Abstract
In 2016, disseminated tuberculosis caused by Mycobacterium caprae was diagnosed in a captive Borneo elephant in Japan. The bacterium was initially identified from clinical isolates. An isolate collected during a relapse showed isoniazid monoresistance and a codon 315 katG mutation.
Keywords: Borneo elephant, disseminated tuberculosis, Mycobacterium caprae, Tuberculosis and other mycobacterial infections, regions of difference, whole genome sequence, bacteria, Japan, zoonoses
Elephants are susceptible to infection by some members of the Mycobacterium tuberculosis complex (MTBC) (1). The MTBC comprises several genetically homogeneous species that have a wide range of hosts and can cause tuberculosis in humans and in animals. Infection in elephants is presumed to originate from human caretakers who have tuberculosis; however, transmission between elephants or from other animals is also possible (2–4).
Phylogenetic events during the divergence of MTBC species are represented within the genomes of MTBC species (5). Whole-genome sequencing has shown that single-nucleotide polymorphism (SNP) microevolution occurs in MTBC strains in the host.
We describe mycobacterial infection in an elephant that was caused by a relatively uncommon species of MTBC. M. caprae infection, a species of the MTBC, was diagnosed in a captive Borneo elephant (Elephas maximus borneensis) that was brought directly from Borneo island after being orphaned.
The Study
In February 2016, an ≈17-year-old female Borneo elephant in the Fukuyama Zoo (Fukuyama, Japan) had low-grade fever (99.9°F), anorexia, progressive weight loss, and cough with sputum. The elephant had been housed alone in a facility with a roofed room and an open-air enclosure and had no contact with other animals. She was seropositive by Chembio DPP VetTB Assay for Elephants (Chembio Diagnostic Systems, Inc., Medford, NY, USA), which detects antibodies to CFP10/ESAT-6 and MPB83 antigens (6,7).
Submitted specimens to the National Hospital Organization Kinki-chuo Chest Medical Center (Sakai, Japan) were sputum, feces, urine, and vaginal discharge recovered from the floor. Acid-fast bacilli were visualized on Ziehl−Neelsen staining performed according to standard methods (8). Isolates with smooth to greasy, domed, nonchromogenic colonies were recovered from all submitted samples using both 7H11 agar and MGIT broth (Becton Dickinson, Fukushima, Japan) and identified as MTBC using loop-mediated isothermal amplification (Eiken Chemical, Tokyo, Japan) and TaqMan PCR (Roche Diagnostics, Tokyo, Japan) (9).
A single colony was sequenced using MiSeq (Illumina, Inc., Tokyo, Japan). Raw reads were trimmed by base quality and were mapped to the M. tuberculosis reference genome, H37Rv (GenBank accession no. CP003248). The initial isolate, EPDC01, was characterized by the presence and absence of genomic regions of difference, the mutation (G to A) of oxyR285, and M. caprae–specific SNPs in lepA (10) (Table 1). These results unexpectedly suggested that the causative agent of tuberculosis in this elephant was M. caprae. The regions of difference analysis suggested that EPDC01 belonged to the Allgäu type of M. caprae found in red deer (Cervus elaphus) (11). However, when we used kSNP3 (12), a kmer-based method, to compare SNPs from the entire genome of EPDC01 to previously published Allgäu and Lechtal types of M. caprae and other MTBC species, the results showed this isolate was not closely related to either Allgäu or Lechtal types (Figure; Technical Appendix Figure) (13).
Table 1. Summary of MTBC PCR-typing panel results in a study of Mycobacterium caprae infection in a Borneo elephant, Japan, 2016*.
| Organism | RD9 | TbD1 | katG 463 | gyrA 95 | RD7 | RD8 | RD9 | RD10 | mmpL6 551 | RD12oryx | RD1mic | RDcan | RDseal | oxyR 285 | RD12bov | RD13 | RD4 | pncA 57 | RD1BCG | lepA | katG 315 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| “Ancentral” M. tuberculosis | + | + | CTG | AGC | + | + | + | + | + | + | + | + | + | G | + | + | + | C | + | – | AGC |
| “Modern” M. tuberculosis | + | – | CTG/CGG† | AGC/ACC† | + | + | + | + | + | + | + | + | + | G | + | + | + | C | + | – | AGC |
| M. africanum subtype I (WA-1) | – | + | CTG | AGC | + | + | + | + | + | + | + | + | + | G | + | + | + | C | + | – | AGC |
| M. africanum subtype II (WA-2) | – | + | CTG | AGC | – | + | + | – | + | + | + | + | + | G | + | + | + | C | + | – | AGC |
| M. orygis | – | + | CTG | AGC | – | + | + | – | – | – | + | + | + | G | + | + | + | C | + | – | AGC |
| M. microti | – | + | CTG | AGC | – | + | + | – | – | + | – | + | + | G | + | + | + | C | + | – | AGC |
| M. canettii | – | + | CTG | AGC | – | + | + | – | – | + | + | – | + | G | + | + | + | C | + | – | AGC |
| M. pinnipedii | – | + | CTG | AGC | – | + | + | – | – | + | + | + | – | G | + | + | + | C | + | – | AGC |
| M. caprae | – | + | CTG | AGC | – | + | + | – | – | – | + | + | + | A | – | – | + | C | + | + | AGC |
| M. bovis | – | + | CTG | AGC | – | + | + | – | – | – | + | + | + | A | – | – | – | G | + | – | AGC |
| M. bovis BCG | – | + | CTG | AGC | – | + | + | – | – | – | + | + | + | A | – | – | – | G | – | – | AGC |
| EPCD01 in this study | – | + | CTG | AGC | – | + | + | – | – | – | + | + | + | A | – | – | + | C | + | + | AGC |
| EPDC02 in this study | – | + | CTG | AGC | – | + | + | – | – | – | + | + | + | A | – | – | + | C | + | + | ACC |
*Includes a differential distribution of known single nucleotide polymorphisms among the MTBC species. MTBC, Mycobacterium tuberculosis complex; RD, region of difference; +, positive; –, negative †”Modern” M, tuberculosis strains are segregated to 2 and 3 in their respective principal genetic group based on the combinations of katG463-gyrA95 polymorphism.
Figure.
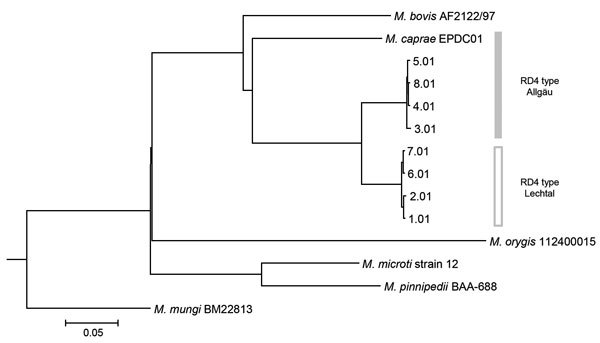
Phylogenetic tree of isolate EPDC01 from a captive Borneo elephant with Mycobacterium caprae infection, Japan, 2016, and 8 Mycobacterium caprae strains (Allgäu and Lechtal types) from a report by Broeckl et al. (13). Short reads of M. caprae strains were assembled by CLC Genomics Workbench version 9.5.1 (https://www.qiagenbioinformatics.com/solutions/functional-genomics/?gclid=EAIaIQobChMIvvGL3L7T2wIVTSOBCh2FAAKtEAAYASAAEgKLWvD_BwE) before analysis. Core single-nucleotide polymorphisms of all 13 strains, including reference M. tuberculosis complex strains (M. bovis, AF2122/97 [GenBank accession no. NC_002945.4]: M. orygis, 112400015 [NZ_APKD00000000.1]: M. pinnipedii, BAA-688 [MWXB00000000.1]: M. microti, strain 12 [CP010333.1]: and M. mungi, BM22813 [NZ_LXTB00000000.1]), were determined and used for tree construction based on neighbor-joining by kSNP3 (12). A tree including all 61 strains described by Broeckl et al. (13) is shown in the Technical Appendix Figure. Scale bar indicates nucleotide substitutions per site.
The isolate was susceptible to isoniazid, rifampin, ethambutol, and levofloxacin according to the broth microdilution method (BrothMIC MTB-1; Kyokuto Pharmaceutical, Inc, Tokyo, Japan). It also was susceptible to pyrazinamide using Bactec MGIT 960 PZA kit (Becton Dickinson).
The infected elephant initially weighed 2,400 kg; isoniazid (4.5–7 mg/kg), pyrazinamide (31–33 mg/kg), and levofloxacin (11 mg/kg) were administered rectally, once a day. When weighing on a scale was not possible, the elephant’s weight was estimated using the chest girth method. After 1 month of treatment, the vaginal discharge disappeared, and the elephant’s sputum culture became negative after 2 months. After 6 months, the multidrug treatment was interrupted for 3 weeks because of severe gastrointestinal disturbance and hepatic dysfunction. Rectal administration of isoniazid and pyrazinamide was resumed for an additional 3 months after recovery from adverse effects. Follow-up trunk wash samples were culture negative, but M. caprae was isolated from a sputum sample collected from the chin in February 2017. This new isolate was resistant to isoniazid. The drug regimen was then changed to oral rifampin (10 mg/kg) and rectal ethambutol (30 mg/kg), levofloxacin (10 mg/kg), and pyrazinamide (30 mg/kg). Under the modified treatment, the recurrence symptoms disappeared, and the routine sputum cultures became negative.
We also sequenced the recurrent isoniazid-resistant isolate (EPDC02) using a MiSeq and detected a mutation in katG where Ser-315 was replaced by 113 Thr (S315T). No mutations were detected in other representative drug resistance–related genes, such as rpoB (rifampin); rrn, gidB, and rpsL (streptomycin); embABC (ethambutol); pncAC (pyrazinamide); and gyrAB (quinolone). The pairwise distance between the initial and relapse isolates (EPDC01 and EPDC02) involved at least 7 single-nucleotide variants found in coding regions (Table 2).
Table 2. Single-nucleotide polymorphism differences between isolates EPDC01 and EPDC02 from a captive Borneo elephant with Mycobacterium caprae infection, Japan, 2016*.
| Genomic position | Gene | Mutation | Amino acid change | Description of function |
|---|---|---|---|---|
| 1,338,135 | Rv1194c | G → frameshift | Gln127 → frameshift | Unknown |
| 1,445,272 | Rv1290A | C → T | Ala72 → Val | Unknown |
| 2,155,168 | katG | C → T | Ser315 → Asn | Catalase; isoniazid resistance |
| 2,577,556 | Rv2306A | G → A | Gly150 → Glu | Unknown |
| 3,200,585 | Rv2891 | T → G | Leu107 → Arg | Unknown |
| 3,777,062 | Rv3365c | C → A | Gly147 → Val | Unknown |
| 4,298,329 | pks2 | G → A | Pro426 → Leu | Lipid metabolism |
| 4,345,372 | Rv3869 | G → C | Ala112 → Pro | Unknown |
*Excluded paralogue genes, which are the descendants of an ancestral gene and underwent a duplication event.
For serologic monitoring of treatment, we tested archived serum samples collected before and during treatment using the DPP assay. Although a sample in November 2003 was reactive on CFP10/ESAT-6 but not on MPB83, at the time of culture-based tuberculosis diagnosis (February 2016), the reaction to CFP10/ESAT-6 was more intense, and the complete band of MPB83 appeared. The intensity to 2 test lines gradually decreased during the initial treatment (September 2016) and disease recrudescence (February 2017). The modified 18-month course of treatment is scheduled to end in October 2018.
Employees at the zoo were assessed for tuberculosis based on symptoms, radiographs, and serology using the QuantiFERON-TB Gold test (QIAGEN, Tokyo, Japan); none had active tuberculosis. Clinical examination, culture, PCR, and tuberculin skin test were used to evaluate as many primates and hoofstock as possible in the collection, and no tuberculosis-positive animals were identified.
Conclusions
A better understanding of tuberculosis in elephants is crucial to improve medical management and reduce risk of transmission to other animals and humans. During the initial treatment of the elephant reported here, treatment was interrupted for 3 weeks because of adverse effects. Reducing the dose of isoniazid was the probable cause of the acquired isoniazid resistance. When treating tuberculosis in elephants, the benefits and adverse effects should be weighed carefully. The DPP provides an indirect measure of infection and disease status (6). In this elephant, declining DPP reactivity was thought to indicate a response to therapy; however, more sensitive biomarkers to monitor therapeutic response are needed.
Previously, epidemiologic observations of elephant tuberculosis by IS6110 restriction fragment-length polymorphism have been based on evidence of local zoonotic risk for transmission to humans or of an epizootic reservoir for transmission to elephants or other animal species (goats and rhinoceros) (1,14). Recently, M. tuberculosis strains in 2 captive elephants in a small traveling circus harbored 3 nucleotide changes, according to whole-genome sequencing (15). M. tuberculosis has been isolated from Asian elephants among regions in southern Asia (3,4). Although the transmission routes have not been defined, our result and those of previous reports indicate that MTBC species may be spilling over into elephants.
Our finding emphasizes the need to identify the species of MTBC when tuberculosis is diagnosed in elephants. Although corroborating epidemiologic evidence of transmission has not been discovered, genomic data of the M. caprae isolates has been registered in the open database (BioProject ID PRJDB6469; BioSample ID SAMD00098240 for EPDC01, SAMD00098241 for EPDC02). Accumulation of genomic data of clinical isolates is expected to be helpful for future comparative studies.
Phylogenetic tree of isolate EPDC01 and 61 Mycobacterium caprae strains.
Acknowledgments
We thank the Fukuyama municipal administrative office, staff veterinarians, animal health and animal care staff, and employees of the Fukuyama Zoo for their participation. We also thank the Infection Control Team staff of National Hospital Organization Kinki-chuo Chest Medical Center for conducting the ventilation assessment.
This work was supported in part by MEXT/JSPS KAKENHI (grant nos. JP16K09120 and JP15H05263), and by the Cooperation Research Program of the Wildlife Research Center, Kyoto University (2017-A-20).
Biography
Ms. Yoshida is a microbiology researcher involved in the Clinical Research Center, National Hospital Organization Kinki-chuo Chest Medical Center, and the Graduate School of Biomedical Sciences, Nagasaki University in Japan. Her research interests include the examination of mycobacterial agents, including those from elephants with tuberculosis.
Footnotes
Suggested citation for this article: Yoshida S, Suga S, Ishikawa S, Mukai Y, Tsuyuguchi K, Inoue Y, et al. Mycobacterium caprae infection in captive Borneo elephant, Japan. Emerg Infect Dis. 2018 Oct [date cited]. https://doi.org/10.3201/eid2410.180018
References
- 1.Murphree R, Warkentin JV, Dunn JR, Schaffner W, Jones TF. Elephant-to-human transmission of tuberculosis, 2009. Emerg Infect Dis. 2011;17:366–71. 10.3201/eid1703.101668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandranaik BM, Shivashankar BP, Umashankar KS, Nandini P, Giridhar P, Byregowda SM, et al. Mycobacterium tuberculosis infection in free-roaming wild Asian elephant. Emerg Infect Dis. 2017;23:555–7. 10.3201/eid2303.161439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obanda V, Poghon J, Yongo M, Mulei I, Ngotho M, Waititu K, et al. First reported case of fatal tuberculosis in a wild African elephant with past human-wildlife contact. Epidemiol Infect. 2013;141:1476–80. 10.1017/S0950268813000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zachariah A, Pandiyan J, Madhavilatha GK, Mundayoor S, Chandramohan B, Sajesh PK, et al. Mycobacterium tuberculosis in wild Asian elephants, southern India. Emerg Infect Dis. 2017;23:504–6. 10.3201/eid2303.161741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–73. 10.1073/pnas.0511240103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyashchenko KP, Greenwald R, Esfandiari J, Mikota S, Miller M, Moller T, et al. Field application of serodiagnostics to identify elephants with tuberculosis prior to case confirmation by culture. Clin Vaccine Immunol. 2012;19:1269–75. 10.1128/CVI.00163-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenwald R, Lyashchenko O, Esfandiari J, Miller M, Mikota S, Olsen JH, et al. Highly accurate antibody assays for early and rapid detection of tuberculosis in African and Asian elephants. Clin Vaccine Immunol. 2009;16:605–12. 10.1128/CVI.00038-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard (M24–A2). Wayne (PA): The Institute; 2011. [PubMed] [Google Scholar]
- 9.Nagai K, Horita N, Yamamoto M, Tsukahara T, Nagakura H, Tashiro K, et al. Diagnostic test accuracy of loop-mediated isothermal amplification assay for Mycobacterium tuberculosis: systematic review and meta-analysis. Sci Rep. 2016;6:39090. 10.1038/srep39090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddington K, O’Grady J, Dorai-Raj S, Niemann S, van Soolingen D, Barry T. A novel multiplex real-time PCR for the identification of mycobacteria associated with zoonotic tuberculosis. PLoS One. 2011;6:e23481. 10.1371/journal.pone.0023481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domogalla J, Prodinger WM, Blum H, Krebs S, Gellert S, Müller M, et al. Region of difference 4 in alpine Mycobacterium caprae isolates indicates three variants. J Clin Microbiol. 2013;51:1381–8. 10.1128/JCM.02966-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–8. 10.1093/bioinformatics/btv271 [DOI] [PubMed] [Google Scholar]
- 13.Broeckl S, Krebs S, Varadharajan A, Straubinger RK, Blum H, Buettner M. Investigation of intra-herd spread of Mycobacterium caprae in cattle by generation and use of a whole-genome sequence. Vet Res Commun. 2017;41:113–28. 10.1007/s11259-017-9679-8 [DOI] [PubMed] [Google Scholar]
- 14.Oh P, Granich R, Scott J, Sun B, Joseph M, Stringfield C, et al. Human exposure following Mycobacterium tuberculosis infection of multiple animal species in a Metropolitan Zoo. Emerg Infect Dis. 2002;8:1290–3. 10.3201/eid0811.020302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson G, Zimmerman R, Shashkina E, Chen L, Richard M, Bradford CM, et al. Mycobacterium tuberculosis infection among Asian elephants in captivity. Emerg Infect Dis. 2017;23:513–6. 10.3201/eid2303.160726 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of isolate EPDC01 and 61 Mycobacterium caprae strains.


