Escherichia coli sequence type 131 (ST131) is currently one of the leading causes of multidrug-resistant extraintestinal infections globally. Here, we analyzed the phenotypic and genotypic characteristics of 169 ST131 isolates from various sources (wildlife, wastewater, companion animals, community, and hospitals) to determine whether wildlife and the environment share similar strains with humans, supporting transmission of ST131 between different ecological niches.
KEYWORDS: Escherichia coli ST131, ESBL, virulence, nosocomial and community-acquired infections, wildlife, environment
ABSTRACT
Escherichia coli sequence type 131 (ST131) is currently one of the leading causes of multidrug-resistant extraintestinal infections globally. Here, we analyzed the phenotypic and genotypic characteristics of 169 ST131 isolates from various sources (wildlife, wastewater, companion animals, community, and hospitals) to determine whether wildlife and the environment share similar strains with humans, supporting transmission of ST131 between different ecological niches. Susceptibility to 32 antimicrobials was tested by disc diffusion and broth microdilution. Antibiotic resistance genes, integrons, plasmid replicons, 52 virulence genes, and fimH-based subtypes were detected by PCR and DNA sequencing. Genomic relatedness was determined by pulsed-field gel electrophoresis (PFGE). The genetic context and plasmid versus chromosomal location of extended-spectrum beta-lactamase and AmpC beta-lactamase genes was determined by PCR and probe hybridization, respectively. The 169 ST131 study isolates segregated predominantly into blaCTX-M-15 H30Rx (60%) and blaCTX-M-27 H30R1 (25%) subclones. Within each subclone, isolates from different source groups were categorized into distinct PFGE clusters; genotypic characteristics were fairly well conserved within each major PFGE cluster. Irrespective of source, the blaCTX-M-15 H30Rx isolates typically exhibited virotype A (89%), an F2:A1:B− replicon (84%), and a 1.7-kb class 1 integron (92%) and had diverse structures upstream of the blaCTX-M region. In contrast, the blaCTX-M-27 H30R1 isolates typically exhibited virotype C (86%), an F1:A2:B20 replicon (76%), and a conserved IS26-ΔISEcp1-blaCTX-M-like structure. Despite considerable overall genetic diversity, our data demonstrate significant commonality between E. coli ST131 isolates from diverse environments, supporting transmission between different sources, including humans, environment, and wildlife.
INTRODUCTION
Escherichia coli sequence type 131 (ST131) currently is one of the most important globally disseminated bacterial lineages, causing severe hospital-acquired and community-onset multidrug-resistant (MDR) infections and contributing to the global spread of blaCTX-M-15 and other extended-spectrum beta-lactamase (ESBL) genes (1–3). A substantial level of virulence, combined with carriage of plasmids with transferable antimicrobial resistance-encoding elements, likely has facilitated this clone's pandemic success (1–4).
Discriminatory molecular typing methods have elucidated the fine clonal structure of ST131 by resolving various epidemiologically important subclones. H30, the most prevalent subclone and the main driver of the recent worldwide ST131 expansion, was so designated based on its association with allele 30 of the type 1 fimbrial adhesin gene fimH (1, 2). Within H30, whole-genome sequencing has resolved two main subclones, H30R1 and H30Rx (also called clades C1 and C2, respectively), which comprise nearly all quinolone-resistant ST131 isolates (1, 5–7). The globally distributed H30Rx subset is closely associated with blaCTX-M-15 and with extensive antimicrobial resistance (5, 8–12). In contrast, although most H30R1 subclone isolates are ESBL negative, some of them harbor blaCTX-M-27 or blaCTX-M-14 (11).
Although ST131 exhibits a narrower range of virulence factor-encoding genes than does the species overall (3, 13), significant within-ST131 variation in virulence gene content is evident, leading to the delineation of specific ST131 virotypes, designated A to D (13). Virotype C is considered to be the most widespread (3), but virotype A has predominated among H30Rx isolates in several recent studies from Europe (8, 10). ST131 also is highly diverse with respect to pulsed-field gel electrophoresis (PFGE) genomic DNA profiles (14). Certain highly prevalent pulsotypes (e.g., 968, 800, and 812) are globally distributed (8, 12, 14).
Mobile genetic elements play a key role in the dissemination of virulence and antibiotic resistance generally, and IncF plasmids in particular likely have contributed to ST131's success (4, 15). IncF plasmids with a complex mosaic structure consisting of multiple plasmid replicons, typically FII, FIA, and/or FIB, dominate within ST131. This is probably due to unique features such as combinations of virulence genes and diverse antibiotic resistance determinants that contribute to bacterial fitness, plus postsegregational killing systems that ensure their maintenance and propagation in the bacterial cell (2–4, 15). ISEcp1-blaCTX-M-orf477 and ISEcp1-blaCTX-M-IS903 are two major genetic platforms described in Enterobacteriaceae in association with blaCTX-M genes (5, 11, 15). These platforms can be inserted at both plasmidic and chromosomal sites, and their structure is frequently interrupted by IS26 elements that are inserted upstream and downstream of the blaCTX-M gene (4, 5, 15).
Although ST131 is known mainly for causing infections in humans, including urinary tract infections, bloodstream infections, and neonatal sepsis, it has also been identified in companion animals, poultry, livestock, wild animals, and food (3, 16). Its presence in food, water, the environment, and other nonhuman sources suggests a minimally explored complexity of potential transmission routes, whereby migrating birds and wastewater may play an important role in ST131's distribution and circulation. To address these unknowns, we compared the phenotypic, genotypic, and clonal characteristics of 169 E. coli ST131 isolates of diverse origins, including human community and nosocomial sources (n = 130), wastewater (n = 19), dogs (n = 3), and wild birds (n = 17), to determine whether wild animals and the environment share highly similar strains with humans, implying exchange of ST131 strains between different ecological niches.
RESULTS
Plasmid-mediated resistance genes, integrons, replicons, fimH30 subtyping, and antimicrobial resistance.
In total, 169 ST131 E. coli isolates were analyzed, including 151 ESBL-positive, one AmpC beta-lactamase-positive, and 17 ESBL-negative, AmpC-negative isolates. All but three (166/169, 98%) were type O25b, and nearly all of the ESBL-positive isolates carried either blaCTX-M-15 (106/169, 63%) or blaCTX-M-27 (42/169, 25%) (Fig. 1; see also Fig. S1 in the supplemental material). blaCTX-M-15-positive isolates frequently also contained blaOXA-1 (92/106, 87%) and aac(6′)-Ib-cr (93/106, 88%), in some instances accompanied by blaTEM-1 (26/106, 25%) and/or the quinolone resistance gene qnrB1 (5/106, 5%), whereas most blaCTX-M-27-positive isolates (33/42, 79%) carried no additional beta-lactam or quinolone resistance genes (see Fig. S1 in the supplemental material). Likewise, a 1.7-kb class 1 integron that contained dfrA17 and aadA5 gene cassettes was significantly more frequent among blaCTX-M-15-positive isolates than those with blaCTX-M-27 (96/106, 91%, versus 20/42, 48%: P < 0.001) (Fig. 1).
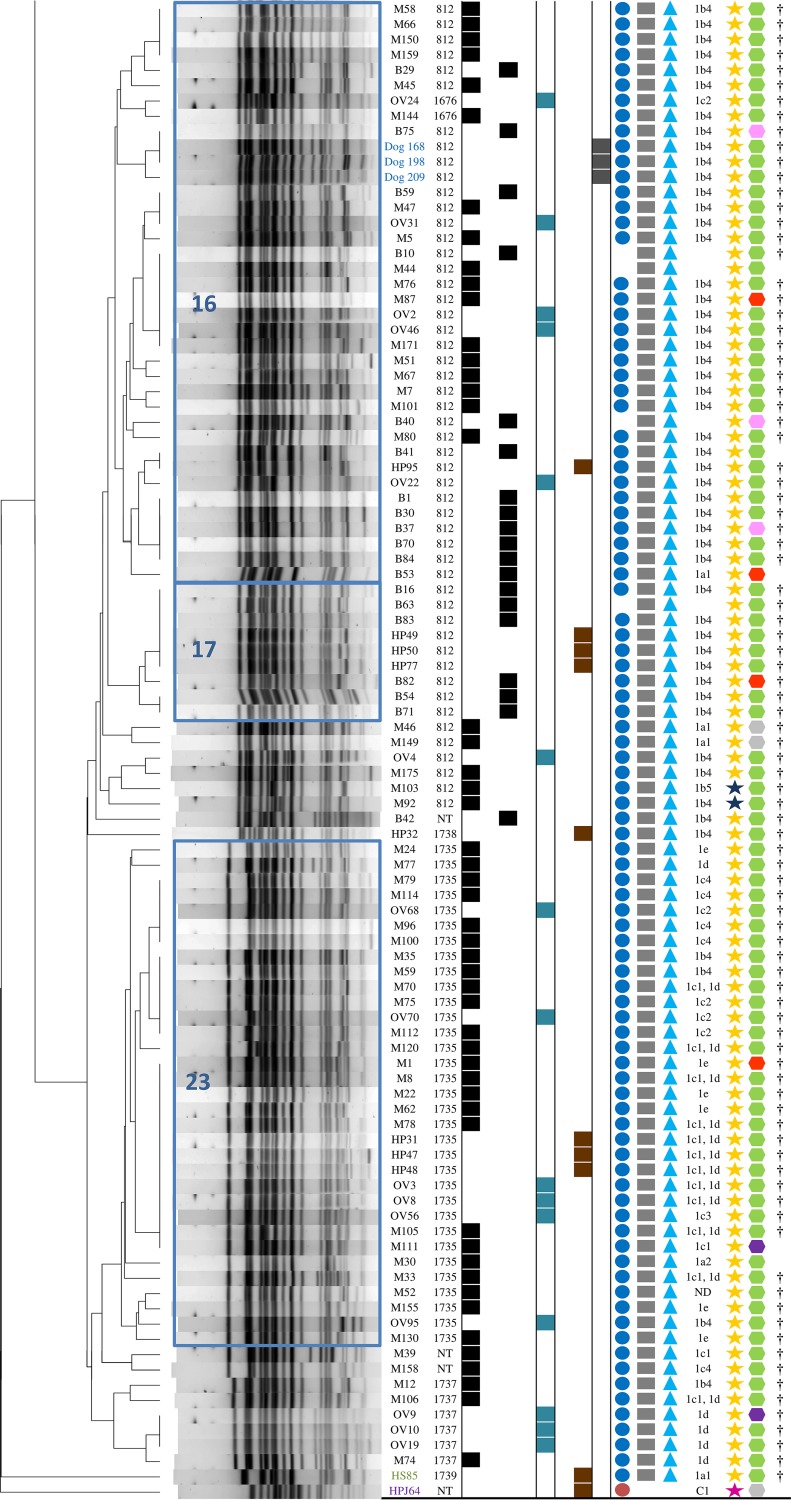
FIG 1.
The clonality and common characteristics of representatives of 169 Escherichia coli ST131 from various sources. The superscript notations in the figure are defined as follows. aNT, not typeable using reference pulsotypes 788, 797, 800, 805, 806, 812, 837, 842, 903, 905, 906, 943, 945, 968, 987, 1067, 1140, 1676, 1733, 1734, 1735, 1737, 1738, and 1739. bHuman clinical isolates: hospital 1, St. Anne's Faculty Hospital, Brno, Czech Republic (CZ); hospital 2, Children's Medical Center, Brno, CZ; hospital 3, University Hospital Motol, Prague, CZ; Community: human community isolate, Karvina, CZ. cWWTP, wastewater treatment plant. d,g,iAn empty box indicates that no corresponding gene was detected. eFQ, fluoroquinolones (an empty box indicates that the isolate was not resistant to fluoroquinolone antibiotics). f,hAn empty box indicates that the isolate could not be assigned to particular subclone or virotype. gThe structure of blaCTX-M and blaCMY genetic environment is displayed in Fig. 2; ND, not determined.
All but one of the study isolates (168/169, 99%) belonged to the ST131-H30 subclone and were accounted for by its H30Rx (109/169, 64%) and H30R1 (59/169, 35%) subsets. blaCTX-M-15 was disproportionately prevalent among H30Rx isolates (102/109 [94%], H30Rx, versus 7 [6%], others: P < 0.001), whereas blaCTX-M-27 was disproportionately prevalent among H30R1 isolates (42/59 [71%], H30R1, versus 17 [29%], others: P < 0.001) (Fig. 1).
Resistance was detected to all individual agents except meropenem and ertapenem, and 140 (83%) of the 169 isolates were MDR (see Fig. S1 in the supplemental material). blaCTX-M-15-positive H30Rx isolates exhibited a significantly higher prevalence of resistance to several antimicrobial agents than did the blaCTX-M-27 H30R1 isolates (Table 1; see also Fig. S1 in the supplemental material).
TABLE 1.
Characteristics of two major subgroups among 144 diverse-source blaCTX-M-positive E. coli ST131 isolates
| Category | Specific characteristic | Prevalence of characteristics, no. of isolates (column %) |
Pa (blaCTX-M-15 vs blaCTX-M-27) | |
|---|---|---|---|---|
| blaCTX-M-15, H30Rx (n = 102) | blaCTX-M-27, H30R1 (n = 42) | |||
| Antimicrobial resistanceb | Ampicillin | 102 (100) | 42 (100) | |
| Piperacillin | 101 (99) | 42 (100) | ||
| Cefazolin | 101 (99) | 42 (100) | ||
| Cefuroxime | 102 (100) | 42 (100) | ||
| Cefoxitin | 4 (4) | 0 (0) | ||
| Ceftazidime | 58 (57) | 4 (10) | <0.001 | |
| Cefotaxime | 102 (100) | 42 (100) | ||
| Cefoperazone | 77 (75) | 31 (74) | ||
| Cefepime | 71 (70) | 4 (10) | <0.001 | |
| Aztreonam | 88 (86) | 15 (36) | <0.001 | |
| Amoxicillin-clavulanate | 93 (91) | 2 (5) | <0.001 | |
| Ampicillin-sulbactam | 79 (77) | 2 (5) | <0.001 | |
| Piperacillin-tazobactam | 32 (31) | 0 (0) | <0.001 | |
| Nalidixic acid | 102 (100) | 42 (100) | ||
| Ciprofloxacin | 101 (99) | 42 (100) | ||
| Ofloxacin | 102 (100) | 42 (100) | ||
| Trimethoprim-sulfamethoxazole | 92 (90) | 21 (50) | <0.001 | |
| Tetracycline | 82 (80) | 20 (48) | <0.001 | |
| Doxycycline | 80 (78) | 20 (48) | <0.001 | |
| Sulfonamides | 95 (93) | 22 (52) | <0.001 | |
| Trimethoprim | 95 (93) | 21 (50) | <0.001 | |
| Azithromycin | 95 (93) | 21 (50) | <0.001 | |
| Streptomycin | 95 (93) | 21 (50) | <0.001 | |
| Tobramycin | 86 (84) | 0 (0) | <0.001 | |
| Gentamicin | 5 (5) | 0 (0) | ||
| Amikacin | 26 (25) | 0 (0) | <0.001 | |
| Nitrofurantoin | 4 (4) | 0 (0) | ||
| Tigecycline | 5 (5) | 2 (5) | ||
| Colistin | 1 (1) | 0 (0) | ||
| Other resistance genes | blaOXA-1 | 88 (86) | 0 (0) | <0.001 |
| blaTEM-1 | 22 (22) | 4 (10) | ||
| aac(6′)-Ib-cr | 89 (87) | 6 (14) | <0.001 | |
| qnrB1 | 1 (1) | 0 (0) | ||
| Integronsc | IntIA (1.7 kb) | 94 (92) | 20 (48) | <0.001 |
| IntIB (0.8 kb) | 0 (0) | 1 (2) | ||
| Replicons | F2:A1:B− | 86 (84) | 0 (0) | <0.001 |
| F1:A2:B20 | 0 (0) | 32 (76) | <0.001 | |
| F1:A2:B− | 0 (0) | 5 (12) | <0.001 | |
| Virulence genotype | Virotype A | 91 (89) | 1 (2) | <0.001 |
| Virotype C | 8 (8) | 36 (86) | <0.001 | |
| Virotype undefined | 3 (3) | 5 (12) | 0.03 | |
| ExPECd | 102 (100) | 37 (88) | <0.001 | |
P values (determined by chi-square test) are shown where P < 0.05.
No isolate was resistant to meropenem, ertapenem, or chloramphenicol.
That is, IntI1A, class 1 integron (1.7 kb), dfrA17-aadA5, and IntI1B (0.8 kb), class 1 integron, dfrA5.
ExPEC, extraintestinal pathogenic E. coli.
Genetic environment of blaCTX-M and blaCMY.
According to PCR mapping and sequencing, the genetic context of the region upstream of blaCTX-M and blaCMY could be divided into several groups (Fig. 1 and 2).
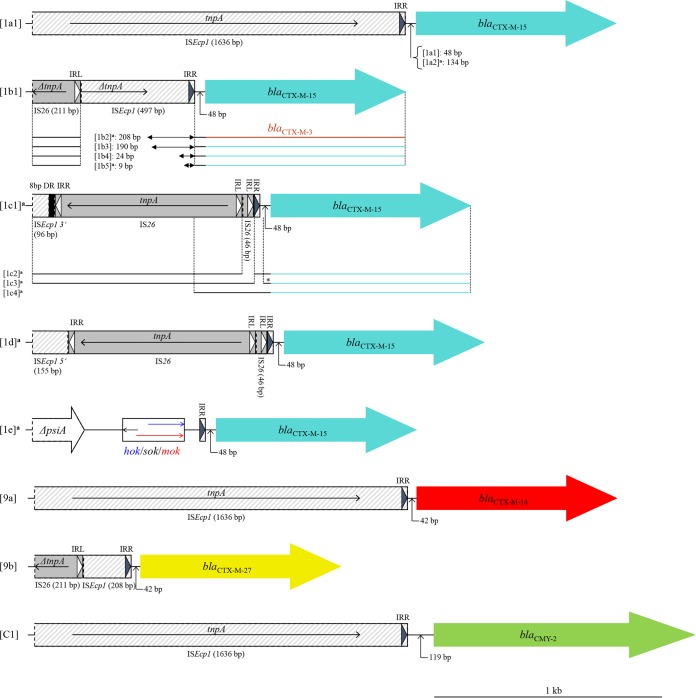
FIG 2.
Genetic environment of blaCTX-M and blaCMY-2 groups among E. coli ST131 isolates. A dotted line in a gene box indicates truncation. IRR, right-hand inverted repeat; IRL, left-hand inverted repeat; DR, direct repeat of partial ISEcp1 inverted repeat; *, shortened spacer (32 bp) in variant 1c3; white triangle, IS26 inverted repeat; black triangle, ISEcp1 inverted repeat. aNovel genetic environment structure found in this study (no identical sequence was deposited in GenBank).
The great majority of blaCTX-M-15-positive isolates (87/106, 82%) had ISEcp1 truncated by IS26 in different positions, with remnant parts of various lengths (Fig. 2). The most prevalent organization, formed by IS26 upstream of a 24-bp remnant of ISEcp1 (group 1b4 in Fig. 2), occurred in 51/106 (48%) isolates (Fig. 1).
Here, the IS26-ΔISEcp1-blaCTX-M-like platform was also very common among blaCTX-M-27-positive isolates. The dominant arrangement consisted of 208-bp remnant of ISEcp1 flanked by IS26 upstream of blaCTX-M-27 (Fig. 1; group 9b in Fig. 2).
The one AmpC-positive ST131 isolate contained an intact ISEcp1 119-bp upstream of blaCMY-2 (Fig. 2).
Plasmid analysis and hybridization of ESBL genes.
All isolates contained at least one IncF replicon, most commonly F2:A1:B− (92/169, 54%), followed by F1:A2:B20 (40/169, 24%) and F1:A2:B− (9/169, 5%). The F2:A1:B− replicon was associated with blaCTX-M-15 (86/92, 93%) and H30Rx (92/92, 100%), whereas the F1:A2:B20 and F1:A2:B− replicons were associated with H30R1 (49/49, 100%) and blaCTX-M-27 (32/40, 80%, and 5/9, 56%, respectively) (Fig. 1).
In total, 49 isolates, representing different sources, subclones, molecular characteristics, plasmid groups, and PFGE clusters, were subjected to S1 PFGE and probe hybridization to determine the plasmidic versus chromosomal location of blaCTX-M (see Table S1 and Fig. S1 in the supplemental material). blaCTX-M was located on an IncF plasmid in 33 isolates, on plasmids other than IncF in 4, on the chromosome in 10, and on both plasmid and chromosome in 2 (see Table S1 and Fig. S1 in the supplemental material). Overall, blaCTX-M-15 was located mostly on F2:A1:B− plasmids in H30Rx strains, while blaCTX-M-27 with F1:A2:B20 plasmids in H30R1 strains (see Table S1 and Fig. S1 in the supplemental material).
Virulence genes.
Based on the presence or absence of specific virulence genes, isolates were classified into established ST131 virotypes. Of these, virotype A was the most common, occurring in 97 isolates (57% of 169), followed by virotype C (60/169, 36%) and virotype D4 (1/169, 1%). Virotype A was associated mainly with H30Rx (95/97, 98%), and virotype C with H30R1 (49/60, 82%) (Fig. 1). Based on the criteria of Dahbi et al. (10), 11 isolates could not be categorized into any of the established virotypes.
In addition, cluster analysis was used to identify associations among all 52 detected virulence genes. Based on unique combinations of virulence genes, the isolates were divided into 20 clusters (according to 100% similarity). The largest such cluster included 74 isolates (44% of 169) with a unique set of 11 virulence genes (afa/dra, iha, fimH, sat, fyuA, iutA, kpsII, traT, usp, ompT, and malX) that corresponded to virotype A. These isolates carried almost exclusively blaCTX-M-15 (71/74; 96%) or were ESBL negative (3/74; 4%), and all represented the H30Rx subclone. In addition, 70 (95% of 74) and 68 (92%) also harbored a 1.7-kb class 1 integron and the F2:A1:B− replicon, respectively (see Fig. S2 in the supplemental material).
The second largest cluster, which comprised 44 H30R1 isolates (26% of 169), contained mainly blaCTX-M-27-positive isolates (34/44, 77%) with a different unique set of 11 virulence genes (iha, fimH, sat, fyuA, iutA, kpsII, kpsM II-K5, traT, usp, ompT, and malX), that corresponded to virotype C. Of these isolates, 27 (61% of 44) harbored an F1:A2:B20 replicon, and 19 (43%) harbored a 1.7-kb class 1 integron (see Fig. S2 in the supplemental material).
Isolates classified as extraintestinal pathogenic E. coli (ExPEC; 153/169, 91%) were distributed in all source groups (see Fig. S1 in the supplemental material), but predominantly among blaCTX-M-15 H30Rx isolates (Table 1; see also Fig. S1 in the supplemental material).
PFGE and distribution of pulsotypes by source group.
PFGE was used for genomic comparison between isolates in relation to source group. Among the 169 study isolates, PFGE analysis yielded 27 unique pulsotypes at the 91% similarity level (Fig. 1). The PFGE profiles of 148 (88%) of the 169 study isolates corresponded with established human-associated international pulsotypes (14), including, in order of descending prevalence (no., percentage of 169), pulsotypes 812 (51, 30%), 1735 (33, 20%), 1733 (20, 12%), 788 (16, 9%), 1676 (9, 5%), 1737 (6, 4%), 1067 (5, 3%), and 945 (3, 2%) comprising >1 isolate (Table 2).
TABLE 2.
Distribution of 169 E. coli ST131 isolates by pulsotype, ecological source, and location
| Pulsotype | Total (%) | Sourcea |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Humans (CZ)b |
WWTPc | Wild birds |
Companion animals (dogs) | ||||||
| H1 | H2 | H3 | CO | Cormorantsd | Corvids | ||||
| 788 | 16 (9) | 15 | 1 (CZ-1) | ||||||
| 812 | 51 (30) | 21 | 18 | 5 (CZ) | 4 (CZ-2) | 3 (KE) | |||
| 943 | 1 (1) | 1 (CZ) | |||||||
| 945 | 3 (2) | 1 | 2 (SB) | ||||||
| 968 | 1 (1) | 1 (CZ) | |||||||
| 1067 | 5 (3) | 1 | 3 | 1 (SL) | |||||
| 1676 | 9 (5) | 7 | 1 (CZ) | 1 (US) | |||||
| 1733 | 20 (12) | 17 | 1 (CZ) | 2 (CZ) | |||||
| 1734 | 1 (1) | 1 (US) | |||||||
| 1735 | 33 (20) | 24 | 6 (CZ) | 3 (CZ-2) | |||||
| 1737 | 6 (4) | 3 | 3 (CZ) | ||||||
| 1738 | 1 (1) | 1 (CZ-2) | |||||||
| 1739 | 1 (1) | 1 (SB) | |||||||
| NTe | 21 (12) | 4 | 1 | 14 | 1 | 1 (PL) | |||
| Total | 169 | 93 | 1 | 35 | 1 | 19 | 2 | 15 | 3 |
CZ, Czech Republic; KE, Kenya; PL, Poland; SB, Serbia; SL, Slovakia; US, United States. CZ-1, Zidlochovice, 17 km from Brno, CZ; CZ-2, Přerov, 67 km from Brno, CZ.
H, Human clinical isolates, all from the Czech Republic: H1, hospital 1, St. Anne's Faculty Hospital, Brno, CZ; H2, hospital 2, Children's Medical Center, Brno, CZ; H3, hospital 3, University Hospital Motol, Prague, CZ; CO, human community isolate, Karvina, CZ.
WWTP, wastewater treatment plant (municipal), Brno-Modrice, CZ.
Nature Reserve Oskovec, Straznice, CZ, 58 km from Brno, CZ.
NT, not typeable using reference pulsotypes 788, 797, 800, 805, 806, 812, 837, 842, 903, 905, 906, 943, 945, 968, 987, 1067, 1140, 1676, 1733, 1734, 1735, 1737, 1738, and 1739.
In the PFGE dendrogram, 123 isolates (73% of 169) were grouped within the five main clusters, arbitrarily labeled 7, 8, 16, 17, and 23. Of these, clusters 16, 17, and 23 accounted collectively for 80 (47% of 169) isolates, all from the H30Rx subclone (Fig. 1). These three clusters had in common multiple accessory traits, including blaCTX-M-15 (76/80, 95%), 1.7-kb class 1 integrons (75/80, 94%), F2:A1:B− replicons (72/80, 90%), and virulence genes that corresponded to virotype A (80/80, 100%) (Fig. 1). The main genetic difference distinguishing clusters 16 and 17 from cluster 23 was in the region upstream of blaCTX-M-15. Specifically, the most common genetic environment for blaCTX-M-15 in clusters 16 and 17 included group 1b4 (41/43, 95%) (Fig. 1 and 2), while in cluster 23 various structures were identified (Fig. 1). In addition, the consensus PFGE profile of clusters 16 and 17 corresponded with international pulsotype 812 (45/47, 96%), and that of cluster 23 with pulsotype 1735 (33/33, 100%). All three clusters contain isolates from multiple sources, including three canine isolates from Kenya, with considerable overlap of sources among clusters (Fig. 1).
In contrast, clusters 7 and 8 comprised collectively 43 isolates, all from to H30R1 subclone (43/43, 100%). These two clusters were characterized by multiple shared traits, including blaCTX-M-27 (42/43, 98%), the F1:A2:B20 replicon (32/43, 74%), a conserved set of virulence genes that corresponded with virotype C (37/43, 86%), and a conserved region upstream of blaCTX-M that represented group 9b (42/43, 98%) (Fig. 1 and 2). Cluster 7 was divided into subclusters 7a and 7b, which had consensus PFGE profiles corresponding with international pulsotypes 1733 (20/20, 100%) and 788 (16/16, 100%), respectively. In contrast to the cluster 7a isolates, which frequently carried 1.7-kb class 1 integron (19/20, 95%) and were MDR, most cluster 7b and 8 isolates carried no integron and were less extensively resistant (Fig. 1; see also Fig. S1 in the supplemental material). All three clusters showed a broad and overlapping source distribution.
DISCUSSION
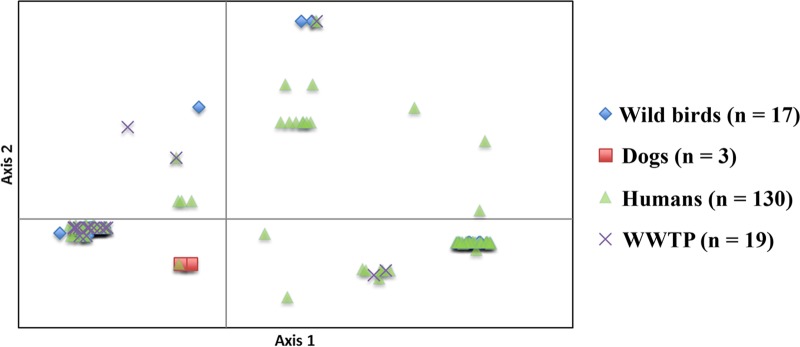
E. coli ST131 represents one of the most medically important and globally widespread clonal lineages and is a major challenge to public health worldwide. However, it remains unknown to what extent and through which pathways these pathogenic bacteria are transmitted between different ecological niches. Here, accessory traits, including virulence gene profiles, plasmid replicons, integron content, and antimicrobial resistance profiles, were fairly well conserved within each major PFGE cluster but differed between clusters. Importantly, genetic and phylogenetic structure of studied ST131 population did not segregate according to source group (Fig. 1 and 3 and Table 2). These broad genetic commonalities across sources strongly suggest an exchange of highly genetically related strains between sources groups (Fig. 3), consistent with the existence of environmental (wastewater) reservoirs and animal (wildlife) vectors for strains of human health importance. Furthermore, the presence of the H30R1 and H30Rx subclones among environmental and animal isolates, which are the most highly prevalent subclones of ST131 among human-source isolates (5), strongly supports our hypothesis of transmission of this clone between different ecological niches.
FIG 3.
Principal-coordinate analysis (PCoA) of virulence genes, O-type, and H30 subclone designation among 169 E. coli ST131 isolates. The PCoA was based on results of all 52 virulence genes, O25b, and H30R1 and H30Rx status. Each isolate is plotted according to its value for PCoA coordinates 1 (horizontal axis) and 2 (vertical axis), where coordinates 1 and 2 capture 55.6% and 18.1% of the total variation, respectively. Based on PCoA, the genotypes and phylogeny structure of ST131 overlap extensively across the source groups.
In addition, isolates obtained from different hospitals (H1 and H3) in different regions of Czech Republic showed indistinguishable pulsotypes and represented the internationally important H30R1 and H30Rx subclones, suggesting the successful spread of these lineages within Czech health care facilities. Here, blaCTX-M-15-positive H30Rx ST131 isolates predominated, as also reported in multiple studies of human-source isolates from the community and health care facilities in different continents (5, 8–10, 12). In contrast, blaCTX-M-27-containing H30R1 isolates, the second largest ST131 subset in our study population, were detected in just one of three sampled Czech hospital. This hospital contributed multiple independent isolates of what proved to be the same strain (hospital H1; 37 H30R1 isolates among 93 total ST131 isolates, each from a different patient), suggesting local spread within this facility. Furthermore, our blaCTX-M-27-containing H30R1 isolates shared extensive genetic commonalities with isolates previously reported from Japanese and Korean hospitals (11, 17), suggesting that Asia might be the original source of this clone.
In the PFGE dendrogram, although the H30Rx and H30R1 isolates generally clustered by subclone, this segregation was incomplete, and certain isolates clustered with different subclones, indicating the limitation of PFGE for strain typing (5). Whole-genome sequence analysis should allow more accurate resolution of phylogenetic relationships (5, 18).
The strong associations we found of blaCTX-M-15 with the F2:A1:B− multireplicon (19, 20) and the H30Rx subclone (4), and of blaCTX-M-27 with the F1:A2:B20 replicon (19, 20) and the H30R1 subclone (4), have been documented in multiple previous studies. This indicates strong association of particular plasmid types and certain ST131 subclones. Since IncF plasmids likely contribute to ST131's success, further plasmid characterization may help to elucidate the relationship between particular plasmids and the epidemic success of high-risk clonal lineages.
In Enterobacteriaceae, blaCTX-M-15 has been reported typically as a part of an ISEcp1-blaCTX-M-15-orf477 transposition unit, nested within a Tn2 element (15), whereas blaCTX-M-27 typically occurs within ISEcp1-blaCTX-M-IS903 (11, 15). The intact ISEcp1 48-bp upstream of blaCTX-M-15 is considered the most common structure distributed globally (11, 15). However, we identified this arrangement only sporadically among our isolates; most blaCTX-M-15-positive isolates had ISEcp1 truncated by IS26 in different positions, with remnant parts of various lengths (Fig. 2). The most prevalent organization was formed by IS26 upstream of a 24-bp remnant of ISEcp1 (group 1b4 in Fig. 2), as identified previously in clinical isolates and in the UK epidemic ST131 “strain A” (20), in E. coli isolates from travelers returning from India (21), and in H30Rx isolates from Japanese hospitals (11).
The IS26-ΔISEcp1-blaCTX-M-like platform was also prevalent among blaCTX-M-27-positive isolates. The dominant organization consisted of 208-bp remnant of ISEcp1 flanked by IS26 upstream of blaCTX-M-27 (group 9b in Fig. 2), as reported previously in most blaCTX-M-27-positive H30 isolates from Japan (11).
Virotype C is considered to be the most broadly distributed virotype within ST131, occurring in both the H30R1 and H30Rx subclones (3, 9, 13). Here, virotype C predominated among blaCTX-M-27-positive H30R1 isolates, as also reported recently from Japan (11). In contrast, virotype A was found by several European studies to predominate among H30Rx isolates in association with human-associated pulsotype 812, corresponding with the UK epidemic “strain A” (8, 12, 13). These findings, in conjunction with our data, suggest that virotype A might be highly prevalent in Europe among blaCTX-M-15-H30Rx isolates, most likely due to the wide dissemination of international pulsotype 812. Here, pulsotype 812 was the most common, observed in isolates from all investigated sources in the Czech Republic. Moreover, we uniquely identified it in stray dogs from Africa and corvids from the United States (Fig. 1 and Table 2), indicating its broad geographical and host species distribution.
Collectively, these findings indicate that ST131 may spread successfully via different pathways from humans to wildlife and the environment, pointing out the capability of ST131 to survive and thrive in diverse hosts and ecological niches without direct antimicrobial selective pressure. Several potential transmission routes can be considered. Surface waters such as rivers and lakes, which contain water originating from diverse sources (e.g., wastewater treatment plants [WWTPs], urban or industrial effluents, and agricultural activities), may represent important reservoirs of antibiotic-resistant bacteria that might further disseminate to diverse ecological niches (22). It was estimated that each day a single WWTP can release into surface waters staggering numbers (>600 billion cells) of ESBL-producing E. coli, including epidemiologically important clones (23). The presence of ST131 isolates containing blaCTX-M-27 has been reported in Europe from diverse aquatic environments such as rivers and lakes (23, 24), and the intestines of freshwater fish (24).
Here, similar PFGE profiles were found among blaCTX-M-27-positive H30R1 isolates from hospital H1, WWTPs, and wild waterfowl (great cormorants). The foraging behavior of such waterfowl is strongly associated with aquatic environments, and all of these sources are located in close proximity to each other within the South Moravian Region in the Czech Republic. Although the direction of transmission is conjectural, these findings support that WWTP effluents contaminated by isolates of human origin may influence the commensal microbiota of water-associated animals such as waterfowl that live, breed, and forage in or near such contaminated aquatic environments.
We also found that corvids, which are synanthropic omnivorous birds that live in close contact with humans in habitats highly influenced by human activities (25, 26), were colonized by the (human-associated) ST131-H30Rx subclone (5). This could result from such birds' foraging behavior, which includes seeking for food in settings that may contain abundant resistant bacteria, e.g., fields that have been fertilized by sewage sludge or different types of animal waste, or various human or animal waste depots such as landfills (25, 26).
Related to this, in France, 7,500 metric tons of sewage sludge containing 2.6 × 105 ESBL-producing E. coli per gram is produced annually by one WWTP and is used as fertilizer on agricultural fields (23). ST131 has been reported from food-producing and companion animals (3, 16). Several studies recovered ESBL/AmpC-producing strains from food-producing animal feces, their housing environment, manure slurry, and fertilized fields near the poultry, pig, and cattle farms (27–29). This application of slurry to farm fields as a fertilizer raised major biosafety concerns, since slurry exhibited the highest detection rate of ESBL/AmpC-producing E. coli among the investigated samples (27, 28), and bacteria with PFGE profiles indistinguishable from those of slurry-derived isolates could be detected on the field surfaces for several months after slurry application (27, 29). Therefore, such a contaminated environment might become a source of MDR bacteria for wildlife that feed in these habitats.
In summary, we found that our ST131 isolates from diverse ecological sources exhibited broad genetic commonality for multiple accessory traits and phylogenetic background, providing strong evidence that this pandemic clone spreads between different ecological niches, including humans, the environment, and wildlife. This presumably creates additional environmental reservoirs and vectors for strains of human medical importance, e.g., H30R1 and H30Rx, and may contribute to the global dissemination of these MDR pathogens. Because the expansion of H30R1 and H30Rx subclones has had a major impact on human health, interruption of such dissemination and elimination of relevant reservoirs should be a public health priority.
MATERIALS AND METHODS
Isolates.
The study population was a convenience sample of 169 E. coli ST131 isolates, some previously published (25, 26, 30–34), as obtained from diverse sources and geographical locations. We compared isolates of human origin (n = 130) with isolates from synanthropic (avian symbionts of humans) wild bird species (n = 17) and companion animals (n = 3), which live in close contact with humans and thus have an elevated likelihood of colonization by human-source bacteria. We also compared them with isolates from WWTP effluent (n = 19) that was discharged into surface waters, which could serve as a connecting link between humans, the environment, and wildlife. Most of the isolates (73%, 123/169) were collected within the South Moravian Region in the Czech Republic; however, international isolates also were included. Most study isolates were obtained by selective cultivation on media supplemented with cefotaxime (2 mg/liter) and/or were identified as ESBL producers. To analyze the commonalities between different ST131 populations, quinolone-resistant or fully susceptible isolates were included. Detailed isolate characteristics (e.g., source, sampling location, isolation method, sampling date, etc.) are listed in Table 3 and also Table S2 in the supplemental material.
TABLE 3.
Source characteristics of the 169 E. coli ST131 study isolates
| Origin | Location | Type of studya | Sampling period | No. of isolates (n = 169) | Source or reference |
|---|---|---|---|---|---|
| Czech Republic | 159 | ||||
| Humansc | |||||
| Hospital 1 (H1) | South Moravian Region: Brno | ESBL E. coli from patients with UTIs | 2008–2011 | 93 | 33 |
| Hospital 2 (H2) | South Moravian Region: Brno | ESBL E. coli from patient with febrile neutropenia | 2009 | 1 | 31 |
| Hospital 3 (H3) | Prague Region: Prague | FQ-R E. coli from urine samples | 2011–2012 | 35 | NPb |
| Community (CO) | Moravian-Silesian Region: Karvina | E. coli from cultivation on MCA | 2012 | 1 | NP |
| WWTPd | |||||
| Treated municipal wastewaters | South Moravian Region: Brno | E. coli from selective cultivation on MCACTX | 2008–2009 | 18 | 30 |
| Wildlife | |||||
| Rooks | South Moravian Region: Prerov | E. coli from selective cultivation on MCACTX | 2010 | 8 | 25 |
| Rooks | South Moravian Region: Zidlochovice | E. coli from selective cultivation on MCACTX | 2012 | 1 | NP |
| Great cormorants | South Moravian Region: Straznicee | E. coli from selective cultivation on MCACTX | 2008 | 2 | 32 |
| International isolates | 10 | ||||
| WWTP | |||||
| Treated municipal wastewaters | Slovakia | E. coli from cultivation on MCA | 2011–2013 | 1 | NP |
| Wildlife | |||||
| Rooks | Poland | E. coli from selective cultivation on MCACTX | 2011 | 1 | 25 |
| Rooks | Serbia | E. coli from selective cultivation on MCACTX | 2011 | 3 | 25 |
| American crows | United States | E. coli from selective cultivation on MCACTX | 2012 | 2 | 26 |
| Companion animals | |||||
| Dogs | Kenya | E. coli from selective cultivation on MCACTX | 2009 | 3 | 34 |
MCA, MacConkey agar; MCACTX, MacConkey agar supplemented with cefotaxime (2 mg/liter); ESBL, extended-spectrum beta-lactamases; UTI, urinary tract infection; FQ-R, fluoroquinolone resistant.
NP, not published.
H, human clinical isolates: H1, St. Anne's Faculty Hospital, Brno, CZ; H2, Children's Medical Center, Brno, CZ; H3, University Hospital Motol, Prague, CZ; CO: human community isolate, Karvina, CZ.
WWTP, wastewater treatment plant.
Nature Reserve Oskovec.
Molecular typing methods.
ST131 status was verified for all isolates by PCR-based detection of ST131-specific SNPs in mdh and gyrB (35), and for selected representatives (43 of 169) of particular PFGE clusters, by multilocus sequence typing (MLST) based on sequence analysis of seven housekeeping genes (http://enterobase.warwick.ac.uk/species/index/ecoli). ST131 clonal subsets were identified by subclone-specific PCR (5, 36). The O25b and O16 rfb variants were detected by O-type-specific PCR (37).
Data regarding antimicrobial resistance-associated phenotypic and genotypic characteristics were available for some isolates, but all characteristics were newly generated here. Genes encoding ESBLs (blaCTX-M, blaTEM, blaOXA, and blaSHV), AmpC beta-lactamases (blaDHA, blaACC-1, blaACC-2, blaMOX, blaCMY, and blaFOX), and plasmid-mediated quinolone resistance [qnrA, qnrB, qnrC, qnrD, qnrS, aac(6′)-Ib-cr, qepA, and oqxAB] were detected by PCR and DNA sequencing (25). The presence and structure of class 1 and class 2 integrons was determined by restriction fragment length polymorphism mapping or sequencing of PCR amplicons (26). Plasmid incompatibility groups were determined by PCR-based replicon typing (38), and IncF plasmids were further analyzed by replicon sequence typing (39).
The genetic environment upstream of blaCMY and blaCTX-M variants representing CTX-M-1 (blaCTX-M-3 and blaCTX-M-15) and CTX-M-9 (blaCTX-M-14 and blaCTX-M-27) groups was mapped by PCR and sequencing by using published primers (30). The novel reverse primers CMY-mapIS (5′-CGATCCTAGCTCAAACAGC-3′) and CTX-M-9-mapIS (5′-CTTTTGCTGCACCGCACTC-3′) were used in combination with previously published forward primers (30) to examine the upstream region of blaCMY and CTX-M group 9 variants, respectively. Isolates were grouped based on identical upstream region PCR results, and selected representatives of each such PCR group underwent Sanger sequencing of the PCR products to define their exact composition.
Virulence genes.
Using multiplex PCR, the isolates were tested for 52 virulence genes associated with ExPEC (40). Isolates were classified as ExPEC if positive for ≥2 of the following genes: papA and/or papC (P fimbriae; counted as one), sfa or focDE (S and F1C fimbriae), afa or draBC (Dr-binding adhesins), kpsMII (group 2 capsule synthesis), and iutA (aerobactin system) (41). The virulence score was estimated as the number of extraintestinal virulence genes detected, adjusted for multiple detection of the pap, sfa, foc, and kps operons. Virulence gene patterns were calculated using BioNumerics (v6.6). Virotypes and subtypes were determined according to the presence of specific gene combinations (10).
Antimicrobial susceptibility.
Susceptibility to 10 and 22 antimicrobial agents was assessed by disc diffusion and broth microdilution, respectively (42, 43). Interpretation of antimicrobial susceptibility was based on inhibition zone diameters or MIC values. Antimicrobial agents and reference strains used for susceptibility testing are listed in Fig. S1 in the supplemental material. Breakpoints were as specified by the Clinical and Laboratory Standards Institute (CLSI) for all antimicrobials excepting colistin and tigecycline (42) (CLSI breakpoints undefined), for which EUCAST breakpoints were used (43), and azithromycin (no defined E. coli breakpoints), for which Streptococcus pneumoniae breakpoints were used (42). Intermediate results were interpreted as resistant. Isolates resistant to at least one representative of ≥3 antimicrobial classes were interpreted as multidrug resistant.
PFGE and hybridization experiments.
Genomic relatedness was defined by XbaI PFGE (25). Macrorestriction patterns were analyzed using BioNumerics v6.6 software (Applied Maths, Ghent, Belgium). Cluster analysis of the Dice similarity indices according to the unweighted-pair group method was used to infer a dendrogram describing the similarity relationships among PFGE profiles. Clusters were defined at an arbitrary 91% similarity level. Macrorestriction profiles of study isolates were compared with international pulsotypes (types 788, 797, 800, 805, 806, 812, 837, 842, 903, 905, 906, 943, 945, 968, 987, 1067, 1140, 1676, 1733, 1734, 1735, 1737, 1738, and 1739) within a large private PFGE profile library (14). In representative isolates, the plasmid versus chromosomal location of blaCTX gene was determined by S1 PFGE and hybridization with DIG-11-dUTP-labeled probes specific for blaCTX-15, blaCTX-27, and IncF replicons (44). The isolates for S1 PFGE were selected to provide broad diversity with respect to source groups, ST131 subclones, molecular characteristics, plasmid types, and PFGE clusters.
Statistical analysis.
Comparisons of proportions were tested for significance by a chi-square test, using MS Excel (Microsoft, Redmond, WA). The significance criterion was P < 0.05. Principal-coordinate analysis (PCoA) was used to reduce the dimensionality of the molecular data set for simplified comparisons (45).
Accession number(s).
The nucleotide sequences of the blaCTX-M genetic environment (variant designation) from strains M46 (1a1), M30 (1a2), B81 (1b1), B100 (1b2), HS115 (1b3), M81 (1b4), M103 (1b5), M111 (1c1), M75 (1c2), OV56 (1c3), M79 (1c4), M77 (1d), M24 (1e), B95 (9a), and M57 (9b) have been deposited in GenBank under accession numbers MH357356, MH357357, MH357358, MH357359, MH357360, MH357361, MH357362, MH357363, MH357364, MH357365, MH357366, MH357367, MH357368, MH357369, and MH357370, respectively. The nucleotide sequence of the blaCMY-2 genetic environment from strain HPJ64 (C1) has been deposited in GenBank under accession number MH357371.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Czech Science Foundation (18-23532S), by CEITEC 2020 (LQ1601), by CETOCOEN PLUS (CZ.02.1.01/0.0/0.0/15_003/0000469) (L.M.), and by the Office of Research Development of the U.S. Department of Veterans Affairs (J.R.J.). The opinions expressed here are strictly those of the authors and not those of their respective institutions or the funders, including the Department of Veterans Affairs.
We thank Petra Siskova, Katerina Albrechtova, Dagmar Tausova, Karel Bubenicek, and Vladimir Kmet for providing the test isolates. We thank Stephen Porter, Alois Cizek, Martina Masarikova, Jana Hofirkova, Eva Suchanova, Iva Kutilova, Jiri Vojtech, Julie C. Ellis, and Veronika Oravcova for their help in the field or laboratory. We thank Constantinos C. Papagiannitsis and Jaroslav Hrabak for providing laboratory equipment for part of the experiments and their help with analysis of hybridization results. We thank George Jacoby (Lahey Clinic, Inc., Burlington, MA), Lina Cavaco and Henrik Hasman (National Food Institute, Technical University of Denmark, Kongens Lyngby, Denmark), and Alessandra Carattoli (Istituto Superiore di Sanità, Rome, Italy) for providing positive-control strains. We thank the team of curators of the Institut Pasteur MLST and whole-genome MLST and pMLST databases for curating the data and making them publicly available at http://bigsdb.web.pasteur.fr/.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00519-18.
REFERENCES
- 1.Banerjee R, Johnson JR. 2014. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathers AJ, Peirano G, Pitout JD. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson TJ, Danzeisen JL, Youmans B, Case K, Llop K, Munoz-Aguayo J, Flores-Figueroa C, Aziz M, Stoesser N, Sokurenko E, Price LB, Johnson JR. 2016. Separate F-type plasmids have shaped the evolution of the H30 subclone of Escherichia coli sequence type 131. mSphere 1:e00121-16. doi: 10.1128/mSphere.00288-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:10. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson JR, Porter S, Thuras P, Castanheira M. 2017. The pandemic H30 subclone of sequence type 131 (ST131) as the leading cause of multidrug-resistant Escherichia coli infections in the United States (2011-2012). Open Forum Infect Dis 4:ofx089. doi: 10.1093/ofid/ofx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347-. doi: 10.1128/mBio.00958-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olesen B, Frimodt-Moller J, Leihof RF, Struve C, Johnston B, Hansen DS, Scheutz F, Krogfelt KA, Kuskowski MA, Clabots C, Johnson JR. 2014. Temporal trends in antimicrobial resistance and virulence-associated traits within the Escherichia coli sequence type 131 clonal group and its H30 and H30-Rx subclones, 1968 to 2012. Antimicrob Agents Chemother 58:6886–6895. doi: 10.1128/AAC.03679-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peirano G, van der Bij AK, Freeman JL, Poirel L, Nordmann P, Costello M, Tchesnokova VL, Pitout JD. 2014. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum beta-lactamases: global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother 58:3762–3767. doi: 10.1128/AAC.02428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahbi G, Mora A, Mamani R, Lopez C, Alonso MP, Marzoa J, Blanco M, Herrera A, Viso S, Garcia-Garrote F, Tchesnokova V, Billig M, de la Cruz F, de Toro M, Gonzalez-Lopez JJ, Prats G, Chaves F, Martinez-Martinez L, Lopez-Cerezo L, Denamur E, Blanco J. 2014. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int J Med Microbiol 304:1247–1257. doi: 10.1016/j.ijmm.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, Ichiyama S. 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. doi: 10.1093/jac/dkv017. [DOI] [PubMed] [Google Scholar]
- 12.Ludden C, Cormican M, Vellinga A, Johnson JR, Austin B, Morris D. 2015. Colonisation with ESBL-producing and carbapenemase-producing Enterobacteriaceae, vancomycin-resistant enterococci, and meticillin-resistant Staphylococcus aureus in a long-term care facility over one year. BMC Infect Dis 15:168. doi: 10.1186/s12879-015-0880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanco J, Mora A, Mamani R, Lopez C, Blanco M, Dahbi G, Herrera A, Marzoa J, Fernandez V, de la Cruz F, Martinez-Martinez L, Alonso MP, Nicolas-Chanoine MH, Johnson JR, Johnston B, Lopez-Cerero L, Pascual A, Rodriguez-Bano J. 2013. Four main virotypes among extended-spectrum-beta-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 51:3358–3367. doi: 10.1128/JCM.01555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson JR, Nicolas-Chanoine MH, DebRoy C, Castanheira M, Robicsek A, Hansen G, Weissman S, Urban C, Platell J, Trott D, Zhanel G, Clabots C, Johnston BD, Kuskowski MA. 2012. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967-2009. Emerg Infect Dis 18:598–607. doi: 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TE, Johnson JR, Didelot X, Walker AS, Crook DW. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162-15. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Platell JL, Johnson JR, Cobbold RN, Trott DJ. 2011. Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet Microbiol 153:99–108. doi: 10.1016/j.vetmic.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Kim SY, Park YJ, Johnson JR, Yu JK, Kim YK, Kim YS. 2016. Prevalence and characteristics of Escherichia coli sequence type 131 and its H30 and H30Rx subclones: a multicenter study from Korea. Diagn Microbiol Infect Dis 84:97–101. doi: 10.1016/j.diagmicrobio.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 18.Johnson JR, Davis G, Clabots C, Johnston BD, Porter S, DebRoy C, Pomputius W, Ender PT, Cooperstock M, Slater BS, Banerjee R, Miller S, Kisiela D, Sokurenko EV, Aziz M, Price LB. 2016. Household clustering of Escherichia coli sequence type 131 clinical and fecal isolates according to whole-genome sequence analysis. Open Forum Infect Dis 3:ofw129. doi: 10.1093/ofid/ofw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumura Y, Yamamoto M, Nagao M, Ito Y, Takakura S, Ichiyama S. 2013. Association of fluoroquinolone resistance, virulence genes, and IncF plasmids with extended-spectrum-beta-lactamase-producing Escherichia coli sequence type 131 (ST131) and ST405 clonal groups. Antimicrob Agents Chemother 57:4736–4742. doi: 10.1128/AAC.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doumith M, Dhanji H, Ellington MJ, Hawkey P, Woodford N. 2012. Characterization of plasmids encoding extended-spectrum beta-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J Antimicrob Chemother 67:878–885. doi: 10.1093/jac/dkr553. [DOI] [PubMed] [Google Scholar]
- 21.Dhanji H, Patel R, Wall R, Doumith M, Patel B, Hope R, Livermore DM, Woodford N. 2011. Variation in the genetic environments of blaCTX-M-15 in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother 66:1005–1012. doi: 10.1093/jac/dkr041. [DOI] [PubMed] [Google Scholar]
- 22.Lupo A, Coyne S, Berendonk TU. 2012. Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Front Microbiol 3:18. doi: 10.3389/fmicb.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brechet C, Plantin J, Sauget M, Thouverez M, Talon D, Cholley P, Guyeux C, Hocquet D, Bertrand X. 2014. Wastewater treatment plants release large amounts of extended-spectrum beta-lactamase-producing Escherichia coli into the environment. Clin Infect Dis 58:1658–1665. doi: 10.1093/cid/ciu190. [DOI] [PubMed] [Google Scholar]
- 24.Muller A, Stephan R, Nuesch-Inderbinen M. 2016. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci Total Environ 541:667–672. doi: 10.1016/j.scitotenv.2015.09.135. [DOI] [PubMed] [Google Scholar]
- 25.Jamborova I, Dolejska M, Vojtech J, Guenther S, Uricariu R, Drozdowska J, Papousek I, Pasekova K, Meissner W, Hordowski J, Cizek A, Literak I. 2015. Plasmid-mediated resistance to cephalosporins and fluoroquinolones in various Escherichia coli sequence types isolated from rooks wintering in Europe. Appl Environ Microbiol 81:648–657. doi: 10.1128/AEM.02459-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamborova I, Dolejska M, Zurek L, Townsend AK, Clark AB, Ellis JC, Papousek I, Cizek A, Literak I. 2017. Plasmid-mediated resistance to cephalosporins and quinolones in Escherichia coli from American crows in the U S A. Environ Microbiol 19:2025–2036. doi: 10.1111/1462-2920.13722. [DOI] [PubMed] [Google Scholar]
- 27.von Salviati C, Laube H, Guerra B, Roesler U, Friese A. 2015. Emission of ESBL/AmpC-producing Escherichia coli from pig fattening farms to surrounding areas. Vet Microbiol 175:77–84. doi: 10.1016/j.vetmic.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Laube H, Friese A, von Salviati C, Guerra B, Rosler U. 2014. Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. Vet Microbiol 172:519–527. doi: 10.1016/j.vetmic.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Hartmann A, Locatelli A, Amoureux L, Depret G, Jolivet C, Gueneau E, Neuwirth C. 2012. Occurrence of CTX-M-producing Escherichia coli in soils, cattle, and farm environment in France (Burgundy region). Front Microbiol 3:83. doi: 10.3389/fmicb.2012.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolejska M, Frolkova P, Florek M, Jamborova I, Purgertova M, Kutilova I, Cizek A, Guenther S, Literak I. 2011. CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J Antimicrob Chemother 66:2784–2790. doi: 10.1093/jac/dkr363. [DOI] [PubMed] [Google Scholar]
- 31.Dolejska M, Brhelova E, Dobiasova H, Krivdova J, Jurankova J, Sevcikova A, Dubska L, Literak I, Cizek A, Vavrina M, Kutnikova L, Sterba J. 2012. Dissemination of IncFII(K)-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from children in hospital paediatric oncology wards. Int J Antimicrob Agents 40:510–515. doi: 10.1016/j.ijantimicag.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Tausova D, Dolejska M, Cizek A, Hanusova L, Hrusakova J, Svoboda O, Camlik G, Literak I. 2012. Escherichia coli with extended-spectrum beta-lactamase and plasmid-mediated quinolone resistance genes in great cormorants and mallards in Central Europe. J Antimicrob Chemother 67:1103–1107. doi: 10.1093/jac/dks017. [DOI] [PubMed] [Google Scholar]
- 33.Micenkova L, Siskova P, Bosak J, Jamborova I, Cernohorska L, Smajs D. 2014. Characterization of human uropathogenic ESBL-producing Escherichia coli in the Czech Republic: spread of CTX-M-27-producing strains in a University Hospital. Microb Drug Resist 20:610–617. doi: 10.1089/mdr.2014.0013. [DOI] [PubMed] [Google Scholar]
- 34.Albrechtova K, Dolejska M, Cizek A, Tausova D, Klimes J, Bebora L, Literak I. 2012. Dogs of nomadic pastoralists in northern Kenya are reservoirs of plasmid-mediated cephalosporin- and quinolone-resistant Escherichia coli, including pandemic clone B2-O25-ST131. Antimicrob Agents Chemother 56:4013–4017. doi: 10.1128/AAC.05859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother 53:2733–2739. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR. 2013. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis 57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JR, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, Junka AF, Maczynska B, Denamur E. 2014. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J Clin Microbiol 52:1358–1365. doi: 10.1128/JCM.03502-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 40.Johnson JR, Porter S, Johnston B, Kuskowski MA, Spurbeck RR, Mobley HL, Williamson DA. 2015. Host characteristics and bacterial traits predict experimental virulence for Escherichia coli bloodstream isolates from patients with urosepsis. Open Forum Infect Dis 2:ofv083. doi: 10.1093/ofid/ofv133.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, Smith KE. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother 47:2161–2168. doi: 10.1128/AAC.47.7.2161-2168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. Document M100-S24. CLSI, Wayne, PA. [Google Scholar]
- 43.European Committee on Antimicrobial Susceptibility Testing. g. 2015. Breakpoint tables for interpretation of MICs and zone diameters, version 5.0 European Committee on Antimicrobial Susceptibility Testing, Vaxjo, Sweden: http://www.eucast.org/clinical_breakpoints/. [Google Scholar]
- 44.Studentova V, Dobiasova H, Hedlova D, Dolejska M, Papagiannitsis CC, Hrabak J. 2015. Complete nucleotide sequences of two NDM-1-encoding plasmids from the same sequence type 11 Klebsiella pneumoniae strain. Antimicrob Agents Chemother 59:1325–1328. doi: 10.1128/AAC.04095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.