Abstract
Key message
Fluorescence in situ hybridization with probes for 45 cDNAs and five tandem repeats revealed homoeologous relationships of Agropyron cristatum with wheat. The results will contribute to alien gene introgression in wheat improvement.
Abstract
Crested wheatgrass (Agropyron cristatum L. Gaertn.) is a wild relative of wheat and a promising source of novel genes for wheat improvement. To date, identification of A. cristatum chromosomes has not been possible, and its molecular karyotype has not been available. Furthermore, homoeologous relationship between the genomes of A. cristatum and wheat has not been determined. To develop chromosome-specific landmarks, A. cristatum genomic DNA was sequenced, and new tandem repeats were discovered. Their distribution on mitotic chromosomes was studied by fluorescence in situ hybridization (FISH), which revealed specific patterns for five repeats in addition to 5S and 45S ribosomal DNA and rye subtelomeric repeats pSc119.2 and pSc200. FISH with one tandem repeat together with 45S rDNA enabled identification of all A. cristatum chromosomes. To analyze the structure and cross-species homoeology of A. cristatum chromosomes with wheat, probes for 45 mapped wheat cDNAs covering all seven chromosome groups were localized by FISH. Thirty-four cDNAs hybridized to homoeologous chromosomes of A. cristatum, nine hybridized to homoeologous and non-homoeologous chromosomes, and two hybridized to unique positions on non-homoeologous chromosomes. FISH using single-gene probes revealed that the wheat-A. cristatum collinearity was distorted, and important structural rearrangements were observed for chromosomes 2P, 4P, 5P, 6P and 7P. Chromosomal inversions were found for pericentric region of 4P and whole chromosome arm 6PL. Furthermore, reciprocal translocations between 2PS and 4PL were detected. These results provide new insights into the genome evolution within Triticeae and will facilitate the use of crested wheatgrass in alien gene introgression into wheat.
Electronic supplementary material
The online version of this article (10.1007/s00122-018-3148-9) contains supplementary material, which is available to authorized users.
Introduction
Among wild relatives of cultivated Triticeae, the genus Agropyron is a promising donor of important traits for improvement of bread wheat (Triticum aestivum L., 2n = 6x = 42, genome AABBDD). The most widely distributed species in the genus, Agropyron cristatum (L.) Gaertn (Yang et al. 2014), known as crested wheatgrass, deserves particular attention because of its high crossability with wheat and other Triticeae and because it is a source of a number of genes controlling resistance to biotic and abiotic stress, such as resistance to barley yellow dwarf virus (Sharma et al. 1984; Shukle et al. 1987), wheat streak mosaic virus (Sharma et al. 1984; Shukle et al. 1987; Brettell et al. 1988; Friebe et al. 1991), yellow rust, leaf rust and stem rust (Knott 1964, 1968; Cauderon and Rhind 1976), and powdery mildew (Wu et al. 2006; Luan et al. 2010; Han et al. 2014; Ye et al. 2015; Zhang et al. 2015; Song et al. 2016), as well as cold tolerance (Limin and Fowler 1990), salinity tolerance and drought tolerance (McGuire and Dvorak 1981), and genes affecting yield (Song et al. 2013).
As alien gene transfer by interspecific hybridization is affected by chromosome collinearity, it is important to establish syntenic relationships between chromosomes of the donor alien species and wheat (Friebe et al. 1996; Qi et al. 2007; Feuillet et al. 2008). If collinearity between the donor and recipient genomes is broken down due to evolutionary chromosome rearrangements, gene transfer by homoeologous chromosome recombination may result in progenies with non-balanced genomes (Devos et al. 1993; Zhang et al. 1998). Altered structures of the donor chromosomes may interfere with recombination and hamper attempts to reduce the size of introgressed chromatin and eliminate undesirable traits (Nasuda et al. 1998).
Crested wheatgrass is a perennial, self-compatible, facultative allogamous species comprising a complex of polyploid series based on the P genome: diploid (2n = 2x = 14, PP), tetraploid (2n = 4x = 28, PPPP), and hexaploid (2n = 6x = 42, PPPPPP) (Dewey 1983; Asay et al. 1992). The species is distributed from central Europe and the Middle East across Central Asia to Siberia, China, and Mongolia (Dewey and Asay 1982). To date, the knowledge of the A. cristatum genome remains poor and, as yet, the homoeology between A. cristatum and wheat chromosomes has not been investigated in detail. Only a few attempts were made to characterize the A. cristatum karyotype on the basis of chromosome morphology (Asghari et al. 2007; Yang et al. 2014). Han et al. (2014) found that the A. cristatum chromosome 6P differs from wheat homoeologs by large rearrangements. This observation underlines the need for detailed analysis of the structures of all A. cristatum chromosomes. The ability to identify A. cristatum chromosomes would open avenues for the detection of introgressed chromosomes and their segments in the genetic background of other Triticeae species.
Fluorescence in situ hybridization (FISH) is a widely used cytogenetic method to study chromosome structural alterations (Alkhimova et al. 1999; Benavente et al. 2008; Fu et al. 2013). The utility of FISH depends on the availability of probes that provide chromosome-specific labeling patterns. Only a few repetitive DNA sequences have been applied as FISH probes to A. cristatum chromosomes. Apart from chromosome-specific 45S and 5S ribosomal DNA repeats (Smit et al. 2007), Svitashev et al. (1998) isolated repeats pAgc1 (350 bp) and pAgc30 (458 bp) from A. cristatum and used them to identify the P genomes involved in the evolution of the Elymus species. Wu et al. (2010) cloned three A. cristatum-specific sequences and applied them to detect the P genome in wheat-A. cristatum addition lines. FISH with subtelomeric tandem repeats pSc119.2 and pSc200 isolated from rye (Secale cereale L.) (Bedbrook et al. 1980; Appels et al. 1981) was applied to A. cristatum (Badaeva et al. 1996, 2004; Schneider et al. 2005; Linc et al. 2017). To date, a standard FISH karyotype of A. cristatum on the basis of homoeologous relationships with other species from Triticeae has not been established. The lack of standard karyotype of A. cristatum hampers a wider use of chromosome mediated gene transfer into wheat.
While FISH with repetitive sequences has been useful in chromosome identification in numerous plant species (He et al. 2015; Tang et al. 2016), it is not reliable for detecting a cross-species homoeology (Danilova et al. 2014). The presence and copy number of repetitive elements varies among Triticeae species (Kato et al. 2004). Moreover, other types of probes, such as, telomeric, subtelomeric or centromeric probes do not permit a complete identification of all chromosomes and will not facilitate a precise comparative analysis in most species of Triticeae (Tang et al. 2008, 2014; Mirzaghaderi et al. 2014; Fu et al. 2015). In contrast, genic sequences and their order are generally more conserved in grass genomes (Gale and Devos 1998). Devos and Gale (2000) and Paterson et al. (2000), utilized the existence of conserved sequences, usually cDNAs, to study the homoeologous relationship between related species from Triticeae.
Visualization of single-genes on mitotic metaphase chromosomes allows to investigate chromosome rearrangements and cross-species homoeology (Danilova et al. 2012, 2014, 2017; Karafiátová et al. 2013). To visualize low-copy sequences or small genomic targets on chromosomes, different types of probes have been used. Jiang et al. (1995) and Kim et al. (2005) used genomic BAC clones as probes for physical mapping of rice and sorghum genomes. This strategy works successfully in plant species with small gene-rich genomes. In plant species with bigger genomes, the application of cDNA sequences as FISH probes allowed chromosome identification, comparative analysis of chromosome structure between species, and identification of gene order along the chromosomes. The method called single-gene FISH was successfully used in rice (Kharb et al. 2001), maize (Wang et al. 2006), barley (Karafiátová et al. 2013), wheat and its wild relatives; i.e., Aegilops caudata, Ae. markgrafii, and Ae. umbellulata (Danilova et al. 2014, 2017). Danilova et al. (2014, 2017) used wheat cDNA sequences from Kawaura et al. (2009) as FISH probes to develop a wheat single-gene FISH map and analyze homoeologous relationships and chromosomal rearrangements within the Triticeae. Development of a single-gene FISH map for A. cristatum could help in chromosome identification and study the evolutionary genome rearrangements of this species in relation to wheat.
The objective of the present study was to develop a molecular karyotype and analyze the homoeology of the diploid A. cristatum chromosomes with those of bread wheat. During the course of the work, P genome-specific repetitive sequences were isolated, and their chromosome distribution patterns were explored. A set of wheat cDNA sequences were localized on A. cristatum chromosomes, and the cross-genome homoeology was established.
Materials and methods
Plant material and genomic DNA isolation
The seeds of diploid A. cristatum cv. Parkway (2n = 2x = 14, PP), accession number PI 415799, released by the Canada Department of Agriculture in 1969, were kindly provided by Dr. Joseph Robins, ARS Forage and Range Research Laboratory, USDA, Logan, USA. The seeds were germinated on moistened filter paper in a Petri dish and transferred to the soil. Plants were grown in a greenhouse, and genomic DNA was isolated from young leaf tissues using the NucleoSpin Plant II kit (Macherey–Nagel GmbH & Co. KG, Düren, Germany) following the manufacturer’s recommendations. DNA content analysis was performed according to Doležel et al. (2007) using Secale cereale cv. Dankovske (2C = 16.19 pg DNA) as internal reference standard.
Illumina sequencing
Libraries for sequencing were prepared from genomic DNA using the TruSeq DNA PCR-Free Library Preparation Kits (Illumina, San Diego, USA). Two micrograms of DNA dissolved in 100 μl of deionized water were fragmented with Bioruptor Plus (Diagenode, Denville, USA) with the following settings: 5 cycles, 30/90 (On/Off) seconds. DNA was sheared to 500–700-bp fragments, purified, end-repaired, adenylated, size-selected to 1000 bp and ligated with adapters according to the manufacturer’s protocol (Illumina). Libraries were diluted 1:10,000, and their concentration was estimated by real-time PCR using the KAPA Library Quantification Kit (Kapa Biosystems, Woburn, USA). The libraries were then pooled and diluted to a final concentration of 17 pM. Samples were paired-end sequenced on an Illumina MiSeq instrument for 600 cycles using the MiSeq Reagent Kit v3 (Illumina).
Sequence data analysis and identification of tandem organized repeats
Illumina reads were trimmed for adapters and for quality using the FASTX-toolkit [-q 20 -p 90] (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Chloroplast DNA sequences were identified and filtered out from both datasets prior to further analysis using the ERNE-FILTER program (Del Fabbro et al. 2013), and then, a stand-alone version of the Repeat Explorer pipeline (Novák et al. 2013) was used for de novo repeat identification and characterization. In the first step, a data set of 1,393,600 250-bp long paired-end reads were randomly selected and used for the reconstruction of repetitive DNA elements using the graph-based method according to Novák et al. (2010). This dataset represented 0.05 × coverage of the A. cristatum genome. Characterization and annotation of the assembled sequences were completed using similarity searches by RepeatMasker (http://www.repeatmasker.org) and its Viridiplantae databases, which were augmented with other repeats of selected plant species (Novák et al. 2010), and by BLASTX and BLASTN (Altschul et al. 1990) against the GenBank NR (Benson et al. 2010) and protein database created specifically for the Repeat explorer pipeline (Novák et al. 2010). All putative tandem organized repeats were identified using Dotter (Sonnhammer and Durbin 1995), and contigs with tandem repeats were then used for specific primer design using the Primer3 program (Untergasser et al. 2012). Newly characterized tandem repeats of A. cristatum were deposited in the GenBank/EMBL data libraries under accession numbers MG323511–MG323515.
Preparation of cDNA probes
We selected 45 cDNA probes previously mapped by single-gene FISH to bread wheat chromosomes by Danilova et al. (2014). The cDNA clones were developed by the National BioResource Project-Wheat, Japan. One sequence; the Acc2 cDNA pooled probe included in this study was developed from wheat RNA by Danilova et al. (2012). All cDNA clones were kindly supplied by the Department of Plant Pathology, Wheat Genetics Resource Center, Kansas State University. cDNA sequences were amplified using PCR with T3/T7 primers and LongAmp DNA Polymerase (New England Biolabs, Massachusetts, USA) following the manufacturer’s recommendations. The PCR products were purified with the Invitrogen PCR purification kit (Life Technologies, Carlsbad, USA) according to the manufacturer’s instructions.
Probe labeling
Probes for newly identified tandem repeats were labeled by PCR either with biotin-dUTP or digoxigenin-dUTP (Roche, Mannheim, Germany) using primers listed in Table S1 and A. cristatum DNA as a template. The PCR mix contained 30 ng of genomic DNA, reaction buffer (consisting of 10 mM Tris–HCl pH 8, 50 mM KCl, 0.1% Triton X-100 and 1.5 mM MgCl2), 200 μM dNTPs including biotin-dUTP or digoxigenin-dUTP, 1 μM primers and 0.4 U of Taq polymerase (New England Biolabs). The thermocycler conditions were set as follows: initial denaturation step at 94 °C for 5 min, followed by 35 cycles of denaturation (94 °C/50 s), annealing at 54 °C for 50 s and extension (72 °C/50 s). The final extension was allowed for 5 min at 72 °C. Probes for rye pSc119.2 and pSc200 repeats (Bedbrook et al. 1980 and Appels et al. 1981) were labeled with biotin-dUTP. The 120-bp unit of pSc119.2 inserted into the plasmid pBR322 was amplified by PCR using the M13 universal primers following Nagaki et al. (1995) and Contento et al. (2005). Genomic rye DNA was used as a template to amplify pSc200 by PCR according to Tang et al. (2008).
Probes for rDNA sequence were labeled either with biotin-dUTP, digoxigenin-dUTP, or with a fluorochrome. In all cases, template rice DNA for the 5S rDNA probe was amplified by PCR according to Fukui et al. (1994), while the plasmid pTa71 (45S rDNA) containing 9-kb fragment from T. aestivum with 18S-5.8S-26S rDNA and intergenic spacers (Gerlach and Bedbrook 1979) was labeled by nick translation following Kato et al. (2004, 2006). The 45S rDNA was directly labeled with ChromaTide® Alexa Fluor® 546-14-dUTP (Thermo Fisher Scientific, Waltham, USA), while 5S rDNA was directly labeled with ChromaTide® Fluorescein-12-dUTP (Thermo Fisher Scientific). Out of the newly identified tandem repeats, ACRI_CL78 was also directly labeled with ChromaTide® Fluorescein-12-dUTP (Thermo Fisher Scientific).
All cDNA probes were labeled with ChromaTide® Texas Red®-12-dUTP (Thermo Fisher Scientific). Direct probe labeling for cDNA and 45S rDNA by nick translation using 2 μg DNA was done as described by Kato et al. (2004, 2006). The probe quality was checked on 1.5% agarose gel. To increase probe concentration and remove non-incorporated nucleotides, probes were precipitated and purified by adding 173 μl 1 × TE buffer (pH 7.5), 20 μl NaAc (3 M, pH 5.2) and 500 μl 100% ethanol; 3 μg herring sperm DNA (Promega, Wisconsin, USA) was added to block DNA. After overnight precipitation at − 20 °C, the probes were centrifuged for 30 min at 4 °C at 14,000 × g, rinsed in 70% ethanol and air-dried for 7–15 min. Subsequently, probes were dissolved in 20 μl of 2 × SSC and 1 × TE buffer (pH 7.6) in a 1:1 ratio at 65 °C for 10 min.
Mitotic chromosome preparations
Seeds of A. cristatum were germinated on moistened filter paper in a glass Petri dish in the dark at 25 °C for 3–4 days. Root tips were transferred to distilled water and incubated overnight at 1 °C in a box filled with ice-water. Subsequently, the root tips were fixed in ice-cold 90% acetic acid for 10 min followed by 3 washes in 70% ethanol and stored in 70% ethanol at − 20 °C. Chromosome preparations were prepared using the drop technique according to Kato et al. (2004, 2006), with minor modifications as described in Danilova et al. (2012). The quality of chromosome spreads was checked under a microscope, and the best slides were selected for FISH.
Fluorescence in situ hybridization (FISH)
All FISH experiments were done with two or more labeled probes which were hybridized simultaneously. New repetitive DNA sequences, pSc119.2, pSc200 and rDNA were localized following the protocol of Cabrera et al. (2002) with modifications. Briefly, digoxigenin-labeled probes were detected using anti-digoxigenin fluorescein isothiocyanate (Roche). Biotin-labeled probes were detected with Cy3-conjugated streptavidin (Invitrogen). Hybridization mixture per slide (total volume = 10 μl) contained 50 ng labeled probe DNA, 50% v/v formamide, 2 × SSC (0.15 mol/l NaCl plus 0.015 mol/l sodium citrate), 10% w/v dextran sulfate, 0.4 μg salmon sperm DNA and 0.1% w/v sodium dodecyl sulfate. Low-copy sequences were localized in combination with 45S rDNA and one tandem repeat (ACRI_CL78) or 5S rDNA by FISH following Karafiátová et al. (2013) with minor modifications. Hybridization mixture (20 μl) consisting of 10 μl formamide, 2 × SSC, 0.1 M Tris–HCl pH 8, 0.05 M EDTA, 1 μg/μl salmon sperm DNA, 300 ng cDNA probe, 50 ng DNA for 45S rDNA and 50 ng of tandem repeat (ACRI_CL78) or 5S rDNA applied onto slides, and the cover slides were taped up. The chromosomes and probes were denatured together at 80 °C for 3 min under high moisture conditions. The hybridization was carried out overnight at 37 ºC; then, the slides were washed in 2 × SSC for 1 min at RT, 2 × SSC for 20 min at 55 °C and 2 × SSC for 1 min at RT, followed by dehydration in 70, 90, 100% ethanol, with each step lasting 2 min. The chromosomes were mounted and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield media (Vector Laboratories, Burlingame, USA).
Microscopy, software, signal capture and image analysis
Chromosome preparations were examined using an Axio Imager Z.2 Zeiss microscope (Zeiss, Oberkochen, Germany) equipped with a Cool Cube 1 (Metasystems, Altlussheim, Germany) camera and appropriate filter sets. The signal capturing and picture processing were performed using ISIS software (Metasystems). The final image adjustment was done in Adobe Photoshop CS5 (Adobe Systems Incorporated, San Jose, USA). Chromosome measurements and determination of the position of FISH signals were measured using ISIS software (Metasystems).
Chromosome measurements
To establish the karyotype and idiogram, ten best images of metaphase spreads obtained at 100× magnification were selected and mean chromosome arm lengths were determined in micrometers (μm). The centromeric index was calculated by dividing the length of the shorter of the two chromosome arms by the length of the whole chromosome and expressed in percent. Arm ratio was calculated by dividing the length of the longer arm of the chromosome by the length of the shorter arm. Relative length was determined by dividing the length of a particular chromosome by the total length (H) of chromosomes in the haploid set and expressed in per cent (Table S2). Individual chromosomes were classified according to centromeric index. The relative positions of cDNA FISH sites were measured from the centromere. The positions given are average values determined from five measurements on different metaphase spreads. The average relative distance from the centromere, standard deviation and confidence intervals with a significance level of 0.05 were calculated using Microsoft Office Excel 2010 functions. The cDNA positions are summarized in Table S3. The cDNA positions on wheat chromosomes and the wheat idiogram were drawn according to the data from Gill et al. (1991) and Danilova et al. (2014).
Accession numbers
Plant material; diploid A. cristatum cv. Parkway (2n = 2x = 14, PP), accession number PI 415799. Newly characterized tandem repeats of A. cristatum can be found in the GenBank/EMBL data libraries under accession numbers MG323511–MG323515.
Results
Identification and genomic distribution of tandem repeats
5S and 45S rDNA
FISH with a probe for 45S rDNA showed four hybridization sites on the short arm of two chromosome pairs corresponding to two pairs of nucleolus organizing regions (NOR). The 5S rDNA probe showed signals in a subterminal position of the short arms of one of the chromosome pairs carrying 45S rDNA locus. Later, this chromosome pair was identified based on cDNA mapping as 5P, while the other pair was identified as 1P, which is characterized by secondary constriction and a satellite on the short arm. Interestingly, another chromosome with a secondary constriction and satellite on the short arm was observed but had no 5S or 45S rDNA signals (Supplementary Fig. S1). Using cDNA probes from wheat, this chromosome pair was identified as 2P. Thus, two satellite chromosome pairs (1P and 2P) were characterized by secondary constriction and satellites on the short arm, but only 1P corresponds to the NOR position confirmed by the 45S rDNA signal. The chromosome 5P carried 45S and 5S signals on terminal and subterminal positions, respectively. The use of 5S and 45S rDNA probes simultaneously enabled identification of two out of the seven pairs of A. cristatum chromosomes.
The pSc119.2 and pSc200 sequences
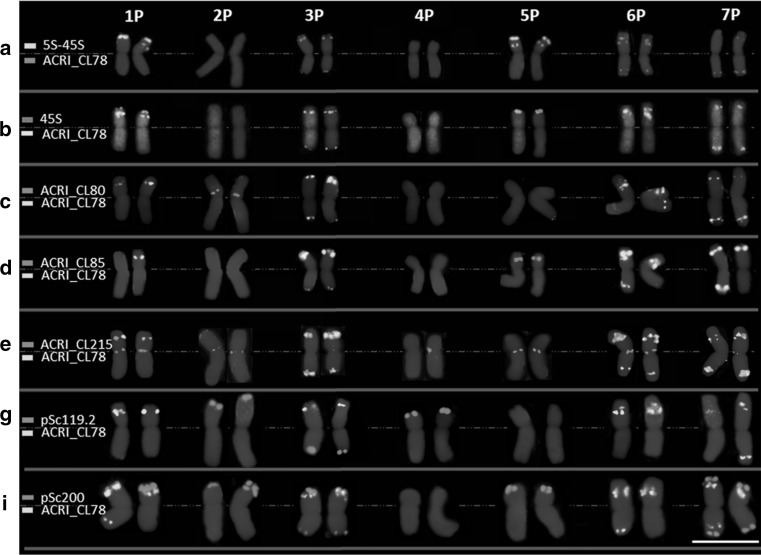
Three of the seven chromosome pairs possessed pSc119.2 repeat clusters. Chromosomes 2P and 4P carried pSc119.2 signals on short arms, while 3P carried them on the long arm. Five chromosome pairs carried pSc200 signals. FISH with the pSc200 probe gave FISH signals on the short arms of chromosomes 1P, 2P, 3P and 5P, while on 7P, the signals were on both short and long arms. In all cases, pSc119.2 and pSc200 probes gave strong telomeric signals (Figs. 1 and 2, and Supplementary Fig. S2). Two chromosomes, 3P and 7P, could be distinguished from the remaining A. cristatum chromosomes by characteristic signals of pSc119.2 and pSc200, respectively. The chromosome 3P was identified by a unique and strong signal of pSc119.2 on the tip of the long arm, while 7P was characterized by clear pSc200 signals on the terminal regions of both arms.
Fig. 1.
FISH on mitotic chromosomes of diploid A. cristatum cv. Parkway with probes for 5S and 45S rDNA, newly identified tandem repeats ACRI_CL78, ACRI_CL80, ACRI_CL85 and ACRI_CL215, and rye tandem repeats pSc119.2 and pSc200. Bar = 10 μm
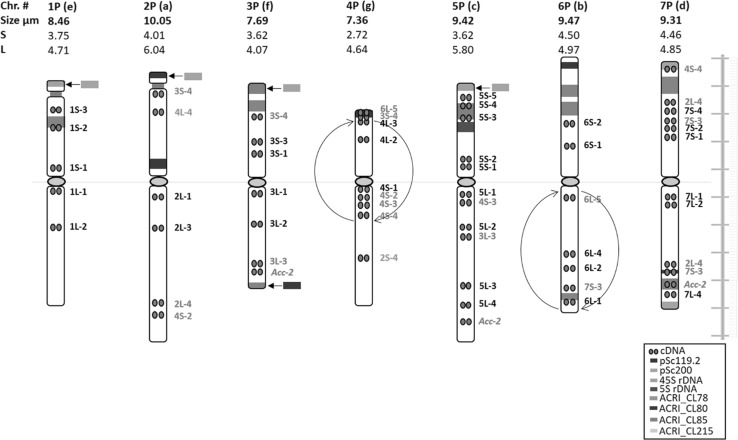
Fig. 2.
Idiogram for chromosomes of diploid A. cristatum cv. Parkway showing the distribution of 5S and 45S rDNA gene clusters, newly identified tandem repeats ACRI_CL78, ACRI_CL80, ACRI_CL85 and ACRI_CL215, rye tandem repeats pSc119.2, pSc200, and cDNAs. The names of cDNA probes that hybridized to more than one chromosome are highlighted in red. The names of cDNA probes that hybridized only to a non-homoeologous chromosome are highlighted in green. Inversions on chromosomes 4P and 6PL are indicated by circular arrows. The letters in parentheses show the order of the chromosome by size from the longest (a) to the shortest (g). S and L letters refer to the short and long arm, respectively. The color scheme (bottom right) shows the color of each probe as represented in this idiogram. The scale on the right side, consists of 10 units, and each unit represents 1 μm
New tandem repeats isolated from A. cristatum
Using flow cytometry, we estimated 2C DNA amount of 12.99 ± 0.11 pg DNA (mean ± SD) for A. cristatum. Its 1C genome size was determined as 6352 Mbp considering 1 pg DNA = 0.978 Mbp (Doležel et al. 2003). After quality filtering and trimming, the sequences to 250 bp, partial Illumina sequencing of A. cristatum resulted in a cumulative length of ~ 7639 Mb, corresponding to ~ 1x coverage of the nuclear genome. A set of randomly selected paired-end reads representing 0.05× genome coverage was used for repeat reconstruction and characterization using the Repeat Explorer pipeline. In general, Ty3/gypsy elements (38.10%) were found to be more abundant than Ty1/copia (18.57%), while non-LTR elements and DNA transposons accounted for only 0.15 and 3.28% of the genome, respectively (Table 1). Among the Ty3/gypsy superfamily, the elements from the Athila lineage were the most common and one particular element of the Athila lineage accounted for more than 20% of the nuclear genome. The Ty1/copia superfamily was mostly represented by retroelements from the Angela lineage (~ 13.5%) and the Sire/Maximus lineage (~ 4.4%). The elements representing other lineages of Ty1/copia were rare (Table 1). Dotter analysis was applied to all assembled contigs from clusters with layouts indicating tandem characteristics (Novák et al. 2010) and revealed fourteen different types of tandem repeats in A. cristatum. The most abundant repeat, ACRI_CL20, had a 379-bp long unit and comprised ~ 0.48% of the nuclear genome. A comparison of ACRI_CL20 and the pSc200 satellite (GenBank accession number Z54189) revealed 93% sequence identity. Two other tandem repeats showed similarities to known plant satellites; ACRI_CL78 was 80% similar to the tandem repeat 4P6 (AY249985) from Aegilops tauschii Coss. (2n = 2x = 14, genome DD) (Zhang et al. 2004). Tandem repeat ACRI_CL215 had 87% similarity to tandem repeat AB016970 of the pLrTail family, first characterized in Leymus racemosus (GB code: AB016970). BLAST searches with 9 putative tandemly organized repeats identified in A. cristatum revealed homology to some parts (usually non-annotated) of wheat 3B chromosome pseudomolecule, and two repeats (ACRI_CL85 and ACRI_CL198) did not show any homology with sequences deposited in the GenBank database.
Table 1.
Proportion of repetitive DNA sequences of A. cristatum cv. Parkway identified after partial Illumina sequencing
| Repeat | Lineage/class | Alternative names | Proportion in the analyzed data [%] | |
|---|---|---|---|---|
| LTR retroelements | Ty1/copia | Maximus-SIRE | 4.42 | |
| Angela | 13.51 | |||
| TAR | Tont | 0.56 | ||
| Tork | Tnt | 0.01 | ||
| Ale | Hopscotch | 0.05 | ||
| Bianca | 0.02 | |||
| Total Ty1/copia | 18.57 | |||
| Ty3/gypsy | Athila | 22.96 | ||
| Chromovirideae | 7.16 | |||
| Ogre-Tat | 7.98 | |||
| Total Ty3/gypsy | 38.10 | |||
| Other | LINE | 0.15 | ||
| DNA transposons | 3.28 | |||
| Tandem repeats | 1.62 | |||
| rRNA genes | 0.21 | |||
| Unclassified LTR elements | 3.94 | |||
| Unclassified repeats | 6.71 | |||
| Non-annotated sequences | 8.36 |
Out of the fourteen tandem repeats, eleven were used as FISH probes, and five of them showed specific distribution patterns. For one of them (ACRI_CL187), dispersed labeling was observed on all chromosomes, except in centromeric and most pericentromeric regions. Repeat ACRI_CL215 showed a preferential location in primary constrictions of all chromosomes. FISH with a probe for the ACRI_CL78 repeat identified the seven pairs of chromosomes, and it is thus recommended for karyotyping. Although there was a slight difference in the signals pattern of the repeat sequence ACRI_CL78 between the homologous chromosomes from different plants of the diploid A. cristatum. However, these small polymorphisms did not hamper the precise characterization of the individual chromosomes. Two tandem repeats gave a fewer number of FISH signals, and each of them showed characteristic patterns on two different chromosome pairs; for instance, ACRI_CL80 showed a strong signal on the pericentromeric region of 2PS and the subterminal region of 6PS, while ACRI_CL85 distinguished other two chromosomes by clear signals on the terminal region of the short arm of chromosome 3P and the subterminal region of the short arm of 5P. (Figs. 1 and 2, and Supplementary Fig. S2). The five new tandem repeats enabled the precise characterization of all the individual chromosomes; the labeling patterns were always chromosome specific. The remaining six new tandem repeats did not show any FISH signals on chromosomes of A. cristatum.
Karyotype analysis
The measurement and chromosome classification is summarized in Supplementary Table S2. All examined mitotic metaphase plates showed standard diploid constitution (2n = 14), characterized by seven pairs of chromosomes, two pairs being satellite chromosomes. During the preparation of the karyotype, the homologous chromosomes were tentatively paired based on tandem repeat ACRI_CL78 distribution, morphology, size, and the presence of 5S and 45S rDNA loci. In the karyotype, each chromosome was identified by a specific number and small letter (Fig. 2). The numbers ranged from 1 to 7, indicating the correspondence of each chromosome to its homoeolog in bread wheat according to the cDNA data. The letters differed between ‘a’ and ‘g’. The letter ‘a’ indicates the largest chromosome, while the letter ‘g’ indicates the smallest one. The chromosome sizes ranged from 7.36 to 10.05 μm. The relative A. cristatum chromosome length ranged from 16.27% for the largest chromosome, 2P, to 11.92% for the smallest, 4P (Supplementary Table S2). Following Naranjo et al. (1983) and based on the data in Supplementary Table S2, the chromosomes of A. cristatum were classified into two different types based on centromere position and arm ratio. Four chromosomes were metacentric (1P, 3P, 6P and 7P), while the rest were submetacentric (Fig. 2). Identification of the A. cristatum chromosomes using FISH pattern of the tandem repeat ACRI_CL78 together with the chromosome size and centromere position, facilitated the co-localization of the remaining tandem repeats and further mapping of cDNAs (Fig. 1 and 2 and Supplementary Fig. S2). The results are summarized in the idiogram representing chromosomes, arranged according to their homoeology to wheat based on cDNA data (Fig. 2).
Chromosome structure and rearrangements as revealed by cDNA FISH
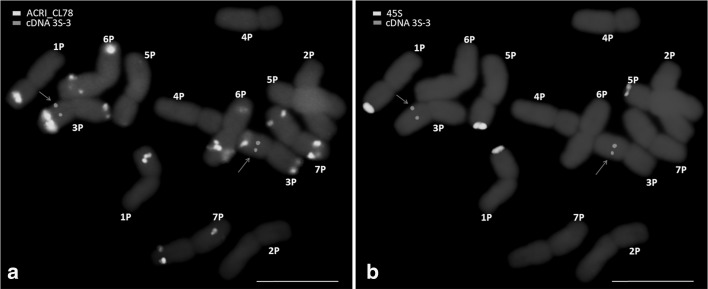
A set of 45 cDNA probes which were mapped to bread wheat chromosomes (Danilova et al. 2014) was used. FISH with the cDNA probes produced clear signals on chromosomes of A. cristatum (Fig. 3). The forty-five cDNA probes hybridized to fifty-eight positions on A. cristatum chromosomes (Fig. 2 and Supplementary Fig. S3, and Supplementary Table S4). Apart from the hybridization of thirty-four probes (75.6%) to homoeologous chromosomes on thirty-four positions, nine (20%) probes hybridized to non-homoeologous chromosomes of A. cristatum on twenty-two positions: probe 2L-4 hybridized to chromosome arms 2PL, 7PS and 7PL; probe 3S-4 hybridized to 2PS, 3PS and 4PS; probe 3L-3 hybridized to 3PL and 5PL; probe Acc-2 hybridized to 3PL, 5PL and 7PL; probe 4S-2 hybridized to 2PL and 4PL; probe 4S-3 hybridized to 4PL and 5PL; probe 4S-4 hybridized to 4PL and 7PS; probe 6L-5 hybridized to 4PS and 6PL; and probe 7S-3 hybridized to 6PL, 7PS and 7PL. Some of the cDNA probes hybridized to more than one position per chromosome. For example, probes 2L-4 and 7S-3 produced signals on both arms of chromosome 7P, such case was not reported in wheat. While the number of the cDNA probes located on multiple loci (Acc-2, 2L-4, 3S-4, 3L-3, 4S-2, 4S-3, 4S-4, 6L-5, 7S-3) on A. cristatum genome was higher, and representing 20% of the used cDNAs, in wheat genomes, only about 7% of these cDNAs (Acc-2, 4S-3 and 4S-4) were located on more than one locus, on chromosome arms 3L, 4AL, and 5L; 4S, 4AL, 5L, and 7AL; 2S, 4S and 4AL, respectively. Besides the hybridization of the forty-three cDNA probes described here, two (4.4%) cDNA probes (2S-4 and 4L-4) hybridized to unique sites on non-homoeologous chromosomes (4PL and 2PS, respectively) of A. cristatum.
Fig. 3.
FISH on mitotic chromosomes of diploid A. cristatum cv. Parkway with probes for the tandem repeat ACRI_CL78, cDNAs (red arrows) and 45S rDNA. a Tandem repeat ACRI_CL78 (green) and cDNAs (red); b 45S rDNA (green) and cDNAs (red). The chromosomes were counterstained with DAPI (blue). Bars = 10 μm
A comparison between the cDNA positions on diploid A. cristatum cv. Parkway and the homoeologous groups of hexaploid wheat cv. Chinese Spring are shown in Supplementary Fig. S4. Most of the probes hybridized to corresponding homoeologous chromosome arms of A. cristatum, generally in the same order, indicating preserved cross-genome collinearity. No major collinearity distortion was detected on chromosomes 1P, 3P and 5P, though additional probes need to be applied to 1PL to verify its structure. However, many cDNA hybridization sites indicated the collinearity distortion for the rest of the A. cristatum genome. For example, signals of cDNA probes 2S-4 on 4PL and probe 4L-4 on 2PS indicate reciprocal translocation between chromosome arms 2PS and 4PL. Moreover, all of the group-4 short arm-specific cDNA probes hybridized to the long arm, while all of the group-4 long arm-specific cDNA probes hybridized to the short arm, thus suggesting pericentric inversion on 4P. On the long arm of chromosome 6P, the probes hybridized in an inverted order as compared to their order on homoeologous wheat chromosome arm, which points to a paracentric inversion of 6PL. The presence of signals of nine cDNAs in multiple loci may indicate either a gene or a chromosome segment duplication.
Discussion
Secondary constrictions and nucleolus organizing regions (NORs)
We found four 45S rDNA sites and two 5S rDNA site in the diploid chromosome set of A. cristatum. The 45S sites were located at the terminal position of the short arms of 1P and 5P, 1P carry a secondary constriction, while on 5P no secondary constriction was observed. The chromosome 2P carries a secondary constriction, but the 45S rDNA signals were absent. Two pairs of unidentified satellite chromosomes in A. cristatum were observed in previous studies (Hsiao et al. 1986, 1989). Linc et al. (2017) observed one chromosome pair with a clear secondary constriction in two accessions of diploid A. cristatum, but the constriction on the second chromosome pair was visible only in the genotype MvGB1521. Moreover, they reported that two chromosome pairs carrying 45S signals on the short arms were found in one accession MvGB1521, but in the accession MvGB1509, the signals were detected only on one chromosome pair, while the other chromosome pair showed pSc119.2 site at the short arm instead. The 45S rDNA on the terminal position of the A. cristatum chromosome arm 5PS was detected by Jubault et al. (2006) in the wheat-A. cristatum chromosome addition line. Secondary constrictions or NOR sites are commonly located in the terminal regions of the chromosomes (Sousa et al. 2011). In many cases, the 45S rDNA sites were observed to be restricted to the NORs, although in various species other smaller or less active sites have also been detected outside the NORs (Sato et al. 1980; Guerra et al. 1996; De Carvalho and Guerra 2002). Our FISH results for chromosome 1P and 5P are in agreement with these observations. However, it seems that secondary constrictions in this species are not always related to the rDNA unit, and this observation deserves further investigation.
To date, there was no information about genomic distribution of 5S rDNA sites in A. cristatum. We located 5S rDNA locus in subterminal region of the short arm of chromosome 5P. In most Triticeae, the 5S rDNA sites are located in homoeologous groups 1 and 5 (Dvorák et al. 1989; Reddy and Appels 1989). In Thinopyrum ponticum, which is related to A. cristatum, the 45S rDNA locus is distally located in relation to the 5S rDNA locus (Brasileiro-Vidal et al. 2003). This disposition is similar to our results on 5P and to that found in homoeologous group 5 of the D genome of wheat (Mukai et al. 1990, 1991) and Aegilops umbellulata (Castilho and Heslop-Harrison 1995).
Chromosome morphology
Relative chromosome length can be used as a rough estimate of the proportion of the genome that a given chromosome represents (Jabeen et al. 2012). We found that chromosome 2P is the longest, while 4P is the shortest chromosome in the karyotype of A. cristatum (Table S2). The data on chromosome size are of special importance if these chromosomes are intended to be sequenced. We observed that four chromosomes of diploid A. cristatum were metacentric, while the rest were submetacentric, which is in agreement with data of Linc et al. (2017). In contrast, Yang et al. (2014) found karyotypes of A. cristatum from several populations to be variable, as the chromosomes of some diploids were all metacentric, while other di-, tetra- and hexaploids showed a combination of metacentric and submetacentric chromosomes. We compared the size of A. cristatum chromosomes with those of wheat in Gill et al. (1991). The chromosomes of A. cristatum were in general shorter than their homoeologous chromosome groups from bread wheat (Supplementary Fig. S4). The chromosome morphology of A. cristatum is of special importance for genomic studies and the quick identification of alien chromosomes in the substitution or addition lines.
FISH pattern of tandem repeats allows identification of all A. cristatum chromosomes
With the aim to achieve reliable identification of all A. cristatum chromosomes, we determined chromosome distribution of two tandem repeats pSc119.2 and pSc200. Linc et al. (2017) performed FISH with the same repeats in two A. cristatum accessions and found that one of the two accessions (MvGB1509) showed the same number of pSc119.2 signals as we found on A. cristatum cv. Parkway. Previous studies on Triticeae showed that combination of pSc119.2 and other FISH probes in most species does not permit identification of all chromosomes (Mirzaghaderi et al. 2014; Li et al. 2018). These tandem repeats were present in genomes of several species forming large telomeric blocks, especially in genera Aegilops and Secale, and were used to distinguish between their chromosomes (Tang et al. 2008, 2014; Mirzaghaderi et al. 2014; Fu et al. 2015). In bread wheat, the pSc119.2 probes hybridized to B genome chromosomes in addition to chromosomes 4A, 5A, 2D, 3D, and 4D, while the signals of pSc200 were not observed (Tang et al. 2008, 2014; Fu et al. 2015). However, pSc200 presented 93% sequence identity to ACRI_CL20, the most abundant tandem repeat in the A. cristatum genome, and was similar to two other tandem repeats. After comparing hybridization patterns of pSc119.2 and pSc200 in A. cristatum as observed in our study and in wheat (Schneider et al. 2003; Contento et al. 2005; Tang et al. 2008, 2014; Fu et al. 2015), we concluded that each of the two probes produced characteristic patterns on chromosomes of A. cristatum, but only the use of pSc200 facilitates detection of the P genome in wheat introgression lines. For instance, using pSc200, five chromosomes (1P, 2P, 3P, 5P and 7P) could be distinguished. Two chromosomes 3P and 7P can be easily distinguished from the rest of A. cristatum chromosomes based on the pattern of pSc119.2 and pSc200 hybridization.
To distinguish all A. cristatum chromosomes using FISH, new tandem repeats were needed. Han et al. (2017) found that some A. cristatum repetitive sequences were distributed all over the P genome or at centromeres, pericentromeric regions, distal regions, and terminal regions. The sequences located at telomeres or subtelomeres were mostly tandem repeats, similar to pSc200 and pSc250 from rye (Vershinin et al. 1995), pAesKB52 from Ae. speltoides (Anathawat-Jonsson and Heslop-Harrison 1993), and pHvMWG2315 from barley (Busch et al. 1995). Although the sequences of Han et al. (2017) enabled identification of wheat-A. cristatum introgression lines, they did not permit identification of individual A. cristatum chromosomes. We obtained sequences of tandem repeats located at centromeric, telomeric, subtelomeric and pericentric regions by whole-genome sequencing, which were found to give specific patterns for individual A. cristatum chromosomes and enabled discrimination among all of the chromosomes. In addition, the region-specific repeats could be helpful in deducing the part of a P genome chromosome in wheat-A. cristatum translocation and introgression lines, as was shown by Li et al. (2012) for Dasypyrum villosum and Han et al. (2017) for A. cristatum introgression lines.
In our work, five tandem repeat sequences with different distribution patterns on the A. cristatum genome were identified by FISH. We found that the distribution patterns as well as the signal intensity of the tandem repeats were specific to different chromosomes. Thus, A. cristatum chromosomes can be identified using the hybridization pattern together with chromosome size and arm ratio. In addition, tandem repeats could be used to study the chromosome rearrangements during polyploidization (Han et al. 2005, 2017; Zhang et al. 2013). Hence, future studies on differences in genomic distribution of specific repetitive sequences between several populations from A. cristatum could be used to reveal their different roles during A. cristatum genome evolution (Han et al. 2017). The absence of FISH signals for six out of the eleven tandem repeats could be attributed to the fact that these sequences do not form clusters long enough to be detected by fluorescence microscopy. Similar findings were reported by Han et al. (2017), who isolated repetitive sequences from chromosome 6PS of tetraploid A. cristatum and only fourteen out of 48 sequences were detectable by FISH. In the present work, the slight differences observed in the FISH pattern of the repeat ACRI_CL78 between the homologous chromosomes of different plants from the same genotype can be attributed to the open pollinated nature of A. cristatum. This phenomenon was also reported for other open pollinated species such as rye (Szakács and Molnár-Láng 2008) as well as in other genotypes of diploid A. cristatum (Linc et al. 2017).
cDNA revealed chromosome structure and evolutionary rearrangements in the P genome
The fact that a considerable percentage (24.4%) of the wheat cDNA sequences localized to both homoeologous and non-homoeologous chromosomes and that other mapped to non-homoeologous chromosomes indicates that some A. cristatum chromosomes did not preserve cross-species collinearity. Distorted cross-species collinearity was observed in chromosomes 2P, 4P, 5P, 6P and 7P. The order of cDNA probes in the pericentric region of chromosome 4P is inverted as compared to homoeologous regions of wheat chromosomes 4B and 4D shown in Danilova et al. (2014), but in accordance with their order on the homoeologous region of 4A. Chromosome 4A of wheat is known to be rearranged as a result of a 4AL/5AL translocation followed by a 7BS translocation and one paracentric and two pericentric inversions (Naranjo et al. 1987; Devos et al. 1995; Mickelsonyoung et al. 1995; Miftahudin et al. 2004). Thus, the A. cristatum chromosome 4P underwent paracentric inversion similar to wheat 4A. The inverted order of cDNA probes on the long arm of chromosome 6P when compared to their order on homoeologous wheat chromosomes (Danilova et al. 2014) indicates the presence of a P genome-specific paracentric inversion. A similar observation on chromosome 6P from the tetraploid A. cristatum was made by Han et al. (2014). This phenomenon of rearrangements can be found in the genomes of wild relatives of wheat (Kishii et al. 2004; Said and Cabrera 2009; Wang et al. 2010; Wang 2011; Hu et al. 2012; McArthur et al. 2012; Said et al. 2012; Han et al. 2014). Although our data showed chromosomal rearrangements in A. cristatum with respect to wheat, it seems that this does not hamper alien chromosome introgression into wheat. For instance, a series of disomic addition and translocation lines have been obtained after hybridization of bread wheat with A. cristatum and have shown high resistance to powdery mildew and leaf rust, longer grain length and higher thousand-grain weight (Li et al. 1998; Song et al. 2013; Ochoa et al. 2015; Li et al. 2016).
Transferring desirable alien genes and improving their utilization efficiency in wheat improvement can be hampered by linkage drag and insufficient compensation for the substituted wheat chromatin. To overcome this difficulty, it is necessary to identify the alien chromosomal regions of target genes and to analyze their homoeologous relationships with wheat chromosomes. Only well-compensating translocations or introgressions produced by induced homoeologous recombination are beneficial for wheat improvement (Qi et al. 2007; Han et al. 2014). To date, all useful alien genes that significantly contributed to wheat improvement are, without exception, compensating translocations (Friebe et al. 1996; Gill et al. 2006; Han et al. 2014). Thus, understanding the genetic structure of the A. cristatum chromosome set may be helpful for producing introgressions or compensating translocations for gene transfer into the wheat genome. In this work, the cDNA FISH probes were distributed unequally along the arms of most chromosomes of A. cristatum, leaving some parts uncovered. For instance, the arms 1PL and 6PS have markers only at proximal regions. Thus, more markers are required at certain genomic positions to cover whole chromosome arms of A. cristatum. These results provide resources for further studies of the A. cristatum genome structure and evolution and contribute to the identification and utilization of wheat-A. cristatum derivatives for wheat improvement.
General discussion, conclusions and future prospects
Prior to this study, several attempts were made to establish a standard karyotype for A. cristatum. Generally, the chromosomes of A. cristatum were arranged according to their size and centromere position (Asghari et al. 2007; Yang et al. 2014), without considering cross-genome homoeology to Triticeae. In our study, the chromosomes of A. cristatum were morphologically characterized and arranged using accurate data on the homoeology between A. cristatum and wheat. The molecular karyotype developed in this work enables the identification of an introduced A. cristatum chromosome or its segment in the wheat background. Moreover, with the knowledge of genome collinearity with wheat, breeders may predict the probability of induced homoeologous recombination with wheat or other Triticeae. A. cristatum was believed to be a species complex of high genetic isolation from other genera of Triticeae (Martin et al. 1999). Nevertheless, our data show a considerable synteny (75.6%) between diploid A. cristatum and wheat. Hybridization between wheat and tetraploid A. cristatum can be easily done (Chen et al. 1989). To date, all the introgressions from A. cristatum into wheat were made using tetraploid A. cristatum that originates from derivatives of hybridizations between diploid Agropyron cristatum and Agropyron mongolicum (Han et al. 2014). Although the diploids A. cristatum and A. mongolicum contain the same basic P genome, their P genomes exhibited rearrangements and variation (Hsiao et al. 1989; Wu et al. 2006). The two P genomes exhibit segmental autonomy in the tetraploid A. cristatum (Stebbins 1947) and are distinguished from each other by structural rearrangements (Hsiao et al. 1989). Schulz-Schaeffer et al. (1963) proposed the segmental alloploid nature of tetraploid and hexaploid A. cristatum. Studies of genetic diversity showed that there are genetic variations among different individuals within one population (Liu et al. 2010; Wang et al. 2010); thus, there might be large structural chromosome rearrangements and genetic differences between diploid and tetraploid A. cristatum. Further analysis of chromosome structure and rearrangements of tetraploid A. cristatum, which is commonly used and successfully hybridizes with wheat, is required. The development of the A. cristatum karyotype and comparative cDNA chromosome maps in this study will support alien chromosome introgressions into wheat and may facilitate the use of crested wheatgrass in alien introgression breeding by enabling the transfer of compensating segments or translocations from the targeted alien chromosome into wheat.
Author contribution statement
The work presented here was carried out in collaboration between all authors. JD, JV and MS conceived and defined the research theme. BF, BG, MS and TD designed the experiments. EH performed the sequencing of DNA, bioinformatics analysis and development of tandem repeat sequences. JČ and TD also worked on laboratory experiments. MK, MS and TD discussed FISH with cDNA methods. EH, JD, MS and TD discussed the results. MS designed the methods, carried out the experiments, analyzed the data and wrote the manuscript. All authors have contributed to, seen, read and approved the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Dr. Joseph Robins from ARS Forage and Range Research Laboratory, USDA, Logan, USA, for providing the plant material used in this study. We are grateful to Dr. Shuhei Nasuda, Laboratory of Plant Genetics, Graduate School of Agriculture, Kyoto University, Japan, for a permission to the use the cDNA clones. We acknowledge Marie Kubaláková and Dr. Jan Šafář from the Institute of Experimental Botany, Czech Republic, for their continuous help and assistance with the plasmid transformation. Special thanks are due to Dr. Adoración Cabrera from the Department of Genetics at the University of Cordoba in Spain, for feedback, support and encouragement. We are also grateful to Dr. Istvan Molnar for critical reading of the manuscript. This work has been supported by the Grant Agency of the Czech Republic (award No. 16-16992S) and by the Czech Ministry of Education, Youth and Sports (award LO1204 from the National Program of Sustainability I).
Compliance with ethical standards
Conflict of interest
All the authors declare that they have no conflict of interest.
References
- Alkhimova AG, Heslop-Harrison JS, Shchapova AI, Vershinin AV. Rye chromosome variability in wheat-rye addition and substitution lines. Chromosome Res. 1999;7:205–212. doi: 10.1023/a:1009299300018. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Anathawat-Jonsson K, Heslop-Harrison JS. Isolation and characterization of genome-specific DNA-sequences in Triticeae species. Mol Gen Genet. 1993;240:151–158. doi: 10.1007/bf00277052. [DOI] [PubMed] [Google Scholar]
- Appels R, Dennis ES, Smyth DR, Peacock WJ. 2 Repeated DNA-sequences from the heterochromatic regions of rye (Secale cereale) chromosomes. Chromosoma. 1981;84:265–277. doi: 10.1007/bf00399137. [DOI] [Google Scholar]
- Asay KH, Jensen KB, Hsiao C, Dewey DR. Probable origin of standard crested wheatgrass, Agropyron desertorum Fisch ex Link, Schultes. Can J Plant Sci. 1992;72:763–772. doi: 10.4141/cjps92-092. [DOI] [Google Scholar]
- Asghari A, Agayev Y, Fathi SAA. Karyological study of four species of wheat grass (Agropyron sp.) Pak J Biol Sci. 2007;10:1093–1097. doi: 10.3923/pjbs.2007.1093.1097. [DOI] [PubMed] [Google Scholar]
- Badaeva ED, Friebe B, Gill BS. Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome. 1996;39:293–306. doi: 10.1139/g96-040. [DOI] [PubMed] [Google Scholar]
- Badaeva ED, Amosova AV, Samatadze TE, et al. Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Syst Evol. 2004;246:45–76. doi: 10.1007/s00606-003-0072-4. [DOI] [Google Scholar]
- Bedbrook JR, Jones J, Odell M, et al. Molecular description of telomeric heterochromatin in Secale species. Cell. 1980;19:545–560. doi: 10.1016/0092-8674(80)90529-2. [DOI] [PubMed] [Google Scholar]
- Benavente E, Cifuentes M, Dusautoir JC, David J. The use of cytogenetic tools for studies in the crop-to-wild gene transfer scenario. Cytogenetic Genome Res. 2008;120:384–395. doi: 10.1159/000121087. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasileiro-Vidal AC, Cuadrado A, Brammer SP, et al. Chromosome characterization in Thinopyrum ponticum (Triticeae, Poaceae) using in situ hybridization with different DNA sequences. Genet Mol Biol. 2003;26:505–510. doi: 10.1590/s1415-47572003000400014. [DOI] [Google Scholar]
- Brettell RIS, Banks PM, Cauderon Y, et al. A single wheatgrass chromosome reduces the concentration of barley yellow dwarf virus in wheat. Ann Appl Biol. 1988;113:599–603. doi: 10.1111/j.1744-7348.1988.tb03337.x. [DOI] [Google Scholar]
- Busch W, Herrmann RG, Martin R. Refined physical mapping of the Sec-1 locus on the satellite of chromosome 1R of rye (Secale cereale) Genome. 1995;38:889–893. doi: 10.1139/g95-117. [DOI] [PubMed] [Google Scholar]
- Cabrera A, Martin A, Barro F. In situ comparative mapping (ISCM) of Glu-1 loci in Triticum and Hordeum. Chromosome Res. 2002;10:49–54. doi: 10.1023/a:1014270227360. [DOI] [PubMed] [Google Scholar]
- Castilho A, Heslop-Harrison JS. Physical mapping of 5S and 18S-25S rDNA and repetitive DNA-sequences in Aegilops umbellulata. Genome. 1995;38:91–96. doi: 10.1139/g95-011. [DOI] [PubMed] [Google Scholar]
- Cauderon Y, Rhind JM. Effect on wheat of an Agropyron chromosome carrying stripe rust resistance. Ann De L Amelior Des Plantes. 1976;26:745–749. [Google Scholar]
- Chen Q, Jahier J, Cauderon Y. Production and cytogenetical studies of hybrids between Triticum aestivum and Agropyron cristatum Gaertn. Comptes Rendus De L Academie Des Sciences Serie Iii-Sciences De La Vie-Life Sciences. 1989;308:425–430. [Google Scholar]
- Contento A, Heslop-Harrison JS, Schwarzacher T. Diversity of a major repetitive DNA sequence in diploid and polyploid Triticeae. Cytogenet Genome Res. 2005;109:34–42. doi: 10.1159/000082379. [DOI] [PubMed] [Google Scholar]
- Danilova TV, Friebe B, Gill BS. Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma. 2012;121:597–611. doi: 10.1007/s00412-012-0384-7. [DOI] [PubMed] [Google Scholar]
- Danilova TV, Friebe B, Gill BS. Development of a wheat single gene FISH map for analyzing homoeologous relationship and chromosomal rearrangements within the Triticeae. Theor Appl Genet. 2014;127:715–730. doi: 10.1007/s00122-013-2253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova TV, Akhunova AR, Akhunov ED, et al. Major structural genomic alterations can be associated with hybrid speciation in Aegilops markgrafii (Triticeae) Plant J. 2017;92:317–330. doi: 10.1111/tpj.13657. [DOI] [PubMed] [Google Scholar]
- De Carvalho R, Guerra M. Cytogenetics of Manihot esculenta Crantz (cassava) and eight related species. Hereditas. 2002;136:159–168. doi: 10.1034/j.1601-5223.2002.1360212.x. [DOI] [PubMed] [Google Scholar]
- Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS ONE. 2013 doi: 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Gale MD. Genome relationships: the grass model in current research. Plant Cell. 2000;12:637–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, et al. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet. 1993;85:673–680. doi: 10.1007/bf00225004. [DOI] [PubMed] [Google Scholar]
- Devos KM, Dubcovsky J, Dvorak J, et al. Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor Appl Genet. 1995;91:282–288. doi: 10.1007/bf00220890. [DOI] [PubMed] [Google Scholar]
- Dewey DR. Historical and current taxonomic perspectives of Agropyron, Elymus, and related genera. Crop Sci. 1983;23:637–642. doi: 10.2135/cropsci1983.0011183X002300040009x. [DOI] [Google Scholar]
- Dewey DR, Asay KH. Cytogenetic and taxonomic relationships among three diploid crested wheat grasses. Crop Sci. 1982;22:645–650. doi: 10.2135/cropsci1982.0011183X002200030052x. [DOI] [Google Scholar]
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry. 2003;51:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Dvořák J, Zhang HB, Kota RS, Lassner M. Organization and evolution of the 5S-ribosomal RNA gene family in wheat and related species. Genome. 1989;32:1003–1016. doi: 10.1139/g89-545. [DOI] [Google Scholar]
- Feuillet C, Langridge P, Waugh R. Cereal breeding takes a walk on the wild side. Trends Genet. 2008;24:24–32. doi: 10.1016/j.tig.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Friebe B, Mukai Y, Dhaliwal HS, et al. Identification of alien chromatin specifying resistance to wheat streak mosaic and greenbug in wheat germ plasm by C-banding and in situ hybridization. Theor Appl Genet. 1991;81:381–389. doi: 10.1007/BF00228680. [DOI] [PubMed] [Google Scholar]
- Friebe B, Jiang J, Raupp WJ, et al. Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica. 1996;91:59–87. doi: 10.1007/bf00035277. [DOI] [Google Scholar]
- Fu S, Lv Z, Guo X, et al. Alteration of terminal heterochromatin and chromosome rearrangements in derivatives of wheat-rye hybrids. J Genet Genom. 2013;40:413–420. doi: 10.1016/j.jgg.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Fu S, Chen L, Wang Y, et al. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci Rep. 2015;5:10552. doi: 10.1038/srep10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K, Kamisugi Y, Sakai F. Physical mapping of 5S rDNA loci by direct-cloned biotinylated probes in barley chromosomes. Genome. 1994;37:105–111. doi: 10.1139/g94-013. [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Plant comparative genetics after 10 years. Science. 1998;282:656–659. doi: 10.1126/science.282.5389.656. [DOI] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. Cloning and characterization of ribosomal-RNA genes from wheat and barley. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill BS, Friebe B, Endo TR. Standard karyotype and nomenclature system for description of chromosome bands and structural-aberrations in wheat (Triticum aestivum) Genome. 1991;34:830–839. doi: 10.1139/g91-128. [DOI] [Google Scholar]
- Gill BS, Friebe B, Raupp WJ, et al. Wheat Genetics Resource Center: the first 25 years. Adv Agron. 2006;89:73–136. doi: 10.1016/s0065-2113(05)89002-9. [DOI] [Google Scholar]
- Guerra M, Kenton A, Bennett MD. rDNA sites in mitotic and polytene chromosomes of Vigna unguiculata (L) Walp and Phaseolus coccineus L revealed by in situ hybridization. Ann Bot. 1996;78:157–161. doi: 10.1006/anbo.1996.0108. [DOI] [Google Scholar]
- Han FP, Fedak G, Guo WL, Liu B. Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGcIR-1a) in newly synthesized wheat allopolyploids. Genetics. 2005;170:1239–1245. doi: 10.1534/genetics.104.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Bai L, Su J, et al. Genetic rearrangements of six wheat-Agropyron cristatum 6P addition lines revealed by molecular markers. PLoS ONE. 2014 doi: 10.1371/journal.pone.0091066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Liu W, Lu Y, et al. Isolation and application of P genome-specific DNA sequences of Agropyron Gaertn. in Triticeae. Planta. 2017;245:425–437. doi: 10.1007/s00425-016-2616-1. [DOI] [PubMed] [Google Scholar]
- He Q, Cai Z, Hu T, et al. Repetitive sequence analysis and karyotyping reveals centromere-associated DNA sequences in radish (Raphanus sativus L.) BMC Plant Biol. 2015 doi: 10.1186/s12870-015-0480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C, Wang RRC, Dewey DR. Karyotype analysis and genome relationships of 22 diploid species in the tribe Triticeae. Can J Genet Cytol. 1986;28:109–120. doi: 10.1139/g86-015. [DOI] [Google Scholar]
- Hsiao C, Asay KH, Dewey DR. Cytogenetic analysis of interspecific hybrids and amphiploids between 2 diploid crested wheatgrasses, Agropyron mongolicum and A. cristatum. Genome. 1989;32:1079–1084. doi: 10.1139/g89-557. [DOI] [Google Scholar]
- Hu L-J, Liu C, Zeng Z-X, et al. Genomic rearrangement between wheat and Thinopyrum elongatum revealed by mapped functional molecular markers. Genes Genom. 2012;34:67–75. doi: 10.1007/s13258-011-0153-7. [DOI] [Google Scholar]
- Jabeen R, Iftikhar T, Mengal T, Khattak MI. A comparative chromosomal count and morphological karyotyping of three indigenous cultivars of kalongi (Nigella sativa L.) Pak J Bot. 2012;44:1007–1012. [Google Scholar]
- Jiang J, Gill BS, Wang GL, et al. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA. 1995;92:4487–4491. doi: 10.1073/pnas.92.10.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubault M, Tanguy AM, Abelard P, et al. Attempts to induce homoeologous pairing between wheat and Agropyron cristatum genomes. Genome. 2006;49:190–193. doi: 10.1139/g05-074. [DOI] [PubMed] [Google Scholar]
- Karafiátová M, Bartoš J, Kopecký D, et al. Mapping nonrecombining regions in barley using multicolor FISH. Chrom Res. 2013;21:739–751. doi: 10.1007/s10577-013-9380-x. [DOI] [PubMed] [Google Scholar]
- Kato A, Lamb JC, Birchler JA. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA. 2004;101:13554–13559. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Albert PS, Vega JM, Birchler JA. Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech Histochem. 2006;81:71–78. doi: 10.1080/10520290600643677. [DOI] [PubMed] [Google Scholar]
- Kawaura K, Mochida K, Enju A, et al. Assessment of adaptive evolution between wheat and rice as deduced from full-length common wheat cDNA sequence data and expression patterns. BMC Genom. 2009 doi: 10.1186/1471-2164-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharb P, Dong JJ, Islam-Faridi MN, et al. Fluorescence in situ hybridization of single copy transgenes in rice chromosomes. In Vitro Cellular Dev Biol Plant. 2001;37:1–5. doi: 10.1007/s11627-001-0001-6. [DOI] [Google Scholar]
- Kim JS, Islam-Faridi MN, Klein PE, et al. Comprehensive molecular cytogenetic analysis of Sorghum genome architecture distribution of euchromatin heterochromatin, genes and recombination in comparison to rice. Genetics. 2005;171:1963–1976. doi: 10.1534/genetics.105.048215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishii M, Yamada T, Sasakuma T, Tsujimoto H. Production of wheat-Leymus racemosus chromosome addition lines. Theor Appl Genet. 2004;109:255–260. doi: 10.1007/s00122-004-1631-y. [DOI] [PubMed] [Google Scholar]
- Knott DR. Effect on wheat of Agropyron chromosome carrying rust resistance. Can J Genet Cytol. 1964;6:500–507. doi: 10.1139/g64-064. [DOI] [PubMed] [Google Scholar]
- Knott DR. Translocations involving Triticum chromosomes and Agropyron chromosomes carrying rust resistance. Can J Genet Cytol. 1968;10:695–696. doi: 10.1139/g68-087. [DOI] [PubMed] [Google Scholar]
- Li L, Yang X, Zhou R, et al. Establishment of wheat-Agropyron cristatum alien addition lines: II. Identification of alien chromosomes and analysis of development approaches. Acta Genet Sin. 1998;25:538–544. [Google Scholar]
- Li G-R, Liu C, Wei P, et al. Chromosomal distribution of a new centromeric Ty3-gypsy retrotransposon sequence in Dasypyrum and related Triticeae species. J Genet. 2012;91:343–348. doi: 10.1007/s12041-012-0181-3. [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Y, Pan C, et al. Characterization of a novel wheat-Agropyron cristatum 2P disomic addition line with powdery mildew resistance. Crop Sci. 2016;56:2390–2400. doi: 10.2135/cropsci2015.10.0638. [DOI] [Google Scholar]
- Li D, Li T, Wu Y, et al. FISH-Based markers enable identification of chromosomes derived from tetraploid Thinopyrum elongatum in hybrid lines. Front Plant Sci. 2018;9:526. doi: 10.3389/fpls.2018.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limin AE, Fowler DB. An interspecific hybrid and amphiploid produced from Triticum aestivum crosses with Agropyron cristatum and Agropyron desertorum. Genome. 1990;33:581–584. doi: 10.1139/g90-085. [DOI] [Google Scholar]
- Linc G, Gaal E, Molnar I, et al. Molecular cytogenetic (FISH) and genome analysis of diploid wheatgrasses and their phylogenetic relationship. PLoS ONE. 2017 doi: 10.1371/journal.pone.0173623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Liu W, Wu J, et al. Analysis of genetic diversity in natural populations of Psathyrostachys huashanica Keng using microsatellite (SSR) markers. Agric Sci China. 2010;9:463–471. doi: 10.1016/s1671-2927(09)60118-8. [DOI] [Google Scholar]
- Luan Y, Wang X, Liu W, et al. Production and identification of wheat-Agropyron cristatum 6P translocation lines. Planta. 2010;232:501–510. doi: 10.1007/s00425-010-1187-9. [DOI] [PubMed] [Google Scholar]
- Martin A, Cabrera A, Esteban E, et al. A fertile amphiploid between diploid wheat (Triticum tauschii) and crested wheatgrass (Agropyron cristatum) Genome. 1999;42:519–524. doi: 10.1139/gen-42-3-519. [DOI] [PubMed] [Google Scholar]
- McArthur RI, Zhu XW, Oliver RE, et al. Homoeology of Thinopyrum junceum and Elymus rectisetus chromosomes to wheat and disease resistance conferred by the Thinopyrum and Elymus chromosomes in wheat. Chrom Res. 2012;20:699–715. doi: 10.1007/s10577-012-9307-y. [DOI] [PubMed] [Google Scholar]
- McGuire PE, Dvorak J. High salt tolerance potential in wheatgrasses. Crop Sci. 1981;21:702–705. doi: 10.2135/cropsci1981.0011183X002100050018x. [DOI] [Google Scholar]
- Mickelsonyoung L, Endo TR, Gill BS. A cytogenetic ladder-map of the wheat homoeologous group-4 chromosomes. Theor Appl Genet. 1995;90:1007–1011. doi: 10.1007/BF00222914. [DOI] [PubMed] [Google Scholar]
- Miftahudin Ross K, Ma XF, et al. Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics. 2004;168:651–663. doi: 10.1534/genetics.104.034827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaghaderi G, Houben A, Badaeva ED. Molecular-cytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Mol Cytogenet. 2014 doi: 10.1186/s13039-014-0091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai Y, Endo TR, Gill BS. Physical mapping of the 5S ribosomal-RNA multigene family in common wheat. J Hered. 1990;81:290–295. doi: 10.1093/oxfordjournals.jhered.a110991. [DOI] [Google Scholar]
- Mukai Y, Endo TR, Gill BS. Physical mapping of the 18S.26S ribosomal-RNA multigene family in common wheat: identification of a new locus. Chromosoma. 1991;100:71–78. doi: 10.1007/bf00418239. [DOI] [Google Scholar]
- Nagaki K, Tsujimoto H, Isono K, Sasakuma T. Molecular characterization of a tandem repeat, AFA family, and distribution among Triticeae. Genome. 1995;38:479–486. doi: 10.1139/g95-063. [DOI] [PubMed] [Google Scholar]
- Naranjo CA, Poggio L, Brandham PE. A practical method of chromosome classification on the basis of centromere position. Genetica. 1983;62:51–53. doi: 10.1007/BF00123310. [DOI] [Google Scholar]
- Naranjo T, Roca A, Goicoechea PG, Giraldez R. Arm homoeology of wheat and rye chromosomes. Genome. 1987;29:873–882. doi: 10.1139/g87-149. [DOI] [Google Scholar]
- Nasuda S, Friebe B, Gill BS. Gametocidal genes induce chromosome breakage in the interphase prior to the first mitotic cell division of the male gametophyte in wheat. Genetics. 1998;149:1115–1124. doi: 10.1093/genetics/149.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinform. 2010 doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, et al. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics. 2013;29:792–793. doi: 10.1093/bioinformatics/btt054. [DOI] [PubMed] [Google Scholar]
- Ochoa V, Madrid E, Said M, et al. Molecular and cytogenetic characterization of a common wheat-Agropyron cristatum chromosome translocation conferring resistance to leaf rust. Euphytica. 2015;201:89–95. doi: 10.1007/s10681-014-1190-5. [DOI] [Google Scholar]
- Paterson AH, Bowers JE, Burow M, et al. Comparative genomics of plant chromosomes. Plant Cell. 2000;12:1523–1539. doi: 10.1105/tpc.12.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Friebe B, Zhang P, Gill BS. Homoeologous recombination, chromosome engineering and crop improvement. Chrom Res. 2007;15:3–19. doi: 10.1007/s10577-006-1108-8. [DOI] [PubMed] [Google Scholar]
- Reddy P, Appels R. A 2nd locus for the 5S multigene family in Secale L.: sequence divergence in 2 lineages of the family. Genome. 1989;32:456–467. doi: 10.1139/g89-469. [DOI] [PubMed] [Google Scholar]
- Said M, Cabrera A. A physical map of chromosome 4Hch from H. chilense containing SSR, STS and EST-SSR molecular markers. Euphytica. 2009;167:253–259. doi: 10.1007/s10681-009-9895-6. [DOI] [Google Scholar]
- Said M, Recio R, Cabrera A. Development and characterisation of structural changes in chromosome 3 Hch from Hordeum chilense in common wheat and their use in physical mapping. Euphytica. 2012;188:429–440. doi: 10.1007/s10681-012-0712-2. [DOI] [Google Scholar]
- Sato S, Hizume M, Kawamura S. Relationship between secondary constrictions and nucleolus organizing regions in Allium sativum chromosomes. Protoplasma. 1980;105:77–85. doi: 10.1007/bf01279851. [DOI] [Google Scholar]
- Schneider A, Linc G, Molnar-Lang M. Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breed. 2003;122:396–400. doi: 10.1046/j.1439-0523.2003.00891.x. [DOI] [Google Scholar]
- Schneider A, Linc G, Molnar I, Molnar-Lang M. Molecular cytogenetic characterization of Aegilops biuncialis and its use for the identification of 5 derived wheat-Aegilops biuncialis disomic addition lines. Genome. 2005;48:1070–1082. doi: 10.1139/g05-062. [DOI] [PubMed] [Google Scholar]
- Schulz-Schaeffer J, Allderdice PW, Creel GC. Segmental allo-polyploidy in tetraploid and hexaploid Agropyron species of the crested wheatgrass complex (Section Agropyron) Crop Sci. 1963;3:525–530. doi: 10.2135/cropsci1963.0011183X000300060021x. [DOI] [Google Scholar]
- Sharma HC, Gill BS, Uyemoto JK. High levels of resistance in Agropyron species to barley yellow dwarf and wheat streak mosaic viruses. Phytopathol Z J Phytopathol. 1984;110:143–147. doi: 10.1111/j.1439-0434.1984.tb03402.x. [DOI] [Google Scholar]
- Shukle RH, Lampe DJ, Lister RM, Foster JE. Aphid feeding behavior: relationship to barley yellow dwarf virus-resistance in Agropyron species. Phytopathology. 1987;77:725–729. doi: 10.1094/Phyto-77-725. [DOI] [Google Scholar]
- Smit S, Widmann J, Knight R. Evolutionary rates vary among rRNA structural elements. Nucl Acids Res. 2007;35:3339–3354. doi: 10.1093/nar/gkm101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Jiang L, Han H, et al. Efficient induction of wheat-Agropyron cristatum 6P translocation lines and GISH detection. PLoS ONE. 2013 doi: 10.1371/journal.pone.0069501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Lu Y, Zhang J, et al. Physical mapping of Agropyron cristatum chromosome 6P using deletion lines in common wheat background. Theor Appl Genet. 2016;129:1023–1034. doi: 10.1007/s00122-016-2680-8. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene-Combis. 1995;167:1–10. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- Sousa A, Barros e Silva AE, Cuadrado A, et al. Distribution of 5S and 45S rDNA sites in plants with holokinetic chromosomes and the “chromosome field” hypothesis. Micron. 2011;42:625–631. doi: 10.1016/j.micron.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Types of polyploids: their classification and significance. Adv Genet Incorporating Mol Genet Med. 1947;1:403–429. doi: 10.1016/s0065-2660(08)60490-3. [DOI] [PubMed] [Google Scholar]
- Svitashev S, Bryngelsson T, Li XM, Wang RRC. Genome-specific repetitive DNA and RAPD markers for genome identification in Elymus and Hordelymus. Genome. 1998;41:120–128. doi: 10.1139/gen-41-1-120. [DOI] [PubMed] [Google Scholar]
- Szakács É, Molnár-Láng M. Fluorescent in situ hybridization polymorphism on the 1RS chromosome arms of cultivated Secale cereale species. Cereal Res Commun. 2008;36:247–255. doi: 10.1556/CRC.36.2008.2.5. [DOI] [Google Scholar]
- Tang Z-X, Fu S-L, Ren Z-L, et al. Variations of tandem repeat, regulatory element, and promoter regions revealed by wheat-rye amphiploids. Genome. 2008;51:399–408. doi: 10.1139/g08-027. [DOI] [PubMed] [Google Scholar]
- Tang Z, Yang Z, Fu S. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet. 2014;55:313–318. doi: 10.1007/s13353-014-0215-z. [DOI] [PubMed] [Google Scholar]
- Tang S, Qiu L, Xiao Z, et al. New oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes. 2016;7:118. doi: 10.3390/genes7120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, et al. Primer3-new capabilities and interfaces. Nucl Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershinin AV, Schwarzacher T, Heslop-Harrison JS. The large-scale genomic organization of repetitive dna families at the telomeres of rye chromosomes. Plant Cell. 1995;7:1823–1833. doi: 10.1105/tpc.7.11.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RRC. Agropyron and Psathyrostachys. Wild Crop Relat Genom Breed Res Cereals. 2011 doi: 10.1007/978-3-642-14228-4_2. [DOI] [Google Scholar]
- Wang CJR, Harper L, Cande WZ. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell. 2006;18:529–544. doi: 10.1105/tpc.105.037838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Xiang J, Gao A, et al. Analysis of chromosomal structural polymorphisms in the St, P, and Y genomes of Triticeae (Poaceae) Genome. 2010;53:241–249. doi: 10.1139/g09-098. [DOI] [PubMed] [Google Scholar]
- Wu J, Yang X, Wang H, et al. The introgression of chromosome 6P specifying for increased numbers of florets and kernels from Agropyron cristatum into wheat. Theor Appl Genet. 2006;114:13–20. doi: 10.1007/s00122-006-0405-0. [DOI] [PubMed] [Google Scholar]
- Wu M, Zhang JP, Wang JC, et al. Cloning and characterization of repetitive sequences and development of SCAR markers specific for the P genome of Agropyron cristatum. Euphytica. 2010;172:363–372. doi: 10.1007/s10681-009-0033-2. [DOI] [Google Scholar]
- Yang C-T, Fan X, Wang X-L, et al. Karyotype analysis of Agropyron cristatum (L.) Gaertner. Caryologia. 2014;67:234–237. doi: 10.1080/0144235x.2014.974351. [DOI] [Google Scholar]
- Ye X, Lu Y, Liu W, et al. The effects of chromosome 6P on fertile tiller number of wheat as revealed in wheat-Agropyron cristatum chromosome 5A/6P translocation lines. Theor Appl Genet. 2015;128:797–811. doi: 10.1007/s00122-015-2466-4. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jia J, Gale MD, Devos KM. Relationships between the chromosomes of Aegilops umbellulata and wheat. Theor Appl Genet. 1998;96:69–75. doi: 10.1007/s001220050710. [DOI] [Google Scholar]
- Zhang P, Li WL, Fellers J, et al. BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma. 2004;112:288–299. doi: 10.1007/s00412-004-0273-9. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bian Y, Gou X, et al. Intrinsic karyotype stability and gene copy number variations may have laid the foundation for tetraploid wheat formation. Proc Natl Acad Sci USA. 2013;110:19466–19471. doi: 10.1073/pnas.1319598110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu W, Han H, et al. De novo transcriptome sequencing of Agropyron cristatum to identify available gene resources for the enhancement of wheat. Genomics. 2015;106:129–136. doi: 10.1016/j.ygeno.2015.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.