Clostridium difficile infection (CDI) is the leading cause of antibiotic-associated diarrhea and has gained worldwide notoriety due to emerging hypervirulent strains and the high incidence of recurrence. We previously reported protection of mice from CDI using the antigerminant bile salt analog CamSA.
KEYWORDS: C. difficile, microbiome, antigermination, CamSA, hamster
ABSTRACT
Clostridium difficile infection (CDI) is the leading cause of antibiotic-associated diarrhea and has gained worldwide notoriety due to emerging hypervirulent strains and the high incidence of recurrence. We previously reported protection of mice from CDI using the antigerminant bile salt analog CamSA. Here we describe the effects of CamSA in the hamster model of CDI. CamSA treatment of hamsters showed no toxicity and did not affect the richness or diversity of gut microbiota; however, minor changes in community composition were observed. Treatment of C. difficile-challenged hamsters with CamSA doubled the mean time to death, compared to control hamsters. However, CamSA alone was insufficient to prevent CDI in hamsters. CamSA in conjunction with suboptimal concentrations of vancomycin led to complete protection from CDI in 70% of animals. Protected animals remained disease-free at least 30 days postchallenge and showed no signs of colonic tissue damage. In a delayed-treatment model of hamster CDI, CamSA was unable to prevent infection signs and death. These data support a putative model in which CamSA reduces the number of germinating C. difficile spores but does not keep all of the spores from germinating. Vancomycin halts division of any vegetative cells that are able to grow from spores that escape CamSA.
INTRODUCTION
Clostridium difficile is an anaerobic, spore-forming pathogen that colonizes the gut of susceptible individuals and causes Clostridium difficile infection (CDI) (1). CDI is recognized as the leading cause of antibiotic-associated diarrhea in nosocomial environments. CDI poses a large burden for the health care system, costing the United States >3 billion dollars annually (2). An emergent hypervirulent C. difficile strain (BI/NAP1/027) is associated with even more severe disease (3, 4). The morbidity and mortality rates associated with CDI have prompted the CDC to upgrade C. difficile to an urgent antimicrobial resistance threat (5).
C. difficile spores are transmitted via the fecal-oral route, and CDI most often occurs after a patient has been treated with antibiotics. In fact, most broad-spectrum antibiotics can render a person susceptible to C. difficile (6). Antibiotic use disrupts the natural gut microbiota that normally resists colonization by C. difficile (7). In this new favorable environment, C. difficile spores germinate; the resulting vegetative cells proliferate in the gut and release two exotoxins, toxin A and toxin B. These toxins cause injury to and inflammation of the colonic lining, resulting in disease (8, 9). Because of the intrinsic antibiotic resistance of C. difficile spores and the inability of current antibiotic treatments for CDI to completely clear the pathogen from the intestinal tract, nonantibiotic strategies should be investigated (10–14). Continued antibiotic use to treat CDI prevents commensal microbiota from repopulating the gut, allowing persistent C. difficile spores to germinate and to reestablish disease. As a result, 25 to 30% of CDI patients experience repeated bouts of relapse (15–17).
C. difficile spores must germinate to establish disease. The activation of dormant C. difficile spores requires binding of the primary bile salt taurocholate and amino acids (18, 19). We showed that a synthetic bile salt analog, CamSA, was a competitive inhibitor of taurocholate-induced C. difficile spore germination in vitro (18). Furthermore, CamSA prevents CDI in a murine model in a dose-dependent manner (20).
In the current study, we assessed the effectiveness of CamSA in the more susceptible hamster CDI model. As in mice, CamSA showed no toxicity and did not cause large-scale disruption of the hamster intestinal microbiota. CamSA alone was unable to prevent CDI in hamsters but resulted in a delay in the onset of disease. Interestingly, CamSA in combination with a suboptimal dose of vancomycin, administered at the same time as C. difficile spores, prevented CDI in 70% of challenged hamsters. However, CamSA-vancomycin combinations were unable to prevent CDI recurrence when vancomycin was administered 24 h postchallenge with C. difficile spores. These results suggest that antigermination therapies have the potential to be used in combination with current antibiotic treatments to reduce CDI progression. This study has also revealed limitations of CamSA alone as prophylaxis for CDI in hamsters.
RESULTS
CamSA toxicity.
Hamsters treated for 30 consecutive days with the highest CamSA dose (300 mg/kg) used in previous studies (20) showed no obvious adverse physiological or behavioral effects. Similarly, gross anatomical examination of treated animals showed no lesions or abnormalities. Microscopic examinations of colons showed no histological anomalies, and findings were indistinguishable from those for untreated animals (see Fig. S1 in the supplemental material).
Microbiome effects.
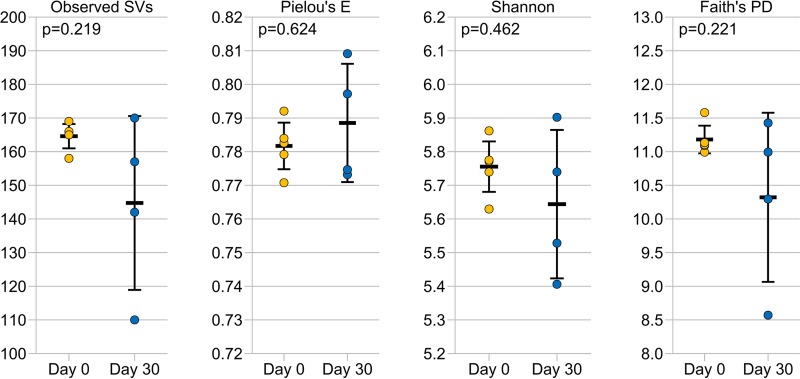
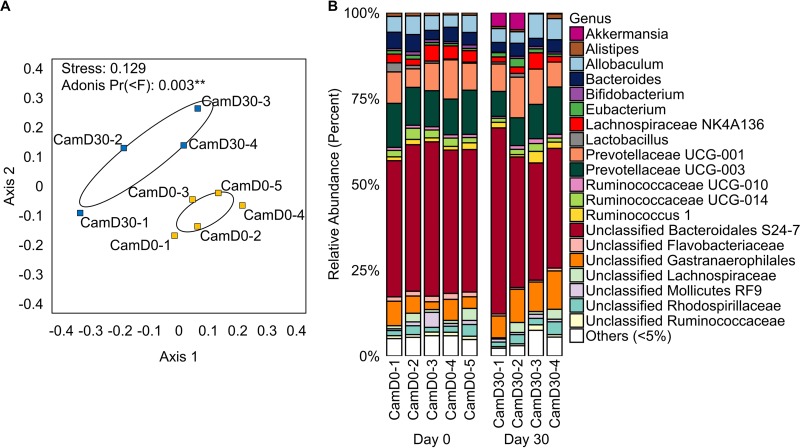
CamSA (300 mg/kg) treatment for 30 days did not significantly alter the richness, evenness, or diversity of the hamster intestinal microbiome (Fig. 1). However, nonmetric multidimensional scaling (NMDS) analysis based on Bray-Curtis dissimilarity, along with a corresponding Adonis analysis, showed that treatment with CamSA for 30 days led to a statistically significant change in the community composition of hamster gut microbiomes (Fig. 2A). Bar graphs showing the relative abundance of microbial taxa before and after CamSA treatment, along with a corresponding SIMPER analysis, showed these shifts to be small in comparison to broad-spectrum antibiotic treatment (Fig. 2B; also see Fig. S2 and Table S1 in the supplemental material) (5, 6, 21). Taxa that contributed most to the variance between microbiomes before and after CamSA treatment included a relative increase in Akkermansia (Verrucomicrobia) and shifts in populations of unclassified Gastranaerophilales (Cyanobacteria), Prevotellaceae (Bacteroidetes), Lachnospiraceae (Firmicutes), and Bacteroidetes.
FIG 1.
Alpha diversity metrics comparing hamster fecal microbiomes before (day 0, n = 5) (yellow circles) and after (day 30, n = 4) (blue circles) a 30-day CamSA treatment. The data for 1 hamster after CamSA treatment failed quality control standards and was removed from the data set. Bars represent means and 95% confidence intervals. No values were significantly different before versus after CamSA treatment (pairwise Kruskal-Wallis test, P > 0.05). Observed SVs, observed sequence variants; Pielou's E, Pielou's evenness; Shannon, Shannon diversity index; Faith's PD, Faith's phylogenetic diversity.
FIG 2.
(A) NMDS analysis based on Bray-Curtis dissimilarity, demonstrating that gut microbiomes were significantly different before (CamD0) (yellow squares) versus after (CamD30) (blue squares) CamSA treatment. Numbers after the hyphens refer to specific hamsters. Ellipses represent the standard errors of the mean (95% confidence). (B) Bar plot showing the relative abundance of gut microbiota at the genus level in each hamster before (day 0) and after (day 30) CamSA treatment. Uncultivated and unnamed genera are listed according to the lowest named taxonomic level, as categorized by SILVA. In cases in which the SILVA taxonomy was flagged as misclassified, the Ribosomal Database Project (RDP) taxonomy was used.
Delay of CDI onset by CamSA.
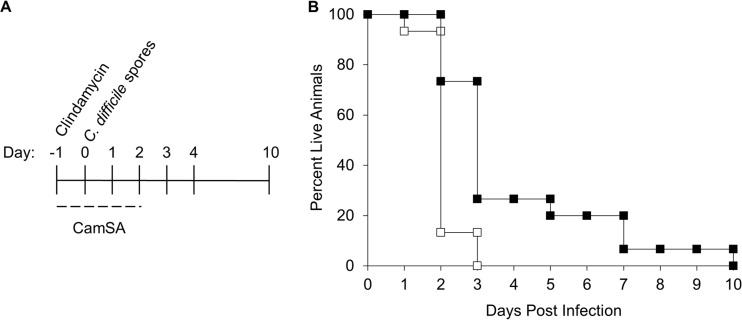
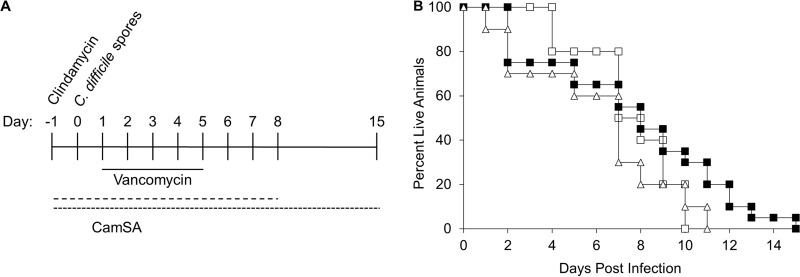
Hamsters challenged with C. difficile spores became symptomatic rapidly and were culled within 3 days postchallenge (Fig. 3A and B). Challenged hamsters treated with 300 mg/kg CamSA for 4 days developed CDI signs similar to those in untreated animals and were culled, but the onset was delayed up to 7 days, leading to a statistically significant difference (P = 0.0006) in survival times between CamSA-treated and untreated hamsters. Under these conditions, CamSA alone increased the mean time to death from 1.73 to 3.77 days.
FIG 3.
Effects of CamSA on hamsters infected with C. difficile spores. (A) Timeline of experiments. All hamsters were treated with a single 30-mg/kg dose of clindamycin (day −1). On day 0, animals were challenged with 100 C. difficile spores. Animals were treated with either 0 or 300 mg/kg CamSA in DMSO for 4 consecutive days beginning at day −1. Animals were scored for signs of CDI twice daily. (B) Percent survival, presented as a Kaplan-Meier survival plot, which shows a significant difference between CamSA-treated (■) and untreated (□) animals (P = 0.0006).
Effects of CamSA in combination with vancomycin.
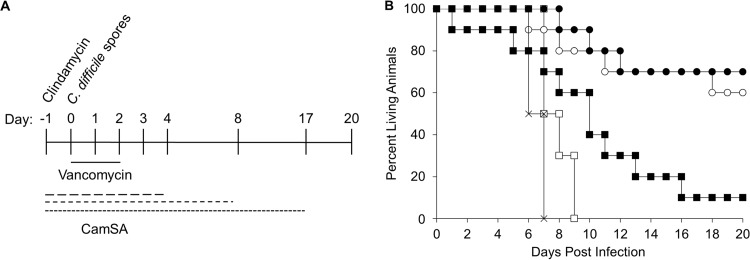
To determine the effects of combining vancomycin and CamSA treatments, animals were challenged with C. difficile spores and treated with a suboptimal vancomycin dose (5 mg/kg) (22, 23) and CamSA (300 mg/kg) (Fig. 4A). As expected, animals treated with vancomycin alone become moribund within 9 days postchallenge. Combining vancomycin with CamSA for 19 consecutive days resulted in asymptomatic survival of 70% of the animals (Fig. 4B). Similar results were obtained when the CamSA dosage was stopped after 10 consecutive days. In contrast, limiting CamSA treatment to 6 doses resulted in survival of only 10% of infected animals. The increases in survival rates for hamsters treated with vancomycin and CamSA for 6 (P = 0.04), 10 (P = 0.001), or 19 (P = 5.18 × 10−5) consecutive days were significant, compared to hamsters treated with vancomycin alone. When the CamSA dosage was reduced to 50 mg/kg for 10 days, C. difficile-challenged animals developed CDI signs, with scores indistinguishable from those of untreated animals.
FIG 4.
Protection of hamsters from CDI with combined administration of a suboptimal vancomycin dose and CamSA. (A) Timeline of experiments. All hamsters were treated with a single 30-mg/kg dose of clindamycin (day −1). Also starting at day −1, animals were treated daily with 300 mg/kg CamSA for 0, 6, 10, or 19 consecutive days. Another group of animals was treated with 50 mg/kg CamSA for 10 consecutive days. On day 0, all animals were challenged with 100 C. difficile spores. Starting at day 0, all animals were also treated daily with 5 mg/kg vancomycin for 3 consecutive days. (B) Percent survival, presented as a Kaplan-Meier survival plot, which shows significant increases in animal survival rates with 300-mg/kg CamSA treatment for 6 days (P = 0.04) (■), 10 days (P = 0.001) (○), and 19 days (P = 5.18 × 10−5) (●), compared to untreated animals (□). Survival of animals treated with 50 mg/kg CamSA for 10 days (×) was undistinguishable from that of untreated animals.
Vancomycin-CamSA-protected animals showed intact colonic mucosal lining, with no distinct tissue damage or separation of the submucosal region, and were indistinguishable from control animals. In contrast, histological examination of animals showing signs of CDI presented extreme intestinal tissue damage, including disruption or loss of the colonic mucosal lining, loss of integrity of epithelial cells, and substantial separation of the submucosa (Fig. S3).
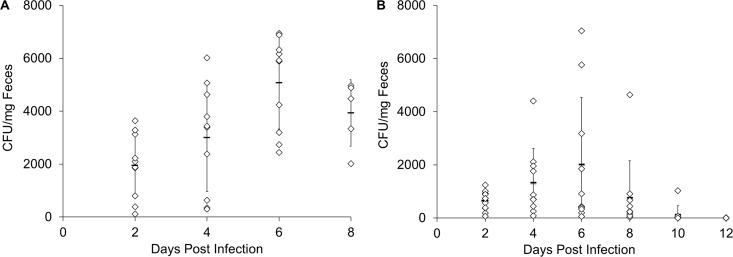
Fecal samples from challenged animals showed only C. difficile vegetative cells; no spores were recovered. Samples were heterogeneous with regard to C. difficile cells shed and the timing needed to clear C. difficile from the gastrointestinal tract. Most vancomycin-treated control animals showed maximum shedding between 4 and 6 days postchallenge but continued shedding until culling (Fig. 5A). CamSA-treated animals that did not develop CDI started shedding as early as 2 days and continued until 10 days postchallenge (Fig. 5B). Animals treated with only vancomycin showed a statistically significant (P = 0.005) increase in C. difficile shedding over time. In contrast, shedding in vancomycin-CamSA-treated animals remained constant (P = 0.2). These differences in shedding behavior resulted in statistically significant decreases in fecal C. difficile loads in vancomycin-CamSA-treated animals, compared to animals treated with only vancomycin, at days 2 (P = 0.005), 4 (P = 0.042), 6 (P = 0.005), and 8 (P = 0.001) postchallenge. Fecal samples were collected up to day 30 postchallenge; however, C. difficile was undetectable in fecal samples after day 12. CamSA-treated animals that developed CDI shed C. difficile up to culling or 10 days postchallenge (data not shown). All data collected correspond to total C. difficile found in feces. Heat treatment of feces to obtain only C. difficile spores was unsuccessful. Surviving animals from all groups did not develop any CDI signs, even 30 days after the final CamSA dose.
FIG 5.
Box-and-whisker plots showing total numbers of C. difficile in hamster feces, including spores and vegetative cells (mean ± standard deviation). C. difficile-challenged animals were treated with 5 mg/kg vancomycin only (A) or with 5 mg/kg vancomycin and 300 mg/kg CamSA (B) for 19 consecutive days. Two-way mixed ANOVA was used to assess the significance of differences for groups treated with vancomycin only or CamSA plus vancomycin on days 2 (P = 0.005), 4 (P = 0.042), 6 (P = 0.005), and 8 (P = 0.001). Within groups, animals treated with vancomycin only showed significant differences in C. difficile CFU per milligram of feces over time (P = 0.005), but no significant differences were found for the CamSA-vancomycin group (P = 0.2) over time.
Effects of CamSA on CDI in hamsters with delayed vancomycin treatment.
To test for synergistic effects of antibiotic and antigermination treatments on CDI, vancomycin treatment was started 1 day after spore challenge, as described previously (24–26) (Fig. 6A). This delay in vancomycin dosing negated the synergistic effects of vancomycin-CamSA combinations (Fig. 6B). There was no statistically significant difference in survival times between hamsters treated with vancomycin and CamSA (10-day treatment, P = 0.67; 17-day treatment, P = 0.25) and hamsters given vancomycin alone. Even animals that were continuously administered CamSA daily developed CDI within 15 days after C. difficile challenge, resulting in only 17 consecutive CamSA doses. The calculated mean time to death for infected untreated animals was 7.2 days. The calculated mean times to death for animals treated with CamSA and vancomycin for 10 and 19 consecutive CamSA doses were 6.0 days and 7.5 days, respectively.
FIG 6.
Effects of delayed treatment on CDI with combined administration of a suboptimal vancomycin dose and CamSA. (A) Timeline of experiments. All hamsters were treated with a single 30-mg/kg dose of clindamycin (day −1). Starting at day −1, animals were treated daily with 300 mg/kg CamSA for 0, 10, or 17 consecutive days. On day 0, all animals were challenged with 100 C. difficile spores. The beginning of vancomycin treatment was delayed for 24 h (day 1) after challenge, and treatment was continued daily for 5 consecutive days. Animals were scored for signs of CDI twice daily. (B) Percent survival, presented as a Kaplan-Meier survival plot. No difference in survival rate was conferred with 10 days of CamSA treatment (P = 0.67) (△) or 17 days of CamSA treatment (P = 0.25) (■), compared to untreated animals (□).
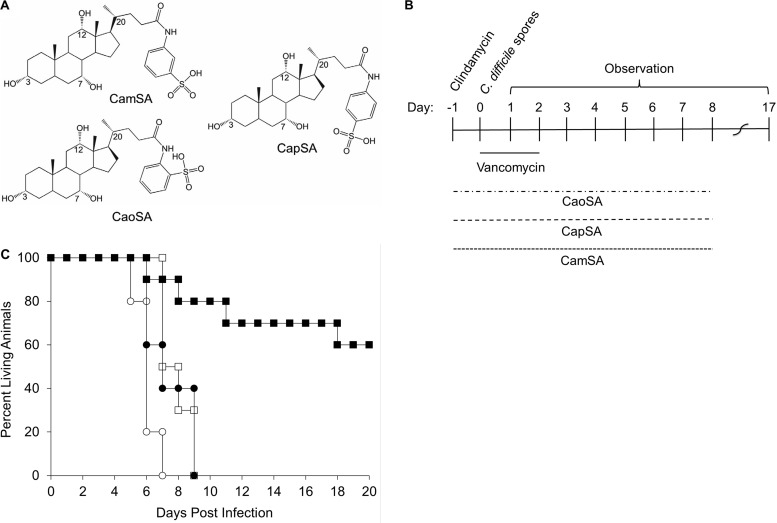
Effects of CamSA analogs on CDI in hamsters.
The structure of CamSA contains a sulfonic acid moiety at the meta-position of the aromatic ring conjugated to cholic acid (Fig. 7A). We showed previously that analogs of CamSA with the sulfonic acid moiety in either the para- or ortho-position negated inhibition of C. difficile spore germination in vitro, but in vivo studies were not performed (18). To test the effects of CamSA structural isomers in CDI protection, hamsters were treated with suboptimal concentrations of vancomycin and either CapSA (para-analog) or CaoSA (ortho-analog) for 10 consecutive days or until they were moribund (Fig. 7B). As shown above, CamSA-treated hamsters showed a significant increase in survival times (P = 0.001), compared to hamsters given vancomycin alone (Fig. 7C). In contrast, CapSA did not offer protection (P = 0.75), and CaoSA-treated animals showed decreased survival times (P = 0.001), compared to animals administered vancomycin alone (Fig. 7C).
FIG 7.
Effects of CapSA and CaoSA on CDI prevention in hamsters. (A) Structures of CamSA, CapSA, and CaoSA. (B) Timeline of experiments. Animals were treated with clindamycin (day −1) and challenged with C. difficile spores (day 0), as described above. Vancomycin (5 mg/kg) was administered for 3 consecutive days beginning at day 0. Starting at day −1, CaoSA, CapSA, or CamSA at 300 mg/kg was administered daily to animals for 10 consecutive days or until the animals became moribund. Animals treated with vancomycin only served as controls. (C) Percent survival, presented as a Kaplan-Meier survival plot, demonstrating that CamSA (■) offered protection (P = 0.001), compared to untreated control animals (□), whereas CapSA (●) did not (P = 0.75) and CaoSA (○) led to more rapid deaths (P = 0.001).
DISCUSSION
C. difficile spore germination is required for the establishment of CDI (11) and thus can be used as a target for CDI prophylaxis. Using the antigerminant CamSA, we reported the complete prevention of CDI in mice (20, 27). In this study, we extended antigermination therapy to the more stringent hamster model of CDI.
Reportedly, mice can be quite resistant to CDI and require an aggressive regimen of antibiotics to make them susceptible to infection (28–30). Others showed that mice can be quite susceptible to various C. difficile strains under different treatment strategies (21, 31, 32). Even so, mice must be challenged with a nonphysiologically large number of C. difficile spores. Murine CDI signs develop slowly and follow a predictable pattern (29, 33). Furthermore, in our experience, most mice recover within a few days (20, 27). The murine model used previously to test CamSA mimics human CDI in showing gradual increases in CDI signs, and the murine relapse model has been refined to resemble human relapse more closely (28, 34).
Due to their high susceptibility to CDI, hamsters have been the standard CDI model. Hamster CDI is readily fulminant after challenge with a few dozen spores (35). These extreme symptoms are rarely seen in primary human CDI (26, 35). Indeed, hamster CDI progresses so rapidly that it more closely resembles an intoxication than a developing infection. However, suboptimal vancomycin treatment of infected hamsters results in delayed sign onset, similar to human CDI relapse (23, 35). Testing the effects of CamSA in the hamster model of CDI and CDI after delayed vancomycin treatment allowed us to determine the limits of antigermination therapy. CamSA prevented CDI in mice in a dose-dependent manner, even when mice were challenged with 1 million times more C. difficile spores than in the current study (20). In the more susceptible hamster model, antigermination therapy delayed the onset of disease and prevented CDI in only 70% of hamsters when administered for ≥10 doses and in the presence of a suboptimal concentration of vancomycin.
In our hands, the antigerminant CamSA has lower efficacy in protecting hamsters from C. difficile strain 630, compared to mice; this finding is not unexpected. Previous reports have consistently shown that CDI treatment in hamsters results in worse outcomes than in a corresponding mouse model (36–39). Furthermore, treatment efficacy is dependent on the C. difficile strain used in the hamster model (40). The most recently approved CDI treatment, fidaxomicin, was able to protect 100% of hamsters from C. difficile strain ATCC 9689 (ribotype 001) (41) and 80% of animals from C. difficile strain BI1 (ribotype 027) but protected only 40% of animals from the same C. difficile strain 630 (ribotype 012) used in our experiments (25).
The role of the microbiome in preventing CDI has been used to develop probiotic treatments (42). A recent article reported that specific normal gut bacterial species, such as Clostridium scindens, can ameliorate CDI in mice (43). Differences in the microbiome before and after antibiotic disruption have been reported to produce changes in bile salt composition, which can affect C. difficile spore germination (44). Here we provide evidence, using deep sequencing of the V4 region of the 16S rRNA gene, that CamSA treatment for 30 days induced small but statistically significant changes in the gut microbiota. The small increase in Akkermansia is not suggestive of harmful dysbiosis. Akkermansia is an abundant gut inhabitant that has been linked to gut health and resistance to obesity (45, 46). The effects of minor shifts in unclassified Gastranaerophilales, Prevotellaceae, Lachnospiraceae, and Bacteroidetes species are difficult to interpret, since no close relatives of these organisms have been isolated and characterized. In addition, our study does not rule out other causes for changes in the microbiome during CamSA treatment. It is possible that changes were caused by aging, stress induced by oral gavage, or the dimethyl sulfoxide (DMSO) vehicle, rather than CamSA. Together, these data suggest that CamSA does not induce large-scale dysbiosis in the gut microbiome; however, more detailed work may be needed to understand more fully the effects of CamSA on the hamster gut microbiome.
In this study, CamSA alone was unable to prevent CDI development in hamsters even at high doses. Interestingly, CDI onset in CamSA-treated animals was delayed, suggesting that CamSA provides some antigermination effects. However, spore germination is an irreversible process and, even if CamSA inhibits the germination of most spores, residual spore germination into toxin-producing vegetative cells may be sufficient to cause colitis in hamsters. In contrast, due to their intrinsic resistance to CDI, potentially due to colonization resistance of natural microbiota, mice can tolerate the germination of a few spores without developing CDI signs (30, 42). It is important to note that in this study we were unable to test the direct effect of CamSA on spore germination in the hamsters, as was conducted in the murine study (20, 27). We were unable to recover C. difficile spores from fecal samples, likely due to the small number of C. difficile spores used for challenge (100 spores). Hypotheses and extrapolations based on the antigermination effects of CamSA in the hamster model are based on previous studies in vitro and in the murine model (18, 20, 27).
Vancomycin has been shown to temporarily protect hamsters from CDI, and the onset and severity of disease can vary, depending on whether vancomycin is administered at the time of infection or 1 day following infection (23, 24, 47). Furthermore, lower concentrations of vancomycin show suboptimal protection and result in delayed CDI onset (23, 47). Treatment of C. difficile-challenged hamsters with low-dose vancomycin failed to prevent CDI but delayed the onset of disease. Combining 300 mg/kg CamSA with suboptimal concentrations of vancomycin administered prophylactically protected most hamsters from both immediate and delayed CDI onset. In fact, surviving animals did not develop CDI even 30 days after CamSA treatment ended. In mice, 50 mg/kg CamSA was sufficient to protect animals from CDI (20), but this same dose did not prevent CDI in hamsters when administered alone (data not shown) or in combination with suboptimal doses of vancomycin. We postulate that the two treatments work synergistically. CamSA prevents most C. difficile spores from germinating, as seen previously in mice (27). Indeed, animals cotreated with CamSA and vancomycin showed significant reductions in C. difficile loads, compared to animals treated with vancomycin alone (compare Fig. 5A and B). C. difficile shedding throughout CamSA-vancomycin treatment remained constant over the course of 10 days, while animals treated with only vancomycin showed an increase in shedding of C. difficile between day 2 and day 6. CamSA treatment may also allow the excretion of ungerminated spores, thus significantly reducing the infective load of C. difficile (27). The number of spores required to initiate disease in hamsters, i.e., 100 spores, is 6 orders of magnitude lower than the number needed to infect mice (27, 29, 48). The greater spore load in mice allowed easy recovery of C. difficile from feces, for calculation of spore versus vegetative cell excretion (20, 27). Unfortunately, in this study we were unable to separate spores from vegetative cells; therefore, C. difficile found in hamster feces is reported as total C. difficile.
Vancomycin can prevent germinated spores from proliferating in the hamster gut. Indeed, there is a strong correlation between the transit time of C. difficile spores in the hamster gut and the length of CamSA treatment necessary to protect animals. Animals stopped shedding C. difficile cells by day 10 postchallenge, and antigermination therapy must be continued for at least the same amount of time to be effective. This correlation is strengthened by the finding that delaying vancomycin treatment for 24 h (day 1) eliminated CDI protection (Fig. 6B), with all animals succumbing by day 15, compared to 70% survival in animals treated with vancomycin on day 0 (Fig. 4B). This delay in vancomycin treatment potentially allows a few spores to germinate into vegetative cells, which could start producing toxins (49). At that point, hamsters' intestines are irrevocably compromised.
CamSA is a meta-aminosulfonate cholate derivative that has been shown to inhibit C. difficile spore germination effectively in vitro and in mice (18, 20) and to reduce the incidence of CDI in hamsters in combination with vancomycin. In contrast, the ortho-isomer (CaoSA) and the para-isomer (CapSA) of CamSA did not inhibit C. difficile spore germination in vitro (18). However, due to the highly dynamic nature of in vivo testing (50), these isomers were assessed for activity in hamster CDI. As expected, both CaoSA and CapSA were ineffective in the prevention of CDI in hamsters, even when combined with vancomycin. These data support earlier hypotheses that putative C. difficile spore germination receptors are very selective in binding germinants and inhibitors such as CamSA (19, 51, 52).
In conclusion, CamSA is a nontoxic, highly valuable tool for the investigation of C. difficile spore germination in rodent models of CDI. Importantly, CamSA does not cause major changes to the gut microbiota. CamSA reduces the incidence of CDI in hamsters when administered in conjunction with low doses of vancomycin. In this study, CamSA was unable to prevent CDI recurrence in hamsters. Antigermination treatments continue to prove to be a novel method worth further exploration for the prevention of CDI and the elucidation of this disease in vivo.
MATERIALS AND METHODS
Materials.
Bile salts were purchased from Sigma-Aldrich (St. Louis, MO) or were synthesized in the Abel-Santos laboratory (18). Clostridium difficile selective agar (CDSA) and brain heart infusion (BHI) medium were purchased from BD Biosciences (Franklin Lakes, NJ). ChromID C. difficile agar was purchased from bioMérieux (Marcy l'Etoile, France), and the PRO disc kit was purchased from Remel (Lenexa, KS). C. difficile strain 630 (ATCC BAA-1382) was purchased from ATCC (Manassas, VA). Reagents for DNA isolation were obtained from Qiagen (product no. 51504; Qiagen, Germantown, MD). Reagents for PCR were obtained from Quantabio (product no. 2200410; Quantabio, Beverly, MA).
Animals.
The institutional animal care and use committee at the University of Nevada, Las Vegas, approved the animal protocol (protocol R0411-266) used in this study. All experiments were performed according to the National Institutes of Health guidelines in the Guide for Care and Use of Laboratory Animals. Weaned female golden Syrian hamsters (3 to 4 weeks of age) were purchased from Harlan Laboratories (Indianapolis, IN) and Charles River (Wilmington, MA). Animals were housed individually and fed ad libitum. Water, food, and bedding were autoclaved prior to use. Animals were allowed to acclimate in the animal facility for 1 week. The following protocols involved animals 5 to 8 weeks of age. Animals were divided into experimental groups of 5, and each experiment was repeated at least twice, with the exception of the CamSA analog experiment (described below).
CamSA toxicity.
Three groups of 5 hamsters each were analyzed for CamSA-mediated toxicity. The first group was treated with 135 μl of DMSO, by oral gavage, once daily for 30 days, the second group was treated with 300 mg/kg CamSA dissolved in DMSO, by oral gavage, once daily for 30 days, and the third group received water ad libitum. Animals were weighed daily and observed for external signs of toxicity. Fecal samples were collected and analyzed as discussed below. On day 30, hamsters were sacrificed and their colons were removed for histological studies.
Histology.
Colon tissue was sectioned and prepared for histological analysis as described previously (24, 53). Briefly, tissues were snap-frozen in Cryo-OCT (Thermo Scientific, Waltham, MA) and stored at −80°C. Ultrathin sections were prepared using a Vibratome UltraPro 5000 cryostat set at 50 nm. Tissue samples were fixed for 15 min in cold acetone and stored at −80°C. Samples were stained using the Rapid Chrome hematoxylin and eosin (H & E) kit (Thermo Scientific, Waltham, MA) and visualized by light microscopy. Colon samples were blindly evaluated for epithelial damage, integrity of the mucosal lining, and separation of the submucosa.
Effects of CamSA on the hamster microbiome.
Fecal samples for microbiome analysis were collected aseptically, within 3 h after deposition, on day 0 and day 30 from a cohort of 5 hamsters treated with either DMSO or 300 mg/kg CamSA. Fecal samples from DMSO-treated hamsters and CamSA-treated hamsters were resuspended in Qiagen ASL buffer, flash frozen, and archived at −80°C. DNA was harvested from freshly thawed samples using the QIAamp DNA stool minikit and was quantified using a NanoDrop 2000 spectrophotometer. Amplification and sequencing of the V4 region of 16S rRNA genes of bacteria and archaea were performed as described by Kozich et al. (54), with the following modifications: (i) to be more inclusive of several archaeal lineages, forward primer 515F was modified to contain a C or T at the 4th position from the 5′ end (5′-GTGYCAGCMGCCGCGGTAA) (55), and a corresponding modification was made to the read 1 sequencing primer; (ii) 5 Prime HotMasterMix DNA polymerase was used; (iii) 33 cycles were used for PCR; and (iv) sequencing on the Illumina MiSeq platform was performed at Micro-Seq Enterprises (Las Vegas, NV). Paired-end Illumina MiSeq reads were quality filtered, aligned, and analyzed using Qiime2 (56). A single sample (CamSA treatment, day 30, hamster 5) yielded fewer than 1,000 quality sequences and was removed from analysis. The remaining reads were truncated at the first position with a PHRED score of <30, quality filtered, and clustered into sequence variants (SVs) using the q2-dada2 plugin (57) in Qiime2 version 2017.6 (56). Each SV was classified under the SILVA NR 99 reference database (version 128), using the classify-sklearn function of the q2-feature-classifier plugin (56). SVs identified as mitochondrial, chloroplastic, or from an unknown domain were removed. SVs were aligned using mafft (58) through the q2-alignment plugin, with default settings. Samples were rarified to contain 13,318 features. Alpha diversity metrics (richness, Shannon diversity, Faith's phylogenetic diversity, and Pielou's evenness) were calculated using the q2-diversity plugin (56). Kruskal-Wallis tests were performed in R version 3.4.1 (56). Proportions of taxa at each taxonomic rank were calculated using the R package phyloseq version 1.20.0 (59) and were visualized using ggplot2 version 2.2.1 (60). Bray-Curtis dissimilarity, Adonis, and SIMPER analyses were performed using phyloseq and vegan version 2.4.4 (61), and results were visualized with NMDS using vegan. Adonis partitions the variance similar to analysis of variance (ANOVA) and tests whether the variation within a category is smaller or larger than the variation between categories; it calculates a pseudo-F value, a P value, and a correlation coefficient (R2). Data categories with Adonis P values of <0.05 were considered significantly different.
Preparation of C. difficile spores.
Spores were purified following the method described by Howerton et al. (20). Briefly, C. difficile 630 was plated on BHI agar supplemented with 1% yeast extract, 0.1% l-cysteine HCl, and 0.05% sodium taurocholate, to yield single-cell clones. Individual C. difficile colonies were grown in BHI broth and replated to confluence. Plates were incubated anaerobically for 7 days at 37°C. Bacterial lawns were harvested by washing with ice-cold nanopure water and gentle scraping. The cell mass was pelleted and washed three times. Pellets were then centrifuged through a Histodenz gradient. Purified spores were washed and analyzed using the Shaeffer-Fulton staining method (62), to ensure >95% pure spores.
Before infection, C. difficile spores were heat activated at 68°C for 30 min and washed. Purified spores were resuspended in water to an optical density at 580 nm (OD580) of 1.0. Spore aliquots were serially diluted onto supplemented BHI agar to enumerate CFU.
Effects of CamSA on CDI.
To determine the effects of CamSA on hamster CDI, 30 animals were orogastrically dosed with 30 mg/kg clindamycin 1 day prior to infection (day −1). On day 0, animals were challenged with 100 C. difficile spores (25, 48). A test group (n = 15) was treated daily with CamSA at 300 mg/kg, by oral gavage, for 4 consecutive days starting at day −1. An infected control group (n = 15) was treated with DMSO, also by oral gavage. Fecal samples were collected using sterilized spatulas, at preselected time points (see below). Animals were observed twice daily and scored for CDI sign progression based on the following rubric: mild lethargy (score of 1), severe lethargy (score of 2), wet/sticky feces (score of 1), runny diarrhea (score of 2), wet tail and pink anogenital area (score of 1), wet anogenital area (score of 2), hunched posture (score of 2). Animals scoring ≥5 were considered moribund and were euthanized. Animals scoring ≤1 were indistinguishable from uninfected controls.
Enumeration of C. difficile from hamster feces.
Fecal samples were weighed and homogenized in sterile water to a concentration of 0.05 mg/ml. An aliquot of the fecal suspension was heated to 68°C for 15 min. Heated and unheated fecal samples were serially diluted and plated on CDSA and ChromID C. difficile selective agar (63, 64). Plates were incubated anaerobically for 48 h, and CFU were counted. CFU obtained from unheated samples represent the sum of C. difficile vegetative cells and spores. CFU obtained from heated samples represent the number of C. difficile spores only. The PRO disc kit was used to confirm C. difficile identity (63, 65).
Effects of coadministration of CamSA and vancomycin.
Animals were treated with 30 mg/kg clindamycin, by oral gavage, 1 day prior to challenge (day −1). On day 0, animals were challenged with 100 C. difficile spores, by oral gavage. Vancomycin (5 mg/kg) was administered once daily for 3 consecutive days starting at day 0, also by oral gavage (22–24). Starting at day −1, animals were treated once daily with 0 mg/kg (n = 10) or 300 mg/kg CamSA for 6 (n = 10), 10 (n = 10), or 19 (n = 10) consecutive days. Another group of animals (n = 4) was treated with a lower dose of CamSA (50 mg/kg) for 10 consecutive days or until the animals were moribund. CamSA or DMSO (control) was administered by oral gavage. CDI signs were scored up to 30 days post-CamSA treatment. Fecal samples were analyzed as described above. Selected animals from control groups and the 19-day CamSA treatment group were necropsied after euthanasia, their gastrointestinal tracts were removed and observed for visual signs of disease, and the colons were evaluated by histological staining.
Effects of coadministration of CamSA analogs CaoSA and CapSA on hamster CDI.
CamSA is a meta-aminosulfonate cholate analog. To determine whether the para- or ortho-isomers of CamSA prevent CDI in hamsters, C. difficile-challenged hamsters were treated with 5 mg/kg vancomycin, as described above. Starting at day −1, 300 mg/kg of the meta-analog CamSA (n = 10), the ortho-analog CaoSA (n = 5), or the para-analog CapSA (n = 5) (18) was administered once daily by oral gavage for 10 consecutive days or until animals became moribund. CDI signs were scored as described above.
Effects of coadministration of CamSA and vancomycin in a CDI delayed-treatment model.
Following previous models for CDI relapse (23, 24, 26, 66), clindamycin-treated hamsters were challenged with C. difficile spores, as described above. The beginning of vancomycin treatment (5 mg/kg) was delayed for 24 h after the challenge (day 1), and vancomycin was administered once daily for 5 days. Starting at day −1, animals were also administered 300 mg/kg CamSA once daily for 10 days (n = 10) or 17 days (n = 20) or until the animals became moribund. The remaining challenged animals (n = 10) were treated with DMSO. All treatments were administered by oral gavage. CDI signs were scored as described above.
Statistical analysis.
Microbiome data were analyzed statistically using pairwise Kruskal-Wallis tests (alpha diversity) or Adonis (beta diversity), as described in more detail above. Hamster survival was assessed by Kaplan-Meier analysis. Statistical comparisons of survival curves were calculated using a log-rank test in R. The scores for the severity of signs were analyzed as box-and-whisker plots. Data were expressed as the mean ± standard deviation in box-and-whisker plots. Standard deviations represent at least 3 independent measures. Student's unpaired t test was used to determine the significance of differences of means. The data on CFU per milligram of feces were analyzed as box-and-whisker plots. A two-way mixed ANOVA was performed to assess the significance of the differences of between-group means (CFU per milligram of feces for untreated hamsters versus CamSA-treated hamsters per day) and within-group means (changes in CFU per milligram of feces over time within experimental groups). The 5 hamsters that died by day 8 in the control group were excluded from this analysis because of the criteria for the ANOVA calculation.
Accession number(s).
Files containing the original unfiltered sequences are available from the NCBI Sequence Read Archive under project no. PRJNA376248. The following Qiime2-compatible supplemental files are available at https://github.com/hedlundb/MACamSA: metadata, unfiltered feature table, representative sequences, and SILVA taxonomy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grant R01-AI109139). A.H. was supported by the Nevada State College Provost-sponsored SEED grant program. B.P.H. was supported by a Faculty Opportunity Award from the University of Nevada, Las Vegas.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02251-17.
REFERENCES
- 1.Bartlett JG. 1992. Antibiotic-associated diarrhea. Clin Infect Dis 15:573–581. doi: 10.1093/clind/15.4.573. [DOI] [PubMed] [Google Scholar]
- 2.O'Brien JA, Lahue BJ, Caro JJ, Davidson DM. 2007. The emerging infectious challenge of Clostridium difficile-associated disease in Massachusetts hospitals: clinical and economic consequences. Infect Control Hosp Epidemiol 28:1219–1227. doi: 10.1086/522676. [DOI] [PubMed] [Google Scholar]
- 3.Spigaglia P, Barbanti F, Mastrantonio P, Brazier JS, Barbut F, Delmee M, Kuijper E, Poxton IR. 2008. Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. J Med Microbiol 57:784–789. doi: 10.1099/jmm.0.47738-0. [DOI] [PubMed] [Google Scholar]
- 4.Awad MM, Johanesen PA, Carter GP, Rose E, Lyras D. 2014. Clostridium difficile virulence factors: insights into an anaerobic spore-forming pathogen. Gut Microbes 5:579–593. doi: 10.4161/19490976.2014.969632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 6.Bartlett JG. 1981. Antimicrobial agents implicated in Clostridium difficile toxin-associated diarrhea of colitis. Johns Hopkins Med J 149:6–9. [PubMed] [Google Scholar]
- 7.Bartlett JG, Moon N, Chang TW, Taylor N, Onderdonk AB. 1978. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 75:778–782. [PubMed] [Google Scholar]
- 8.Savidge TC, Pan W-H, Newman P, O'Brien M, Anton PM, Pothoulakis C. 2003. Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology 125:413–420. doi: 10.1016/S0016-5085(03)00902-8. [DOI] [PubMed] [Google Scholar]
- 9.Pothoulakis C, Lamont JT. 2001. Microbes and microbial toxins: paradigms for microbial-mucosal interactions. II. The integrated response of the intestine to Clostridium difficile toxins. Am J Physiol Gastrointest Liver Physiol 280:G178–G183. doi: 10.1152/ajpgi.2001.280.2.G178. [DOI] [PubMed] [Google Scholar]
- 10.Peláez T, Alcalá L, Alonso R, Rodríguez-Créixems M, García-Lechuz JM, Bouza E. 2002. Reassessment of Clostridium difficile susceptibility to metronidazole and vancomycin. Antimicrob Agents Chemother 46:1647–1650. doi: 10.1128/AAC.46.6.1647-1650.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 12.Debast SB, Bauer MP, Kuijper EJ. 2014. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect 20(Suppl 2):1–26. doi: 10.1111/1469-0691.12418. [DOI] [PubMed] [Google Scholar]
- 13.Crobach MJT, Dekkers OM, Wilcox MH, Kuijper EJ. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin Microbiol Infect 15:1053–1066. doi: 10.1111/j.1469-0691.2009.03098.x. [DOI] [PubMed] [Google Scholar]
- 14.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. 2013. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol 108:478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 15.Fekety R. 1997. Guidelines for the diagnosis and management of Clostridium difficile-associated diarrhea and colitis. Am J Gastroenterol 92:739–750. [PubMed] [Google Scholar]
- 16.Wilcox MH, Fawley WN, Settle CD, Davidson A. 1998. Recurrence of symptoms in Clostridium difficile infection: relapse or reinfection? J Hosp Infect 38:93–100. doi: 10.1016/S0195-6701(98)90062-7. [DOI] [PubMed] [Google Scholar]
- 17.Surawicz CM, Alexander J. 2011. Treatment of refractory and recurrent Clostridium difficile infection. Nat Rev Gastroenterol Hepatol 8:330–339. doi: 10.1038/nrgastro.2011.59. [DOI] [PubMed] [Google Scholar]
- 18.Howerton A, Ramirez N, Abel-Santos E. 2011. Mapping interactions between germinants and Clostridium difficile spores. J Bacteriol 193:274–282. doi: 10.1128/JB.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howerton A, Patra M, Abel-Santos E. 2013. A new strategy for the prevention of Clostridium difficile infection. J Infect Dis 207:1498–1504. doi: 10.1093/infdis/jit068. [DOI] [PubMed] [Google Scholar]
- 21.Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. 2011. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2:145–158. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babcock GJ, Broering TJ, Hernandez HJ, Mandell RB, Donahue K, Boatright N, Stack AM, Lowy I, Graziano R, Molrine D, Ambrosino DM, Thomas WDJ. 2006. Human monoclonal antibodies directed against toxins A and B prevent Clostridium difficile-induced mortality in hamsters. Infect Immun 74:6339–6347. doi: 10.1128/IAI.00982-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong MY, Chang TW, Gorbach SL. 1987. Treatment of Clostridium difficile colitis in hamsters with a lipopeptide antibiotic, LY146032. Antimicrob Agents Chemother 31:1135–1136. doi: 10.1128/AAC.31.7.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kokkotou E, Moss AC, Michos A, Espinoza D, Cloud JW, Mustafa N, O'Brien M, Pothoulakis C, Kelly CP. 2008. Comparative efficacies of rifaximin and vancomycin for treatment of Clostridium difficile-associated diarrhea and prevention of disease recurrence in hamsters. Antimicrob Agents Chemother 52:1121–1126. doi: 10.1128/AAC.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sattar A, Thommes P, Payne L, Warn P, Vickers RJ. 2015. SMT19969 for Clostridium difficile infection (CDI): in vivo efficacy compared with fidaxomicin and vancomycin in the hamster model of CDI. J Antimicrob Chemother 70:1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anton PM, O'Brien M, Kokkotou E, Eisenstein B, Michaelis A, Rothstein D, Paraschos S, Kelly CP, Pothoulakis C. 2004. Rifalazil treats and prevents relapse of Clostridium difficile-associated diarrhea in hamsters. Antimicrob Agents Chemother 48:3975–3979. doi: 10.1128/AAC.48.10.3975-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howerton A, Patra M, Abel-Santos E. 2013. Fate of ingested Clostridium difficile spores in mice. PLoS One 8:e72620. doi: 10.1371/journal.pone.0072620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Wang H, Zhang Y, Chen K, Davis B, Feng H. 2011. Mouse relapse model of Clostridium difficile infection. Infect Immun 79:2856–2864. doi: 10.1128/IAI.01336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, Kelly CP. 2008. A mouse model of Clostridium difficile-associated disease. Gastroenterology 135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, Mastroeni P, Scott P, Raisen C, Mottram L, Fairweather NF, Wren BW, Parkhill J, Dougan G. 2009. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun 77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, Cheknis A, Figueroa I, Johnson S, Gerding D, Rood JI, Dougan G, Lawley TD, Lyras D. 2015. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio 6:e00551. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon SA, Hutton ML, Rood JI, Cheung JK, Lyras D. 2016. CdtR regulates TcdA and TcdB production in Clostridium difficile. PLoS Pathog 12:e1005758. doi: 10.1371/journal.ppat.1005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erikstrup LT, Aarup M, Hagemann-Madsen R, Dagnaes-Hansen F, Kristensen B, Olsen KEP, Fuursted K. 2015. Treatment of Clostridium difficile infection in mice with vancomycin alone is as effective as treatment with vancomycin and metronidazole in combination. BMJ Open Gastroenterol 2:e000038. doi: 10.1136/bmjgast-2015-000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douce G, Goulding D. 2010. Refinement of the hamster model of Clostridium difficile disease. Methods Mol Biol 646:215–227. doi: 10.1007/978-1-60327-365-7_14. [DOI] [PubMed] [Google Scholar]
- 36.Butler MM, Shinabarger DL, Citron DM, Kelly CP, Dvoskin S, Wright GE, Feng H, Tzipori S, Bowlin TL. 2012. MBX-500, a hybrid antibiotic with in vitro and in vivo efficacy against toxigenic Clostridium difficile. Antimicrob Agents Chemother 56:4786–4792. doi: 10.1128/AAC.00508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Locher HH, Seiler P, Chen X, Schroeder S, Pfaff P, Enderlin M, Klenk A, Fournier E, Hubschwerlen C, Ritz D, Kelly CP, Keck W. 2014. In vitro and in vivo antibacterial evaluation of cadazolid, a new antibiotic for treatment of Clostridium difficile infections. Antimicrob Agents Chemother 58:892–900. doi: 10.1128/AAC.01830-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghose C, Eugenis I, Sun X, Edwards AN, McBride SM, Pride DT, Kelly CP, Ho DD. 2016. Immunogenicity and protective efficacy of recombinant Clostridium difficile flagellar protein FliC. Emerg Microbes Infect 5:e8. doi: 10.1038/emi.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghose C, Eugenis I, Edwards AN, Sun X, McBride SM, Ho DD. 2016. Immunogenicity and protective efficacy of Clostridium difficile spore proteins. Anaerobe 37:85–95. doi: 10.1016/j.anaerobe.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Secore S, Wang S, Doughtry J, Xie J, Miezeiewski M, Rustandi RR, Horton M, Xoconostle R, Wang B, Lancaster C, Kristopeit A, Wang S-C, Christanti S, Vitelli S, Gentile M-P, Goerke A, Skinner J, Strable E, Thiriot DS, Bodmer J-L, Heinrichs JH. 2017. Development of a novel vaccine containing binary toxin for the prevention of Clostridium difficile disease with enhanced efficacy against NAP1 strains. PLoS One 12:e0170640. doi: 10.1371/journal.pone.0170640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson RN, Hardy DJ, Shipkowitz NL, Hanson CW, Ramer NC, Fernandes PB, Clement JJ. 1991. In vitro and in vivo evaluation of tiacumicins B and C against Clostridium difficile. Antimicrob Agents Chemother 35:1108–1111. doi: 10.1128/AAC.35.6.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theriot CM, Koenigsknecht MJ, Carlson PEJ, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li ZJ, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giel JL, Sorg JA, Sonenshein AL, Zhu J. 2010. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One 5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KCH, Aalvink S, Martinez LO, Dumas M-E, Maiter D, Loumaye A, Hermans MP, Thissen J-P, Belzer C, de Vos WM, Cani PD. 2017. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 46.Belzer C, de Vos WM. 2012. Microbes inside—from diversity to function: the case of Akkermansia. ISME J 6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Browne RA, Fekety RJ, Silva JJ, Boyd DI, Work CO, Abrams GD. 1977. The protective effect of vancomycin on clindamycin-induced colitis in hamsters. Johns Hopkins Med J 141:183–192. [PubMed] [Google Scholar]
- 48.Buckley AM, Spencer J, Candlish D, Irvine JJ, Douce GR. 2011. Infection of hamsters with the UK Clostridium difficile ribotype 027 outbreak strain R20291. J Med Microbiol 60:1174–1180. doi: 10.1099/jmm.0.028514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. 1985. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun 47:349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saeidnia S, Manayi A, Abdollahi M. 2015. From in vitro experiments to in vivo and clinical studies: pros and cons. Curr Drug Discov Technol 12:218–224. doi: 10.2174/1570163813666160114093140. [DOI] [PubMed] [Google Scholar]
- 51.Ramirez N, Liggins M, Abel-Santos E. 2010. Kinetic evidence for the presence of putative germination receptors in Clostridium difficile spores. J Bacteriol 192:4215–4222. doi: 10.1128/JB.00488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goulding D, Thompson H, Emerson J, Fairweather NF, Dougan G, Douce GR. 2009. Distinctive profiles of infection and pathology in hamsters infected with Clostridium difficile strains 630 and B1. Infect Immun 77:5478–5485. doi: 10.1128/IAI.00551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou W, Wang S, Dong H, Jiang H, Briggs BR, Peacock JP, Huang Q, Huang L, Wu G, Zhi X, Li W, Dodsworth JA, Hedlund BP, Zhang C, Hartnett HE, Dijkstra P, Hungate BA. 2013. A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS One 8:e53350. doi: 10.1371/journal.pone.0053350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP.. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 61.Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P, Stevens HH, Wangner H. 2009. vegan: community ecology package, version 2.2-1. https://CRAN.R-project.org/package=vegan.
- 62.Hamouda T, Shih AY, Baker JRJ. 2002. A rapid staining technique for the detection of the initiation of germination of bacterial spores. Lett Appl Microbiol 34:86–90. doi: 10.1046/j.1472-765x.2002.01047.x. [DOI] [PubMed] [Google Scholar]
- 63.Park KS, Ki C-S, Lee NY. 2015. Isolation and identification of Clostridium difficile using chromID C. difficile medium combined with Gram staining and PRO disc testing: a proposal for a simple culture process. Ann Lab Med 35:404–409. doi: 10.3343/alm.2015.35.4.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han SB, Chang J, Shin SH, Park KG, Lee GD, Park YG, Park Y-J. 2014. Performance of chromID Clostridium difficile agar compared with BBL C. difficile selective agar for detection of C. difficile in stool specimens. Ann Lab Med 34:376–379. doi: 10.3343/alm.2014.34.5.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fedorko DP, Williams EC. 1997. Use of cycloserine-cefoxitin-fructose agar and l-proline-aminopeptidase (PRO discs) in the rapid identification of Clostridium difficile. J Clin Microbiol 35:1258–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peterfreund GL, Vandivier LE, Sinha R, Marozsan AJ, Olson WC, Zhu J, Bushman FD. 2012. Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile. PLoS One 7:e46966. doi: 10.1371/journal.pone.0046966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.