A total of 108 meropenem-resistant Enterobacteriaceae isolates were obtained from 1,658 rectal swabs collected from 15 unrelated commercial chicken farms in China between 2014 and 2016. These samples yielded 16 Escherichia coli and 2 Klebsiella pneumoniae isolates of diverse sequence types carrying a blaNDM-5-bearing IncX3 plasmid.
KEYWORDS: Escherichia coli, Klebsiella pneumoniae, blaNDM-5, IncX3 plasmid
ABSTRACT
A total of 108 meropenem-resistant Enterobacteriaceae isolates were obtained from 1,658 rectal swabs collected from 15 unrelated commercial chicken farms in China between 2014 and 2016. These samples yielded 16 Escherichia coli and 2 Klebsiella pneumoniae isolates of diverse sequence types carrying a blaNDM-5-bearing IncX3 plasmid. K. pneumoniae strain sequence type 709 (ST709) has two blaNDM-5-carrying plasmids that were transferred together to E.coli.
TEXT
Multidrug-resistant organisms, including those that are carbapenemase-producing Enterobacteriaceae, are becoming a challenging threat to public health worldwide (1). Gene blaNDM-5, a variant of blaNDM, was first identified in 2011 in an Escherichia coli sequence type 648 (ST648) isolate from a patient in the United Kingdom (2). Since then, it has been reported in various parts of the world, including South Korea (3), Denmark (4), Algeria (5), and China (6–8). The cooccurrence of NDM-5 and other carbapenemase enzyme isolates from the same patient is extremely worrisome because it might lead to therapeutic failure and death. The first cooccurrence of NDM-4- and NDM-5-producing Klebsiella pneumoniae in the same patient was reported in 2016 (9). In 2017, a carbapenem-resistant K. pneumoniae ST147 isolate harboring blaNDM-5 and blaOXA-181 from a hospitalized patient was found in the United States (10). In 2018, a blaNDM-5- and blaOXA-48-like-coproducing E. coli strain was first isolated in South Korea (11). Meanwhile, the first case of a clinical Klebsiella michiganensis isolate producing KPC-2, NDM-1, and NDM-5 was reported in China (12). A fusion plasmid (IncX3 and IncFIB) recoverable from an NDM-5-producing clinical E. coli isolate was recently characterized, and these types of recombination events presumably play a potential role in the development of new plasmids with extended resistance profiles (13). In this study, we identified the presence of various sequence types of Enterobacteriaceae carrying blaNDM-5 in chickens from multiple farms across seven Chinese provinces.
A total of 108 nonrepeated meropenem-resistant Enterobacteriaceae (97 E. coli and 11 K. pneumoniae) isolates (6.5%) were obtained from 1,658 rectal swabs collected from 15 unrelated commercial chicken farms in China between 2014 and 2016 (see Fig. S1 in the supplemental material). All of the Enterobacteriaceae isolates were selected on MacConkey agar plates supplemented with 2 μg/ml meropenem. Species identification was performed with the BD Phoenix-100 system (Becton Dickinson) and confirmed by 16S rRNA gene sequencing. Antimicrobial susceptibility testing was performed on Mueller-Hinton agar plates testing for 16 antimicrobials according to CLSI guidelines (14), except polymyxin B, for which European Committee on Antimicrobial Susceptibility Testing breakpoints were used (15).
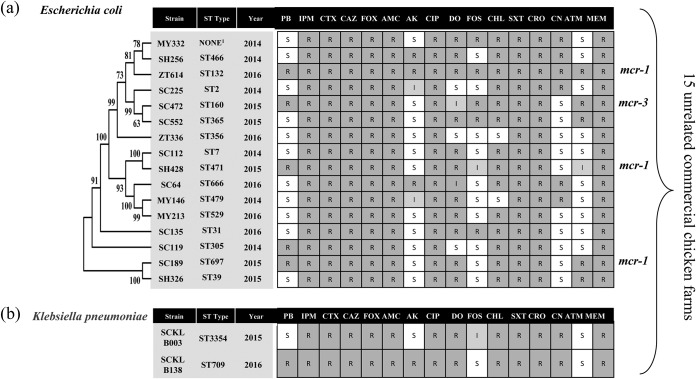
These Enterobacteriaceae isolates were then subjected to screening for the presence of blaNDM and mcr genes by PCR assay (see Table S1 in the supplemental material) as previously described (16). Of 108 meropenem-resistant Enterobacteriaceae strains, 52 (48 E. coli and 4 K. pneumonia; 3.14%) were found to harbor the blaNDM-5 gene. Multilocus sequence typing (MLST) was performed as previously described (http://bigsdb.pasteur.fr/); 16 different sequence types of E. coli isolates and 2 sequence types of K. pneumoniae isolates were found. Three of the 16 strains of E. coli were found to coharbor the mcr-1 gene. Multidrug-resistant E. coli strain ZT614 ST132 was resistant to all 16 antimicrobials. Phylogenetic relationship, antimicrobial resistance phenotypes, and sequence types of the strains are shown in Fig. 1.
FIG 1.
Phylogenetic relationship, antimicrobial resistance phenotypes, source of isolate, and sequence types of the 18 different Enterobacteriaceae: (a) Escherichia coli and (b) Klebsiella pneumoniae. A phylogenetic tree based on a maximum-likelihood method was built by MEGA6 with nucleotide sequences of 8 MLST genes to reveal a more detailed relationship among the analyzed strains. Bootstrap values (percentages of 1,000 replications) of >50% are shown at each node. 1, Failure to find any corresponding ST type with MLST database blasting. S, susceptible; I, intermediate; R, resistant; PB, polymyxin B; IPM, imipenem; CTX, cefotaxime; CAZ, ceftazidime; FOX, cefoxitin; AMC, amoxicillin-clavulanate; AK, amikacin; CIP, ciprofloxacin; DO, doxycycline; FOS, fosfomycin; CHL, chloramphenicol; SXT, trimethoprim- sulfamethoxazole; CRO, ceftriaxone; CN, gentamicin; ATM, aztreonam; MEM, meropenem.
Conjugation experiments were performed between 18 different isolates (Fig. 1) and E. coli J53 Azr as the recipient. Transconjugants were selected on Mueller-Hinton agar (MHA; Oxoid) plates that contained 200 μg/ml sodium azide with 2 μg/ml imipenem. All of them could successfully transfer their carbapenem resistance genes to the recipient strain E. coli J53 Azr. There was no cotransfer of carbapenem and colistin resistance phenotype transconjugant. The total plasmid DNA from 18 transconjugants was extracted using a Qiagen plasmid minikit following manufacturer's recommendations (Qiagen, Hilden, Germany). Whole-genome sequencing was performed on the Illumina MiSeq platform (Majorbio, Shanghai) using a 350-bp paired-end TruSeq library with a 2 × 300 run. A draft assembly of the plasmids was made with plasmidSPAdes (17). Predicted gaps were closed by PCR and Sanger sequencing the using specifically designed primers listed in Table S1. Identification of antibiotic resistance genes was done by ResFinder 3.0 (http://www.genomicepidemiology.org/), and plasmid replicon types were determined by using the PlasmidFinder tool (http://genomicepidemiology.org/).
Sequence analysis revealed that all of the transconjugants harbored a 46-kb blaNDM-5-bearing IncX3 plasmid. BLASTN results showed that all 18 IncX3 plasmids had almost 100% identity to the 46,161-bp plasmid pBJ114-46 (GenBank accession no. MF679143) with only 1 to 4 single-base changes (data not shown), indicating that the blaNDM-5-bearing IncX3 plasmid was an important vector responsible for the dissemination of NDM-5 among Enterobacteriaceae isolates originating from chicken farms in China. Recently, a study identified the occurrence of similar IncX3 plasmids carrying blaNDM-5 in pigs originating from multiple farms across China (18), confirming that this mobile NDM vector is widespread in animal production.
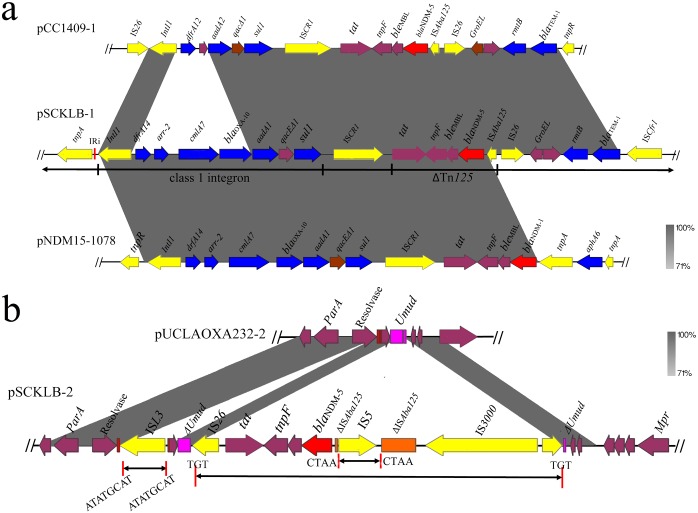
Interestingly, a transconjugant of the ST709 K. pneumoniae strain named SCKLB138 harbored two blaNDM-5-bearing plasmids, pSCKLB-1 and pSCKLB-2 (see Fig. S2 in the supplemental material). Plasmid pSCKLB-1 was found to comprise the IncFIB and IncFII replicons; the blaNDM-5, blaOXA-10, rmtB, aadA1, and blaTEM-1 genes; and some other resistance gene cassettes bounded by various insertion sequences. blaNDM-5 together with the bleomycin resistance gene bleMBL, tat, and tnpF form part of the transposon Tn125 (ΔTn125). Upstream of the ISAba125, IS26 and ISCfr1 were identified bracketing blaTEM-1 and the 16S rRNA methylase gene rmtB, conferring resistance to aminoglycosides. The same arrangement of ΔTn125 and rmtB was found in ST147 K. pneumoniae plasmid pCC1409-1 (GenBank accession no. KT725789), except that the upstream of the blaTEM-1 gene of pCC1409-1 was truncated by Tn2 resolvase rather than ISCfr1 (Fig. 2a). The blaOXA-10 gene was localized downstream of blaNDM-5 in a class 1 integron with the dfrA14-arr-2-cmlA7-blaOXA-10-aadA1 cassette array and the ISCR1 element behind the 3′-conserved segments. The region bracketed by blaNDM-1 and tnpR in plasmid pHN-NDM0711 exhibited 99% identity to the corresponding region of pSCKLB-2 (Fig. 2a). Plasmid pSCKLB-2 was a 46-kb IncX3 blaNDM-5-bearing plasmid. An IS5 was inserted with ISAba125 upstream of blaNDM-5 and the ble, trpF, and tat genes downstream from blaNDM-5. Comparison of the genetic characteristics of pSCKLB-2 and pUCLAOXA232-2 (GenBank accession no. NZ_CP012563) showed that an ISL3 was inserted downstream of the resolvase gene, leading to the flanking 8-bp direct repeats (ATATGCAT). The blaNDM-5-carrying region bracketed by IS26 and tnpA was inserted into the umuD gene, resulting in a pair of 3-bp direct repeats (TGT). The ISAba125 gene was interrupted by IS5 and split into two fragments, resulting in a pair of 4-bp direct repeats (CTAA) (Fig. 2b).
FIG 2.
The genetic context of blaNDM-5 on pSCKLB138-1 and pSCKLB138-2 compared with other plasmids. (a) Genetic structure of blaNDM-5 gene on pSCKLB138-1 compared with pCC1409-1 and pNDM15-1078. (b) Genetic structure of blaNDM-5 gene on pSCKLB138-2 compared with pUCLAOXA232-2.
In conclusion, this study identified a self-transmissible IncX3 plasmid carrying blaNDM-5 that was an important vector responsible for the dissemination of NDM-5 among Enterobacteriaceae isolates originating from chicken farms in China. The cooccurrence of blaNDM-5 and other resistance rmtB and mcr-1 genes in Enterobacteriaceae isolated in chicken farms strongly suggests a potential food chain dissemination pathway, which warrants further attention. To the best of our knowledge, this is the first report of two blaNDM-5-carrying plasmids coexisting in a K. pneumoniae strain isolated from commercial chicken farms in China. The results highlight that the chicken farms are an important reservoir of Enterobacteriaceae carrying blaNDM-5 gene.
Accession number(s).
The complete nucleotide sequences of plasmids pSCKLB-1 and pSCKLB-2 characterized in this study were submitted to the GenBank database and assigned accession numbers MH161191 and MH161192.
Supplementary Material
ACKNOWLEDGMENTS
We thank the team of the curators of the Institut Pasteur MLST system (Paris, France) for importing novel MLST profiles at http://bigsdb.web.pasteur.fr.
This work was supported by China Agriculture Research System (CARS-40) National System for Layer Production Technology (CARS-40-K14), National Natural Science Fund of China (31772769), the general program of National Natural Science Foundation of China (31572547), and Special Fund for Agro-Scientific Research in the Public Interest of China (201403054).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00779-18.
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho SY, Huh HJ, Baek JY, Chung NY, Ryu JG, Ki CS, Chung DR, Lee NY, Song JH. 2015. Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg Infect Dis 21:1088–1089. doi: 10.3201/eid2106.150048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammerum AM, Hansen F, Olesen B, Struve C, Holzknecht BJ, Andersen PS, Thye AM, Jakobsen L, Roder BL, Stegger M, Hansen DS. 2015. Investigation of a possible outbreak of NDM-5-producing ST16 Klebsiella pneumoniae among patients in Denmark with no history of recent travel using whole-genome sequencing. J Glob Antimicrob Resist 3:219–221. doi: 10.1016/j.jgar.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Yousfi M, Mairi A, Bakour S, Touati A, Hassissen L, Hadjadj L, Rolain JM. 2015. First report of NDM-5-producing Escherichia coli ST1284 isolated from dog in Bejaia, Algeria. New Microbes New Infect 8:17–18. doi: 10.1016/j.nmni.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang F, Xie L, Wang X, Han L, Guo X, Ni Y, Qu H, Sun J. 2016. Further spread of blaNDM-5 in Enterobacteriaceae via IncX3 plasmids in Shanghai, China. Front Microbiol 7:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang LP, Xue WC, Meng DY. 2016. First report of New Delhi metallo-beta-lactamase 5 (NDM-5)-producing Escherichia coli from blood cultures of three leukemia patients. Int J Infect Dis 42:45–46. doi: 10.1016/j.ijid.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Yang P, Xie Y, Feng P, Zong Z. 2014. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother 58:7548–7552. doi: 10.1128/AAC.03911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalifa HO, Soliman AM, Ahmed AM, Shimamoto T, Shimamoto T. 2016. NDM-4- and NDM-5-producing Klebsiella pneumoniae coinfection in a 6-month-old infant. Antimicrob Agents Chemother 60:4416–4417. doi: 10.1128/AAC.00479-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rojas LJ, Hujer AM, Rudin SD, Wright MS, Domitrovic TN, Marshall SH, Hujer KM, Richter SS, Cober E, Perez F, Adams MD, van Duin D, Bonomo RA. 2017. NDM-5 and OXA-181 β-lactamases, a significant threat continues to spread in the Americas. Antimicrob Agents Chemother 61:e00454-. doi: 10.1128/AAC.00454-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jhang J, Wang HY, Yoo G, Hwang GY, Uh Y, Yoon KJ. 2018. NDM-5 and OXA-48 co-producing uropathogenic Escherichia coli isolate: first case in Korea. Ann Lab Med 38:277–279. doi: 10.3343/alm.2018.38.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng B, Xu H, Yu X, Lv T, Jiang X, Cheng H, Zhang J, Chen Y, Huang C, Xiao Y. 2018. Identification and genomic characterization of a KPC-2-, NDM-1- and NDM-5-producing Klebsiella michiganensis isolate. J Antimicrob Chemother 73:536–538. doi: 10.1093/jac/dkx415. [DOI] [PubMed] [Google Scholar]
- 13.Xie M, Li R, Liu Z, Chan EWC, Chen S. 2018. Recombination of plasmids in a carbapenem-resistant NDM-5-producing clinical Escherichia coli isolate. J Antimicrob Chemother 73:1230–1234. doi: 10.1093/jac/dkx540. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; 28th informational supplement. CLSI document M100-S28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.European Committee on Antimicrobial Susceptibility Testing. 2017. Breakpoint tables for interpretation of MICs and zone diameters, version 7.1. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf.
- 16.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 66:2002–2005. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 17.Laczny CC, Galata V, Plum A, Posch AE, Keller A. 2017. Assessing the heterogeneity of in silico plasmid predictions based on whole-genome-sequenced clinical isolates. Brief Bioinform. doi: 10.1093/bib/bbx162. [DOI] [PubMed] [Google Scholar]
- 18.Ho PL, Wang Y, Liu MC, Lai EL, Law PY, Cao H, Chow KH. 2018. IncX3 epidemic plasmid carrying blaNDM-5 in Escherichia coli from swine in multiple geographic areas in China. Antimicrob Agents Chemother 62:e02295-17. doi: 10.1128/AAC.02295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.