Abstract
Despite various hypothesized benefits of dietary isoflavone genistein (GEN) from soy-based products, many questions surrounding GEN’s immunotoxic effects, especially during perinatal exposure, have yet to be answered. The objective of the study was to determine if there existed a sex-specific effect of GEN on type 1 diabetes (T1D) following perinatal exposure. We exposed offspring of non-obese diabetic (NOD) mice to GEN per oral at a physiological dose (20 mg/kg body weight) from embryonic day 7 to postnatal day (PND) 21. In female offspring, perinatal GEN dosing significantly increased the incidence of T1D at early time points, and the exacerbation was associated with decreased serum levels of interleukin (IL)-10, IgG2a, and IgM. In male offspring dosed with GEN, a decrease in serum IgG1 was also observed. Flow cytometric analysis in females suggested an increased pro-inflammatory splenic CD5+CD24− and CD4−CD8+ cell counts, while both %T cells and %CD4+ T cells were significantly decreased in males, suggesting an anti-inflammatory effect. Gut microbiota (GMB) analysis indicated that fecal microbiota from PND 90 female offspring exhibited an increased level of Enterobacteriales (suggesting a pro-inflammatory response), while the similar changes were not found in PND 30 females. Moreover, RNA sequencing showed that intestinal α-defensin expression was down-regulated in GEN-treated females, supporting a pro-inflammatory response. However, perinatal GEN administration perturbed GMB toward an anti-inflammatory response in PND 90 males. Taken together, a strong sex-specific effect was found in the perinatal GEN exposure window, and the T1D exacerbation in NOD females was associated with GMB-related immunomodulatory mechanisms.
Keywords: genistein, gut microbiota, flow cytometry, antibody, cytokine/chemokine, type 1 diabetes
Among children in the United States, the prevalence of type 1 diabetes (T1D) has increased from 1.48 to 2.32 per 1000 during 2002–2013 (Li et al., 2016a), and several hypotheses have been brought into attention, such as pathogen, vitamin D deficiency, and food (Egro, 2013). Soy products are of particular importance since they are an intrinsic part of Asian cuisine, and the market of soy-based formula is increasing in the United States (about 20% of the formula market). A cross-sectional study found that the urinary genistein (GEN) concentration in infants consuming soy-based formula was 500 times higher than in those consuming milk-based formula (Cao et al., 2009). Adult GEN dosing is reported to have anti-diabetic function on both T1D (Guo et al., 2014) and type 2 diabetes (Gilbert and Liu, 2013) due to its immunomodulatory properties and protective effects on pancreatic β-cells in murine models while epidemiological evidences are limited to provide a solid association between GEN exposure and T1D. However, the effects of GEN on immune regulation and the onset of T1D and glucose regulation during perinatal stages have not been fully understood. Perinatal GEN treatment was found to have immunomodulatory effects in animal studies. Our group has reported changes in immune response (such as increased cytotoxic T cells activity and decreased regulatory T cells) following perinatal GEN dosing in female B6C3F1 mice, and the response depends largely on sex, exposure duration, as well as litter order (Guo et al., 2005).
The association of high soy consumption during perinatal period and risk of T1D is uncertain and intricate. Among the limited epidemiological reports, one study showed that for infants aged 4–6 and 7–12 months, soy milk formula consumption was associated with a two-fold increase of T1D risk in China (Strotmeyer et al., 2004), while information on other exposure windows is largely unknown. Another epidemiological study suggested twice as many diabetic children consumed soy-based formula in infancy as healthy children (Fort et al., 1986). However, the mechanism underlying the effect of soy intake on T1D in perinatal stages is largely unknown, and studies on the gender effect are lacking.
In the fetus, early-life gut bacterial colonization plays an important role in metabolic tissue development and in influencing the risk of autoimmune diseases (Kozyrskyj and Sloboda, 2016) because intestinal immune system development starts as early as 11 weeks of gestation in humans (Mejia-Leon and de la Barca, 2015). Bacterial DNA can transfer from mother to fetus through amniotic fluid, umbilical cord (Kozyrskyj and Sloboda, 2016), and placenta (Romano-Keeler and Weitkamp, 2015). There is evidence to show that modulation of gut microbiota (GMB) by probiotics (Lactobacillus rhamnosus or Bifidobacterium lactis) during pregnancy alters infant immune responses (Prescott et al., 2008). The mode of delivery, the antibiotic used after birth, and the infant formula consumption could all help shape the infant microbiota (Mejia-Leon et al., 2015; Mueller et al., 2015; Romano-Keeler and Weitkamp, 2015) and further modulate the immune system. In mice, the colonization of thymus starts from day 10 to day 11 of gestation (Holladay and Smialowicz, 2000). To understand the effects of perinatal GEN intake on T1D, which represented a major deficit in our present knowledge, we investigated the potential links between GEN exposure at the physiological level (20 mg/kg body weight, BW) and increased risk of developing T1D in non-obese diabetic (NOD) mice, a model that closely resembles human T1D (Anderson and Bluestone, 2005), with regards to different exposure windows (perinatal period in conjunction with our previous findings on adults) and sex from perspectives of immunomodulation and GMB.
MATERIALS AND METHODS
Perinatal study
A total of 24 time-mated (male: female = 1:1) NOD female mice with normal blood glucose level (BGL) were used and randomized into vehicle (VH) and GEN groups (12 per group) based on body weight and baseline BGL. From gestational day (GD) 7, the mice in VH group received a 0.1 ml/10 g BW sodium carbonate (Na2CO3, 25 mM) daily because of GEN’s low water solubility, and the mice in GEN group were gavaged with 0.1 ml/10 g BW GEN dissolved and sonicated in VH (2 mg GEN/ml [Guo et al., 2014] till PND 21). Thus, the perinatal dosing included both in utero and lactational exposures. The daily dosing was performed with 18 G gavage needles. After parturition, the pups were housed in the same cage with dams with one litter per cage. Animal husbandry is included in the Supplementary Material. As the pups on 5K96 diet were usually smaller than the ones on regular diet, they were separated from the dams on PND 28 and housed up to five same-sex littermates per cage. Each litter was treated as an experimental unit.
In the second study, a total of 8 NOD females with normal BGL were randomized into VH (N = 4) and GEN (N = 4) groups, time-mated (male: female = 1:2) and received the same dosing regimen as above. Gestational day 0 was defined as the plug day. These 8 females gave birth to 8 litters. This separate study was conducted for GMB analysis, transcriptomic analysis, and to verify previous results. Each litter was treated as an experimental unit. All animal procedures in the lab were approved by the UGA Institutional Animal Care and Use Committee (IACUC).
Body weight (bw) and blood glucose level (bgl) measurement
The BW and BGL were monitored every week for the NOD offspring. The non-fasting BGL was measured using a blood glucose meter (Bayer Contour, Ascensia Diabetes Care, Parsippany, New Jersey) with a small sample of venous blood sample. T1D was defined as a BGL higher than or equal to 250 mg/dl (13.9 mmol/l) (Guo et al., 2014). Mice with BGL of 600 mg/dl or higher (33.3 mmol/l) in two consecutive weeks or at the end of the study were euthanized with carbon dioxide (CO2) inhalation. Individual organs were collected and weighted during necropsy. In addition, glucose and insulin tolerance tests were performed (see Supplementary Material).
Serum antibody measurement
The enzyme-linked immunosorbent assay (ELISA) was performed to detect the total antibody levels (IgG2a, IgG2b, IgM, and IgG1) in mouse sera according to the reported procedures (Guo et al., 2014). Briefly, the plates were added with blocking solution (5% milk powder in PBS with Tween 20) for 1 h and washed with PBST (Tween 20 in PBS). The diluted serum was then added in the plates and incubated in room temperature for 2 h. Then the secondary antibody was added and incubated for 1 h. Substrate TMB was added at 100 μl/well and the plate was read at the wavelength of 405 nm. Serum insulin and anti-insulin antibody were also measured and listed in the Supplementary Material.
Serum cytokine and chemokine
A total of 9 cytokines (IFN-γ, IL-1α, IL-1β, IL-2, IL-6, IL-10, IL-12p40, IL-12p70, TNF-α, both pro-inflammatory and anti-inflammatory) and 1 chemokine (MCP-1) were measured using Luminex Mouse Cytokine/Chemokine Magnetic Bead Panel (EMD Millipore Corporation, Billerica, Massacheussts) following manufacturer’s protocols. Sera and beads were added, and the plates were incubated at 4°C overnight on a shaker. Then detection antibody was added and followed by streptavidin phycoerythrin. Plates were run on a Bio-Plex MAGPIXTM Multiplex Reader with Bio-Plex ManagerTM MP Software (Luminex, Austin, Texas). Each biomarker concentration was calculated as pg/ml.
Flow cytometry
The spleen was subject to flow cytometric analysis following mashing between two slides with frosted ends (Guo et al., 2006). For male offspring, different cell surface markers were labeled with a cocktail of monoclonal antibodies conjugated with a fluorescent molecule (Phycoerythrin onjugated antibody for CD4, Peridinin ChlorophyII Protein Complex [PerCP] for CD8, fluorescein isothiocyanate [FITC] for IgM, and PerCP for CD3), for visualization. For female offspring (Huang et al., 2017), the fluorochrome-labeled antibody cocktails for cell staining were: CD4 (V450), CD8 (APC-H7), CD3 (PerCP-Cy5.5; total T cells) and CD45R (PE-CF594; B cells), and CD24 and CD5 (FITC and PE, respectively). All antibodies were obtained from BD PharMingen (San Diego, California). Isotype-matched irrelevant antibodies were used as controls. Following the addition of the reagents, the cells were incubated at 4°C in the dark for 30 min. The cells were washed, and enumeration performed on a Becton Dickinson LSRII Flow Cytometer in which log fluorescence intensity was read and a forward scatter threshold high enough to eliminate red blood cells. Ten thousand cells were counted for each sample.
16s rRNA taxonomy and diversity
The DNA extraction and sequencing are included in Supplementary Material. Bioinformatics analysis was performed as described previously with fecal DNA amplicons (Huang et al., 2017; Lefever et al., 2016). In brief, Read 1 and Read 2 sequence files were merged with Mr_Demuxy 1.2.0 (available from https://pypi.python.org/pypi/Mr_Demuxy), a python program designed for demultiplexing sequences with dual index, allowing a maximum difference of 15. The multiple_split_libraries.py script was then run with a default Phred offset and a Phred quality threshold of 20, allowing for 1% incorrect base call probability. The fasta file then was analyzed with Quantitative Insights into Microbial Ecology (QIIME) version 1.7.0 (Caporaso et al., 2010). The workflows included pick_de_novo_otus.py, which picked operational taxonomic units (OTUs) based on 97% similarity, assigned taxonomy by uclust, aligned representative sequences with PyNAST, built a phylogenetic tree, and then made the OTU table. The sequences were rarified at a depth of 6000 reads, then α-diversity and β-diversity were computed with core_diversity_analysis.py. The metrics for α-diversity include PD whole tree, observed OTUs, and chao1. β-diversity was represented by both phylogenetic-based (unweighted Unifrac and weighted Unifrac) and non-phylogenetic-based (Jaccard distance and Bray-Curtis distance) (Caporaso et al., 2010). In addition, linear discriminant analysis effect size (LEfSe) was applied to identify the most biologically informative microbial features differentiating between two treatment groups (Segata et al., 2011) on the website: http://huttenhower.sph.harvard.edu/galaxy. For Illumina output, a total of 140 479 reads were obtained from PND 30 female fecal samples with a mean of 419.8 nucleotides (NTs) per sample; after filtering, 122 899 reads remained. For PND 90 female fecal samples, a total of 136 001 reads were obtained with a mean of 417.0 NTs per sample; after filtering, 122 781 reads still left. Among PND 90 male fecal samples, a total of 538 291 reads were obtained and after filtering, 446 814 reads left. The Illumina Miseq provides excellent quality with 90% reads reaching a Phred Quality Score > 30.
RNA-sequencing (RNA-seq)
As Paneth cells in ileum section can sense bacterial stimuli and initiate anti-microbial actions through peptides, the ileum tissues from NOD female offspring were harvested upon euthanasia at the end of study (PND 206), emptied for intestinal content, transferred to liquid nitrogen and later −80°C freezer for storage. RNA was extracted using RNeasy Mini Kit (QIAGEN), and the extracted RNA was measured for concentration, quality (as indicate by A260/280 and A260/230), and integrity (RNA integrity number greater than 6). The RNA was then submitted to Genewiz (South Plainfield, New Jersey) for quality control, DNA removal, library preparation, and sequencing with Illumina Hi-seq (San Diego, CA), which yielded 2X150 pair-end reads (total = 372 034 607 with a mean quality score of 37.48). The subsequent steps were performed in Georgia Advanced Computing Resource Center (GACRC): First, the data passed the quality control via FastQC; then the low-quality reads and adapters were trimmed by Trimmomatic (Bolger et al., 2014), with >90% pair-end reads survived. The reads were aligned with the mouse reference genome mm10, available at https://genome.ucsc.edu/cgi-bin/hgGateway? db=mm10) using Tophat 2.1.1 (Trapnell et al., 2009) allowing for a minimum intron length of 20 and a maximum intron length of 500 000. Subsequently the mapped reads were searched for differentially expressed genes (DEGs) in transcript expression using Cuffdiff (Trapnell et al., 2010). The DEGs were identified with a q value less than 0.05.
Quantitative real-time PCR (qRT-PCR) for mRNA expression
To confirm RNA-seq results that there were actual changes in α-defensin mRNA, qRT-PCR was used. RNA was extracted as described in RNA-seq section, and RNA isolates were reverse transcribed to cDNA with the iScriptTM cDNA synthesis kit (Bio-Rad). The cDNA obtained was used for real-time PCR analysis. The qRT-PCR assays were performed in 1 μl reactions containing 5 μl SYBR Green I Master and 1 μl gene-specific primers (described in Supplementary Table 1). Samples were analyzed in technical triplicate using a Stratagene Mx3005 qPCR thermocycler (Agilent Technologies, La Jolla, California).
Histology
The histology of pancreas was evaluated by a board-certified veterinary pathologist (Dr Nagy). The details of histologic analysis were described in the Supplementary Material.
Statistical analysis
Dunnett’s test was used for comparisons with VH as reference group when the equal variance assumption was met; otherwise, Wilcoxon test was performed. Likelihood ratio was performed for comparing diabetes incidence between groups. In LEfSe analysis, Kruskal-Wallis and Wilcoxon tests were used for logarithmic LDA scores >2.0 with a critical value of 0.05. JMP Pro 12 (SAS Inc., Cary, North Carolina), GraphPad Prism 7 (GraphPad Software Inc., La Jolla, California), and R 3.3.1 (R Core Team, 2013) were used for statistical analysis and data visualization.
RESULTS
T1D Incidence Was Increased in Female, but Not in Male Offspring, Following Perinatal GEN Dosing
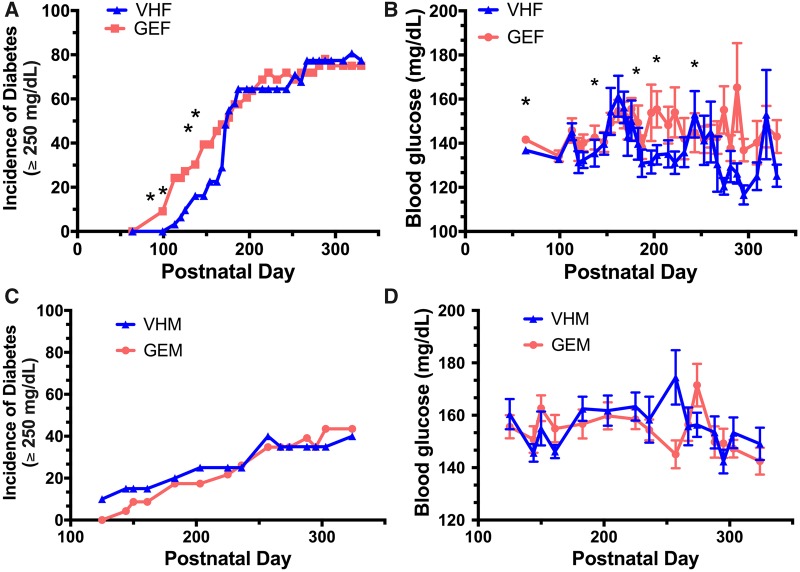
Female offspring perinatally dosed with GEN (from GD7 to PND 21) had a significant earlier onset of T1D (Figure 1A), and the incidence of T1D in GEN-treated group was twice as much as that in VH group from PND 100 to PND 175; significant increases in the incidence of T1D were observed at PND 113 (24% vs 3%), 120 (24% vs 6%), 147 (39% vs 16%), and 162 (45% vs 23%). The incidence of severe T1D (BGL ≥ 400 mg/dl) was also increased at PND 113 (33% vs 4%), 120 (33% vs 8%), 125 (38% vs 12%), 147 (54% vs 20%), 162 (63% vs 28%), and 168 (67% vs 36%) (Supplementary Figure 1). In addition to T1D incidence, perinatal GEN treatment exacerbated BGL in non-diabetic female offspring on PND 64, 203, and 274 (Figure 1B). However, the T1D incidence in males (Figure 1C) and the BGL in non-diabetic males (Figure 1D) were not significantly affected by the GEN treatment. Taken together, administration of GEN during early postnatal development caused a detrimental effect in female offspring (both the onset of T1D and the BGL), but not in the male offspring.
Figure 1.
Perinatal GEN treatment increased T1D incidence in female, but not in male, offspring. (A) The incidence of T1D (BGL ≥ 250 mg/dl) among female offspring perinatally exposed to GEN. (B) The BGL for non-diabetic female offspring. (C) The incidence of T1D among male offspring. (D) The BGL for non-diabetic male offspring. VHF, females exposed to vehicle control (N = 36); GEF, females exposed to GEN (N = 33); VHM, males exposed to vehicle control (N = 32), GEM, males exposed to GEN (N = 35). * indicates p < .05. BGL, blood glucose level; GEN, genistein; T1D, type 1 diabetes.
Perinatal GEN Dosing Elicited a Pro-inflammatory Response in Females While an Anti-inflammatory Response in Males
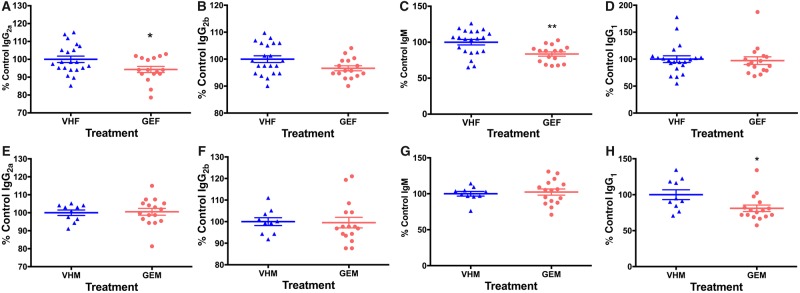
Serum antibody levels were altered following perinatal GEN dosing (Figure 2): Among the four antibodies measured (IgG1, IgG2a, IgG2b, and IgM), IgG2a was decreased by 5.7% (p < .05, Figure 2A), and IgM was decreased by 16.4% (p < .01, Figure 2C) in female offspring. Among male offspring, serum IgG1 level was reduced by 13.6% (p < .05, Figure 2H). No significant alterations in other antibodies were observed (Figure 2).
Figure 2.
The serum antibody levels in NOD female (A–D) and male offspring (E–H). (A, E) IgG2a, (B, F) IgG2b, (C, G) IgM, and (D, H) IgG1 levels in the sera of NOD female and male offspring were measured at dilutions of 1:50, 1:50, 1:500, and 1:5000 (v/v), respectively, following titration by serial dilution using ELISA. The sera were obtained when the mice were euthanized. *, p < .05 and **, p < .01. Each of the serum antibody concentration was calculated as the percent control. VHF, females exposed to vehicle control (N = 21); GEF, females exposed to GEN (N = 16); VHM, males exposed to vehicle control (N = 10); GEM, males exposed to GEN (N = 16). GEN, genistein; ELISA, enzyme-linked immunosorbent assay.
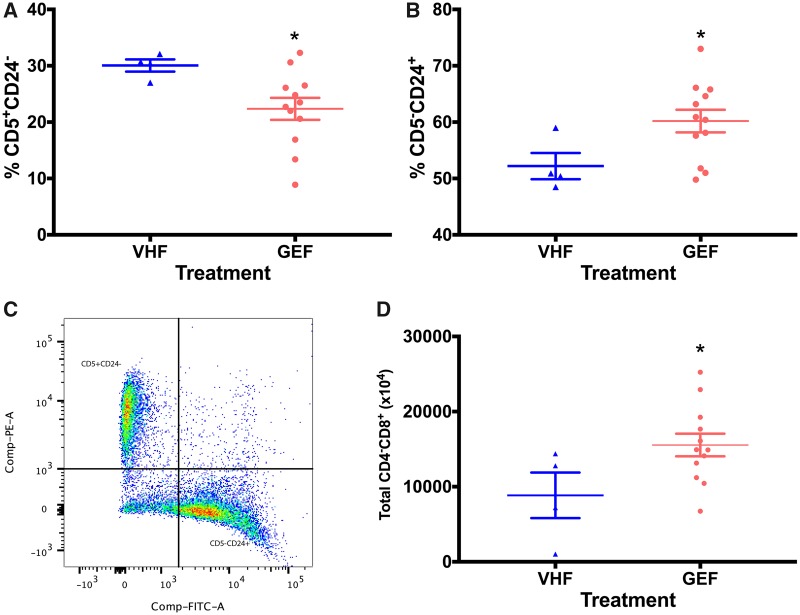
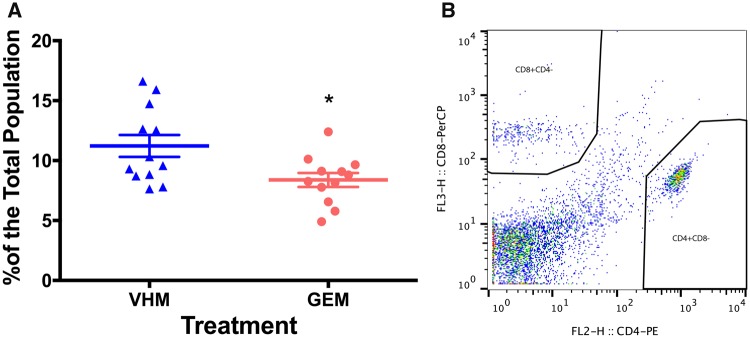
Flow cytometric analysis of splenic immune cells also demonstrated a sex-specific effect following perinatal GEN dosing (Figs. 3 and 4). For female offspring, the percentage of CD5+CD24− splenic subpopulation was significantly reduced (Figure 3A), which was accompanied by an increase in CD5−CD24+ subpopulation (Figure 3B). In addition, an increase in total CD4−CD8+ subpopulation (representing cytotoxic T cells; Figure 3D) was observed. The alterations of total T cells and B cells did not achieve statistical significances (p > .05; data not shown). Splenic T cell and its subpopulation were also analyzed in male offspring for comparison. Following perinatal GEN dosing, both the percentages of CD3+IgM− (representing total T cells, data not shown) and CD4+CD8− cells (representing helper T cells, Figure 4A) were significantly reduced, while the CD4−CD8+ subpopulation was not altered (data not shown). B cell population and its subtypes did not show a difference (data not shown).
Figure 3.
GEN treatment caused alteration in subpopulations of splenic immune cells in female offspring. (A) The percentage of splenic CD5+CD24−. (B) The percentage of splenic CD5−CD24+. (C) The gating strategy for CD5CD24 markers. X-axis represents CD24, and y-axis represents CD5. (D) The total number of splenic CD4−CD8+ cells. VHF, female exposed to vehicle control (N = 4); GEF, females exposed to genistein (N = 12). * indicates p < .05.
Figure 4.
Flow cytometric analysis of splenic T cell subpopulation in male offspring. (A) The percentage of helper T cell population. (B) The gating strategy for helper T cell population (CD4+CD8−). VHM, males exposed to vehicle control (N = 12); GEM, males exposed to genistein (N = 12). * indicates p < .05.
Among the 9 cytokines (IFN-γ, IL-1α, IL-1β, IL-2, IL-6, IL-10, IL-12p40, IL-12p70, and TNF-α) and 1 chemokine (MCP-1) analyzed in this study, there was no significant difference in most cytokines/chemokines except for serum IL-10, which was noticeably reduced (by 8.9%) in female offspring following perinatal GEN dosing (Table 1). However, no significant changes were observed in male offspring in either the cytokines or chemokine measured (Table 1).
Table 1.
The Cytokine/Chemokine Levels Measured by Multiplex Mouse Cytokine/Chemokine Magnetic Bead Panel Kit 96 Well Plate Assay in Sera of NOD Male and Female Offspring
| IFN-γ | IL-1 α | IL-1 β | IL-2 | IL-6 | |
|---|---|---|---|---|---|
| VHF | 6.21±0.81 | 82.34±2.60 | 33.48±1.21 | 22.23±1.71 | 22.52±1.13 |
| GEF | 5.15±0.25 | 78.66±6.23 | 31.38±0.00 | 21.90±1.09 | 22.74±2.85 |
| VHM | 5.22±0.73 | 88.85±7.47 | 31.34±0.83 | 21.70±1.39 | 22.92±1.84 |
| GEM | 5.35±0.60 | 79.10±2.41 | 31.71±0.33 | 21.24±0.83 | 21.87±0.82 |
| MCP-1 | TNF- α | IL-10 | IL-12p40 | IL-12p70 | |
| VHF | 79.96±1.27 | 26.67±3.52 | 14.46±0.46 | 13.86±0.79 | 22.13±0.93 |
| GEF | 76.86±1.34 | 25.92±0.63 | 13.17±0.36* | 13.44±0.98 | 22.17±0.40 |
| VHM | 79.13±2.21 | 26.11±0.51 | 13.76±0.35 | 15.54±1.51 | 21.68±0.46 |
| GEM | 79.94±0.86 | 26.47±0.53 | 14.45±0.49 | 17.73±2.85 | 22.33±0.66 |
Note: All values represent mean ± S.E. (pg/ml). IL = interleukin, MCP-1 = monocyte chemoattractant protein-1, TNF-α = tumor necrosis factor-α, VHM = NOD males dosed with vehicle (N = 10); VHF = NOD females dosed with vehicle (N = 17); GEM = NOD males dosed with genistein (N = 16); and GEF = NOD females dosed with genistein (N = 8). *, p < .05.
Gut Microbiota Was Perturbed Toward a Pro-inflammatory Response Among Female Offspring, But Toward an Anti-inflammatory Response Among Male Offspring at PND 90
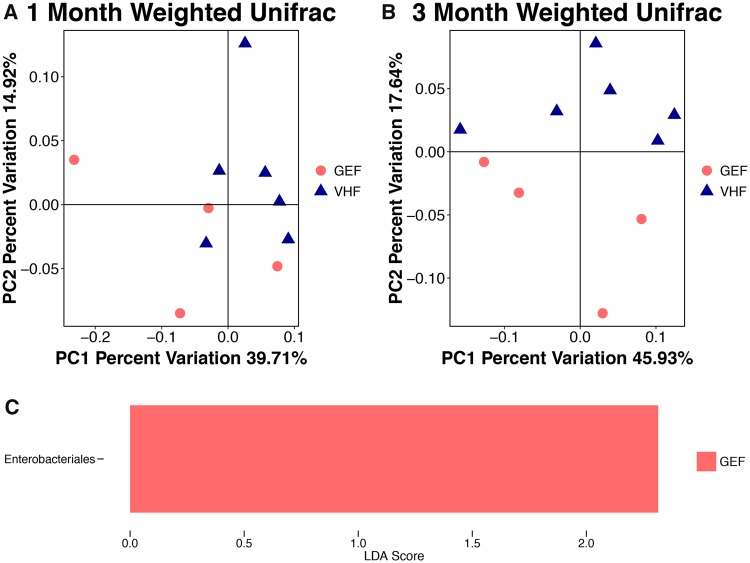
Fecal samples from both PND 30 and PND 90 female offspring were used for 16s rRNA sequencing. At the phylum level, the ratio of Bacteroides/Firmicutes was numerically decreased (p > .05) following GEN (GEF) dosing compared with VH (VHF) at PND 30 in female offspring (2.63 in GEF and 2.70 in VHF; Supplementary Figure 2A). No alteration in both α diversity and taxonomy at either order or genus level was observed (data not shown). Furthermore, principal coordinate analysis (PCoA) did not show a significant difference in the bacterial community composition at PND 30 between GEN and VH-treated females with both unweighted Unifrac metric (data not shown) and weighted Unifrac (Figure 5A).
Figure 5.
The composition of gut microbiota (GMB) based on 16S ribosomal RNA sequencing in NOD female offspring treated with genistein perinatally (GEF, N = 4) or vehicle (VHF, N = 6). The β-diversity based on the weighted Unifrac index at (A) postnatal day (PND) 30 and (B) PND 90. Each of the symbols (closed circles and triangles) represents one animal and is illustrated by principal coordinate component (PCoA), and the primary principal component (PC1) and secondary principal component (PC2) are shown. (C) Linear discriminant analysis Effect Size (LefSe) showing the microbial species with differential abundance at the order level at PND 90.
At PND 90, the Bacteroides/Firmicutes ratio was non-significantly decreased from 2.38 in VHF to 1.47 in GEF in female offspring (GEF vs VHF at PND 90: p = .088 by Wilcoxon test; Supplementary Figure 2B), indicating that the taxonomy at phylum level was not significantly affected by GEN treatment within first 3 months. In addition, α-diversity remained unchanged following perinatal GEN dosing in PND 90 females (p > .05, data not shown). However, at PND 90, GEN-treated mice could be well separated by weighted Unifrac, a phylogenetic-based metric that takes bacterial abundance into account (Figure 5B, p = .045 by non-parametric test). In addition, Enterobacterials at the order level (Figure 5C) was significantly increased following perinatal GEN dosing at PND 90 (LDA score > 2.0, p < .05 by non-parametric test). The non-phylogenetic-based metrics did not show a difference (data not shown).
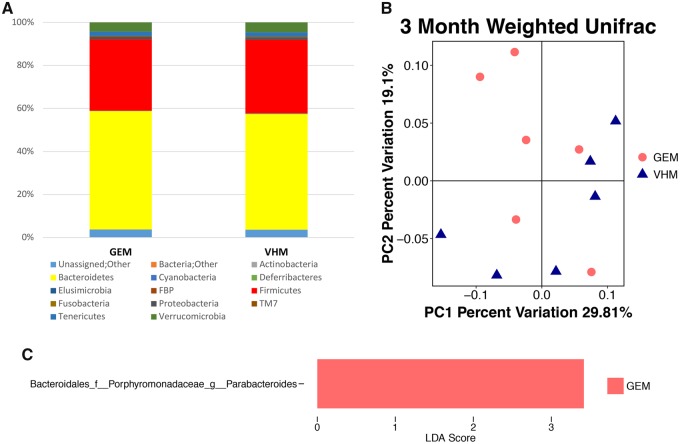
Fecal samples from PND 90 male offspring were also used for 16s rRNA sequencing. In male offspring, taxonomy at phylum level (Figure 6A), unweighted (data not shown) and weighted Unifrac (Figure 6B) did not show a difference at PND 90, while an increased genus Parabacteroides was found in males perinatally dosed with GEN (Figure 6C). At order level, no alteration in taxonomy was observed, and the non-phylogenetic-based metrics did not show a difference (data not shown).
Figure 6.
The composition of gut microbiota (GMB) based on 16S ribosomal RNA sequencing in NOD male offspring at postnatal day (PND) 90. (A) The taxonomy of the GMB at the phylum level when the individual animal data were combined for analysis according to the treatment. (B) The β-diversity based on the weighted Unifrac index. Each of the symbols (closed circles and triangles) represents one animal and is illustrated by principal coordinate component (PCoA), and the primary principal component (PC1) and secondary principal component (PC2) are shown. (C) Linear discriminant analysis Effect Size (LefSe) showing Parabacteroides with differential abundance at the order at the genus level. VHM, males exposed to VH (N = 6); GEM, males exposed to GEN (N = 6).
RNA-seq Identified Down-Regulation of Multiple Defensin Subclasses and Other Inflammatory Markers
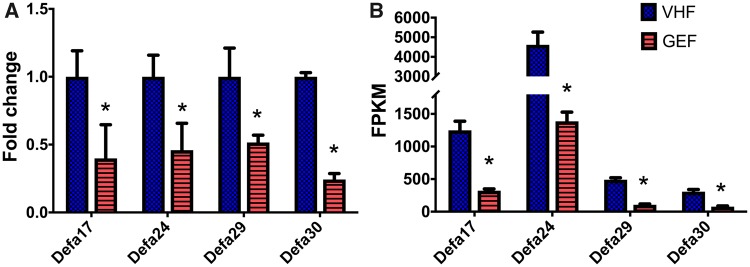
To explore further the mechanism of pro-inflammatory immune response following perinatal GEN dosing in female offspring, a transcriptomic study was conducted. A total of 124 out of 35 607 genes were identified with a p-value less than 0.05. Significantly up-regulated and down-regulated genes from RNA-seq were shown in Supplementary Table 2. Noticeably and consistently, perinatal GEN dosing in NOD female offspring down-regulated Defa17, Defa24, Defa29, and Defa30, which were confirmed by examining their mRNA levels using quantitative PCR analyses (p = .01, Figs. 7A and 7B).
Figure 7.
The expression of mRNA transcripts. (A) qRT-PCR (N = 4/group), and (B) RNA-seq. Error bar = standard error of mean. * indicates p < .05. FPKM, fragment per kilobase of transcript per million mapped reads.
DISCUSSION
T1D, a devastating and expensive organ-specific autoimmune disease, is increasing in incidence in U.S. adults (once mainly in juveniles) with 40% excess risk of death in women (Huxley et al., 2015; Li et al., 2016a). Environmental exposure (eg endocrine disruptors) and genetic predisposition may be the underlying causes (Jobling et al., 2013). Soy milk formula consumption during infancy was associated with a two-fold higher risk of T1D (Fort et al., 1986; Strotmeyer et al., 2004) and a significant increase in use of asthma or allergy drugs in young women (Strom et al., 2001). Our studies have identified unique windows of T1D susceptibility in female NOD mice for GEN: perinatal GEN dosing (from GD 7 to PND 21) accelerated the onset of T1D. In contrast, similar detrimental effect was not found in males, and adult GEN dosing were protective in both sexes (Huang et al., 2017). Although the mechanisms underlying a fetal basis of adult T1D are currently unclear, sex differences in immunoregulation in early life (Bao et al., 2002) and GMB-driven estrogen metabolism (Markle et al., 2013) might be responsible for our observations that female offspring dosed with GEN perinatally showed an exacerbation of T1D. The dose selected in this study is physiologically relevant because soy formula can provide a 4-month-old infant with approximately 6–9 mg/kg of isoflavones (Irvine et al., 1998). The dose of 20-mg/kg GEN in mice is much lower than a clinical human supplement dose (approximately 100 mg/day [Djuric et al., 2001]) in terms of milligrams per square meter of body surface, which often gives more accurate interspecies extrapolation (Hodgson and Levi, 2010). The serum level of GEN (1.4–7.5 μM) in rodents that have been fed a 1000 ppm GEN-containing diet (about 80-mg/kg BW) was equivalent to that in men who received 100-mg GEN/day (Bhandari et al., 2003; Djuric et al., 2001; Yellayi et al., 2002).
Evidences in both humans and animal models increasingly suggest that T1D is originated in the gut (70% of the body’s immune system dwells here) and associated with a profound dysbiosis during the perinatal period and in adulthood, favoring pro-inflammatory microbial communities (Kostic et al., 2015; Markle et al., 2013; Vaarala et al., 2008). In NOD mice, differences in sex hormones and GMB underlie its sexual dimorphism of T1D (Yurkovetskiy et al., 2013). Thus, an intricate interplay between of perinatal GEN intake, GMB, immune dysregulation, and sex hormone imbalance (Gourdy et al., 2016) may play critical roles in the T1D exacerbation. Our current study revealed that perinatal GEN dosing perturbed the GMB toward a pro-inflammatory response in females at PND 90, but not at PND 30, in addition to altering the β-diversity. In males, however, perinatal GEN dosing perturbed the GMB toward an anti-inflammatory response at PND 90. Our study is the first to identify the unique window of effect for GMB alteration. For inflammatory status following perinatal GEN dosing, the increase in Enterobacterials at the order level and Enterobacteriaceae at the genus level in females were associated with a pro-inflammatory status (Ellis et al., 2011), while an increased genus Parabacteroides was found in males, indicating a modulating effect on immune system and possibly regulatory T cells (Li et al., 2016b). To date, the only study investigating GMB alteration induced by perinatal endocrine disruptor exposure found bisphenol A perturbed β-diversity in rabbits on PND 42 (Reddivari et al., 2017).
With regards to antibody and cytokine alterations, there was evidence that IgG2a demonstrated an anti-inflammatory effect and thus protected female NOD mice from T1D (Todd et al., 1998). In our studies, we observed a decrease in IgG2a in NOD females following perinatal GEN dosing, suggesting a pro-inflammatory status. Moreover, the pro-inflammatory response in female offspring perinatally dosed with GEN was supported by a decreased IgM level, as IgM has been shown to protect against autoimmune diseases (Hampe, 2012). A lack of IgM was also shown to correlate with increases in pathogenic IgG (Hampe, 2012). This is in agreement with a reduced IL-10 level in females perinatally dosed with GEN, as IL-10 producing B cells suppress inflammation in various mouse models of autoimmune diseases (Hampe, 2012). It is interesting to speculate that the IL-10 producing B cells are CD5+ as discussed later (Yanaba et al., 2009). In addition, IL-10-deficient mice experience severe T1D accompanied with an increase in Th1 cytokines (Tian et al., 2001), and the decrease in IgG1 among males perinatally dosed with GEN implies a decreased Th2 response. In this study, no statistical significances in the insulin level, insulin autoantibody level, and histopathology were observed (data not shown), possibly due to the fact that the sera and tissue samples were collected at the end of study when most animals developed T1D.
For splenic cell subpopulation, CD24 is a marker for mature T cell, while CD5 is a common marker for regulatory B cells (Yanaba et al., 2009). In recent studies, it was found that CD5+ cells were negatively associated with pro-inflammatory status in autoimmune disease (Baglaenko et al., 2015). In our study, a decrease in CD5+CD24− and an increase in CD5−CD24+ splenic cells were identified in females following perinatal dosing with GEN, and consequently, a weakened regulation of autoimmune responses was observed as reflected by the exacerbation of T1D. This is in agreement with our finding that IL-10 was decreased in GEN exposed female offspring as the IL-10 producing regulatory B cell subset with a unique CD1dhiCD5+ phenotype controls T cell-dependent inflammatory responses (Vighi et al., 2008). In contrast, among male offspring, the decreases of both helper T cell and total T cell populations would suggest an anti-inflammatory status, and thus were associated with a protective effect against T1D. At the same time, serum level of IL-1α was numerically reduced in GEN-exposed male mice. IL-1 receptor deficiency can slow the progression to diabetes in the NOD mouse (Thomas et al., 2004), suggesting that GEN reduction of IL-1α might be a novel mechanism of action.
In infants, diets play a major role in the alteration of GMB and metabolites (Del Chierico et al., 2015). In the current study, perinatal GEN exposure in female offspring led to an alteration in GMB at PND 90. Importantly, the down-regulated mRNA expression of defensins, an antimicrobial peptide that is an essential component of intestinal innate immunity (Coretti et al., 2017), has been observed in these mice. Interestingly, the production of antimicrobial peptides is decreased in NOD mouse compared with their mutated strain with lower susceptibility (Mullaney et al., 2018). Defensin can stabilize the GMB (Coretti et al., 2017, Salzman 2010) while the reduction of α-defensin is associated with gut dysbiosis (Wehkamp et al., 2005). Among the few studies investigating ileum α-defensin mRNA expression, NOD mice showed a reduced expression as compared with the NOD-resistant mice (Daft et al., 2011). The anti-inflammatory effect of defensin was associated with an increased IL-10 production in humans (Kanda et al., 2011). Consistently, a decreased IL-10 level in female offspring perinatally exposed to GEN was observed in our study, which might explain the pro-inflammatory response following perinatal GEN dosing in females. Moreover, IL-10 deficiency leads to an activation of immune system (Sellon et al., 1998) and dysbiosis, which can be restored by certain bacterial strains (eg Lactobacillus spp.) (Madsen et al., 1999). The down-regulated expression of defensin in intestinal tissues in female offspring perinatally exposed to GEN might be related to the epidemiological evidence that soy-based formula induced a higher T1D risk than milk-based formula (Strotmeyer et al., 2004). After birth, the infant formula consumption, compared with breast feeding, was found to be detrimental to neonatal immune system development (Innis, 2007), as breast milk can provide defensins that enables infants to ward off environmental pathogens (Walker and Iyengar, 2015).
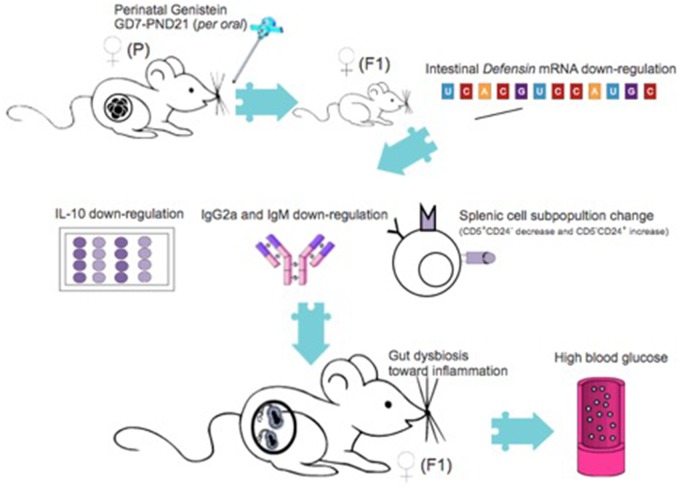
Our previous studies have identified that the immunomodulatory effect of adult GEN treatment toward protection against T1D in NOD mice is associated with the alteration of GMB (Guo et al., 2015). The hypothesized mechanism in perinatal GEN modulation of T1D in female NOD offspring is depicted in Figure 8. Despite some epidemiological studies suggest a potential detrimental effect of perinatal soy consumption on autoimmune diseases such as T1D, this current study is the first to: (1) reveal the susceptible window of exposure, (2) discover the sex dimorphic effect, (3) use the “omics” technique to explore the GMB-immune-transcriptomics consortium of T1D development, and provide solid data for future risk assessment of infant diet on increasing autoimmune diseases. This study can also serve as a model of studying immune-related diseases for other environmental chemicals (such as dietary supplement and endocrine disruptors) and contribute to the discovery of novel biomarkers for early disease detection.
Figure 8.
The hypothesized mechanism for increased type 1 diabetes incidence in females that are perinatally exposed with genistein.
This study, however, also has some limitations. For instance, the harvest of terminal blood and organs has forbidden us to detect a histopathological alteration of disease progression. More time points in sample collection would enable a better trajectory of biomarkers (such as GMB) and could be included in future studies. In addition, the causal effect of GEN-induced mRNA expression, GMB, and intestinal immunological changes are not examined and require future investigation.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH (R21ES24487) and in part by USDA-NIFA and NIH (R41AT009523). The authors declare no conflict of interest.
REFERENCES
- Anderson M. S., Bluestone J. A. (2005). The NOD mouse: a model of immune dysregulation. Ann. Rev. Immunol. 23, 447–485. [DOI] [PubMed] [Google Scholar]
- Baglaenko Y., Manion K. P., Chang N. H., Loh C., Lajoie G., Wither J. E. (2015). Suppression of autoimmunity by CD5(+) IL-10-producing B cells in lupus-prone mice. Genes Immun. 16, 311–320. [DOI] [PubMed] [Google Scholar]
- Bao M., Yang Y., Jun H. S., Yoon J. W. (2002). Molecular mechanisms for gender differences in susceptibility to T cell-mediated autoimmune diabetes in nonobese diabetic mice. J. Immunol. 168, 5369–5375. [DOI] [PubMed] [Google Scholar]
- Bhandari A., Crawford S. E., Huang L., Reenstra W. W. (2003). Effects of oral genistein in mice. Pediatr. Pathol. Mol. Med. 22, 131–141. [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Calafat A. M., Doerge D. R., Umbach D. M., Bernbaum J. C., Twaddle N. C., Ye X., Rogan W. J. (2009). Isoflavones in urine, saliva, and blood of infants: data from a pilot study on the estrogenic activity of soy formula. J. Expo. Sci. Environ. Epidemiol. 19, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I.. et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coretti L., Natale A., Cuomo M., Florio E., Keller S., Lembo F., Chiariotti L., Pero R. (2017). The interplay between defensins and microbiota in Crohn’s disease. Mediat. Inflamm. 2017, 1. Artn 8392523, doi:10.1155/2017/8392523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daft J. G., Wolf K. J., Lorenz R. G. (2011). Alterations in intestinal antimicrobial peptides are associated with increased susceptibility to type 1 diabetes. Gastroenterology 140, S-500. [Google Scholar]
- Del Chierico F., Vernocchi P., Petrucca A., Paci P., Fuentes S., Pratico G., Capuani G., Masotti A., Reddel S., Russo A.. et al. (2015). Phylogenetic and metabolic tracking of gut microbiota during perinatal development. PLoS One 10, e0137347.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuric Z., Chen G., Doerge D. R., Heilbrun L. K., Kucuk O. (2001). Effect of soy isoflavone supplementation on markers of oxidative stress in men and women. Cancer Lett. 172, 1–6. [DOI] [PubMed] [Google Scholar]
- Egro F. M. (2013). Why is type 1 diabetes increasing? J. Mol. Endocrinol. 51, R1–R13. [DOI] [PubMed] [Google Scholar]
- Ellis C. L., Ma Z.-M., Mann S. K., Li C.-S., Wu J., Knight T. H., Yotter T., Hayes T. L., Maniar A. H., Troia-Cancio P. V.. et al. (2011). Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J. Acquir. Immune Defic. Syndr. 57, 363.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Lanes R., Dahlem S., Recker B., Weyman-Daum M., Pugliese M., Lifshitz F. (1986). Breast feeding and insulin-dependent diabetes mellitus in children. J. Am. Coll. Nutr. 5, 439–441. [DOI] [PubMed] [Google Scholar]
- Gilbert E. R., Liu D. (2013). Anti-diabetic functions of soy isoflavone genistein: mechanisms underlying its effects on pancreatic beta-cell function. Food Funct. 4, 200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourdy P., Bourgeois E. A., Levescot A., Pham L., Riant E., Ahui M. L., Damotte D., Gombert J. M., Bayard F., Ohlsson C.. et al. (2016). Estrogen therapy delays autoimmune diabetes and promotes the protective efficiency of natural killer T-cell activation in female nonobese diabetic mice. Endocrinology 157, 258–267. [DOI] [PubMed] [Google Scholar]
- Guo T. L., Chi R. P., Germolec D. R., White K. L. (2005). Stimulation of the immune response in B6C3F1 mice by genistein is affected by exposure duration, gender, and litter order. J. Nutr. 135, 2449–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T. L., Chi R. P., Zhang X. L., Musgrove D. L., Weis C., Germolec D. R., White K. L. (2006). Modulation of immune response following dietary genistein exposure in F-0 and F-1 generations of C57BL/6 mice: evidence of thymic regulation. Food Chem. Toxicol. 44, 316–325. [DOI] [PubMed] [Google Scholar]
- Guo T. L., Germolec D. R., Zheng J. F., Kooistra L., Auttachoat W., Smith M. J., White K. L., Elmore S. A. (2015). Genistein protects female nonobese diabetic mice from developing type 1 diabetes when fed a soy- and alfalfa-free diet. Toxicol. Pathol. 43, 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T. L., Wang Y. B., Xiong T., Ling X., Zheng J. F. (2014). Genistein modulation of streptozotocin diabetes in male B6C3F1 mice can be induced by diet. Toxicol. Appl. Pharmacol. 280, 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe C. S. (2012). B cells in autoimmune diseases. Scientifica. 2012, Artn 215308, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson E., Levi P. E. (2010). A Textbook of Modern Toxicology. Wiley Online Library. [Google Scholar]
- Holladay S. D., Smialowicz R. J. (2000). Development of the murine and human immune system: differential effects of immunotoxicants depend on time of exposure. Environ. Health Perspect. 108, 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Xu J., Lefever D. E., Glenn T. C., Nagy T., Guo T. L. (2017). Genistein prevention of hyperglycemia and improvement of glucose tolerance in adult non-obese diabetic mice are associated with alterations of gut microbiome and immune homeostasis. Toxicol. Appl. Pharmacol. 332, 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley R. R., Peters S. A., Mishra G. D., Woodward M. (2015). Risk of all-cause mortality and vascular events in women versus men with type 1 diabetes: a systematic review and meta-analysis. Lancet Diabetes Endocrinol. 3, 198–206. [DOI] [PubMed] [Google Scholar]
- Innis S. M. (2007). Human milk: maternal dietary lipids and infant development. Proc. Nutr. Soc. 66, 397–404. [DOI] [PubMed] [Google Scholar]
- Jobling S., Bjerregaard P., Blumberg B., Brandt I., Brian J. V., Casey S. C., Frouin H., Giudice L. C., Heindel J. J., Iguchi T.. et al. (2013). Evidence for endocrine disruption in humans and wildlife State of the Science of Endocrine Disrupting Chemicals—(2012), 23–186. [Google Scholar]
- Irvine C. H. G., Fitzpatrick M. G., Alexander S. L. (1998). Phytoestrogens in soy-based infant foods: concentrations, daily intake, and possible biological effects. Pro. Soc. Exp. Biol. Med. 217, 247–253. [DOI] [PubMed] [Google Scholar]
- Kanda N., Kamata M., Tada Y., Ishikawa T., Sato S., Watanabe S. (2011). Human beta-defensin-2 enhances IFN-gamma and IL-10 production and suppresses IL-17 production in T cells. J. Leukoc. Biol. 89, 935–944. [DOI] [PubMed] [Google Scholar]
- Kostic A. D., Gevers D., Siljander H., Vatanen T., Hyotylainen T., Hamalainen A. M., Peet A., Tillmann V., Poho P., Mattila I.. et al. (2015). The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyrskyj A. L., Sloboda D. M. (2016). Perinatal programming of gut microbiota and immunity. J. Dev. Orig. Health Dis. 7, 2–4. [DOI] [PubMed] [Google Scholar]
- Lefever D. E., Xu J., Chen Y., Huang G., Tamas N., Guo T. L. (2016). TCDD modulation of gut microbiome correlated with liver and immune toxicity in streptozotocin (STZ)-induced hyperglycemic mice. Toxicol. Appl. Pharmacol. 304, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sung C. Y. J., Lee N., Ni Y. Q., Pihlajamaki J., Panagiotou G., El-Nezami H. (2016). Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc. Natl. Acad. Sci. U.S.A. 113, E1306–E1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Jick S., Breitenstein S., Michel A. (2016). Prevalence of diabetes and diabetic nephropathy in a large US commercially insured pediatric population, 2002–2013. Diabetes Care 39, 278–284. [DOI] [PubMed] [Google Scholar]
- Madsen K. L., Doyle J. S., Jewell L. D., Tavernini M. M., Fedorak R. N. (1999). Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology 116, 1107–1114. [DOI] [PubMed] [Google Scholar]
- Markle J. G., Frank D. N., Mortin-Toth S., Robertson C. E., Feazel L. M., Rolle-Kampczyk U., von Bergen M., McCoy K. D., Macpherson A. J., Danska J. S. (2013). Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088. [DOI] [PubMed] [Google Scholar]
- Mejia-Leon M. E., de la Barca A. M. C. (2015). Diet, microbiota and immune system in type 1 diabetes development and evolution. Nutrients 7, 9171–9184. Review). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller N. T., Bakacs E., Combellick J., Grigoryan Z., Dominguez-Bello M. G. (2015). The infant microbiome development: mom matters. Trends Mol. Med. 21, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney J. A., Stephens J. E., Costello M.-E., Fong C., Geeling B. E., Gavin P. G., Wright C. M., Spector T. D., Brown M. A., Hamilton-Williams E. E. (2018). Type 1 diabetes susceptibility alleles are associated with distinct alterations in the gut microbiota. Microbiome 6, 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott S. L., Wickens K., Westcott L., Jung W., Currie H., Black P. N., Stanley T. V., Mitchell E. A., Fitzharris P., Siebers R.. et al. (2008). Supplementation with Lactobacillus rhamnosus or Bifidobacterium lactis probiotics in pregnancy increases cord blood interferon-gamma and breast milk transforming growth factor-beta and immunoglobin A detection. Clin. Exp. Allergy 38, 1606–1614. [DOI] [PubMed] [Google Scholar]
- R Core Team (2013). R: A language and environment for statistical computing.
- Reddivari L., Veeramachaneni D. N. R., Walters W. A., Lozupone C., Palmer J., Hewage M. K. K., Bhatnagar R., Amir A., Kennett M. J., Knight R.. et al. (2017). Perinatal bisphenol A exposure induces chronic inflammation in rabbit offspring via modulation of gut bacteria and their metabolites. mSystems 2, e00093-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano-Keeler J., Weitkamp J. H. (2015). Maternal influences on fetal microbial colonization and immune development. Pediatr. Res. 77, 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman N. H. (2010). Paneth cell defensins and the regulation of the microbiome: detente at mucosal surfaces. Gut Microbes 1, 401.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., Huttenhower C. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellon R. K., Tonkonogy S., Schultz M., Dieleman L. A., Grenther W., Balish E., Rennick D. M., Sartor R. B. (1998). Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 66, 5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom B. L., Schinnar R., Ziegler E. E., Barnhart K. T., Sammel M. D., Macones G. A., Stallings V. A., Drulis J. M., Nelson S. E., Hanson S. A. (2001). Exposure to soy-based formula in infancy and endocrinological and reproductive outcomes in young adulthood. JAMA 286, 807–814. [DOI] [PubMed] [Google Scholar]
- Strotmeyer E. S., Yang Z., LaPorte R. E., Chang Y. F., Steenkiste A. R., Pietropaolo M., Nucci A. M., Shen S., Wang L., Wang B.. et al. (2004). Infant diet and type 1 diabetes in China. Diabetes Res. Clin. Pract. 65, 283–292. [DOI] [PubMed] [Google Scholar]
- Thomas H. E., Irawaty W., Darwiche R., Brodnicki T. C., Santamaria P., Allison J., Kay T. W. (2004). IL-1 receptor deficiency slows progression to diabetes in the NOD mouse. Diabetes 53, 113–121. [DOI] [PubMed] [Google Scholar]
- Tian J., Zekzer D., Hanssen L., Lu Y., Olcott A., Kaufman D. L. (2001). Lipopolysaccharide-activated B cells down-regulate Th1 immunity and prevent autoimmune diabetes in nonobese diabetic mice. J. Immunol. 167, 1081–1089. [DOI] [PubMed] [Google Scholar]
- Todd I., Davenport C., Topping J. H., Wood P. J. (1998). IgG2a antibodies non-specifically delay the onset of diabetes in NOD mice. Autoimmunity 27, 209–211. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaarala O., Atkinson M. A., Neu J. (2008). The “perfect storm” for type 1 diabetes - the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 57, 2555–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vighi G., Marcucci F., Sensi L., Di Cara G., Frati F. (2008). Allergy and the gastrointestinal system. Clin. Exp. Immunol. 153(Suppl 1), 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker W. A., Iyengar R. S. (2015). Breast milk, microbiota, and intestinal immune homeostasis. Pediatr. Res. 77, 220–228. [DOI] [PubMed] [Google Scholar]
- Wehkamp J., Salzman N. H., Porter E., Nuding S., Weichenthal M., Petras R. E., Shen B., Schaeffeler E., Schwab M., Linzmeier R.. et al. (2005). Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. U.S.A. 102, 18129–18134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaba K., Bouaziz J. D., Haas K. M., Poe J. C., Fujimoto M., Tedder T. F. (2008). A regulatory B cell subset with a unique CD1dhiCD5+ phenotype controls T cell-dependent inflammatory responses. J. Immunol. 285, 639–650. [DOI] [PubMed] [Google Scholar]
- Yellayi S., Naaz A., Szewczykowski M. A., Sato T., Woods J. A., Chang J., Segre M., Allred C. D., Helferich W. G., Cooke P. S. (2002). The phytoestrogen genistein induces thymic and immune changes: a human health concern? Proc. Natl. Acad. Sci. U.S.A. 99, 7616–7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Burrows M., Khan A. A., Graham L., Volchkov P., Becker L., Antonopoulos D., Umesaki Y., Chervonsky A. V. (2013). Gender bias in autoimmunity is influenced by microbiota. Immunity 39, 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.