for, respectively, 5 or 7 days and then treated three times at days 5, 7, and 9 (PHHs) or days 7, 9, and 11 (dHepaRG cells) postinfection. Cells and supernatants were harvested at day 12 or 14 postinfection (i.e., 7 days posttreatment).
Analysis of viral parameters.
From cell culture supernatants, secreted HBe and HBs antigens were quantified by ELISA, using a chemiluminescence immunoassay kit (Autobio, China) according to manufacturer's instructions. Extracellular viral DNA and RNA were extracted from cell culture supernatant using MagMAx kit (Thermo Scientific) and MagNAPure (Roche), respectively, according to the manufacturer's protocols and treated either with DNase I or RNase A. Intracellular total HBV DNA or RNA were extracted from infected cells using, respectively, NucleoSpin 96 Tissue or NucleoSpin 96 RNA kits (Macherey-Nagel). RNAs were transcribed into cDNA using the SuperScript III reverse transcriptase (Invitrogen). Real-time PCR for total intracellular HBV DNA and RNA was performed using a LightCycler 480 (Roche) and normalized to PRP, a housekeeping gene, with the following primers: 5′-GCTGACGCAACCCCCACT-3′ (HBV-sense), 5′-AGGAGTTCCGCAGTATGG-3′ (HBV-antisense), 5′-TGCTGGGAAGTGCCATGAG-3′ (PRP-sense), and 5′-CGGTGCATGTTTTCACGATAGTA-3′ (PRP-antisense). An HBV standard was used for quantification of extracellular viral DNA and RNA. For cccDNA quantification, total DNA was digested by using Plasmid-Safe DNase (New England BioLabs) for 6 h at 37°C and then subjected to qPCR using a LightCycler 480 Probes master kit (Roche) with a home-made probe-primer mix. cccDNA was normalized to β-globin quantification. For PreC/pgRNA quantification, qPCR was performed using TaqMan Fast Advanced master mix (Life Technologies) using a home-made probe primer mix. PreC/pgRNA levels were normalized to GUSB using a commercial probe primer mix (Life Technologies, catalog no. 4448491; GUSB: Hs99999908_m1).
ELISA.
Apolipoprotein B (ApoB) was quantified in cell culture supernatants using a total human apolipoprotein B ELISA (Alerchek, Inc.) according to the manufacturer's recommendations. The amounts of HBeAg and HBsAg in the supernatant and sera were quantified by commercially available ELISA (Autobio, Ltd., China) according to the manufacturer's instructions. An intracellular HBeAg ELISA was performed after cell lysis in a buffer containing 400 mM potassium acetate, 25 mM KHEPES (pH 7.2), 15 mM magnesium acetate, 1% (vol/vol) NP-40, and 0.5% (wt/vol) deoxycholate supplemented with 1 mM phenylmethylsulfonyl fluoride and 1× protease inhibitor cocktail.
Native agarose gel migration assay and immunoblotting.
The intracellular formation/accumulation of HBV nucleocapsid in infected dHepaRG cells was performed from cells lysed in a buffer containing 50 mM Tris-HCl (pH 8), 1 mM EDTA, and 1% (wt/vol) NP-40. Cell lysates were centrifuged for 1 min at 10,000 rpm and 4°C. The supernatants were recovered and incubated at least 2 h at 37°C with 10 μg/ml of a DNase I/RNase A mix in buffer containing 20 mM Mg(CH3COO)2. Lysates were loaded into a native 1.2% agarose gel, and electrophoresis was performed in 1× Tris-HCl acetate EDTA (TAE) buffer. Capillary transfer of proteins to ECL membrane was performed in 1× Tris-HCl/NaCl/EDTA (TNE) buffer. Immunoblotting was conducted according to a standard protocol with an antibody against HBcAg (Dako, catalog no. B0586) at 1:1,000.
For the detection of HBeAg in cell supernatants, supernatants were 10× concentrated by trichloroacetic acid (Sigma) precipitation and subjected to immunoblotting with Dako B0586 antibody at 1:1,000. An antibody directed against β-actin (Sigma-Aldrich, catalog no. A1978) diluted at 1:5,000 was used for the normalization of results.
Immunofluorescence staining.
dHepaRG cells were fixed with 2% formaldehyde in 1× phosphate-buffered saline (PBS) for 20 min at room temperature and permeabilized/blocked with 0.1% (vol/vol) Triton X-100 (Sigma), 1% (wt/vol) bovine serum albumin (BSA; Euromedex), and 10% (vol/vol) fetal bovine serum (Thermo Scientific) in 1× PBS at room temperature for 30 min. Primary antibodies against HBcAg (Ab8637; used at 1:500), which only recognizes assembled capsid, promyelocytic leukemia protein (PML; Abcam, catalog no. ab31101; used at 1:200), and the proteasomal 20S subunit (Pierce, catalog no. PA1-977; used at 1:200) were diluted in 1× PBS and incubated 1 h at room temperature. Cells were washed four times with wash buffer (1× PBS with 0.1% Triton X-100) and subsequently incubated at room temperature for 1 h with either Alexa Fluor 488-conjugated goat anti-mouse antibody (Invitrogen) for HBcAg detection or Alexa Fluor 594-conjugated goat anti-rabbit antibody (Invitrogen) for PML and 20S subunit detections, both diluted at 1:1,000 in 1× PBS. Counterstaining for the detection of nucleic acid was performed using Hoechst 33258 (Life Technologies) diluted at 1:5,000 in the Alexa Fluor mix. Cells were washed four times with wash buffer, and coverslips were mounted using mounting medium (Dako). Images were obtained with a confocal microscope Leica TSC-SP5X.
Statistical analysis.
Statistical analyses were conducted using nonparametric Mann-Whitney tests using GraphPad Prism software 6.0. Significance is denoted in the figures as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maud Michelet and Jennifer Molle for help with the isolation of primary human hepatocytes and the staff from Michel Rivoire's surgery room for providing assistance with liver resection.
This study was supported by research grants from Janssen, ANRS (French National Agency for Research on AIDS and Viral Hepatitis; grants from CSS4), FINOVI (Foundation for Innovation in Infectiology), and INSERM. This study was also indirectly supported by the DEVweCAN LABEX (ANR-10-LABX-0061) of the Université de Lyon, within the program Investissements d'Avenir (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR). This study was mainly sponsored by Janssen. INSERM U1052 also received institutional funding from INSERM, as well as external grants from ANRS, and DEVweCAN LABEX (ANR-10-LABX-0061) of the Université de Lyon, within the program Investissements d'Avenir (ANR-11-IDEX-0007) operated by the ANR.
T.L., A.F., F.Z., and D.D. have nothing to declare. J.M.B., K.V., K.V., and F.P. are employees of Janssen.
Author contributions were as follows: research design, J.M.B., F.P., F.Z., and D.D.; supervision, J.M.B. and D.D.; experiments, T.L., K.V., A.F., K.V., and D.D.; data analysis, T.L. and D.D.; statistics, T.L.; writing of the manuscript, T.L. and D.D.; and editing and approval of the final manuscript, all authors.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00835-18.
REFERENCES
- 1.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479-480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan SL, Wong VW, Qin S, Chan HL. 2016. Infection and cancer: the case of hepatitis B. J Clin Oncol 34:83–90. doi: 10.1200/jco.2016.34.26_suppl.83. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. 2012. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142:1264–1273. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 3. doi: 10.7554/eLife.00049. [DOI] [PubMed] [Google Scholar]
- 5.Blondot ML, Bruss V, Kann M. 2016. Intracellular transport and egress of hepatitis B virus. J Hepatol 64:S49–S59. doi: 10.1016/j.jhep.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Nassal M. 2015. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 7.Katen S, Zlotnick A. 2009. The thermodynamics of virus capsid assembly. Methods Enzymol 455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zlotnick A. 2005. Theoretical aspects of virus capsid assembly. J Mol Recognit 18:479–490. doi: 10.1002/jmr.754. [DOI] [PubMed] [Google Scholar]
- 9.Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, Endres D. 1999. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry 38:14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- 10.Tuttleman JS, Pourcel C, Summers J. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Sheng Q, Ding Y, Chen R, Sun X, Chen X, Dou X, Lu F. 2017. HBV RNA virion-like particles produced under nucleos(t)ide analogues treatment are mainly replication deficient. J Hepatol doi: 10.1016/j.jhep.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 12.Jansen L, Kootstra NA, van Dort KA, Takkenberg RB, Reesink HW, Zaaijer HL. 2016. Hepatitis B virus pregenomic RNA is present in virions in plasma and is associated with a response to pegylated interferon alfa-2a and nucleos(t)ide analogues. J Infect Dis 213:224–232. doi: 10.1093/infdis/jiv397. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, Zhang R, Chen R, Li T, Zhang T, Yuan Q, Li PC, Huang Q, Colonno R, Jia J, Hou J, McCrae MA, Gao Z, Ren H, Xia N, Zhuang H, Lu F. 2016. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 65:700–710. doi: 10.1016/j.jhep.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 14.Locarnini S, Hatzakis A, Chen DS, Lok A. 2015. Strategies to control hepatitis B: public policy, epidemiology, vaccine, and drugs. J Hepatol 62:S76–S86. doi: 10.1016/j.jhep.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Zoulim F, Durantel D. 2015. Antiviral therapies and prospects for a cure of chronic hepatitis B. Cold Spring Harb Perspect Med 5:a021501. doi: 10.1101/cshperspect.a021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diab A, Foca A, Zoulim F, Durantel D, Andrisani O. 2018. The diverse functions of the hepatitis B core/capsid protein (HBc) in the viral life cycle: implications for the development of HBc-targeting antivirals. Antiviral Res 149:211–220. doi: 10.1016/j.antiviral.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. 2001. Structural organization of the hepatitis B virus minichromosome. J Mol Biol 307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez M, Quiroga JA, Carreno V. 2003. Hepatitis B virus downregulates the human interferon-inducible MxA promoter through direct interaction of precore/core proteins. J Gen Virol 84:2073–2082. doi: 10.1099/vir.0.18966-0. [DOI] [PubMed] [Google Scholar]
- 19.Gruffaz M, Testoni B, Luangsay S, Fusil F, Malika AG, Mancip J, Petit MA, Javanbakht H, Cosset FL, Zoulim F, Durantel D. 2013. The nuclear function of hepatitis B capsid (HBc) protein is to inhibit IFN response very early after infection of hepatocytes. Hepatology 58:276A. [Google Scholar]
- 20.Guo YH, Li YN, Zhao JR, Zhang J, Yan Z. 2011. HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics 6:720–726. doi: 10.4161/epi.6.6.15815. [DOI] [PubMed] [Google Scholar]
- 21.Kwon JA, Rho HM. 2003. Transcriptional repression of the human p53 gene by hepatitis B viral core protein (HBc) in human liver cells. Biol Chem 384:203–212. doi: 10.1515/BC.2003.022. [DOI] [PubMed] [Google Scholar]
- 22.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Xiang A, Ren F, Lei X, Zhang J, Guo R, Lu Z, Guo Y. 2015. The hepatitis B virus (HBV) core protein enhances the transcription activation of CRE via the CRE/CREB/CBP pathway. Antiviral Res 120:7–15. doi: 10.1016/j.antiviral.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Feld JJ, Colledge D, Sozzi V, Edwards R, Littlejohn M, Locarnini SA. 2007. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res 76:168–177. doi: 10.1016/j.antiviral.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Perni RB, Conway SC, Ladner SK, Zaifert K, Otto MJ, King RW. 2000. Phenylpropenamide derivatives as inhibitors of hepatitis B virus replication. Bioorg Med Chem Lett 10:2687–2690. doi: 10.1016/S0960-894X(00)00544-8. [DOI] [PubMed] [Google Scholar]
- 26.Campagna MR, Liu F, Mao R, Mills C, Cai D, Guo F, Zhao X, Ye H, Cuconati A, Guo H, Chang J, Xu X, Block TM, Guo JT. 2013. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J Virol 87:6931–6942. doi: 10.1128/JVI.00582-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick A. 2005. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc Natl Acad Sci U S A 102:8138–8143. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stray SJ, Zlotnick A. 2006. BAY 41-4109 has multiple effects on Hepatitis B virus capsid assembly. J Mol Recognit 19:542–548. doi: 10.1002/jmr.801. [DOI] [PubMed] [Google Scholar]
- 29.Bourne C, Lee S, Venkataiah B, Lee A, Korba B, Finn MG, Zlotnick A. 2008. Small-molecule effectors of hepatitis B virus capsid assembly give insight into virus life cycle. J Virol 82:10262–10270. doi: 10.1128/JVI.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang XY, Wei ZM, Wu GY, Wang JH, Zhang YJ, Li J, Zhang HH, Xie XW, Wang X, Wang ZH, Wei L, Wang Y, Chen HS. 2012. In vitro inhibition of HBV replication by a novel compound, GLS4, and its efficacy against adefovir-dipivoxil-resistant HBV mutations. Antivir Ther 17:793–803. doi: 10.3851/IMP2152. [DOI] [PubMed] [Google Scholar]
- 31.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, Rubsamen-Waigmann H. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 32.Weber O, Schlemmer KH, Hartmann E, Hagelschuer I, Paessens A, Graef E, Deres K, Goldmann S, Niewoehner U, Stoltefuss J, Haebich D, Ruebsamen-Waigmann H, Wohlfeil S. 2002. Inhibition of human hepatitis B virus (HBV) by a novel non-nucleosidic compound in a transgenic mouse model. Antiviral Res 54:69–78. doi: 10.1016/S0166-3542(01)00216-9. [DOI] [PubMed] [Google Scholar]
- 33.Brezillon N, Brunelle MN, Massinet H, Giang E, Lamant C, DaSilva L, Berissi S, Belghiti J, Hannoun L, Puerstinger G, Wimmer E, Neyts J, Hantz O, Soussan P, Morosan S, Kremsdorf D. 2011. Antiviral activity of Bay 41-4109 on hepatitis B virus in humanized Alb-uPA/SCID mice. PLoS One 6:e25096. doi: 10.1371/journal.pone.0025096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Billioud G, Pichoud C, Puerstinger G, Neyts J, Zoulim F. 2011. The main hepatitis B virus (HBV) mutants resistant to nucleoside analogs are susceptible in vitro to non-nucleoside inhibitors of HBV replication. Antiviral Res 92:271–276. doi: 10.1016/j.antiviral.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Berke JM, Tan Y, Verbinnen T, Dehertogh P, Vergauwen K, Vos A, Lenz O, Pauwels F. 2017. Antiviral profiling of the capsid assembly modulator BAY41-4109 on full-length HBV genotype A-H clinical isolates and core site-directed mutants in vitro. Antiviral Res 144:205–215. doi: 10.1016/j.antiviral.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 36.Lam AM, Ren S, Espiritu C, Kelly M, Lau V, Zheng L, Hartman GD, Flores OA, Klumpp K. 2017. Hepatitis B virus capsid assembly modulators, but not nucleoside analogs, inhibit the production of extracellular pregenomic RNA and spliced RNA variants. Antimicrob Agents Chemother 61:e00680-17. doi: 10.1128/AAC.00680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berke JM, Dehertogh P, Vergauwen K, Van Damme E, Mostmans W, Vandyck K, Pauwels F. 2017. Capsid assembly modulators have a dual mechanism of action in primary human hepatocytes infected with hepatitis B virus. Antimicrob Agents Chemother 61:e00560-17. doi: 10.1128/AAC.00560-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo F, Zhao Q, Sheraz M, Cheng J, Qi Y, Su Q, Cuconati A, Wei L, Du Y, Li W, Chang J, Guo JT. 2017. HBV core protein allosteric modulators differentially alter cccDNA biosynthesis from de novo infection and intracellular amplification pathways. PLoS Pathog 13:e1006658. doi: 10.1371/journal.ppat.1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlicksup CJ, Wang JC, Francis S, Venkatakrishnan B, Turner WW, Van Nieuwenhze M, Zlotnick A. 2018. Hepatitis B virus core protein allosteric modulators can distort and disrupt intact capsids. Elife 7:e31473. doi: 10.7554/eLife.31473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belloni L, Palumbo GA, Lupacchini L, Li L, Chirapu SR, Calvo L, Finn MG, Lopatin U, Zlotnick A, Levrero M. 2015. Anti-capsid drugs HAP12 and AT130 target HBV core protein nuclear functions. J Hepatol 62:S513–S514. doi: 10.1016/S0168-8278(15)30736-4. [DOI] [Google Scholar]
- 41.Katen SP, Chirapu SR, Finn MG, Zlotnick A. 2010. Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol 5:1125–1136. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michalak T, Nowoslawski A. 1982. Crystalline aggregates of hepatitis B core particles in cytoplasm of hepatocytes. Intervirology 17:247–252. doi: 10.1159/000149295. [DOI] [PubMed] [Google Scholar]
- 43.Sharma RR, Dhiman RK, Chawla Y, Vasistha RK. 2002. Immunohistochemistry for core and surface antigens in chronic hepatitis. Trop Gastroenterol 23:16–19. [PubMed] [Google Scholar]
- 44.Diab A, Foca A, Fusil F, Lahlali T, Jalaguier P, Amirache F, N′Guyen L, Isorce N, Cosset FL, Zoulim F, Andrisani O, Durantel D. 2017. Polo-like-kinase 1 is a proviral host factor for hepatitis B virus replication. Hepatology 66:1750–1765. doi: 10.1002/hep.29236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lallemand-Breitenbach V, de The H. 2010. PML nuclear bodies. Cold Spring Harb Perspect Biol 2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marion MJ, Hantz O, Durantel D. 2010. The HepaRG cell line: biological properties and relevance as a tool for cell biology, drug metabolism, and virology studies. Methods Mol Biol 640:261–272. doi: 10.1007/978-1-60761-688-7_13. [DOI] [PubMed] [Google Scholar]
- 47.Schulze A, Schieck A, Ni Y, Mier W, Urban S. 2010. Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction. J Virol 84:1989–2000. doi: 10.1128/JVI.01902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klumpp K, Shimada T, Allweiss L, Volz T, Lutgehetmann M, Hartman G, Flores OA, Lam AM, Dandri M. 2018. Efficacy of NVR 3-778, alone and in combination with pegylated interferon, versus entecavir in uPA/SCID mice with humanized livers and HBV infection. Gastroenterology 154:652–662. doi: 10.1053/j.gastro.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 49.Ning X, Nguyen D, Mentzer L, Adams C, Lee H, Ashley R, Hafenstein S, Hu J. 2011. Secretion of genome-free hepatitis B virus: single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog 7:e1002255. doi: 10.1371/journal.ppat.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huber AD, Wolf JJ, Liu D, Gres AT, Tang J, Boschert KN, Puray-Chavez MN, Pineda DL, Laughlin TG, Coonrod EM, Yang Q, Ji J, Kirby KA, Wang Z, Sarafianos SG. 2018. The heteroaryldihydropyrimidine BAY38-7690 induces hepatitis B virus core protein aggregates associated with promyelocytic leukemia nuclear bodies in infected cells. mSphere 3:e00131-18. doi: 10.1128/mSphereDirect.00131-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lang T, Lo C, Skinner N, Locarnini S, Visvanathan K, Mansell A. 2011. The hepatitis B e antigen (HBeAg) targets and suppresses activation of the Toll-like receptor signaling pathway. J Hepatol 55:762–769. doi: 10.1016/j.jhep.2010.12.042. [DOI] [PubMed] [Google Scholar]
- 52.Visvanathan K, Skinner NA, Thompson AJ, Riordan SM, Sozzi V, Edwards R, Rodgers S, Kurtovic J, Chang J, Lewin S, Desmond P, Locarnini S. 2007. Regulation of Toll-like receptor-2 expression in chronic hepatitis B by the precore protein. Hepatology 45:102–110. doi: 10.1002/hep.21482. [DOI] [PubMed] [Google Scholar]
- 53.Chen MT, Billaud JN, Sallberg M, Guidotti LG, Chisari FV, Jones J, Hughes J, Milich DR. 2004. A function of the hepatitis B virus precore protein is to regulate the immune response to the core antigen. Proc Natl Acad Sci U S A 101:14913–14918. doi: 10.1073/pnas.0406282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milich DR, Chen MK, Hughes JL, Jones JE. 1998. The secreted hepatitis B precore antigen can modulate the immune response to the nucleocapsid: a mechanism for persistence. J Immunol 160:2013–2021. [PubMed] [Google Scholar]
- 55.DiMattia MA, Watts NR, Stahl SJ, Grimes JM, Steven AC, Stuart DI, Wingfield PT. 2013. Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein. Structure 21:133–142. doi: 10.1016/j.str.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watts NR, Conway JF, Cheng N, Stahl SJ, Steven AC, Wingfield PT. 2011. Role of the propeptide in controlling conformation and assembly state of hepatitis B virus e-antigen. J Mol Biol 409:202–213. doi: 10.1016/j.jmb.2011.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schodel F, Peterson D, Zheng J, Jones JE, Hughes JL, Milich DR. 1993. Structure of hepatitis B virus core and e-antigen. A single precore amino acid prevents nucleocapsid assembly. J Biol Chem 268:1332–1337. [PubMed] [Google Scholar]
- 58.Bourne CR, Finn MG, Zlotnick A. 2006. Global structural changes in hepatitis B virus capsids induced by the assembly effector HAP1. J Virol 80:11055–11061. doi: 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katen SP, Tan Z, Chirapu SR, Finn MG, Zlotnick A. 2013. Assembly-directed antivirals differentially bind quasiequivalent pockets to modify hepatitis B virus capsid tertiary and quaternary structure. Structure 21:1406–1416. doi: 10.1016/j.str.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Z, Hu T, Zhou X, Wildum S, Garcia-Alcalde F, Xu Z, Wu D, Mao Y, Tian X, Zhou Y, Shen F, Zhang Z, Tang G, Najera I, Yang G, Shen HC, Young JA, Qin N. 2017. Heteroaryldihydropyrimidine (HAP) and sulfamoylbenzamide (SBA) inhibit hepatitis B virus replication by different molecular mechanisms. Sci Rep 7:42374. doi: 10.1038/srep42374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A 99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lecluyse EL, Alexandre E. 2010. Isolation and culture of primary hepatocytes from resected human liver tissue. Methods Mol Biol 640:57–82. doi: 10.1007/978-1-60761-688-7_3. [DOI] [PubMed] [Google Scholar]
- 63.Lucifora J, Bonnin M, Aillot L, Fusil F, Maadadi S, Dimier L, Michelet M, Floriot O, Ollivier A, Rivoire M, Ait-Goughoulte M, Daffis S, Fletcher SP, Salvetti A, Cosset FL, Zoulim F, Durantel D. 2018. Direct antiviral properties of TLR ligands against HBV replication in immune-competent hepatocytes. Sci Rep 8:5390. doi: 10.1038/s41598-018-23525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.