Abstract
Endocannabinoids, an important class of signaling lipids involved in health and disease, are predominantly synthesized and metabolized by enzymes of the serine hydrolase superfamily. Activity-based protein profiling (ABPP) using fluorescent probes, such as fluorophosphonate (FP)-TAMRA and β-lactone-based MB064, enables drug discovery activities for serine hydrolases. FP-TAMRA and MB064 have distinct, albeit partially overlapping, target profiles but cannot be used in conjunction due to overlapping excitation/emission spectra. We therefore synthesized a novel FP-probe with a green BODIPY as a fluorescent tag and studied its labeling profile in mouse proteomes. Surprisingly, we found that the reporter tag plays an important role in the binding potency and selectivity of the probe. A multiplexed ABPP assay was developed in which a probe cocktail of FP-BODIPY and MB064 visualized most endocannabinoid serine hydrolases in mouse brain proteomes in a single experiment. The multiplexed ABPP assay was employed to profile endocannabinoid hydrolase inhibitor activity and selectivity in the mouse brain.
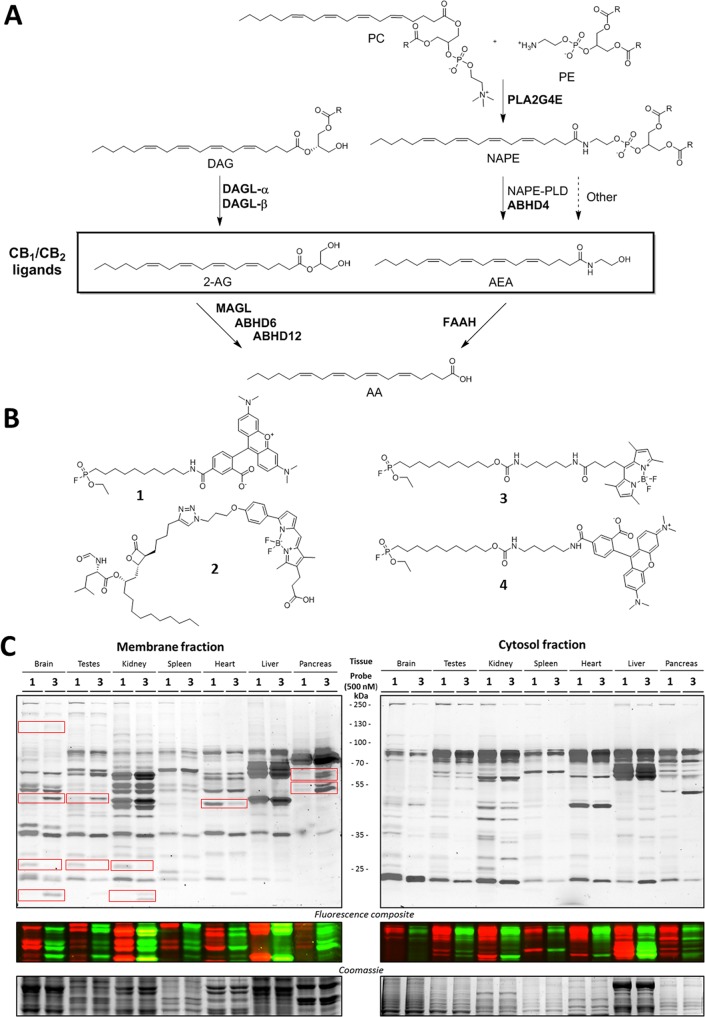
The endocannabinoid system (ECS) influences many physiological processes in the human body, including food intake, energy balance, motor coordination, pain sensation, memory formation, and anxiety.1,2 The ECS has, therefore, been under active investigation for therapeutic exploitation.3,4 There are two main cannabinoid receptors, CB1R and CB2R, which belong to the family of G-protein coupled receptors. They are activated by two endogenous ligands, i.e., anandamide (AEA) and 2-arachidonoyl glycerol (2-AG).5,6 The production and degradation of these endocannabinoids is mainly performed by serine hydrolases (Figure 1A). Diacylglycerol lipase α and β (DAGL-α and -β) are the main enzymes responsible for the biosynthesis of 2-AG through the hydrolysis of diacylglycerol (DAG).7−9 Monoacylglycerol lipase (MAGL) and α,β-hydrolase-domain containing enzymes 6 and 12 (ABHD6 and ABHD12) account for 99% of the 2-AG hydrolysis to arachidonic acid (AA) and glycerol in the brain.10,11 The Ca2+-dependent biosynthesis of endogenous AEA is mediated by the subsequent actions of PLA2G4E12 and N-acylphosphatidylethanolamine-phospholipase D (NAPE-PLD) or ABHD4,13 although other biosynthetic pathways have also been uncovered.3,4,14 Fatty acid amide hydrolase (FAAH) is the key enzyme for the hydrolysis of AEA to AA.15,16 Inhibitors of these enzymes are crucial to investigating the biological role of the hydrolases and may serve as drug candidates to modulate the endocannabinoid levels in human disease.
Figure 1.
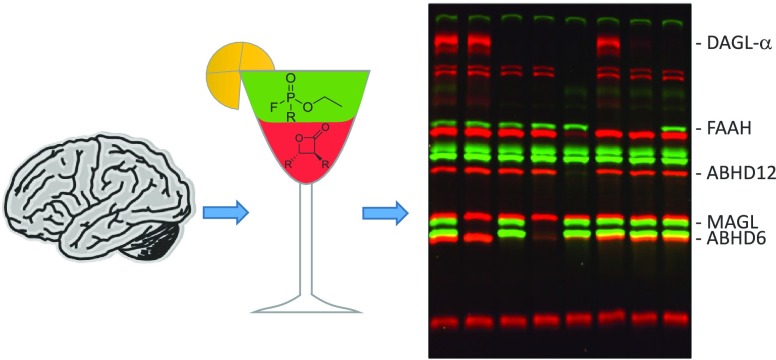
Endocannabinoid system, activity-based probes, and the labeling profiles of FP-TAMRA (1) and FP-BODIPY (3). (A) Schematic overview of the main biosynthetic pathways within the endocannabinoid system. All enzymes except NAPE-PLD belong to the serine hydrolase protein family. PC, phosphatidylcholine; PE, phosphatidylethanolamine; DAG, diacylglycerol; NAPE, N-acylphosphatidylethanolamine; AA, arachidonic acid; PLA2G4E, phospholipase A2 group IVE; DAGL, diacylglycerol lipase; NAPE-PLD, N-acylphosphatidylethanolamine phospholipase D; MAGL, monoacylglycerol lipase; ABHD, α,β-hydrolase-domain containing enzyme; FAAH, fatty acid amide hydrolase. (B) Chemical structures of the four activity-based probes used in this study. (C) Direct comparison of FP-TAMRA (1) and FP-BODIPY (3) labeling patterns of seven mouse tissue lysates.
All endocannabinoid hydrolases except NAPE-PLD belong to the family of serine hydrolases, which consists of over 200 proteins that use a nucleophilic serine to hydrolyze ester-, amide-, or thioesterbonds in small molecules and proteins via a covalent acyl-protein intermediate.17,18 This mode of action is exploited in activity-based protein profiling (ABPP).19,20 Herein, a chemical probe, typically consisting of a reactive “warhead” and a reporter tag, reacts with the catalytically active nucleophilic serine. The reporter tag can be either a fluorophore to visualize the probe-protein adduct by SDS-PAGE and fluorescence scanning21 or a biotin group to enrich proteins from proteomes for identification by LC-MS/MS22 or visualization by Western blotting.23 ABPP is used in drug discovery to efficiently profile activity and selectivity of inhibitors over a protein family in native biological samples.
The archetypical activity-based probe (ABP) for serine hydrolases is the fluorophosphonate (FP) probe (FP-TAMRA (1), Figure 1B), which was introduced by Liu et al. almost 20 years ago.23 This probe is widely used to study serine hydrolases in complex proteomes.24,25 Although the FP-based probes are known for their broad reactivity, they do not react with all serine hydrolases.20 Most notably, DAGL-α is among the enzymes which cannot be visualized by FP-based ABPs.26 To study DAGL-α, MB064 (2, Figure 1B), a tailored chemical probe with a BODIPY-TMR as a fluorophore, was developed.27 In terms of experimental efficiency with respect to time, cost of reagents, and use of valuable biological samples, it would be optimal to combine the commercially available FP-TAMRA (ActivX) and MB064 in the same experiment. However, MB064 cannot be applied in conjunction with FP-TAMRA (1), because the excitation/emission spectra of their fluorophores overlap. Therefore, the aim of the current study was to synthesize, characterize, and apply a new FP-based probe (3) with a different reporter tag (BODIPY-FL) that is compatible with MB064. Such a multiplexed assay, using different activity-based probes, has been shown to work for other enzyme classes in the past.21,28,29 Here, a multiplexed ABPP assay with ABP (3) and MB064 was developed and used to study endocannabinoid hydrolase activity and to profile inhibitors on activity and selectivity in mouse brain proteomes.
Results and Discussion
FP-BODIPY probe (3) was synthesized using a previously described method (Scheme S1).30 In addition, commercially available FP-TAMRA (probe 1) and control compound FP-TAMRA (4), containing the same linker as 3, were synthesized using similar procedures (Schemes S2 and S3, respectively).
To obtain a broad view of serine hydrolase labeling by the FP probes in various tissues, we first incubated FP-TAMRA (1) and FP-BODIPY (3) with membrane and cytosol fractions of a panel of seven mouse tissues (brain, testes, kidney, spleen, heart, liver, and pancreas) at a concentration of 500 nM (Figure 1C). The proteins were resolved using SDS-PAGE, and probe-labeled proteins were visualized by fluorescent scanning of the gel. The overall labeling profile in the various proteomes was comparable between probes 1 and 3, but several differences were observed, denoted with boxes. In the brain, for example, membrane proteome FP-BODIPY (3) labeled additional targets, including in the top left box DAGL-α, the identity of which was confirmed by competition with LEI104 (Figure S1).
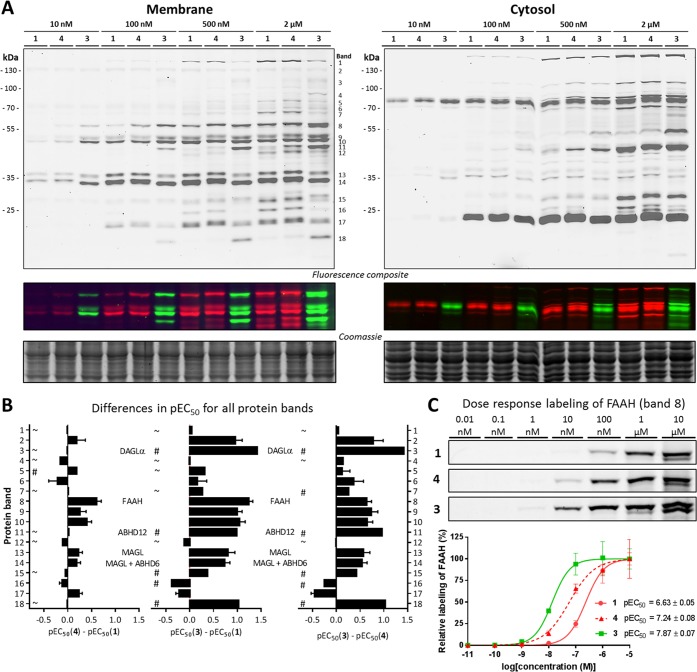
Brain lysates were selected for further profiling of probes 1 and 3, as well as control probe 4, because the brain is the most studied target organ of the ECS. In an initial screen, the three ABPs were incubated with both brain membrane and soluble proteomes (Figure 2A, Supporting Information). While the labeling profile in the soluble proteome was not significantly different between the three probes, FP-BODIPY (3) labeled various proteins at lower concentrations in the membrane proteome. To determine the half-maximum effect (EC50) values of serine hydrolase labeling in the membrane proteome, the probes were dosed at a wide range of concentrations (10 pM to 10 μM), and the fluorescent signal of 18 distinct bands was quantified and corrected for protein loading by coomassie staining (Figures S2 and S3). To study the effect of fluorophore and linker length on serine hydrolase labeling by FP probes in the mouse brain membrane proteome, the change in pEC50 of ABP 3 and 4 relative to ABP 1 was calculated (Figure 2B and Table S1). The increased linker length did not significantly alter the labeling efficiency for FP-TAMRA for most proteins, except for FAAH (left plot, Figure 2C), whereas the change in fluorophore led to a 10-fold increased potency in labeling for several proteins (bands: 3, 8, and 18). Of note, FAAH labeling was already visualized at 10 nM FP-BODIPY 3 and DAGL-α (band 3) at 500 nM (Figure 2C, A, respectively). The third plot in Figure 2B, comparing probes 3 and 4, which only differ in reporter tag, shows that almost all the difference between the commercial FP-TAMRA probe 1 and the newly synthesized FP-BODIPY probe 3 observed in the central plot is due to the change in fluorophore. The most likely explanation for the observed potency increase when changing from TAMRA to BODIPY-FP is the strong increase in lipophilicity. The CLogP of BODIPY-FL is 3.7 points higher than that of TAMRA, which would make it more favorable to stick to proteins and membranes, causing a higher local concentration and thus better labeling. This explanation is in line with the observation that the strongest differences are observed in the membrane fractions and, between organs, in the brain. Finally, the impact of the addition of a reporter tag was visualized by preincubation with “dark” alkyne-FP (5; Figure S4). This competitive labeling shows that alkyne-FP only completely prevented labeling by the fluorescent probes at 5–10 μM, demonstrating the significantly reduced affinity of the fluorophosphonate inhibitor when lacking the reporter tag. All together, these data demonstrate that the choice of fluorophore influences the labeling efficiency of FP-based probes.
Figure 2.
Concentration dependent labeling of probes 1, 3, and 4. (A) Four doses of the FP probes label the mouse brain proteome in a distinct pattern with different affinities. (B) Quantified affinity differences among the evaluated FP probes for the 18 bands denoted in A. A # indicates a pEC50 ≤ 6 for one of the two probes, meaning that the difference is most likely greater than the given value. A ∼ indicates a pEC50 ≤ 6 for both probes, meaning that both probes label these proteins only at high concentrations. All quantifications assumed 100% labeling of protein at 10 μM probe. (C) Example of the labeling pattern of band 8 (FAAH) and corresponding pEC50 curves and values.
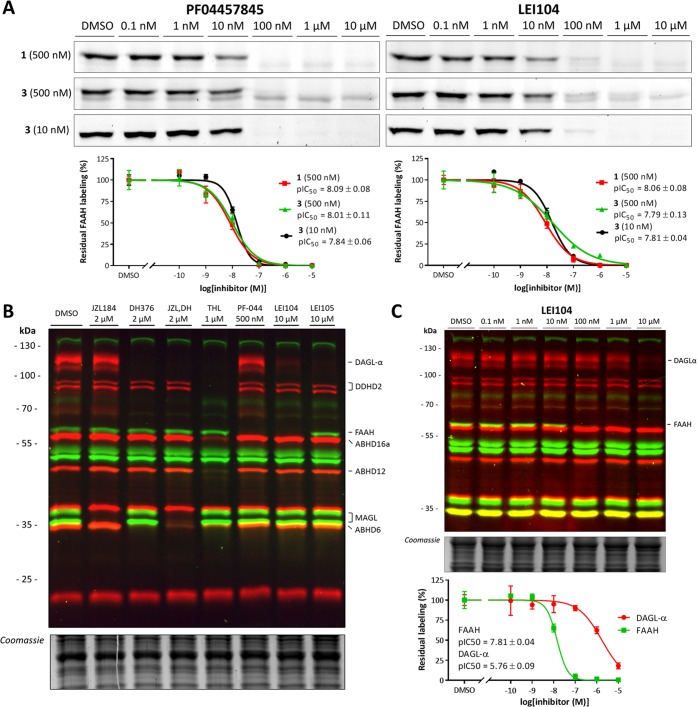
Next, we tested whether the activity and selectivity profile of serine hydrolase inhibitors would be dependent on the reporter group of the activity-based probe. To this end, we tested a covalent irreversible FAAH inhibitor, PF-04457845,31 and a reversible inhibitor, LEI104,27 in a competitive ABPP setting using probe 1 (500 nM) and probe 3 (500 and 10 nM; Figure 3A). Importantly, the pIC50 values of both inhibitors were not dependent on the fluorescent reporter group of the probe, nor the probe concentration. This indicated that FP-BODIPY 3 can be used in a drug discovery setting to profile inhibitor activity using ABPP.
Figure 3.
Illustration of the applicability of the prepared probe cocktail. (A) Dose response inhibition of FAAH using PF-04457845, covalent irreversible, and LEI104, reversible, to test the dependency of the pIC50 determination on probe affinity and concentration. No statistical significant differences have been found between the probe pairs (P > 0.05, two-sided Student’s t test). (B) Seven inhibitors targeted for different endocannabinoid serine hydrolases were shown to inhibit their specific targets using the probe cocktail. (C) Dose response inhibition with LEI104 shows, in one gel, the inhibition of DAGL-α and FAAH. Quantification shows agreement of the pIC50 with literature values.
Having developed two complementary probes (FP-BODIPY (3) and MB064) with different reporter groups and distinct labeling patterns, we tested whether they can be used in a multiplexed ABPP assay to profile the activity and selectivity of compounds inhibiting biosynthetic or metabolic enzymes of the ECS.24,25,32 To this end, a cocktail of FP-BODIPY 3 (10 nM) and MB064 2 (250 nM) was incubated with mouse brain membrane proteomes. This enabled the simultaneous visualization and quantification of DAGL-α, DDHD2, ABHD16a, FAAH, MAGL, ABHD6, and ABHD12 activities in a single experiment (Figure 3B). Bands were identified based on previous studies.27,33 PLA2G4E and ABHD4 can be labeled by FP-BODIPY and MB064, respectively, but their endogenous expression in the brain is too low to be visualized.12 A panel of inhibitors consisting of JZL184 (MAGL),34 DH376 (DAGL-α, ABHD6),32 THL (DAGL-α, ABHD6, ABHD12, ABHD16a, DDHD2),35 PF-04457845 (FAAH),31 LEI104 (DAGL-α, FAAH),27 and LEI105 (DAGL-α)33 (Figure S5) was used to confirm the identity of each fluorescent band (Figure 3B). As a final validation, we confirmed that the inhibitory activity of LEI104 on DAGL-α and FAAH in this new multiplexed ABPP assay was in line with previously reported data (Figure 3C).27
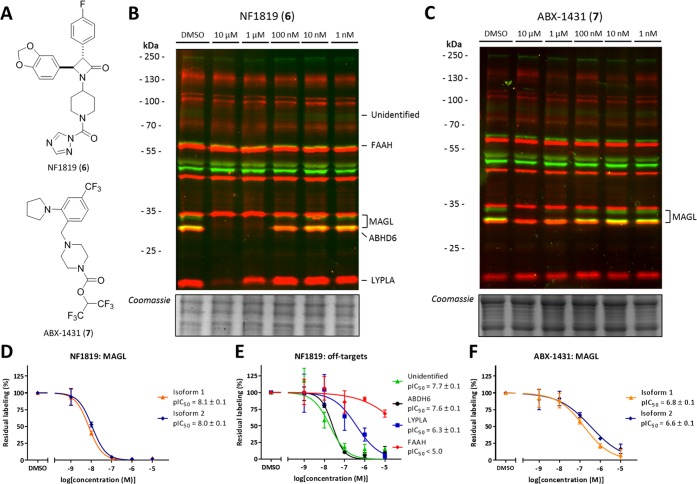
The validated assay was employed to study the selectivity and activity of two MAGL inhibitors. First, we tested the recently published β-lactam based MAGL inhibitor NF1819 (6; Figure 4A), which was active in several animal models of multiple sclerosis, pain, and predator stress-induced long-term anxiety.36,37 The target-interaction profile of NF1819 (6) was compared to the experimental drug ABX-1431 (7), currently in phase 2 clinical trials for the treatment of Tourette syndrome.38−40 To this end, they were incubated at various concentrations with mouse brain membrane proteome (Figure 4B, C and Figure S6). Inhibition of MAGL was confirmed with a pIC50 of 8.1 ± 0.1 for 6 and 6.7 ± 0.1 for 7 (Figure 4D, F), in accordance with previously published data.36,38 Of note, for 6, various off-targets were observed, including ABHD6, LYPLA, and an unidentified protein (Figure 4E). ABHD6 was inhibited at equal potency, whereas LYPLA demonstrated a 50-fold lower potency. FAAH labeling was only slightly reduced at concentrations >10 μM. The target-interaction landscape of 7 is clean; even at 10 μM, no clear off-targets were observed. The relatively small selectivity window of 6 over ABHD6 should be taken into account during the biological evaluations of this inhibitor as it may contribute to the rise of 2-AG levels.
Figure 4.
Off-target profiling of β-lactam based MAGL inhibitor (6) and clinical candidate ABX-1431 in mouse brain membrane proteome. (A) Chemical structure of 6 and 7. (B) Dose response inhibition with 6 shows several off-targets in the mouse brain membrane. (C) Dose response inhibition with 7 shows selective MAGL inhibition in the mouse brain membrane. (D) pIC50 curves and values of 6 against MAGL. (E) pIC50 curves and values of 6 against its off-targets. (F) pIC50 curves and values of 7 against MAGL.
In conclusion, FP-BODIPY (3) was synthesized and characterized as a new ABP, thereby we have extended the chemical toolbox to study serine hydrolase activity in native biological samples. We emphasize that the choice of fluorophore when designing ABPs can be of great influence on labeling patterns, even for broadly reactive probes such as fluorophosphonates. FP-BODIPY (3) in conjunction with MB064 (2) was used to develop a multiplexed ABPP assay, which was validated by profiling inhibitor activity and selectivity on a broad range of endocannabinoid hydrolases in mouse brain tissue in a single experiment. This multiplexed ABPP assay was applied to investigate the specificity of a recently published in vivo active MAGL inhibitor and an experimental drug currently going through clinical trials.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.8b00534.
Figures S1–S6, Table S1, Schemes S1–S4, and synthetic methods (PDF)
Author Contributions
A.P.A.J., D.v.d.V., A.T.B., M.J., and S.H.G. performed the experiments. A.P.A.J., D.v.d.V., A.T.B., M.J., S.H.G., S.B., G.C., and M.v.d.S. designed the experiments and analyzed the results. A.P.A.J., D.v.d.V., and M.v.d.S. wrote the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Mechoulam R.; Parker L. A. (2013) The Endocannabinoid System and the Brain. Annu. Rev. Psychol. 64, 21–47. 10.1146/annurev-psych-113011-143739. [DOI] [PubMed] [Google Scholar]
- Soethoudt M.; Stolze S. C.; Westphal M. V.; van Stralen L.; Martella A.; van Rooden E. J.; Guba W.; Varga Z. V.; Deng H.; van Kasteren S. I.; Grether U.; IJzerman A. P.; Pacher P.; Carreira E. M.; Overkleeft H. S.; Ioan-Facsinay A.; Heitman L. H.; van der Stelt M. (2018) Selective Photoaffinity Probe That Enables Assessment of Cannabinoid CB 2 Receptor Expression and Ligand Engagement in Human Cells. J. Am. Chem. Soc. 140, 6067. 10.1021/jacs.7b11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donvito G.; Nass S. R.; Wilkerson J. L.; Curry Z. A.; Schurman L. D.; Kinsey S. G.; Lichtman A. H. (2018) The Endogenous Cannabinoid System: A Budding Source of Targets for Treating Inflammatory and Neuropathic Pain. Neuropsychopharmacology 43, 52–79. 10.1038/npp.2017.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzo V. (2008) Targeting the Endocannabinoid System: To Enhance or Reduce?. Nat. Rev. Drug Discovery 7, 438–455. 10.1038/nrd2553. [DOI] [PubMed] [Google Scholar]
- Devane W. A.; Hanus L.; Breuer A.; Pertwee R. G.; Stevenson L. A.; Griffin G.; Gibson D.; Mandelbaum A.; Etinger A.; Mechoulam R. (1992) Isolation and Structure of a Brain Constituent That Binds to the Cannabinoid Receptor. Science (Washington, DC, U. S.) 258, 1946–1949. 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- Mechoulam R.; Ben-Shabat S.; Hanus L.; Ligumsky M.; Kaminski N. E.; Schatz A. R.; Gopher A.; Almog S.; Martin B. R.; Compton D. R.; Pertwee R. G.; Griffin G.; Bayewitch M.; Barg J.; Vogel Z. (1995) Identification of an Endogenous 2-Monoglyceride, Present in Canine Gut, That Binds to Cannabinoid Receptors. Biochem. Pharmacol. 50, 83–90. 10.1016/0006-2952(95)00109-D. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Vasilyev D. V.; Goncalves M. B.; Howell F. V.; Hobbs C.; Reisenberg M.; Shen R.; Zhang M.-Y.; Strassle B. W.; Lu P.; Mark L.; Piesla M. J.; Deng K.; Kouranova E. V.; Ring R. H.; Whiteside G. T.; Bates B.; Walsh F. S.; Williams G.; Pangalos M. N.; Samad T. A.; Doherty P. (2010) Loss of Retrograde Endocannabinoid Signaling and Reduced Adult Neurogenesis in Diacylglycerol Lipase Knock-out Mice. J. Neurosci. 30, 2017–2024. 10.1523/JNEUROSCI.5693-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura A.; Yamazaki M.; Hashimotodani Y.; Uchigashima M.; Kawata S.; Abe M.; Kita Y.; Hashimoto K.; Shimizu T.; Watanabe M.; Sakimura K.; Kano M. (2010) The Endocannabinoid 2-Arachidonoylglycerol Produced by Diacylglycerol Lipase α Mediates Retrograde Suppression of Synaptic Transmission. Neuron 65, 320–327. 10.1016/j.neuron.2010.01.021. [DOI] [PubMed] [Google Scholar]
- Reisenberg M.; Singh P. K.; Williams G.; Doherty P. (2012) The Diacylglycerol Lipases: Structure, Regulation and Roles in and beyond Endocannabinoid Signalling. Philos. Trans. R. Soc., B 367, 3264. 10.1098/rstb.2011.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savinainen J. R.; Saario S. M.; Laitinen J. T. (2012) The Serine Hydrolases MAGL, ABHD6 and ABHD12 as Guardians of 2-Arachidonoylglycerol Signalling through Cannabinoid Receptors. Acta Physiol. 204, 267–276. 10.1111/j.1748-1716.2011.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. Z.; Nomura D. K.; Vann R. E.; Walentiny D. M.; Booker L.; Jin X.; Burston J. J.; Sim-Selley L. J.; Lichtman A. H.; Wiley J. L.; Cravatt B. F. (2009) Dual Blockade of FAAH and MAGL Identifies Behavioral Processes Regulated by Endocannabinoid Crosstalk in Vivo. Proc. Natl. Acad. Sci. U. S. A. 106, 20270–20275. 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y.; Parsons W. H.; Kamat S. S.; Cravatt B. F. (2016) A Calcium-Dependent Acyltransferase That Produces N-Acyl Phosphatidylethanolamines. Nat. Chem. Biol. 12, 669–671. 10.1038/nchembio.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G. M.; Cravatt B. F. (2006) Endocannabinoid Biosynthesis Proceeding through Glycerophospho-N-Acyl Ethanolamine and a Role for Alpha/beta-Hydrolase 4 in This Pathway. J. Biol. Chem. 281, 26465–26472. 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- Liu J.; Wang L.; Harvey-White J.; Huang B. X.; Kim H. Y.; Luquet S.; Palmiter R. D.; Krystal G.; Rai R.; Mahadevan A.; Razdan R. K.; Kunos G. (2008) Multiple Pathways Involved in the Biosynthesis of Anandamide. Neuropharmacology 54, 1–7. 10.1016/j.neuropharm.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt B. F.; Giang D. K.; Mayfield S. P.; Boger D. L.; Lerner R. A.; Gilula N. B. (1996) Molecular Characterization of an Enzyme That Degrades Neuromodulatory Fatty-Acid Amides. Nature 384, 83–87. 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- Patricelli M. P.; Lovato M. A.; Cravatt B. F. (1999) Chemical and Mutagenic Investigations of Fatty Acid Amide Hydrolase: Evidence for a Family of Serine Hydrolases with Distinct Catalytic Properties. Biochemistry 38, 9804–9812. 10.1021/bi990637z. [DOI] [PubMed] [Google Scholar]
- Long J. Z.; Cravatt B. F. (2011) The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease. Chem. Rev. 111, 6022–6063. 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahiduzzaman M.; Coombs K. M. (2012) Activity Based Protein Profiling to Detect Serine Hydrolase Alterations in Virus Infected Cells. Front. Microbiol. 3, 1–5. 10.3389/fmicb.2012.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tian Y., Wang M., Wang M., Sun G., and Sun X. (2018) Advanced Activity-Based Protein Profiling Application Strategies for Drug Development. Front. Pharmacol. 9, 10.3389/fphar.2018.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G. M.; Cravatt B. F. (2010) Activity-Based Proteomics of Enzyme Superfamilies: Serine Hydrolases as a Case Study. J. Biol. Chem. 285, 11051–11055. 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli M. P.; Giang D. K.; Stamp L. M.; Burbaum J. J. (2001) Direct Visualization of Serine Hydrolase Activities in Complex Proteomes Using Fluorescent Active Site-Directed Probes. Proteomics 1, 1067–1071. . [DOI] [PubMed] [Google Scholar]
- Jessani N.; Niessen S.; Wei B. Q.; Nicolau M.; Humphrey M.; Ji Y.; Han W.; Noh D.-Y.; Yates J. R.; Jeffrey S. S.; Cravatt B. F. (2005) A Streamlined Platform for High-Content Functional Proteomics of Primary Human Specimens. Nat. Methods 2, 691–697. 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Patricelli M. P.; Cravatt B. F. (1999) Activity-Based Protein Profiling: The Serine Hydrolases. Proc. Natl. Acad. Sci. U. S. A. 96, 14694–14699. 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esbroeck A. C. M.; Janssen A. P. A.; Cognetta A. B.; Ogasawara D.; Shpak G.; van der Kroeg M.; Kantae V.; Baggelaar M. P.; de Vrij F. M. S.; Deng H.; Allarà M.; Fezza F.; Lin Z.; van der Wel T.; Soethoudt M.; Mock E. D.; den Dulk H.; Baak I. L.; Florea B. I.; Hendriks G.; De Petrocellis L.; Overkleeft H. S.; Hankemeier T.; De Zeeuw C. I.; Di Marzo V.; Maccarrone M.; Cravatt B. F.; Kushner S. A.; van der Stelt M. (2017) Activity-Based Protein Profiling Reveals off-Target Proteins of the FAAH Inhibitor BIA 10–2474. Science (Washington, DC, U. S.) 356, 1084–1087. 10.1126/science.aaf7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooden E. J.; Florea B. I.; Deng H.; Baggelaar M. P.; van Esbroeck A. C. M.; Zhou J.; Overkleeft H. S.; van der Stelt M. (2018) Mapping in Vivo Target Interaction Profiles of Covalent Inhibitors Using Chemical Proteomics with Label-Free Quantification. Nat. Protoc. 13, 752–767. 10.1038/nprot.2017.159. [DOI] [PubMed] [Google Scholar]
- Hoover H. S.; Blankman J. L.; Niessen S.; Cravatt B. F. (2008) Selectivity of Inhibitors of Endocannabinoid Biosynthesis Evaluated by Activity-Based Protein Profiling. Bioorg. Med. Chem. Lett. 18, 5838–5841. 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggelaar M. P.; Janssen F. J.; van Esbroeck A. C. M.; den Dulk H.; Allarà M.; Hoogendoorn S.; McGuire R.; Florea B. I.; Meeuwenoord N.; van den Elst H.; van der Marel G. a.; Brouwer J.; Di Marzo V.; Overkleeft H. S.; van der Stelt M. (2013) Development of an Activity-Based Probe and in Silico Design Reveal Highly Selective Inhibitors for Diacylglycerol Lipase-α in Brain. Angew. Chem., Int. Ed. 52, 12081–12085. 10.1002/anie.201306295. [DOI] [PubMed] [Google Scholar]
- Adam G. C.; Sorensen E. J.; Cravatt B. F. (2002) Proteomic Profiling of Mechanistically Distinct Enzyme Classes Using a Common Chemotype. Nat. Biotechnol. 20, 805. 10.1038/nbt714. [DOI] [PubMed] [Google Scholar]
- de Bruin G.; Xin B. T.; Kraus M.; van der Stelt M.; van der Marel G. A.; Kisselev A. F.; Driessen C.; Florea B. I.; Overkleeft H. S. (2016) A Set of Activity-Based Probes to Visualize Human (Immuno)proteasome Activities. Angew. Chem., Int. Ed. 55, 4199. 10.1002/anie.201509092. [DOI] [PubMed] [Google Scholar]
- Tully S. E.; Cravatt B. F. (2010) Activity-Based Probes That Target Functional Subclasses of Phospholipases in Proteomes. J. Am. Chem. Soc. 132, 3264–3265. 10.1021/ja1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. S.; Stiff C.; Lazerwith S. E.; Kesten S. R. S. R. J.; Fay L. K.; Morris M.; Beidler D.; Liimatta M. B.; Smith S. E.; Dudley D. T.; Sadagopan N.; Bhattachar S. N.; Kesten S. R. S. R. J.; Nomanbhoy T. K.; Cravatt B. F.; Ahn K. (2011) Discovery of PF-04457845: A Highly Potent, Orally Bioavailable, and Selective Urea FAAH Inhibitor. ACS Med. Chem. Lett. 2, 91–96. 10.1021/ml100190t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara D.; Deng H.; Viader A.; Baggelaar M. P.; Breman A.; den Dulk H.; van den Nieuwendijk A. M. C. H.; Soethoudt M.; van der Wel T.; Zhou J.; Overkleeft H. S.; Sanchez-Alavez M.; Mori S.; Nguyen W.; Conti B.; Liu X.; Chen Y.; Liu Q. S.; Cravatt B. F.; van der Stelt M. (2016) Rapid and Profound Rewiring of Brain Lipid Signaling Networks by Acute Diacylglycerol Lipase Inhibition. Proc. Natl. Acad. Sci. U. S. A. 113, 26–33. 10.1073/pnas.1522364112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggelaar M. P.; Chameau P. J. P.; Kantae V.; Hummel J.; Hsu K.-L.; Janssen F.; van der Wel T.; Soethoudt M.; Deng H.; den Dulk H.; Allarà M.; Florea B. I.; Di Marzo V.; Wadman W. J.; Kruse C. G.; Overkleeft H. S.; Hankemeier T.; Werkman T. R.; Cravatt B. F.; van der Stelt M. (2015) Highly Selective, Reversible Inhibitor Identified by Comparative Chemoproteomics Modulates Diacylglycerol Lipase Activity in Neurons. J. Am. Chem. Soc. 137, 8851–8857. 10.1021/jacs.5b04883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B.; Wang W.; Long J. Z.; Sun D.; Hillard C. J.; Cravatt B. F.; Liu Q. (2009) Blockade of 2-Arachidonoylglycerol Hydrolysis by Selective Monoacylglycerol Lipase Inhibitor 4-Nitrophenyl 4-(dibenzo[d][1,3]dioxol-5-Yl(hydroxy)methyl)piperidine-1-Carboxylate (JZL184) Enhances Retrograde Endocannabinoid Signaling. J. Pharmacol. Exp. Ther. 331, 591–597. 10.1124/jpet.109.158162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisogno T.; Howell F.; Williams G.; Minassi A.; Cascio M. G.; Ligresti A.; Matias I.; Schiano-Moriello A.; Paul P.; Williams E.-J.; Gangadharan U.; Hobbs C.; Di Marzo V.; Doherty P. (2003) Cloning of the First sn1-DAG Lipases Points to the Spatial and Temporal Regulation of Endocannabinoid Signaling in the Brain. J. Cell Biol. 163, 463–468. 10.1083/jcb.200305129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindisi M.; Maramai S.; Gemma S.; Brogi S.; Grillo A.; Di Cesare Mannelli L.; Gabellieri E.; Lamponi S.; Saponara S.; Gorelli B.; Tedesco D.; Bonfiglio T.; Landry C.; Jung K.-M.; Armirotti A.; Luongo L.; Ligresti A.; Piscitelli F.; Bertucci C.; Dehouck M.-P.; Campiani G.; Maione S.; Ghelardini C.; Pittaluga A.; Piomelli D.; Di Marzo V.; Butini S. (2016) Development and Pharmacological Characterization of Selective Blockers of 2-Arachidonoyl Glycerol Degradation with Efficacy in Rodent Models of Multiple Sclerosis and Pain. J. Med. Chem. 59, 2612–2632. 10.1021/acs.jmedchem.5b01812. [DOI] [PubMed] [Google Scholar]
- Lim J.; Igarashi M.; Jung K.-M.; Butini S.; Campiani G.; Piomelli D. (2016) Endocannabinoid Modulation of Predator Stress-Induced Long-Term Anxiety in Rats. Neuropsychopharmacology 41, 1329–1339. 10.1038/npp.2015.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankman J. L., Clapper J. R., Ezekowitz R. A. B., Fraser I. P., Grice C. A., Jones T. K., O’Neill G. P., Thurston A. W. Jr., and Beals C. R.. Methods of Treating Inflammation or Neuropathic Pain. WO2016183097A1, 2015.
- Cisar J. S., Grice C. A., Jones T. K., Niphakis M. J., Chang J. W., Lum K. M., and Cravatt B.. Carbamate Compounds and of Making and Using Same. US 2015 001 8335A1, January 6, 2012.
- Cisar J. S., Weber O. D., Clapper J. R., Blankman J. L., Henry C. L., Simon G. M., Alexander J. P., Jones T. K., Ezekowitz R. A. B., O’Neill G. P., and Grice C. A.. Identification of ABX-1431, a Selective Inhibitor of Monoacylglycerol Lipase and Clinical Candidate for Treatment of Neurological Disorders. J. Med. Chem. 2018, 10.1021/acs.jmedchem.8b00951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.