Many bacteria are resistant to killing (tolerant) by typically bactericidal antibiotics due to their ability to counteract drug-induced cell damage. Vibrio cholerae, the cholera agent, displays an unusually high tolerance to diverse inhibitors of cell wall synthesis.
KEYWORDS: antibiotic tolerance, beta-lactam, cell wall synthesis, outer membrane, penicillin-binding proteins, peptidoglycan
ABSTRACT
Many bacteria are resistant to killing (tolerant) by typically bactericidal antibiotics due to their ability to counteract drug-induced cell damage. Vibrio cholerae, the cholera agent, displays an unusually high tolerance to diverse inhibitors of cell wall synthesis. Exposure to these agents, which in other bacteria leads to lysis and death, results in a breakdown of the cell wall and subsequent sphere formation in V. cholerae. Spheres readily recover to rod-shaped cells upon antibiotic removal, but the mechanisms mediating the recovery process are not well characterized. Here, we found that the mechanisms of recovery are dependent on environmental conditions. Interestingly, on agarose pads, spheres undergo characteristic stages during the restoration of rod shape. Drug inhibition and microscopy experiments suggest that class A penicillin binding proteins (aPBPs) play a more active role than the Rod system, especially early in sphere recovery. Transposon insertion sequencing (TnSeq) analyses revealed that lipopolysaccharide (LPS) and cell wall biogenesis genes, as well as the sigma E cell envelope stress response, were particularly critical for recovery. LPS core and O-antigen appear to be more critical for sphere formation/integrity and viability than lipid A modifications. Overall, our findings demonstrate that the outer membrane is a key contributor to beta lactam tolerance and suggest a role for aPBPs in cell wall biogenesis in the absence of rod-shape cues. Factors required for postantibiotic recovery could serve as targets for antibiotic adjuvants that enhance the efficacy of antibiotics that inhibit cell wall biogenesis.
INTRODUCTION
The emergence of antibiotic resistance in bacterial pathogens requires the development of new drugs and novel strategies to combat infection. However, antibiotic resistance is not the sole explanation for antibiotic treatment failures. Instead, some infections are caused by fully susceptible pathogens that are thought to survive antibiotic treatment due to a high level of drug tolerance, i.e., the capacity to stay alive in the presence of otherwise bactericidal drugs (1–4). Dormant persister cells, which resist killing by all available antibiotics (4), represent an extreme form of antibiotic tolerance. However, susceptible (nonpersister) bacteria are sometimes capable of surviving severe antibiotic-imposed damage, potentially providing an opportunity to acquire or evolve resistance mechanisms. In addition, surviving bacteria typically exhibit a prolonged lag phase after drug exposure (the postantibiotic effect), during which they can repair antibiotic-induced damage. Currently our knowledge of the molecular processes underlying antibiotic tolerance and the postantibiotic effect is limited. Understanding the mechanistic underpinnings of postantibiotic recovery could yield insights enabling the development of novel approaches to target tolerant organisms.
We and others have previously shown that some Gram-negative bacteria, including Burkholderia pseudomallei, Pseudomonas aeruginosa, and Vibrio cholerae, the causative agent of cholera, exhibit high tolerance to ordinarily bactericidal cell wall-acting antibiotics (e.g., beta lactams) (5–7). In V. cholerae, for example, exposure to beta lactam antibiotics at multiples of the MIC results in cell wall loss, similar to well-studied model organisms, such as Escherichia coli (5). However, in contrast to E. coli, V. cholerae survives as wall-deficient spheres, similar to L-forms (8), with the notable difference that V. cholerae spheres do not divide while in this wall-less state. Remarkably, however, the wall-deficient spherical cells remain viable and have minimal plating defects on medium lacking antibiotics (5). Sphere survival in vivo (in the mouse intestine) and in vitro is enabled by the two-component cell wall stress response system wigKR (also known as vxrAB), which controls several processes, including cell wall (peptidoglycan, or PG) biosynthesis, motility, type VI secretion, and biofilm formation (9–11).
The many steps in PG synthesis include cytoplasmic production of the lipid II precursor, translocation of this precursor into the periplasm, and finally precursor incorporation into the cell wall sacculus via polymerization (transglycosylation, or TG) and intercrosslinking (transpeptidation, or TP) reactions. TG and TP reactions are mediated by two spatiotemporally distinct entities, the Rod system (with RodA as the polymerase and a class B penicillin binding protein [bPBP] as the cross-linking enzyme) and the class A PBPs (aPBPs) that can catalyze both TG and TP reactions (12). In addition, aPBPs require outer membrane (OM)-localized lipoproteins (Lpos) for their activity (13, 14), and the Rod system is associated with (and requires for its activity) the cytoskeletal actin homolog MreB (15–18). Almost the entire V. cholerae PG synthesis pathway is upregulated through wigKR (vxrAB) in response to antibiotics that disrupt cell wall synthesis (9), with the notable exception of components of the Rod system.
We have little knowledge of how V. cholerae cell envelope biogenesis pathways enable recovery from the antibiotic-induced spherical state and if additional factors contribute to survival and recovery from this state. Here, we have characterized the postantibiotic recovery process in V. cholerae. Microscopy using fluorescent protein fusions and cell wall stains revealed that during an ordered sphere recovery process, aPBP1a localizes prominently to the outgrowth area, and its function appears to account for the majority of the initial deposition of new cell wall material. In contrast, the Rod system, which is ultimately required for sphere recovery, plays a minor role in the initial recovery stages. We also used transposon insertion sequencing (TnSeq) to identify the genetic requirements for V. cholerae tolerance to penicillin and found that there is an enrichment in genes important for cell wall and outer membrane biogenesis functions among mutations that confer postantibiotic fitness defects. Collectively, our findings reveal the pleiotropic nature of beta lactam tolerance, provide potential targets for beta lactam adjuvants, and have implications for the role of aPBPs in de novo PG template generation.
RESULTS
Distinct mechanisms of recovery under different growth conditions.
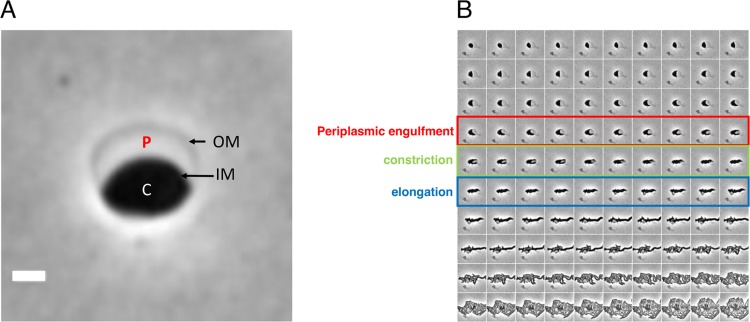
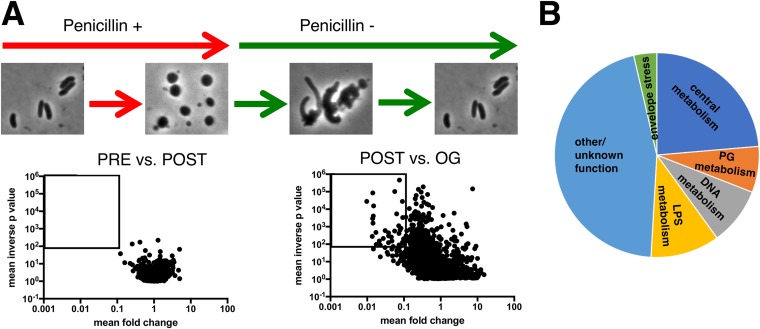
In previous work, we used microscopy to characterize V. cholerae sphere formation following exposure to antibiotics that interfere with cell wall synthesis (5). Here, we used a similar approach to investigate how spheres revert to rod shape. As observed previously, V. cholerae cells grown in minimal medium exposed to penicillin G (100 μg/ml, 10× MIC) form nondividing spheres exhibiting well-defined demarcations between the phase-dark cytoplasm, an enlarged periplasmic space visible as a phase-light bubble, and a clearly visible outer membrane (Fig. 1A). Time-lapse light microscopy was used to monitor cell morphology on agarose pads after removal of the antibiotic by washing. Under these conditions, approximately 10 to 50% of cells fully recovered to form microcolonies (see Movie S1 in the supplemental material for an example). While these conditions were not as favorable for recovery as plating on LB agar (5), they enabled us to discern steps in sphere recovery, which appeared to take place in partially overlapping stages in wild-type (wt) cells (Fig. 1B). Initially, phase-dark material engulfed the periplasmic space (engulfment stage), and then the now elliptically shaped cells reduced their widths (constriction phase), followed by elongation (elongation phase); finally, these elongated cell masses gave rise to rod-shaped cells, which proliferated into a microcolony.
FIG 1.
Recovery of V. cholerae rod morphology on agarose pads. (A) Sphere anatomy after 3 h of treatment with PenG. OM, outer membrane; IM, inner membrane; C, cytoplasm; P, periplasm. Cellular compartments were determined as described in reference 5 using fluorescent protein fusions with known localization patterns. Scale bar, 1 μm. (B) Representative time-lapse images of PenG-generated spheres after removal of the antibiotic on an agarose pad.
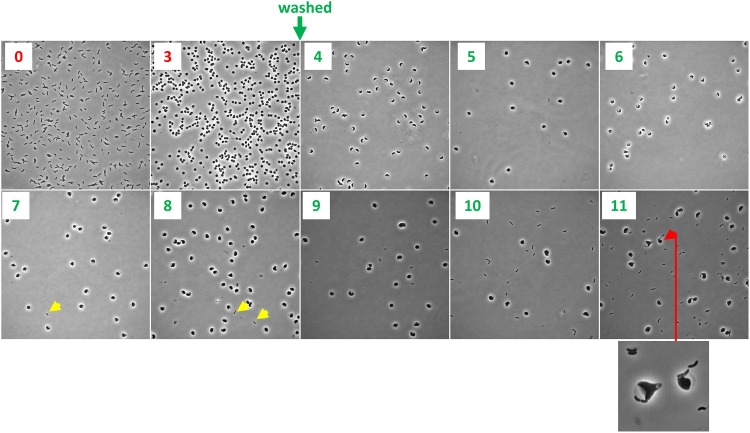
The pattern of recovery of rod shape described above is distinct from that described for osmostabilized, beta lactam-treated E. coli cells (19); however, the latter experiments were conducted in microfluidic chambers rather than agarose pads. Unlike E. coli, V. cholerae does not require osmostabilization for sphere formation; furthermore, V. cholerae spheres retain viability and structural integrity in LB and minimal medium, as well as in rabbit cecal fluid (5). Unlike the conditions in microfluidic chambers, agarose pads may provide external structural support to recovering spheres. Consistent with this idea, we found that the pattern and dynamics of recovery were very different when we repeated recovery experiments in liquid M9 minimal medium. Following exposure to PenG and washing, cells were intermittently removed from the liquid medium and imaged. We did not observe the distinct stages of recovery observed on agarose pads; in general, sphere morphology did not change for the duration of the experiment (12 h), except for a slight increase in volume (Fig. 2). However, normal, rod-shaped cells appeared after ∼4 to 5 h of postantibiotic incubation (Fig. 2, yellow arrow). We surveyed ∼100 cells per time point in each of two biological replicate experiments and did not find any intermediates, suggesting that if such intermediates form, they do so at a frequency of <1/100. The origin of the rod-shaped cells is not clear, but they may have directly budded off spheres from a newly formed pole juxtaposed to the periplasm, similar to the recovery protrusions observed in E. coli after treatment with beta lactams (19) or lysozyme (20). Indeed, we observed some rods that appeared to be budding off spheres (Fig. 2, red arrow). Thus, the morphological transitions and dynamics of sphere-to-rod conversion are dependent on specific culture conditions and may rely on distinct mechanisms.
FIG 2.
Sphere recovery in liquid medium. Cells were grown to a density of ∼2 × 108 CFU/ml (T0) in minimal medium, exposed to penicillin G (100 μg ml−1, 10× MIC) for 3 h (T3), washed twice to remove the antibiotic, and then imaged every hour. Yellow arrowheads show rod-shaped cells, and the red arrowhead (plus enlarged window) shows sphere apparently budding of a rod.
PBP localization dynamics during sphere recovery.
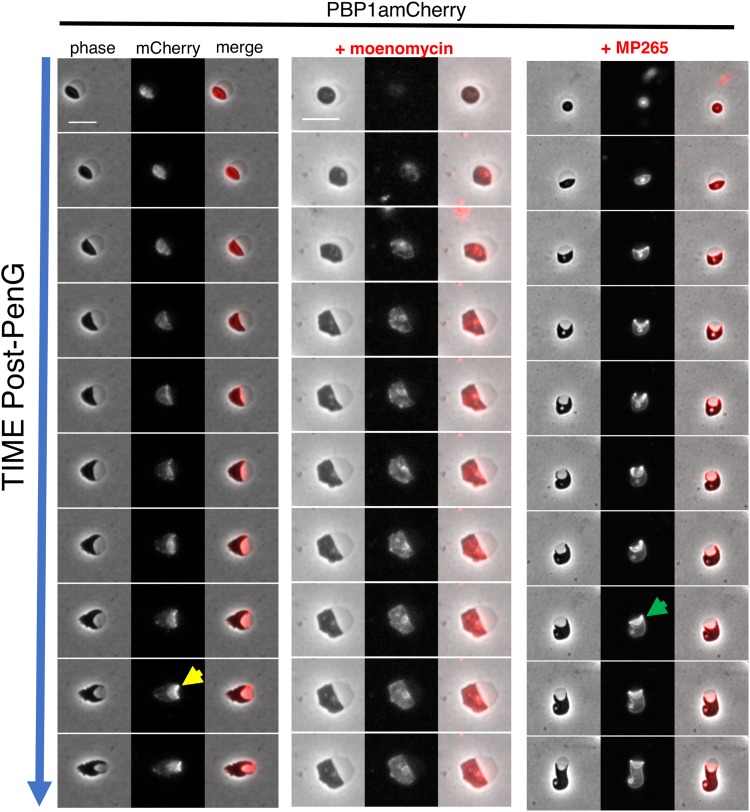
V. cholerae, like the model rod-shaped organisms E. coli and B. subtilis, encodes two distinct cell wall synthesis machineries, namely, the RodAZ-MreBC-PBP2 elongation complex (Rod system) and the aPBPs (15, 21, 22). Nearly the entire cell wall synthesis pathway, including aPBPs, is upregulated by the wigKR cell wall stress response two-component system. Members of the Rod system, however, are conspicuously absent from the wigKR regulon (9). We thus hypothesized that aPBPs were crucial determinants of postantibiotic recovery. To investigate the role of PBP1a, V. cholerae's primary aPBP (23) in the recovery process, we created a functional (Fig. S1) PBP1amCherry translational fusion and tracked its localization in recovering spheres on agarose pads. In the first stages of recovery, PBP1amCherry was diffuse, but then it assumed a striking, band-like pattern along the leading edge of the periplasmic engulfment, migrating ahead of the phase-dark cytoplasmic material (Fig. 3, yellow arrow). Inhibiting PBP1a's TG activity using moenomycin (10 μg ml−1, 10× MIC) arrested sphere recovery in the preengulfment stage and prevented proper PBP1a localization, suggesting that the recovery process depends on PBP1a's PG synthesis capabilities (or at least transglycosylation function). We also tested whether MreB was necessary for PBP1a's leading-edge localization by treating the recovering PBP1amCherry strain with the MreB inhibitor MP265 (24) (200 μM, 10× MIC). Inhibition of MreB suppressed recovery and completion of periplasmic engulfment, establishing that MreB is important for sphere-to-rod recovery, as shown before for E. coli (19). However, engulfment was only partially defective in spheres treated with MP265, and PBP1a still localized in a concentrated, band-like pattern in the presence of MP265 (Fig. 3, green arrow). Thus, while both MreB and PBP1a are important for recovery, aPBPs seems to function earlier than the Rod system in the process.
FIG 3.
PBP1a localizes to the leading edge of engulfment during sphere recovery. Cells were exposed to PenG (100 μg ml−1, 10× MIC) for 3 h, followed by washing and application to agarose pads containing either no antibiotic, the aPBP inhibitor moenomycin, or the MreB inhibitor MP265 (both at 10× MIC). Frames are 10 min apart. Scale bar, 5 μm. Arrows point to examples of ring-like localization of PBP1a in untreated spheres (yellow arrow) or those exposed to MP265 (green arrow).
The aPBPs are important for the initiation of cell wall synthesis early in sphere recovery.
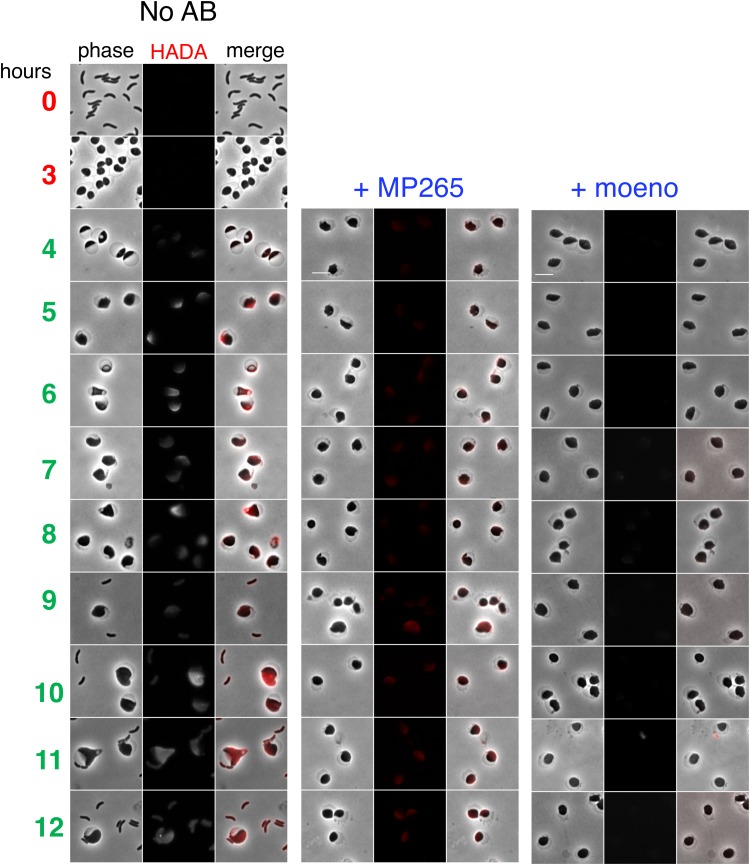
Since PBP1a was concentrated around the leading edge during the engulfment process, we hypothesized that the aPBPs are required for the commencement of cell wall synthesis after antibiotic-induced murein degradation. To test this, we treated cells with PenG, removed the antibiotic by washing, and then used the fluorescent d-amino acid HADA to visualize insertion of new cell wall material. A ΔldtA ΔldtB mutant defective in l,d-transpeptidase activity (25) was used in these experiments to exclude PG synthesis-independent HADA incorporation. In untreated spheres, cell wall deposition generally started at the opposite side of the periplasm (Fig. 4). This is likely the place where aPBPs and their OM activators interact first, as the inner and outer membranes are in close proximity in this area. In the presence of the MreB inhibitor MP265 (at 10× MIC), initial cell wall deposition was reduced compared to that of untreated spheres but remained detectable. In contrast, when cells were incubated with moenomycin (10× MIC), incorporation of HADA-labeled material was drastically reduced (Fig. 4; see Fig. S2 for an image adjusted to lower dynamic range). It follows that while both the aPBPs and MreB are required for sphere recovery, the aPBPs are more active than the Rod system in producing nascent PG in recovering spheres.
FIG 4.
Deposition of new cell wall material in recovering spheres is primarily mediated by aPBPs. N16961 ΔldtA ΔldtB cells were exposed to PenG (100 μg ml−1, 10× MIC) in M9 minimal medium for 3 h (T0 and T3), followed by washing and resuspension in fresh, prewarmed M9 containing the fluorescent d-amino acid analog HADA as a cell wall label (4 to 12 h). Scale bar, 5 μm. The MreB inhibitor MP265 was added at 200 μM (10× MIC) and the aPBP inhibitor moenomycin at 10 μg ml−1 (10× MIC).
Identification of genes required for postantibiotic recovery.
We used transposon insertion site sequencing (TIS; also known as TnSeq) to identify factors required for sphere recovery. Since we observed differences in recovery dynamics on solid (agarose pad) versus in liquid media, we combined both conditions in TIS experiments to uncover a broad array of recovery factors. Cells were exposed to PenG in liquid culture for 4 h, washed, outgrown overnight in the absence of antibiotics, and then plated. The insertion sites in the Tn library were sequenced before addition of the antibiotic (PRE), after incubation in PenG (POST), and after the overnight outgrowth followed by plating (OG). Strikingly, comparison of the insertion profiles under the PRE and POST conditions (Fig. 5A) did not reveal any genes that met stringent criteria for differential fitness (>10-fold difference in insertion abundance; P < 0.01). Thus, no single mutation appears to lead to catastrophic lysis in V. cholerae treated with penicillin G. In contrast, comparing the insertion profiles under the POST versus OG conditions revealed 55 genes which had reduced fitness during postantibiotic outgrowth (>10-fold fewer insertions; P < 0.01).
FIG 5.
Identification of genes required for sphere recovery with TnSeq. (A) Schematic of experimental design and volcano plots of change in relative abundance of insertion mutants between the two conditions (x axis) versus the concordance of independent insertion mutants within each gene (y axis, inverse P value). The square denotes the cutoff criteria applied (>10-fold fitness defect, P < 0.01) for identification of genes contributing to sphere recovery. PRE versus POST is a comparison of insertion frequencies after/before antibiotic exposure, and POST versus OG is a comparison between an outgrowth period and directly after antibiotic exposure (see Materials and Methods for details). (B) Distribution of the main functional categories of gene insertions that confer a postantibiotic fitness defect. Functional categories were assigned manually following annotation of individual proteins in the Kegg database (http://www.genome.jp/kegg/).
Notably, there was an enrichment of several functional pathways among the 55 genes required for robust postantibiotic outgrowth (Fig. 5B). Included among the enriched categories were genes predicted to be required for cell wall synthesis and recycling (pbp1A, vc2153, ampG, pbp5, lpoB, and mltA), lipopolysaccharide (LPS) biosynthesis (core biosynthesis, lipid A acylation, and O-antigen synthesis pathways, vc0212, vc0223, vc0225, vc0236, vc0237, and vc0240), intrinsic stress resistance (superoxide dismutase, rpoE), phosphate uptake (vc0724-0726), and chromosome dynamics (mukBEF) (Fig. S3). Intriguingly, some of the hits (vc2153, rpoE, and PG synthesis factors) were homologs or analogs of factors identified in a recent TnSeq screen for genes that promote tolerance to beta lactams in Burkholderia pseudomallei and B. thailandensis (6), raising the possibility that there are shared tolerance requirements across Gram-negative bacteria.
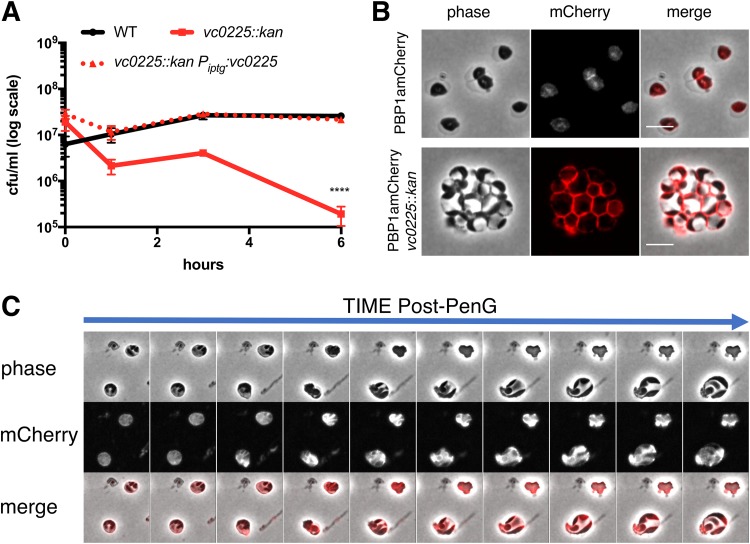
We were particularly interested in the contribution of cell envelope functions to sphere recovery and therefore prioritized genes involved in LPS and cell wall metabolism for further studies. We first focused on the 6 LPS biosynthesis genes that answered our screen. To validate the requirement of LPS core biosynthesis in the recovery process, we created an insertion mutant in vc0225, the gene encoding heptosyltransferase I. This mutation is expected to result in a truncated LPS molecule lacking an outer core and O-antigen, and consistent with this, the mutant strain did not have detectable high-molecular-weight (HMW) LPS in isolated OM material (Fig. S4). wt and vc0225::kan mutant cells were compared in time-dependent viability experiments. Exposing wt cells to penicillin G (at 10× MIC) in minimal medium (unlike LB) in some experiments permitted initial growth (Fig. 6A). We do not know the reason for this initial growth. V. cholerae does not become resistant to PenG in M9, as evidenced by sphere formation (Fig. 6B), but it is possible that antibiotic diffusion through the OM is slower in minimal than in rich medium. Disruption of the LPS core gene vc0225 resulted in the absence of initial growth and a subsequent 100-fold plating defect after exposure to PenG (Fig. 6A), corroborating the TnSeq result. This survival defect could be complemented by expressing vc0225 from a neutral chromosomal locus. Light microscopy revealed that the vc0225 mutant strain still formed spheres (Fig. 6B); however, these spheres were morphologically distinct from wt spheres. In contrast to wt spheres, which were usually seen as single cells, exhibiting well-demarcated separation between the phase-dark cytoplasm and the phase-light periplasm (Fig. 1A), vc0225 mutant cells were mostly grape-like masses showing a checkered pattern of distinct periplasmic enclaves in a sometimes divided cytoplasm (Fig. 6B). Visualizing an inner membrane (IM) marker (PBP1amCherry) also revealed the lack of a clear distinction between the inner and outer membrane in the mutant. Upon removal of the antibiotic, vc0225 spheres were defective in all stages of the recovery process; the spheres underwent modest enlargement without initiating periplasmic engulfment (Fig. 6C). Thus, intact LPS appears necessary for sphere anatomy and internal organization; moreover, these LPS-associated sphere defects seem to impair sphere recovery.
FIG 6.
An LPS core mutant is defective in sphere recovery and organization. (A) Time-dependent viability experiment in the presence of PenG. Strains were exposed to PenG (100 μg ml−1, 10× MIC) at T0 and plated for CFU ml−1 at the indicated times. The vc0225 mutant exhibits a significant plating defect by T3 (****, P < 0.0001). (B) Localization of PBP1a in PenG-treated wt (top) and mutant spheres (bottom). PBP1amCherry was expressed from its native, chromosomal locus. Scale bar, 5 μm. (C) Recovery pattern in a PenG-treated PBP1amCherry vc0225 mutant after exposure to and subsequent removal of PenG. Frames are 10 min apart.
Sphere integrity does not depend on lipid A modifications.
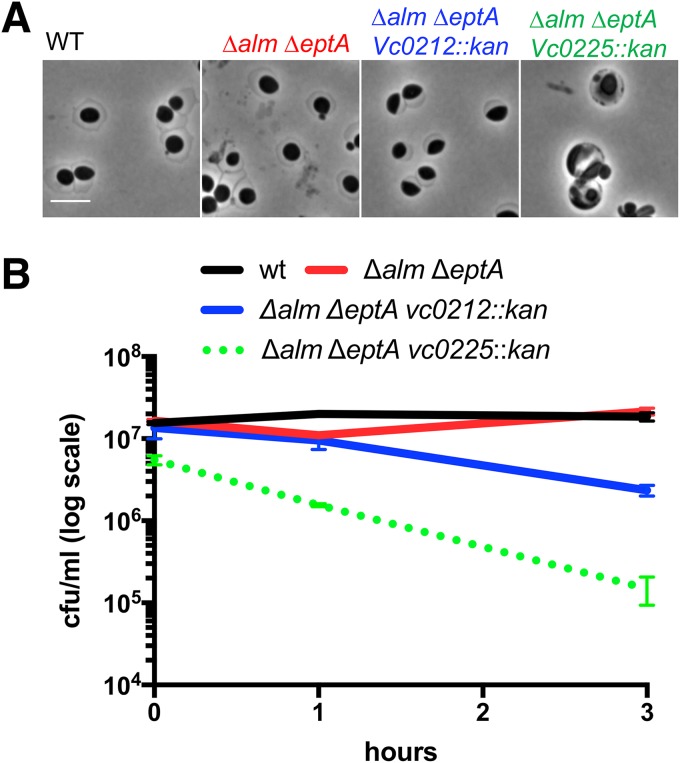
The outer membrane appears to be the principle load-bearing structure in beta lactam-induced spheres, because these cells are largely devoid of detectable cell wall material and are more susceptible to detergents and antimicrobial peptides (5, 7). This is consistent with recent evidence that the Gram-negative OM equals or even surpasses the cell wall in stiffness (26). V. cholerae LPS contains at least two modifications that are not found in E. coli LPS and that potentially stabilize V. cholerae spheres. These modifications, addition of phosphoethanolamine to the 1-phosphate group of lipid A and an unusual glycine addition to a hydroxylauryl chain at the 2′ position of lipid A, both promote resistance to polymyxin (27, 28); glycinylation (by the alm system) is dominant, but the pH-dependent EptA can promote residual polymyxin resistance when the alm system is inactivated (22). We investigated whether these modifications were required for sphere formation and integrity by deleting the alm operon, which encodes the glycine transferase activity, and eptA, which encodes the ethanolamine transferase. As expected, the alm mutation abrogated polymyxin resistance on LB (Fig. S5). When these mutants, either alone (not shown) or in combination, were exposed to PenG they formed spheres that were indistinguishable from wt spheres (Fig. 7A), indicating that these lipid A modifications are not required for sphere generation. The Δalm ΔeptA mutations were then combined with disruptions in vc0225 or vc0212 (encoding the 3-hydroxy laurate transferase LpxN [29]) to test the effect of a core mutation (vc0225) or lipid A underacylation (vc0212) on sphere formation in LPS lacking glycine and ethanolamine modifications. (Note that mutation of lpxN results in the absence of the acyl chain modified by glycine and thus causes polymyxin B sensitivity; Fig. S5). The Δalm ΔeptA vc0212::kan mutant still formed spheres after PenG exposure (Fig. 7A), but these spheres had an ∼5-fold decrease in viability compared to that of the wt (Fig. 7B). The Δalm ΔeptA vc0225::kan mutant also formed spheres but resulted in a more dramatic reduction in viability than the Δalm ΔeptA vc0212::kan mutant (Fig. 7B) and to the single vc0225::kan mutant, where there was less pronounced loss of viability after 3 h (compare with Fig. 6A). Thus, LPS core and O-antigen appear to be more critical for sphere formation/integrity and viability than lipid A modifications. However, glycinylation and/or ethanolamine addition to lipid A appear to promote maintenance of sphere integrity in the absence of LPS core and O antigen, suggesting that these modifications contribute to OM stability in this context.
FIG 7.
Outer membrane modifications are not required for sphere formation. (A) Sphere formation in the wt and mutants defective in glycine (alm) and ethanolamine (eptA) modifications alone and in combination with defects in LPS core biosynthesis (vc0225) and lipid A acylation (vc0212). Scale bar, 5 μm. (B) Time-dependent viability experiment as described for Fig. 4A.
The sigma E cell envelope stress response is required for penicillin tolerance.
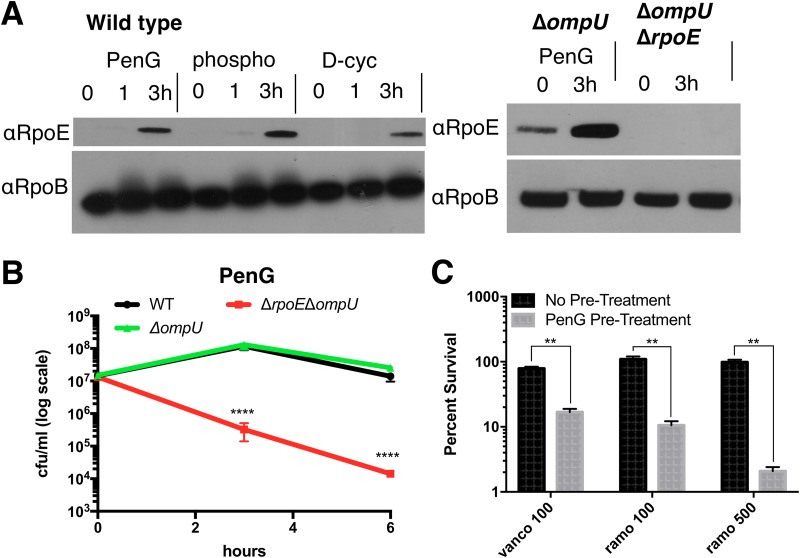
The TnSeq analysis implicated several genes in the sigma E cell envelope stress response as important for sphere viability/recovery (Fig. S3). Misfolding of outer membrane proteins such as OmpU triggers the V. cholerae envelope stress response, wherein sigma E directs the transcription of a set of genes involved in a variety of cell envelope maintenance functions (30, 31). We found that the abundance of RpoE markedly increased several hours after cells were exposed to diverse antibiotics that interrupt cell wall synthesis (PenG, phosphomycin, or d-cycloserine), consistent with the idea that this sigma factor promotes sphere survival (Fig. 8A). Interestingly, PenG treatment increased RpoE abundance independent of OmpU (Fig. 8B). The importance of sigma E for survival after PenG exposure was established by measuring time-dependent viability after antibiotic challenge. Since rpoE is essential, we used an ΔrpoE ΔompU strain (the latter deletion enables rpoE deletion [31]) to investigate rpoE's importance for sphere viability/recovery. Following exposure to PenG, the ΔrpoE ΔompU strain exhibited a drastic (∼1,000-fold) plating defect compared to the wild-type and ΔompU controls (Fig. 8B). Thus, sigma E (and presumably the regulon it controls) is a crucial determinant of V. cholerae beta lactam tolerance.
FIG 8.
Sigma E is induced in response to cell wall-acting antibiotics and is required for beta lactam tolerance. (A) Western blot using anti-RpoE antiserum after exposure to penicillin G (PenG; 100 μg ml−1, 10× MIC), phosphomycin (phospho; 100 μg ml−1, 3× MIC), or d-cycloserine (D-cyc; 100 μg ml−1, 3× MIC). (B) Time-dependent viability of indicated strains after exposure to PenG (100 μg ml−1, 10× MIC). The ompU rpoE mutant exhibits a significant plating defect by T6 (****, P < 0.0001). (C) Pretreatment with PenG sensitizes cells to high-molecular-weight antibiotics (**, P < 0.01). Cells were exposed to either vehicle (no pretreatment) or penicillin G (PenG; 100 μg ml−1) for 3 h, followed by plating on either LB, vancomycin (vanco; 100 μg ml−1), or ramoplanin (ramo; 100 μg ml−1 or 500 μg ml−1). Survival is calculated as a ratio of CFU/ml on antibiotic over CFU/ml on LB within the same pretreatment group.
The importance of the sigma E response for beta lactam tolerance indicated the strong possibility that PenG-treated V. cholerae sustains OM damage; this in turn suggested that beta lactam exposure sensitizes cells to HMW antibiotics that are typically too large to permeate the Gram-negative cell envelope; e.g., vancomycin and ramoplanin. To test this, we plated cells exposed to PenG or a vehicle control on either LB or plates containing vancomycin (100 μg ml−1) or ramoplanin (100 μg ml−1 and 500 μg ml−1) (Fig. 8C). While untreated cultures plated at close to 100% on any of these plates, pretreatment with PenG for 3 h resulted in a 10- to 50-fold plating defect on either HMW antibiotic. Thus, while V. cholerae is tolerant to beta lactam antibiotics, these agents appear to sensitize it against HMW antibiotics.
DISCUSSION
Antibiotic tolerance, the ability to survive and fully recover from exposure to normally lethal doses of bactericidal antibiotics, is a common cause of treatment failure and serves as a stepping stone for the development of antibiotic resistance (32). The mechanisms of antibiotic tolerance and a related phenomenon, the postantibiotic effect (the recovery process of tolerant cells), are understudied and insufficiently understood.
Here, we investigated the postantibiotic effect in V. cholerae, an organism highly tolerant to typically bactericidal inhibitors of cell wall synthesis. Following beta lactam treatment, this pathogen forms viable cell wall-deficient spheres that can reestablish their characteristic rod morphology when antibiotics are no longer present. We found that the process of restoration of rod shape appears to be medium dependent and to differ from that described for the recovery of E. coli spheres induced by cefsulodin (19). At least on agarose pads, 3 successive steps characterized the recovery process. Recovery involves relocalization of PBP1a to the leading edge of a periplasmic engulfment process, and aPBPs, rather than the Rod system, have a primary role, particularly in the early steps of recovery. The characteristic steps and protein localization patterns involved in restoration of rod shape suggest that V. cholerae's ability to recover from a wall-less spherical state is a previously unappreciated type of programmed response to stress. Furthermore, the capacity of wall-less spheres to remain viable and to regain rod shape in a stepwise fashion is not restricted to V. cholerae but found in several other Gram-negative organisms (7, 20, 33).
Not surprisingly, TnSeq analysis revealed that genes required for peptidoglycan biogenesis were essential for sphere recovery; however, additional envelope-related functions, particularly LPS biosynthesis, and the RpoE envelope stress response system were also critical for V. cholerae spheres to recover from beta lactam assault. While this TnSeq data set offers many intriguing leads for future studies of the postantibiotic recovery process (e.g., the role of riboflavin and condensins), here we primarily focused on the role of LPS and PG biosynthesis.
Mutations in LPS core and O-antigen biosynthesis can have at least two potentially detrimental consequences for sphere viability/recovery. First, truncated (rough) LPS has been shown to result in increased OM permeability due to the surface presentation of phospholipid bilayer patches resulting from LPS instability (34). Second, truncated LPS, which cannot be ligated to O-antigen, results in the accumulation of dead-end intermediates of O-antigen fused to the lipid carrier molecule undecaprenol (UNDP) (35). UNDP concentrations are limited, and this carrier is critical for the biosynthesis of a variety of extracellular macromolecules, including peptidoglycan. Thus, in addition to direct effects on OM integrity, LPS core mutations can also impede efficient cell wall synthesis due to the reduced availability of UNDP; the consequences of such inhibition may be particularly severe for PG-deficient spheres. However, LPS core mutants exhibited postantibiotic exposure phenotypes that cannot easily be explained by cell wall precursor depletion alone. The absence of the clear demarcation between the inner and outer membrane in the LPS core (vc0225 mutant)-deficient spheres suggests that IM material (IM phospholipids and associated proteins) is present in the OM. More broadly, our findings demonstrate the importance of LPS integrity for V. cholerae survival of cell wall damage; it is likely that LPS structure/strength modulates susceptibility to beta lactams in other bacteria as well, as has been suggested in E. coli (36).
As expected, we also found cell wall synthesis and recycling factors to be required for sphere recovery. The prominent role of PBP1a, rather than its paralog, PBP1b, in the recovery process supports our previous data showing that PBP1a is the principle aPBP in V. cholerae (23, 37) and is consistent with observations in lysozyme-treated, spherical E. coli, where that organism's principal aPBP (PBP1B) is required for recovery (33). Intriguingly, PBP1a localized as a concentrated ring around the outgrowth area. Moenomycin, an aPBP TG inhibitor, abrogated PBP1a's capacity to localize to the outgrowth area as well as sphere recovery, revealing the essentiality of aPBP enzymatic activity for the recovery process. In contrast, MreB, and by extension likely the associated Rod system, appeared to play a minor role during the early stages of recovery (though MreB was ultimately necessary for full sphere-to-rod conversion). We do not know the exact structure of possible remnant PG in spheres; however, our results suggest that aPBPs are more efficient than the Rod system at starting PG synthesis in the absence of a rod-shaped scaffold. Recent data suggest that MreB determines the directionality of Rod-mediated PG synthesis through its axial, membrane-curvature-induced orientation in the cell (38). Our data are in line with such a model, as axial localization cues are lost in a sphere. Thus, the Rod system might rely on the aPBPs to first mediate some degree of sphere constriction, inducing heterogeneity in membrane curvature that would then enable ordered, Rod-mediated PG deposition resulting in cell elongation.
In summary, we provide here an analysis of factors required for postantibiotic recovery in V. cholerae treated with a beta lactam antibiotic. Our results directly demonstrate a role for OM integrity in beta lactam tolerance and establish a differential role for aPBPs and the Rod system for postantibiotic recovery. The factors identified here could serve as novel targets for antibiotic adjuvants that increase the efficacy of beta lactam antibiotics and other inhibitors of cell wall synthesis toward Gram-negative pathogens.
MATERIALS AND METHODS
Media, chemicals, and growth conditions.
All growth experiments were conducted either in Luria-Bertani broth (LB) or M9 minimal medium supplemented with glucose (0.2%). Antibiotics (suppliers) used were the following: moenomycin (Santa Cruz), MP265 (Santa Cruz), penicillin G (Fisher), and streptomycin (Santa Cruz). Unless otherwise noted, all experiments were conducted in three biological replicates (i.e., experiments conducted on different days). For each experiment, two independent overnight cultures were used; where possible (i.e., for mutants constructed in this study), these cultures were inoculated from two independently isolated clones.
Molecular techniques/strain construction.
For molecular cloning purposes, PCR was conducted using Q5 high-fidelity polymerase (NEB). For diagnostic PCRs, OneTaq mastermix (NEB) was used instead. All cloned constructs were verified using Sanger sequencing of the cloned insert. All plasmid cloning was done using isothermal assembly (ITA) (39). Oligonucleotides are summarized in Table S1 and strains in Table S2 in the supplemental material. Unless otherwise noted, all strains were constructed in the El Tor N16961 background (40).
Unless otherwise noted, mutants were constructed as previously described using the suicide plasmid pCVD442 (41) and homologous recombination to replace genes with the sequence TAATGCGGCCGCACTCGAGTAATAATGATGA. Briefly, the E. coli donor strain Sm10, carrying a pCVD-based deletion plasmid, was mixed 1:1 with the V. cholerae recipient on an LB plate and incubated for at least 2 h at 37°C. The cell mixture was then streaked out on a plate containing carbenicillin (100 μg/ml) and streptomycin (200 μg/ml) to select against the donor strain and for recipients that have integrated the deletion plasmid. To counterselect against pCVD, a single colony was then streaked out on sucrose agar (15 g/liter agar, 10 g/liter tryptone, 5 g/liter yeast extract, filter-sterilized sucrose added after autoclaving to 10% final concentration) plus streptomycin. Plates were incubated at ambient temperature for 1 day and then transferred to 30°C, followed by additional incubation for 1 or 2 days. Successful mutants were verified via colony PCR using primers flanking the gene of interest.
For the vc0225 disruption, sacB-based counterselection did not work due to the inability of LPS core mutants to grow on sucrose agar. We therefore used a kanR (conferring kanamycin resistance) variant of the single-crossover suicide vector pGP704 (42) to create insertion disruption mutants. To this end, a 400-bp internal fragment (nucleotide positions 27 to 427) of vc0225 or a 461-bp internal fragment (nucleotide positions 93 to 554) of vc0212 was cloned into pGPkan and transferred into recipient V. cholerae using the diamino pimelic acid (DAP)-auxotrophic E. coli donor strain MFD lambda pir (43). Insertion mutants were selected on plates containing streptomycin (200 μg/ml) and kanamycin (50 μg/ml).
Procedures for the construction of other knockout plasmids and strains. (i) Pbp1a::pbp1amcherry.
Upstream and downstream regions were amplified using primers TDP 1362/1434 and TDP 1435/232, respectively, and fused with mCherry (amplified using primers 1436/1437) and Xba1-digested pCVD442 using ITA. These primers insert the linker sequence GACATCCTCGAGCTC between PBP1a (no stop codon) and mCherry.
(ii) Δalm ΔeptA plasmids.
For the Δalm plasmid, upstream and downstream homologies were amplified using primers 519/520 and 521/522, respectively. For the ΔeptA plasmid upstream and downstream homology regions were amplified using primers 539/540 and 541/542.
TnSeq.
TnSeq was conducted as described before (44–46); briefly, cultures were subjected to transposon (mariner) mutagenesis in duplicate in LB medium. In the resulting libraries (∼200,000 colonies/replicate), whole-population transposon-chromosome junctions were sequenced (PRE sample, see below); the libraries were then frozen in 30% glycerol (−80°C). For the experiment, libraries were grown to an optical density at 600 nm (OD600) of ∼0.5 and then exposed to penicillin G (100 μg/ml, 10× MIC) for 4 h and sequenced again (POST sample), followed by washing to remove the antibiotic and outgrowth in LB medium overnight, after which the libraries were sequenced again (outgrowth, or OG, sample). Sequencing was performed as follows. Pelleted libraries were lysed and DNA was isolated as described before (43) and then fragmented using NEB fragmentase, followed by blunting (blunting enzyme mix; NEB), A-tailing, and ligation of specific adaptors. Transposon-DNA junctions were then PCR amplified using transposon- and adaptor-specific primers. The libraries were then sequenced on an Illumina MiSeq as described before (43). Data analysis was conducted essentially as described previously (44–46); however, to avoid false negatives that did not pass our stringent cutoff, we also used a candidate-based approach (based on known genetic interactions between cell envelope functions) to visually inspect the TnSeq data set in the genome browser Artemis (this approach yielded, e.g., WigKR and RpoE).
Recovery on agarose pads.
For recovery time lapses, overnight cultures were diluted 100-fold into fresh M9 minimal medium and then grown to an OD600 of 0.3 (3.5 h) at 37°C with shaking. Antibiotic was then added, followed by incubation for another 3 h. Cells were then washed twice in antibiotic-free medium and applied to agarose patches (0.8% agarose in M9) and imaged every 5 min on a Leica DMi8 inverted microscope with an incubated (30°C) stage. For fluorescent readings, exposure time was 500 ms (mCherry), 300 ms (msfGFP), or 1,000 ms (HADA).
Recovery in liquid medium.
Overnight cultures were diluted 100-fold into fresh M9 minimal medium and then grown to an OD600 of 0.3 (3.5 h) at 37°C with shaking. Antibiotic was then added, followed by incubation for another 3 h. Cells were then washed twice in antibiotic-free medium and diluted 10-fold into the same medium containing 100 μM HADA. Cells were withdrawn at the indicated time points, washed once with M9, and imaged as detailed above.
Statistical analyses.
The time-dependent viability experiments in Fig. 6A and 8B were analyzed using a 2-way analysis of variance (ANOVA) on a linear mixed-effects model with fixed effects of strain, time, and the log10 (CFU/ml) and a random effect to control for repeated measurements taken from the same culture over time. Similarly, the plating experiment shown in Fig. 8C was analyzed using a 2-way ANOVA on a linear mixed-effects model with fixed effects of pretreatment, plating antibiotic, and the survival ratio and a random effect to control for repeated measurements of the same culture on multiple plating conditions. P values from all tests were adjusted with a Bonferroni correction, and all error bars represent standard errors of the means (SEM).
Supplementary Material
ACKNOWLEDGMENTS
We thank Hongbaek Cho and Thomas Bernhardt (Harvard Medical School) for the msfGFP template. Carol Gross (UCSF) is acknowledged for her gift of the anti-RpoE antibody.
Research in the Waldor laboratory is supported by the Howard Hughes Medical Institute and the NIH (2R01-AI-042347-23). Research in the VanNieuwenhze laboratory is supported by NIH grant GM113172.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01326-18.
REFERENCES
- 1.Meylan S, Andrews IW, Collins JJ. 2018. Targeting antibiotic tolerance, pathogen by pathogen. Cell 172:1228–1238. doi: 10.1016/j.cell.2018.01.037. [DOI] [PubMed] [Google Scholar]
- 2.Conlon BP, Rowe SE, Lewis K. 2015. Persister cells in biofilm associated infections. Adv Exp Med Biol 831:1–9. doi: 10.1007/978-3-319-09782-4_1. [DOI] [PubMed] [Google Scholar]
- 3.Sharma B, Brown AV, Matluck NE, Hu LT, Lewis K. 2015. Borrelia burgdorferi, the causative agent of lyme disease, forms drug-tolerant persister cells. Antimicrob Agents Chemother 59:4616–4624. doi: 10.1128/AAC.00864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 5.Dorr T, Davis BM, Waldor MK. 2015. Endopeptidase-mediated beta lactam tolerance. PLoS Pathog 11:e1004850. doi: 10.1371/journal.ppat.1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Held K, Gasper J, Morgan S, Siehnel R, Singh P, Manoil C. 2018. Determinants of extreme beta-lactam tolerance in the Burkholderia pseudomallei complex. Antimicrob Agents Chemother 62:e00068-. doi: 10.1128/AAC.00068-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monahan LG, Turnbull L, Osvath SR, Birch D, Charles IG, Whitchurch CB. 2014. Rapid conversion of Pseudomonas aeruginosa to a spherical cell morphotype facilitates tolerance to carbapenems and penicillins but increases susceptibility to antimicrobial peptides. Antimicrob Agents Chemother 58:1956–1962. doi: 10.1128/AAC.01901-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Errington J, Mickiewicz K, Kawai Y, Wu LJ. 2016. L-form bacteria, chronic diseases and the origins of life. Philos Trans R Soc Lond B Biol Sci 371:20150494. doi: 10.1098/rstb.2015.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorr T, Alvarez L, Delgado F, Davis BM, Cava F, Waldor MK. 2016. A cell wall damage response mediated by a sensor kinase/response regulator pair enables beta-lactam tolerance. Proc Natl Acad Sci U S A 113:404–409. doi: 10.1073/pnas.1520333113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng AT, Ottemann KM, Yildiz FH. 2015. Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog 11:e1004933. doi: 10.1371/journal.ppat.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teschler JK, Cheng AT, Yildiz FH. 2017. The two-component signal transduction system VxrAB positively regulates Vibrio cholerae biofilm formation. J Bacteriol 199:e00139-. doi: 10.1128/JB.00139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H, Patel V, Helmann JD, Dorr T. 2017. Don't let sleeping dogmas lie: new views of peptidoglycan synthesis and its regulation. Mol Microbiol 106:847–860. doi: 10.1111/mmi.13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PD, Suh H, Marto JA, Garner EC, Bernhardt TG. 2016. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol 19:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. 2011. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. 2011. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 18.van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. 2011. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci U S A 108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Billings G, Ouzounov N, Ursell T, Desmarais SM, Shaevitz J, Gitai Z, Huang KC. 2014. De novo morphogenesis in L-forms via geometric control of cell growth. Mol Microbiol 93:883–896. doi: 10.1111/mmi.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranjit DK, Young KD. 2013. The Rcs stress response and accessory envelope proteins are required for de novo generation of cell shape in Escherichia coli. J Bacteriol 195:2452–2462. doi: 10.1128/JB.00160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgenstein RM, Bratton BP, Nguyen JP, Ouzounov N, Shaevitz JW, Gitai Z. 2015. RodZ links MreB to cell wall synthesis to mediate MreB rotation and robust morphogenesis. Proc Natl Acad Sci U S A 112:12510–12515. doi: 10.1073/pnas.1509610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee TK, Meng K, Shi H, Huang KC. 2016. Single-molecule imaging reveals modulation of cell wall synthesis dynamics in live bacterial cells. Nat Commun 7:13170. doi: 10.1038/ncomms13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorr T, Moll A, Chao MC, Cava F, Lam H, Davis BM, Waldor MK. 2014. Differential requirement for PBP1a and PBP1b in in vivo and in vitro fitness of Vibrio cholerae. Infect Immun 82:2115–2124. doi: 10.1128/IAI.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. 2010. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J Bacteriol 192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. 2011. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J 30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC. 2018. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559:617–621. doi: 10.1038/s41586-018-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrera CM, Henderson JC, Crofts AA, Trent MS. 2017. Novel coordination of lipopolysaccharide modifications in Vibrio cholerae promotes CAMP resistance. Mol Microbiol 106:582–596. doi: 10.1111/mmi.13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hankins JV, Madsen JA, Giles DK, Brodbelt JS, Trent MS. 2012. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in gram-positive and gram-negative bacteria. Proc Natl Acad Sci U S A 109:8722–8727. doi: 10.1073/pnas.1201313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hankins JV, Madsen JA, Giles DK, Childers BM, Klose KE, Brodbelt JS, Trent MS. 2011. Elucidation of a novel Vibrio cholerae lipid A secondary hydroxy-acyltransferase and its role in innate immune recognition. Mol Microbiol 81:1313–1329. doi: 10.1111/j.1365-2958.2011.07765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathur J, Davis BM, Waldor MK. 2007. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol Microbiol 63:848–858. doi: 10.1111/j.1365-2958.2006.05544.x. [DOI] [PubMed] [Google Scholar]
- 31.Davis BM, Waldor MK. 2009. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoE mutations: one fewer porin is enough. Nucleic Acids Res 37:5757–5767. doi: 10.1093/nar/gkp568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 33.Ranjit DK, Jorgenson MA, Young KD. 2017. PBP1B glycosyltransferase and transpeptidase activities play different essential roles during the de novo regeneration of rod morphology in Escherichia coli. J Bacteriol 199:e00612-. doi: 10.1128/JB.00612-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jorgenson MA, Young KD. 2016. Interrupting biosynthesis of O antigen or the lipopolysaccharide core produces morphological defects in Escherichia coli by sequestering undecaprenyl phosphate. J Bacteriol 198:3070–3079. doi: 10.1128/JB.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao Z, Kahne D, Kishony R. 2012. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol Cell 48:705–712. doi: 10.1016/j.molcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorr T, Lam H, Alvarez L, Cava F, Davis BM, Waldor MK. 2014. A novel peptidoglycan binding protein crucial for PBP1A-mediated cell wall biogenesis in Vibrio cholerae. PLoS Genet 10:e1004433. doi: 10.1371/journal.pgen.1004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussain S, Wivagg CN, Szwedziak P, Wong F, Schaefer K, Izore T, Renner LD, Holmes MJ, Sun Y, Bisson-Filho AW, Walker S, Amir A, Lowe J, Garner EC. 2018. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. Elife 7:e32471. doi: 10.7554/eLife.32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 40.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59:4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrieres L, Hemery G, Nham T, Guerout AM, Mazel D, Beloin C, Ghigo JM. 2010. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaichi Y, Dorr T. 2017. Transposon insertion site sequencing for synthetic lethal screening. Methods Mol Biol 1624:39–49. doi: 10.1007/978-1-4939-7098-8_4. [DOI] [PubMed] [Google Scholar]
- 45.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, Zhang YJ, Rubin EJ, Waldor MK. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet 10:e1004782. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chao MC, Abel S, Davis BM, Waldor MK. 2016. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol 14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.