Abstract
The aryl hydrocarbon receptor (AHR) mediates the toxic effects of dioxin (2, 3, 7, 8-tetrachlorodibenzo-p-dioxin; TCDD), which includes thymic atrophy, steatohepatitis, and a lethal wasting syndrome in laboratory rodents. Although the mechanisms of dioxin toxicity remain unknown, AHR signaling in hepatocytes is necessary for dioxin-induced liver toxicity. We previously reported that loss of TCDD-inducible poly(adenosine diphosphate [ADP]-ribose) polymerase (TIPARP/PARP7/ARTD14), an AHR target gene and mono-ADP-ribosyltransferase, increases the sensitivity of mice to dioxin-induced toxicities. To test the hypothesis that TIPARP is a negative regulator of AHR signaling in hepatocytes, we generated Tiparpfl/fl mice in which exon 3 of Tiparp is flanked by loxP sites, followed by Cre-lox technology to create hepatocyte-specific (Tiparpfl/flCreAlb) and whole-body (Tiparpfl/flCreCMV; TiparpEx3−/−) Tiparp null mice. Tiparpfl/flCreAlb and TiparpEx3−/− mice given a single injection of 10 μg/kg dioxin did not survive beyond days 7 and 9, respectively, while all Tiparp+/+ mice survived the 30-day treatment. Dioxin-exposed Tiparpfl/flCreAlb and TiparpEx3−/− mice had increased steatohepatitis and hepatotoxicity as indicated by greater staining of neutral lipids and serum alanine aminotransferase activity than similarly treated wild-type mice. Tiparpfl/flCreAlb and TiparpEx3−/− mice exhibited augmented AHR signaling, denoted by increased dioxin-induced gene expression. Metabolomic studies revealed alterations in lipid and amino acid metabolism in liver extracts from Tiparpfl/flCreAlb mice compared with wild-type mice. Taken together, these data illustrate that TIPARP is an important negative regulator of AHR activity, and that its specific loss in hepatocytes is sufficient to increase sensitivity to dioxin-induced steatohepatitis and lethality.
Keywords: aryl hydrocarbon receptor, wasting syndrome, ADP-ribosylation, 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin, TCDD-inducible poly-ADP-ribose polymerase
2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin (TCDD; dioxin) is a highly toxic environmental contaminant produced during waste incineration and other high-temperature industrial processes, and it remains a global health concern. The toxic effects of dioxin are mediated through its binding to and activation of the aryl hydrocarbon receptor (AHR), which is a member of the basic helix-loop-helix Period-AHR nuclear translocator (ARNT)-Single-minded family of transcription factors. In the canonical AHR signaling pathway, ligand binding to cytosolic AHR causes its translocation to the nucleus where it dimerizes with ARNT (also known as hypoxia-inducible factor 1β). The AHR: ARNT heterodimer then binds DNA sequence elements (termed AHREs or DREs) located within the regulatory regions of its target genes, which include cytochrome P4501A1 (CYP1A1), CYP1B1, and TCDD-inducible adenosine diphosphate (ADP)-ribose polymerase (TIPARP) (Ma et al., 2001; Whitlock, 1999; Zhang et al., 1998). In addition to its roles in dioxin toxicity, xenobiotic metabolism and vascular development (Stevens et al., 2009), AHR has important roles in T-cell differentiation, in the defense against bacterial infections and in gut homeostasis (Moura-Alves et al., 2014; Quintana et al., 2008). To date, >400 AHR ligands have been identified, including dietary compounds (3, 3'-diindolylmethane), various endogenous ligands (kynurenine, 6-formylindolo(3, 2b)carbazole), and environmental contaminants such as dioxin (Denison and Nagy, 2003).
Dioxin causes diverse toxic effects in laboratory rodents including immunosuppression, steatohepatitis, impaired reproduction, and a lethal wasting syndrome (Birnbaum, 1994, 1995; Poland and Knutson, 1982). A single dose of dioxin induces a lethal starvation-like syndrome, which includes decreased gluconeogenesis, liver damage, steatohepatitis and dyslipidaemia, ultimately leading to lethality (Linden et al., 2010; Seefeld et al., 1984). Acute lethality varies widely both among species and between rodent strains. For example, the median lethal dose (LD50) for guinea pigs is 1–2 μg/kg dioxin, whereas the LD50 for hamsters is >5000 μg/kg (Pohjanvirta and Tuomisto, 1994; Poland and Knutson, 1982). In most strains of mice, lethality occurs only 2–3 weeks after a single dose of 115–300 μg/kg of dioxin (Pohjanvirta and Tuomisto, 1994; Poland and Knutson, 1982). There is a roughly 10-fold difference in susceptibility between the high sensitivity C57BL/6 (Ahrb1 allele) and low sensitivity DBA/2 (Ahrd allele) mouse strains, which is due to polymorphic variations in their ligand-binding domains (Poland et al., 1994). The molecular mechanisms of dioxin-induced wasting syndrome remain obscure, but transgenic mice overexpressing AHR show increased sensitivity to dioxin toxicities, whereas Ahr−/− null and Ahrdbd mice, which express a mutant AHR that does not bind to AHREs, are resistant to the effects of dioxin (Fernandez-Salguero et al., 1996; Lee et al., 2010; Walisser et al., 2005).
TIPARP (PARP7/ARTD14) is an AHR-regulated gene and a member of the poly-adenosine diphosphate (ADP)-ribose polymerase (PARP) family. PARPs are nicotinamide adenine dinucleotide (NAD+)-dependent enzymes that use NAD+ as a substrate to transfer 1 molecule of ADP-ribose, referred to as mono-ADP-ribosylation (MARylation), or several ADP-ribose moieties, referred to as poly-ADP-ribosylation (PARylation), to specific amino acid residues on themselves and on target proteins (Hottiger et al., 2010). Mono- and poly-ADP-ribosylation are reversible posttranslational modifications involved in several biological processes, such as immune cell function, regulation of transcription, protein expression and DNA repair (Kraus and Hottiger, 2013). We previously reported that TIPARP is a mono-ADP-ribosyltransferase that functions as part of a negative feedback loop to repress AHR activity through a mechanism that involves reduced ligand-induced AHR protein levels and that requires TIPARP’s catalytic activity (MacPherson et al., 2013). Moreover, Tiparp−/− mice exhibit increased AHR activity but also increased sensitivity to dioxin-induced toxicities, including steatohepatitis, hepatotoxicity and lethal wasting syndrome (Ahmed et al., 2015). Although the mechanisms of dioxin-induced toxicity remain incompletely understood, AHR expression in hepatocytes is needed to generate the adaptive as well as toxic response to dioxin exposure (Walisser et al., 2005).
Here, we describe the generation of Tiparp conditional mutant mice in which exon 3 of the Tiparp gene is flanked by loxP sites, and the subsequent creation of both hepatocyte-specific (Tiparpfl/flCreAlb) and whole-body (Tiparpfl/flCreCMV; TiparpEx3−/−) TIPARP knockout mice. These mice were used to further investigate the role of TIPARP in AHR signaling and dioxin-induced toxicity.
MATERIALS AND METHODS
Generation of conditional Tiparp null mice
Tiparpfl/fl mice, where exon 3 of Tiparp was flanked by loxP sites, were generated from embryonic stem (ES) cells purchased from European Conditional Mouse Mutagenesis (EUCOMM; Tiparptm1a(EUCOMM)Wtsi). ES cells were expanded by the Toronto Centre for Phenogenomics (TCP). Correct targeted recombination was confirmed in 2 of 5 ES cell clones purchased. Briefly, SphI-digested genomic DNA was used in Southern blotting to produce 8.9 kb (wild-type [WT]) and 12.3 kb (tm1a) fragments spanning the 5’ region of the targeted sequence of Tiparp, and in the targeted allele, including the neomycin-resistance (Neo) cassette. The Neo probe was used to reveal additional or random integrations of the targeting vector in the genomes (fragments of incorrect sizes); these ES clones were excluded. PCR primers used to generate the respective probes are provided in Supplementary Table 1. Based on the positive Southern blot results, ES cell clones D3 and G3 were chosen for aggregation and implantation into pseudopregnant surrogate females (performed by TCP). Chimeric Tiparp+/tm1a mice were bred to C57BL/6 albino females and pups were genotyped to identify those with germline transmission of the conditional mutant Tiparp allele (Tiparp+/tm1a); only ES clone G3 resulted in successful germline transmission. Some Tiparp+/tm1a mice were bred to B6.C-Tg(CMV-cre)1Cgn/J (Jackson Labs, Bar Harbor, Maine) to remove the Neo cassette and the targeted exon and leave the lacZ reporter gene (conversion to tm1b allele) and then outbred once to C57BL/6N to remove Tg-(CMV-cre). Mice heterozygous for the tm1b allele were then intercrossed to make homozygous tm1b mice (Tiparptm1b/tm1b, referred to as TiparpEx3−/−). Other Tiparp+/tm1a mice were bred to B6(C3)-Tg(Pgk1-FLPo)10Sykr/J (Jackson Labs) to remove the lacZ and Neo cassettes and leave the targeted exon flanked by LoxP sites (conversion to tm1c allele) and then outbred once to C57BL/6N to remove Tg-(Pgk1-FLPo) (Tiparp+/tm1c). Tiparp+/tm1c mice were bred to B6.C-Tg(CMV-cre)1Cgn/J to remove the targeted exon (conversion to tm1d allele) and then outbred once to C57BL/6N to remove Tg-(CMV-cre). Mice heterozygous for the tm1d allele were then intercrossed to make homozygous tm1d mice (Tiparptm1d/tm1d; referred to as TiparpEx3−/−). Tiparp+/tm1c mice were also bred to B6N.Cg-Tg(Alb-cre)21Mgn/J (Jackson Labs) to create hepatocyte-specific Tiparp knock-out mice (Tiparptm1c/tm1cCreAlb; referred to as Tiparpfl/flCreAlb. This colony was maintained such that Tiparpfl/fl female mice were paired with Tiparpfl/flCreAlb male mice. Genotypes of all mice were determined by PCR analysis of tail biopsies using PCR primers shown in Supplementary Table 1. The specificity of excision events in Tiparpfl/flCreAlb mice was determined by quantitative real time PCR (qPCR) using primer pairs specific for exon 3 of Tiparp compared with primer pairs specific for intron 1 (Supplementary Table 1). Genotype controls used in experiments for the Tiparpfl/flCreAlb line are Tiparpfl/fl (observed to be phenotypically equivalent to WT mice), and for the TiparpEx3−/− are Tiparp+/+. Both tm1b and tm1d lines were used.
In vivo dioxin treatment studies
All experiments used 8- to 10-week-old male mice. For the acute 6 h exposure studies, TiparpEx3−/− or Tiparpfl/flCreAlb mice and their respective strain-specific WT control mice were treated with a single intraperitoneal (i.p.) injection of 100 μg/kg dioxin, and livers were excised and flash frozen 6 h later. For the subacute dioxin toxicity studies, mice were treated with a single i.p. injection of 10 or 100 μg/kg of dioxin and sacrificed on day 6. The 10 μg/kg dose of dioxin was dissolved in a mixture of corn oil and dimethyl sulfoxide (CO:DMSO; 90:10, referred to as CO), while the 100 μg/kg dose of dioxin was dissolved in pure DMSO. For the survival studies, mice were followed for up to 30 days after a single injection of 10 or 100 μg/kg of dioxin. Control mice received equivalent weight-adjusted volumes of CO or DMSO. The time point for euthanization was determined based on endpoint criteria for our study: a loss of 20% body weight or indications of acute distress. All control mice were euthanized to match the endpoints of dioxin-sensitive mice. For food intake studies, mice were housed individually and provided intact pellets of food that were weighed daily; a baseline was determined for each mouse by monitoring for 1 week prior to treatment as described previously (Ahmed et al., 2015). Whole blood was obtained from the saphenous vein for serum alanine aminotransferase (ALT) analysis as described previously in Ahmed et al. (2015). Hepatic glycogen levels were determined from 10 mg of frozen liver using the Glycogen Assay Kit II (Abcam). Liver, thymus, white and brown adipose tissue (WAT and BAT) were dissected and weighed. Livers from TiparpEx3−/− or Tiparpfl/flCreAlb mice treated with vehicle, 10 or 100 µg/kg dioxin were collected either on day 6 or on the day of euthanization in the survival studies. Care and treatment of animals followed the guidelines set by the Canadian Council on Animal Care, and all protocols were approved by the University of Toronto Animal Care Committee.
Hepatocytes
Tiparpfl/flCreAlb or Tiparpfl/fl male mice (8- to 10-weeks old) were used to isolate primary hepatocytes. Mouse liver was perfused with liver perfusion medium (Invitrogen) for 10 min followed by liver digestion medium for 10 min. Freshly prepared hepatocytes were seeded at a final density of 0.5 × 106 cells/well onto type I collagen coated 6-well plates in attachment medium (William’s E media, 10% dextran-coated charcoal (DCC) stripped fetal bovine serum (FBS), 1× penicillin/streptomycin, and 10-nM insulin). The medium was changed 2 h after plating, and all experiments were performed on the second day. Ligands were added to the cells in M199 media with 5% DCC-FBS and cells were harvested 16 h after ligand treatment for RNA extraction.
RNA extraction and gene expression analysis
Livers were removed, washed in ice-cold PBS, weighed, and flash frozen in liquid nitrogen. Frozen livers were homogenized in TRIZOL reagent (Life Technologies) and total RNA was isolated using the Aurum RNA isolation kits (BioRad) and reverse transcribed as previously described (Ahmed et al., 2015). Primers used to amplify target transcripts are provided in Supplementary Table 1 or described elsewhere (Ahmed et al., 2015). All genes were normalized to TATA-binding protein levels and analyzed using the comparative CT (ΔΔ CT) method.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed as previously described in Lo et al. (2011). Briefly, approximately 100 mg of frozen mouse liver was homogenized in 1% formaldehyde in PBS and incubated for 10 min at room temperature. The homogenate was centrifuged at 8000 × g for 5 min at 4°C. Pellet was washed in ice-cold PBS, centrifuged, and resuspended in 900 μl of TSEI (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate) + 1× Protease Inhibitor Cocktail (Sigma, St Louis, Missouri). Samples were sonicated 10 times for 30 s ON/30 s OFF on the high setting using a Bioruptor (Diagenode). The supernatants were transferred to new microcentrifuge tubes and incubated with rabbit IgG (5 μg; Sigma) and antiAHR (5 μg; SA-210, Enzo) overnight at 4°C under gentle agitation. ChIP samples were washed, the DNA and the ChIP-qPCR was performed as previously described in Lo et al. (2011).
Histology
Hematoxylin and eosin and Oil-Red-O/Hematoxylin staining were performed following standard methods with representative images provided. Paraformaldehyde-fixed or optimal cutting temperature (OCT) compound-embedded tissues were provided to the Histology Core Facility at the Princess Margaret Cancer Centre, Toronto, Ontario, for all histology sample processing, staining and scanning of stained slides.
Western blotting
For hepatic AHR protein detection, whole cell extracts were prepared by homogenizing 100 mg of liver tissue in RIPA lysis buffer. Ten micrograms of total protein were separated by SDS-PAGE and transferred to a nitrocellulose membrane. Membranes were incubated with antiAHR antibody (SA-210) and stripped and then incubated with antiβ-actin antibody (Sigma A-2228).
Metabolomics
Tiparpfl/fl and Tiparpfl/flCreAlb mice were treated with a single intraperitoneal injection of CO: DMSO or 10 μg/kg dioxin. Liver tissue (100 mg) was collected on day 3 and flash frozen in liquid nitrogen. The frozen tissue was extracted and metabolomic analyses were performed by Metabolon (Durham, North Carolina). Raw data received from Metabolon were preprocessed to input missing values and normalized using logarithmic transformation. Shapiro’s test was used to test for normality and Levene’s test was used to test for the homogeneity of variances. Analysis of variance (ANOVA) was used to analyze the differences in group means, followed by Tukey’s-HSD for post hoc correction. The FDR method was used to adjust the p-values for multiple ANOVA tests (one for each metabolite). At an FDR of 10%, all the group comparisons that exhibited a statistically significant difference (post hoc corrected p-values < .05) were considered significant. All the analyses were performed in R version 3.4.1.
Statistical analysis
All data were presented as means and SEM. Two-way ANOVA followed by Sidak’s post hoc test or 1-way ANOVA followed by Tukey’s post hoc test were used to assess statistical significance (p < .05) using GraphPad Prism 6 Software (San Diego, California) or R version 3.4.1.
RESULTS
Generation of Conditional Tiparpfl/fl Mice
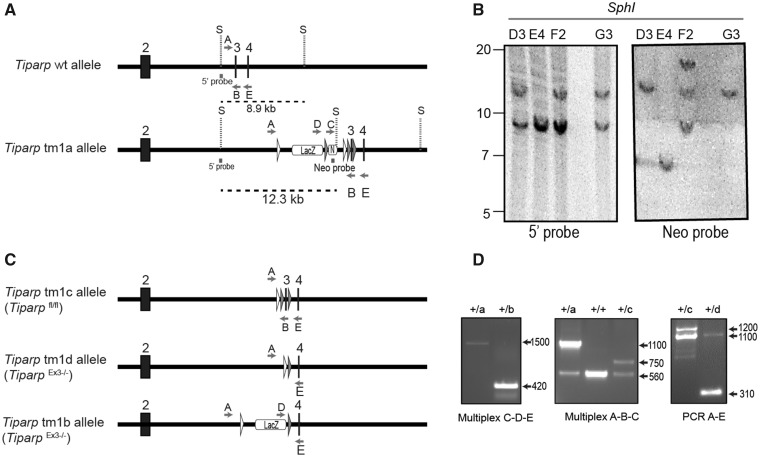
Mice harboring the conditional Tiparpfl allele were generated from Tiparptm1a(EUCOMM)Wtsi ES cells purchased from EUCOMM. A partial map of both the Tiparp WT and Tiparp tm1a alleles is shown in Figure 1A. Southern blotting of genomic DNA isolated from 4 different ES cell clones confirmed the correct integration of the tm1a allele in the ES cell clones D3 and G3 (Figure 1B). Mice generated in this study were from ES cell clone G3. Mice harboring the conditional Tiparpfl allele were generated from the Tiparp tm1a allele by excision of the Neo and LacZ genes through crosses with B6(C3)-Tg(Pgk1-FLPo)10Sykr/J mice, leaving the targeted exon flanked by LoxP sites. Mice heterozygous for the tm1c allele were then intercrossed to make homozygous tm1c mice (Tiparpfl/fl; tm1c allele).
Figure 1.
Generation of the conditional Tiparpfl/fl mice. A, Schematic diagram of Tiparp WT allele and the allele with successful recombination of the tm1 targeting construct (Tiparp tm1a allele) and corresponding Southern blot data. Exon numbers reflect known coding exons; white arrowheads represent FRT sites; gray arrowheads represent LoxP sites; LacZ represents lacZ reporter gene; N represents the Neo resistance cassette; S, SphI site; dashed lines indicate the fragment of DNA generated by SphI digestion that was detected with the radiolabeled probes (5’ probe and Neo probe); letters indicate genotyping primers. B, Southern blots show Tiparp allele fragments detected with the radiolabeled probes: ES clones D3 and G3 show correct homologous recombination of the targeting construct (12.3 kb fragment detected with 5’ probe) without additional random integrations (only the 12.3 kb band detected with Neo probe). C, Schematic diagram of Tiparp tm1 alleles following excision of sequence by CRE or FLP recombinases. D, PCR data from mouse tail biopsies. The Tiparp tm1a allele was converted to tm1b using CRE recombinase to remove the Neo cassette and exon 3 located between the LoxP sites. Primer pair D-E amplify a product spanning the excision sites to show that the floxed sequences were removed (420 bp). The Tiparp tm1a allele was converted to tm1c using FLP recombinase to remove the LacZ and Neo cassettes located between the FRT sites. Primer pair A-B amplifies a product spanning the excision site to show that the sequence was flipped out (750 bp). The Tiparp tm1c allele was converted to tm1d using CRE recombinase to remove the floxed exon 3. Primer pair A-E amplify a product spanning the floxed region to show that the exon is removed (310 bp). Sample genotypes: + indicates the Tiparp WT allele; letters indicate the Tiparp tm1 corresponding allele (tm1a, tm1b, tm1c, and tm1d).
Complete and Hepatocyte-Specific Excision of the Tiparpfl/fl Allele
Using genetrap targeted TIPARP null mice in which a LacZ gene is inserted in front of exon 1 in the Tiparp gene (Schmahl et al., 2007), we reported that loss of TIPARP expression increased the sensitivity of mice to dioxin-induced steatohepatitis and lethality (Ahmed et al., 2015). These findings supported the notion that TIPARP protects against dioxin-induced toxicity by negatively regulating AHR action. Because the type of gene knockout targeting strategy can impact the phenotypes observed from targeting the same gene, we created a complete TIPARP knockout by removal of exon 3 of Tiparp (TiparpEx3−/−) as described in Materials and Methods section. To generate a complete TIPARP null mouse in which exon 3 is removed (TiparpEx3−/−) we used 2 different strategies. In the first approach, mice heterozygous for the tm1c allele were bred to B6.C-Tg(CMV-cre)1Cgn/J mice to remove the targeted exon (TiparpEx3−/−; tm1d allele). In second approach, mice carrying the tm1a allele were bred to B6.C-Tg(CMV-cre)1Cgn/J mice to remove the Neo cassette and the targeted exon and leave the lacZ reporter (TiparpEx3−/−; tm1b allele). A map of the Tiparp tm1c, tm1d, and tm1b alleles is shown in Figure 1C. The correct genotype was confirmed by PCR analysis of genomic DNA (Figure 1D). TiparpEx3−/− mice represent a distinct TIPARP null strain compared with Tiparp targeted knockout using a genetrap approach (Ahmed et al., 2015; Schmahl et al., 2007). To determine whether the loss of TIPARP in hepatocytes is sufficient to increase sensitivity to dioxin-dependent toxicity, we generated mice in which Tiparp was deleted in hepatocytes, Tiparpfl/flCreAlb (Figure 2A). Tiparpfl/flCreAlb mice were created by breeding mice homozygous for the tm1c allele (Tiparpfl/fl) with B6N.Cg-Tg(Alb-cre)21Mgn/J mice to remove the targeted exon specifically in hepatocytes (Walisser et al., 2005). For the Tiparpfl/flCreAlb line, WT mice were referred to as Tiparpfl/fl, whereas for the TiparpEx3−/− line, WT mice were referred to as Tiparp+/+ for simplicity.
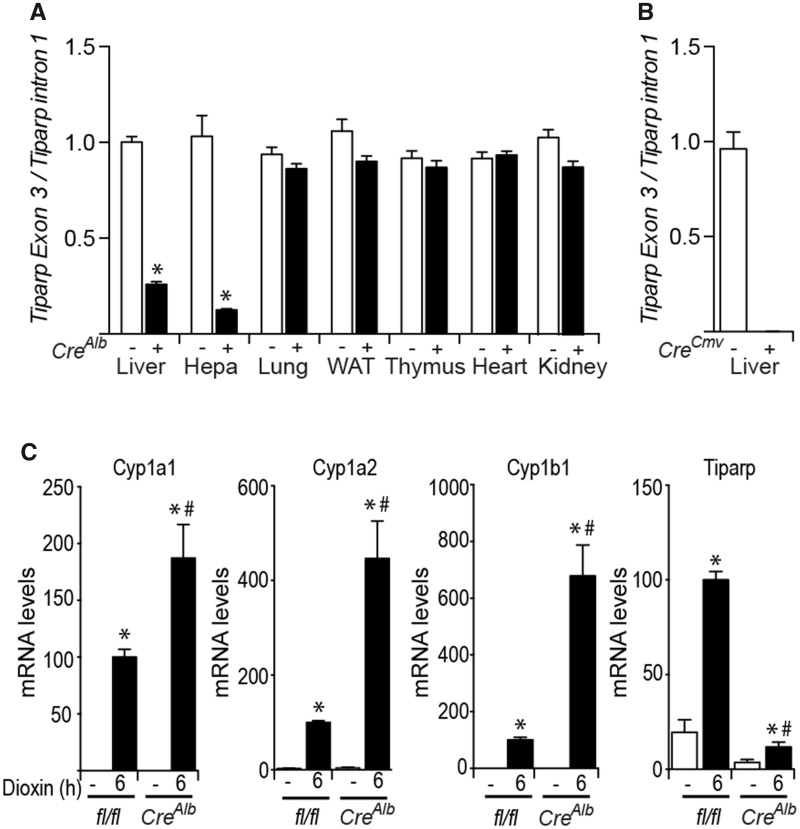
Figure 2.
Specificity of CreAlb-mediated excision of the Tiparpfl allele. A, Specificity of Tiparpfl excision by CreAlb was determined by quantitative real-time PCR-based genotyping for excised and unexcised alleles of Tiparpfl in genomic DNA isolated from various tissues and hepatocytes obtained from Tiparpfl/fl and Tiparpfl/flCreAlb mice. *p < .05 compared with tissue matched Tiparpfl/fl mice. B, Tiparpfl excision by CreCMV was determined from genomic DNA isolated from livers of Tiparp+/+ and TiparpEx3−/− mice. Abbreviations: Hep, hepatocytes; WAT, white adipose tissue. C, Increased AHR regulated Cyp1a1, Cyp1a2, Cyp1b1, and Tiparp mRNA levels in expression in hepatocytes isolated from Tiparpfl/fl and Tiparpfl/flCreAlb mice treated with 10-nM dioxin for 6 h. RNA and qPCR were performed as described in the materials and methods section. *p < .05 compared with genotyped match control treated, #p < .05 compared with dioxin-treated Tiparpfl/fl (n = 3).
To examine the specificity of excision events in Tiparpfl/flCreAlb mice, we analyzed various tissues by determining the relative ratios of Tiparp exon 3 compared with intron 1 using qPCR. Tiparp exon 3 was efficiently excised in liver tissue and in isolated hepatocytes (Hepa) in the presence of CreAlb, but not in the other tissues examined (Figure 2A). Tiparp exon 3 was not detected in liver tissue isolated from TiparpEx3−/− mice (Figure 2B). In the absence of CreAlb, the Tiparpfl/fl mice showed only the Tiparpfl-unexcised allele in all tissues and isolated hepatocytes examined (Figure 2A).
Cloning and DNA sequencing revealed that the conditional inactivation of the Tiparpfl allele accomplished by Cre-mediated deletion of exon 3 (169 bp) results in splicing and fusion between exon 2 (961 bp) and exon 4 (161 bp), leading to the insertion of a premature stop codon. This truncated version of TIPARP contains 311 amino acids, of which the last 4 are the result of a frameshift (the full-length protein is 657 a.a.), and it lacks its tryptophan-tryptophan-glutamate (WWE) and catalytic domains (MacPherson et al., 2013). In agreement with a previous study, cloning and transient transfection of the truncated TIPARP protein failed to inhibit AHR-dependent and dioxin-induced CYP1A1 reporter gene activity (Supplementary Figure 1).
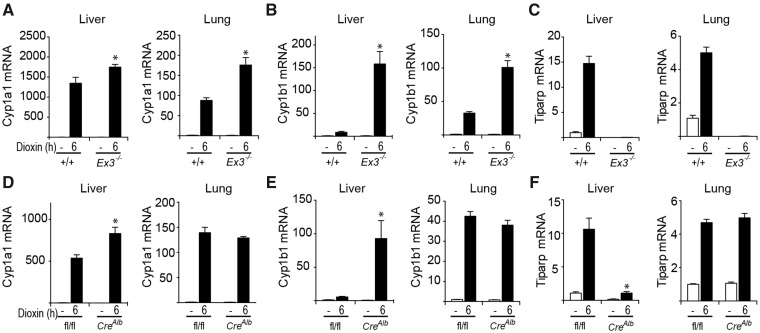
Consistent with TIPARP’s role as a negative regulator of AHR activity, exposure of hepatocytes from Tiparpfl/flCreAlb mice for 6 h to 10 nM dioxin increased mRNA expression levels of the AHR target genes Cyp1a1, Cyp1a2 and Cyp1b1 compared with similarly treated hepatocytes from Tiparpfl/fl mice (Figure 2C). Tiparp mRNA expression levels were markedly decreased in hepatocytes isolated from Tiparpfl/flCreAlb mice compared with Tiparpfl/fl mice. We next examined the effect of TIPARP loss on AHR target gene expression in liver and lung tissues isolated from TiparpEx3−/−, Tiparpfl/flCreAlb, and WT mice 6 h after treatment with 100 μg/kg dioxin. Higher Cyp1a1 and Cyp1b1 mRNA levels were observed in both liver and lung from TiparpEx3−/− mice than in TiparpEx3+/+ mice (Figs. 3A and 3B). Tiparp mRNA levels were not detected in liver or lung from TiparpEx3−/− mice compared with Tiparp+/+ mice (Figure 3C). Significantly increased dioxin-induced Cyp1a1 and Cyp1b1 mRNA levels above those observed in Tiparpfl/fl mice were only observed in liver and lung from Tiparpfl/flCreAlb mice (Figs. 3D and 3E). As expected, TIPARP expression levels were reduced in liver, but not in lung of Tiparpfl/flCreAlb compared with Tiparpfl/fl mice (Figure 3F).
Figure 3.
AHR regulated transcript levels in liver and lung tissue isolate from control and dioxin-treated TiparpEx3−/−, Tiparpfl/flCreAlb, and their respective WT mice. Mice were treated with a single injection of 100 μg/kg dioxin in DMSO, or DMSO vehicle alone, and euthanized 6 h later. RNA and qPCR were performed as described in the Materials and Methods section. *p < .05 compared with genotype matched control-treated; #p < .05 compared with dioxin-treated Tiparp+/+ (A–C) or Tiparpfl/fl (D–F) (n = 4).
TiparpEx3−/− Mice Exhibit Increased Sensitivity to Dioxin-Induced Toxicity and Lethality
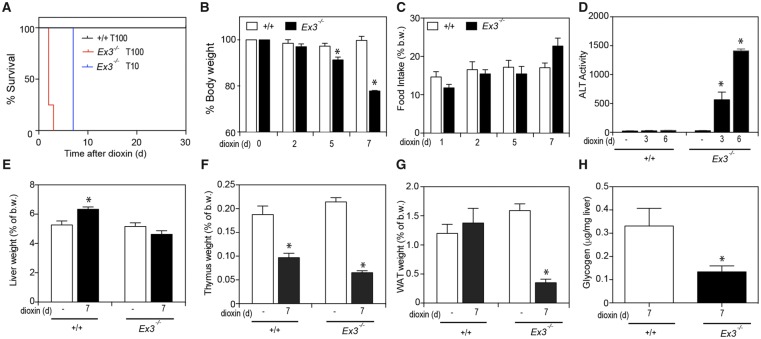
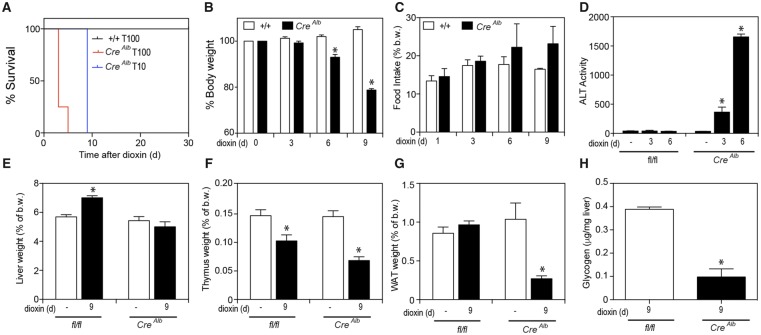
To determine the sensitivity of TiparpEx3−/− mice to dioxin-induced toxicity, these mice and their respective WT controls were injected with a single dose of 10 or 100 μg/kg dioxin and monitored for up to 30 days, as previously described (Ahmed et al., 2015). All TiparpEx3+/+ mice were normal in physical appearance at the end of the 30-day observation period (Figure 4A), while no dioxin-treated TiparpEx3−/− mice (tm1d or tm1b) survived the 30-day experiment (Figure 4A). TiparpEx3−/− mice treated with 100 μg/kg dioxin became weakened and moribund and were humanely euthanized between days 2 and 3, while those treated with 10 μg/kg dioxin were euthanized at day 7. TiparpEx3−/− mice treated with 10 μg/kg dioxin had lost significant body weight by 5 days after treatment (Figure 4B); however, no decrease in food intake was observed (Figure 4C). No changes in food intake or body weight were seen in dioxin-exposed Tiparp+/+ mice. Serum ALT activity, a marker of hepatotoxicity, was significantly increased in dioxin-treated TiparpEx3−/− mice, while no increase above controls was observed in Tiparp+/+ mice (Figure 4D). Increased liver weight was observed in dioxin-treated Tiparp+/+ mice but not in TiparpEx3−/− mice (Figure 4E). Thymic involution, an endpoint associated with dioxin toxicity, responded as expected in both genotypes at day 7 (Figure 4F). A significant decrease in epididymal WAT weight was seen in TiparpEx3−/− mice but not in WT mice (Figure 4G). No differences in BAT weight were observed (data not shown). Hepatic glycogen stores were lower in dioxin-treated TiparpEx3−/− mice than in WT mice (Figure 4H). These data support the importance of TIPARP in regulating AHR action and show that loss of its expression in mice increases their sensitivity to dioxin toxicity.
Figure 4.
Loss of TIPARP increases dioxin-induced hepatotoxicity and lethal wasting syndrome in male mice. A, Kaplan-Meier survival curves for male Tiparp+/+and TiparpEx3−/− mice treated with a single 10 or 100 µg/kg i.p. injection of dioxin and monitored for 30 days. B, Body weight, (C) food intake, (D) serum ALT activity, (E) liver, (F) thymus, and (G) WAT weight expressed as percentage of total body weight, and (H) hepatic glycogen levels were measured from Tiparp+/+ and TiparpEx3−/− mice treated with 10 μg/kg dioxin. Data shown are the mean ± SEM (n = 4–5). For (B–G) *p < .05, 2-way ANOVA followed by Tukey’s post hoc test compared with genotype-matched control treated mice. For (H) *p < .05, Student’s t test.
Hepatocyte-Specific Loss of TIPARP Results in Increased Sensitivity to Dioxin-Induced Toxicity and Lethality
Because AHR expression in hepatocytes is required for dioxin-induced liver toxicity, we hypothesized that the loss of TIPARP expression in hepatocytes would enhance dioxin-dependent liver toxicity. To test this hypothesis, we treated Tiparpfl/flCreAlb and Tiparpfl/fl mice with a single i.p. injection of 10 or 100 μg/kg dioxin and monitored the mice for up to 30 days. As expected, all Tiparpfl/fl mice were normal in physical appearance at the end of the 30-day observation period (Figure 5A). No dioxin-treated Tiparpfl/flCreAlb mice survived the 30-day experiment (Figure 5A). Tiparpfl/flCreAlb mice treated with 100 μg/kg dioxin appeared weakened and moribund and were humanely euthanized between days 3 and 5, while those treated with 10 μg/kg dioxin were humanely euthanized at day 9. Sensitivity to dioxin-induced lethality was significantly different between Tiparpfl/flCreAlb and TiparpEx3−/− mice at both 10 and 100 μg/kg dioxin, suggesting that cells other than hepatocytes also contribute to dioxin lethality in these models. Tiparpfl/flCreAlb mice treated with 10 μg/kg dioxin had lost significant body weight by 6 days after treatment (Figure 5B), while no decrease in food intake was observed (Figure 5C). No change in food intake or body weight was seen in Tiparpfl/fl mice. Serum ALT was significantly increased in dioxin-treated Tiparpfl/flCreAlbmice at days 3 and 6, but no increase was observed in Tiparpfl/fl mice (Figure 5D). Increased liver weight was observed in dioxin-treated Tiparpfl/fl mice but not in Tiparpfl/flCreAlb mice (Figure 5E). Decreased thymus weight was seen in both genotypes at day 9 (Figure 5F). Consistent with dioxin-treated TiparpEx3−/− mice, Tiparpfl/flCreAlb mice had significantly decreased epididymal WAT levels compared with WT mice (Figure 5G). No difference in BAT weight was observed (data not shown). Hepatic glycogen stores were also decreased in dioxin-treated Tiparpfl/flCreAlb mice compared with Tiparpfl/fl mice (Figure 5H). These findings show that hepatocyte-specific deletion of TIPARP is sufficient to increase dioxin-induced toxicity and lethality.
Figure 5.
Hepatocyte-specific loss of TIPARP increases dioxin-induced hepatotoxicity and lethal wasting syndrome in male mice. A, Kaplan-Meier survival curves for male Tiparpfl/fland Tiparpfl/flCreAlb mice treated with a single 10 or 100 µg/kg i.p. injection of dioxin and monitored for 30 days. B, Body weight, (C) food intake, (D) serum ALT activity, (E) liver, (F) thymus, (G) WAT weight expressed as percentage of total body weight, and (H) hepatic glycogen levels were measured from Tiparpfl/fl and Tiparpfl/flCreAlb mice treated with 10 μg/kg dioxin. Data shown are the mean ± SEM (n = 4–5). For (B–G) *p < .05, 2-way ANOVA followed by Tukey’s post hoc test compared with genotype-matched control treated mice. For (H) *p < .05, Student’s t test.
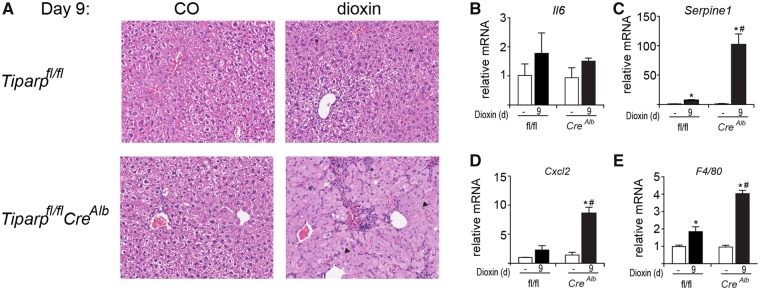
As an independent measure of liver toxicity, livers were sectioned and stained with hematoxylin and eosin. Vehicle-treated Tiparpfl/fl and Tiparpfl/flCreAlb mice had histologically normal liver architecture (Figure 6A). On day 9, dioxin-treated Tiparpfl/fl livers exhibited slight hepatocyte cytoplasmic clearing within periportal regions and inflammatory cell infiltration (Figure 6A). In contrast, day 9 Tiparpfl/flCreAlb livers were characterized by inflammatory infiltration and a predominant microvesicular steatosis. Six days after dioxin treatment, livers from Tiparpfl/fl mice displayed distinct inflammatory cell infiltration and increased clearing of the cytoplasm with the appearance of large vacuoles within hepatocytes. Similar findings were observed in livers isolated from dioxin-treated Tiparp+/+ and TiparpEx3−/− mice (Supplementary Figure 2A).
Figure 6.
Increased liver inflammation and cytokine levels in dioxin-treated Tiparpfl/flCreAlb compared with Tiparpfl/fl mice. A, Representative H&E staining of livers from Tiparpfl/fl and Tiparpfl/flCreAlb mice (n = 4). Control animals were injected with CO and were euthanized on day 9. The asterisks (*) indicate focal inflammatory infiltration, and the arrowheads indicate microvesicular steatosis. All images are to the same scale. Hepatic (B) Il6, (C) Serpine 1, (D) Cxcl2, and (E) F4/80 mRNA levels were determined as described in the methods. Data represent the mean ± SEM (n = 4). *p < .05 2-way ANOVA compared with genotyped-matched control treated mice. #P < 0.05 2-way ANOVA compared with dioxin-treated Tiparpfl/fl mice.
We next determined the mRNA levels of AHR-regulated cytokines and the macrophage marker F4/80 (Casado et al., 2011; Matsubara et al., 2012). Hepatic interleukin 6 (Il6) levels were unaffected by dioxin treatment in both genotypes (Figure 6B). However, dioxin-treated Tiparpfl/flCreAlb mice had increased hepatic expression of Serpine 1 (also known as plasminogen activator inhibitor-1; PAI-1), chemokine (C-X-C motif) ligand 2 (Cxcl2) and F4/80 when compared with Tiparpfl/fl mice (Figs. 6C–E). Similar findings were also seen in livers isolated from dioxin-treated Tiparp+/+ and TiparpEx3−/− mice (Supplementary Figs. 2B–E). The increased cytokine and F4/80 levels indicated increased hepatic inflammation in dioxin-treated Tiparpfl/flCreAlb compared with WT mice.
Hepatocyte-Specific Loss of TIPARP Increases Dioxin-Induced Steatohepatitis
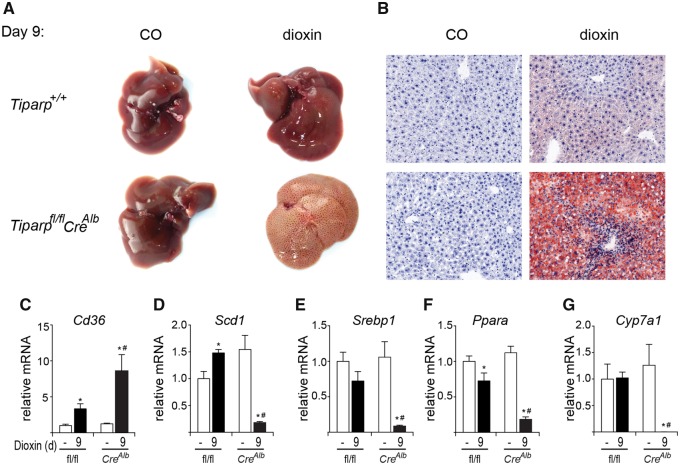
Livers from vehicle-treated Tiparpfl/fl and Tiparpfl/flCreAlb mice were macroscopically normal (Figure 7A). Livers from Tiparpfl/fl mice were enlarged but only slightly pale in color 9 days after dioxin treatment (Figure 7A). Livers from dioxin-treated Tiparpfl/flCreAlb mice were markedly pale in color at 9 days, suggesting a high level of lipid accumulation. We tested for the presence of neutral lipids by Oil-Red-O staining (Figure 7B). Livers from vehicle-exposed Tiparpfl/fl and Tiparpfl/flCreAlb mice were negative for Oil-Red-O staining. On day 9 after dioxin treatment, small droplets of lipid were seen in the livers of Tiparpfl/fl mice, while those from similarly treated Tiparpfl/flCreAlb mice had substantial intracytoplasmic lipid accumulation. Comparable findings were observed in livers isolated from dioxin-treated Tiparp+/+ and TiparpEx3−/− mice (Supplementary Figs. 3A and 3B).
Figure 7.
Dioxin-induced steatosis is increased in Tiparpfl/flCreAlb mice. (A) Livers from male Tiparpfl/fl and Tiparpfl/flCreAlb mice given a single i.p. injection of CO or 10 µg/kg dioxin and euthanized after 9 days (n = 5). B, Oil-Red-O and hematoxylin stained liver sections from Tiparpfl/fl and Tiparpfl/flCreAlb mice. All images are to the same scale. Hepatic mRNA levels of Cd36 (C), Scd1 (D), Srebp1 (E), Ppara (F), and Cyp7a1 (G) were determined as described in the methods. Data represent the mean ± SEM (n = 4). For all data, p < .05 was determined by 2-way ANOVA followed by Tukey’s post hoc test comparison. Significantly different compared with genotype-matched *DMSO- or #dioxin-treated Tiparpfl/fl mice.
We then analyzed the hepatic levels of transcripts encoding genes involved in lipid uptake, lipogenesis and cholesterol/bile acid metabolism. Consistent with previous studies (Lee et al., 2010; Lu et al., 2011), the lipid uptake transporter, scavenger receptor encoded by cluster of differentiation 36 (Cd36), was increased 3-fold by dioxin treatment in Tiparpfl/fl mice and to a greater extent (8-fold) in similarly treated Tiparpfl/flCreAlb mice (Figure 7C). Hepatic expression of lipogenic genes including sterol regulatory element-binding transcription factor 1 (Srebp1), and stearoyl-CoA desaturase (Scd1) were significantly decreased in dioxin treated Tiparpfl/flCreAlb mice compared with Tiparpfl/fl mice (Figs. 7D and 7E). Peroxisome proliferator activating receptor α (Ppara) and Cyp7a1, the rate limiting enzyme bile acid synthesis, were also significantly decreased in Tiparpfl/flCreAlb mice compared with Tiparpfl/fl mice (Figs. 7F and 7G). Similar findings were seen in livers isolated from dioxin-treated Tiparp+/+ and TiparpEx3−/− mice (Supplementary Figs. 3C–G). These data suggest that the increased sensitivity of Tiparpfl/flCreAlb and TiparpEx3−/− mice to dioxin-induced steatohepatitis is due to increased lipid uptake rather than increased hepatic lipogenesis.
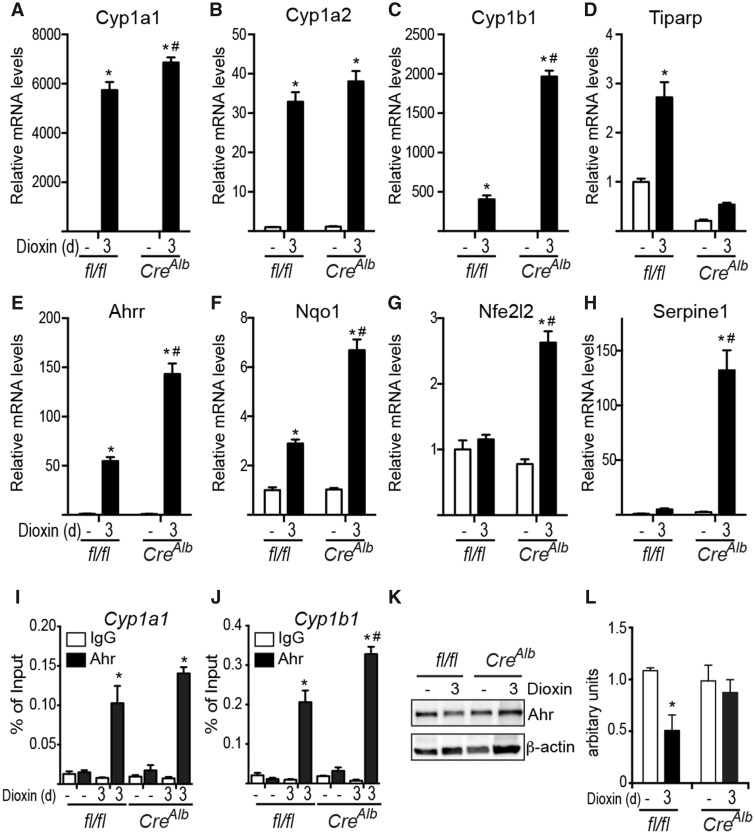
Increased AHR Regulated Gene Expression in Tiparpfl/flCreAlb Mice After Treatment With 10 μg/kg Dioxin
To identify AHR genes and/or changes in metabolites that might provide insight into the molecular mechanisms regulating the increased dioxin sensitivity of Tiparpfl/flCreAlb mice, we analyzed changes in dioxin-induced hepatic mRNA and metabolite levels 3 days after dioxin treatment. Day 3 was chosen because we observed significant increases in serum ALT activity in Tiparpfl/flCreAlb mice, and we reasoned that this time point could be used to identify early changes in AHR target gene expression and/or metabolite levels prior to more severe toxicities that ultimately led to death. Tiparpfl/flCreAlb mice treated with 10 μg/kg dioxin exhibited increased mRNA expression levels of many AHR target genes including Cyp1a1, Cyp1a2, Ahrr, Nqo1, Nfe2l2 and Serpine1 compared with similarly treated Tiparpfl/fl mice (Figs. 8A and 8C–H). Cyp1a2 expression was slightly increased in Tiparpfl/flCreAlb compared with Tiparpfl/fl mice, but this difference was not statistically significant (Figure 8B). No significant increase in AHR recruitment to Cyp1a1 was observed (Figure 8I). Significantly higher levels of AHR were recruited to Cyp1b1 in liver extracts from Tiparpfl/flCreAlb mice compared with Tiparpfl/fl mice (Figure 8J). Consistent with our previous study, we observed reduced dioxin-induced proteolytic degradation of total AHR protein in liver extracts from Tiparpfl/flCreAlb mice compared with Tiparpfl/fl mice (Figure 8K).
Figure 8.
Hepatocyte-specific loss of TIPARP increases dioxin-dependent regulation of hepatic AHR target gene expression. Hepatic mRNA levels of Cyp1a1 (A), Cyp1a2 (B), Cyp1b1 (C), Tiparp (D), Ahrr (E), Nqo1 (F) Nfe2l2 (G) Serpine 1 (H) were determined after 3 days of exposure to CO or 10 μg/kg dioxin as described in experimental procedures (n = 4). Recruitment of AHR to Cyp1a1 (I) and Cyp1b1 (J). Data represent the mean ± SEM. Representative AHR (K), and β-actin protein levels were detected by Western blotting after 3 days of treatment. AHR proteins levels were normalized to β-actin levels, n = 4) (L). For all data, p < .05 was determined by 2-way ANOVA followed by Tukey’s post hoc test comparison. Significantly different compared with genotype-matched *DMSO- or #dioxin-treated Tiparpfl/fl mice.
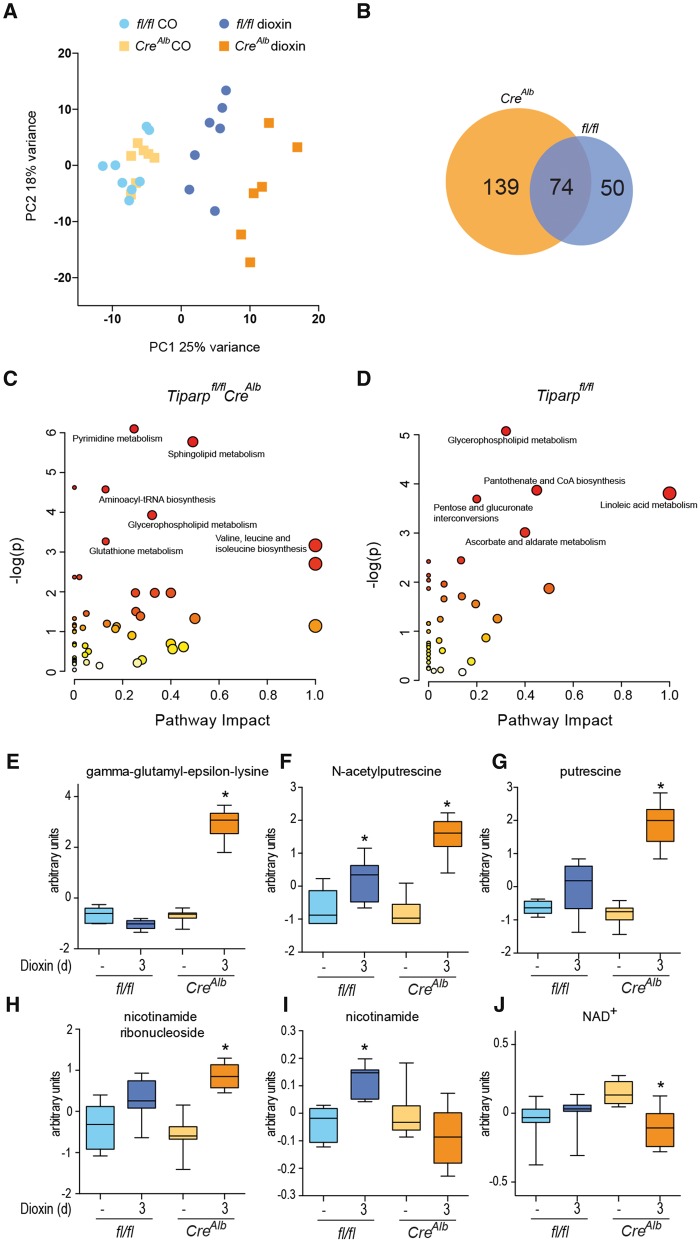
We next did comparative metabolomic analyses on liver extracts from Tiparpfl/flCreAlb and Tiparpfl/fl mice. Principal component analysis (PCA) revealed significant treatment-based separations among the samples, with clear distinctions between the vehicle- and dioxin-treated animals in each genotype. Separations were also apparent between the dioxin-treated Tiparpfl/fl and Tiparpfl/flCreAlb animals (Figure 9A). Of the total of 679 named metabolites examined, 213 were significantly altered (P < 0.05) by dioxin treatment of Tiparpfl/flCreAlb mice compared with 124 in similarly treated Tiparpfl/fl mice (Table 1; Supplementary Tables 2 and 3). Of the 124 metabolites, 74 overlapped with those identified in dioxin-treated Tiparpfl/flCreAlb mice (Figure 9B). Only 7 metabolites were significantly changed in vehicle-treated Tiparpfl/flCreAlb compared with Tiparpfl/fl mice, showing that the loss of TIPARP had little effect on basal liver metabolism (Supplementary Table 4). Consistent with previous studies, dioxin treatment resulted in significant lipidomic changes, with accumulation of several classes of free fatty acids and metabolites linked to complex lipid homeostasis (Nault et al., 2016b). Increased lipids were observed in both the Tiparpfl/fl and Tiparpfl/flCreAlb animals, with a few classes of lipids that were differentially expressed in the 2 groups (Supplementary Tables 2 and 3). These included certain long-chain acylcarnitines, fatty acid dicarboxylates, and complex lipids such as plasmalogens and sphingolipids. However, 129 metabolites were altered in dioxin-treated Tiparpfl/flCreAlb compared with Tiparpfl/fl mice (Table 1; Supplementary Table 5). Altered metabolite levels in Tiparpfl/flCreAlb and Tiparpfl/fl mice were analyzed for pathway overrepresentation (enrichment) and connectivity within related metabolites (impact) using MetaboAnalyst (Xia and Wishart, 2016) (Figs. 9C and 9D). Only glycerophospholipid metabolism was common among the top 5 or 6 significant pathways (Supplementary Tables 6 and 7). Dioxin-treated Tiparpfl/flCreAlb mice also differed with regard to increased γ-glutamyl-ε-lysine levels (12.5-fold; Figure 9E), which may reflect increased transglutaminase activity, and polyamine metabolism (N-acetylputrescine; Figure 9F, and putrescine; Figure 9G). Because dioxin toxicity has been tightly linked with NAD+ levels, its precursors and metabolites, and because TIPARP activity is dependent on NAD+, we examined NAD+ metabolism (Diani-Moore et al., 2010, 2017; He et al., 2013). Dioxin-dependent increases in nicotinamide ribonucleoside (Figure 9H) and nicotinamide (Figure 9I) were observed in Tiparpfl/flCreAlb and Tiparpfl/fl mice, respectively. Significant decreases in intrahepatic NAD+ levels were only observed in dioxin-treated Tiparpfl/flCreAlb mice (Figure 9J).
Figure 9.
Dioxin-induced hepatic metabolomic disruption. A, PCA of hepatic metabolomic analysis after 3-day treatment of Tiparpfl/fl (fl/fl) and Tiparpfl/flCreAlb with CO or 10 μg/kg dioxin. B, Venn diagram of the overlapping metabolites that were significantly altered between dioxin treated Tiparpfl/fl and Tiparpfl/flCreAlb. Metabolic pathway enrichment analysis of altered hepatic metabolites (p < .05) in dioxin-treated Tiparpfl/flCreAlb (C) and Tiparpfl/fl mice (D). Hepatic levels of gamma-glutamyl-epsilon lysine (E), N-acetylputrescine (F), putrescine (G), nicotinamde ribonucleoside (H), nicotinamide (I), and NAD+ (J). Data represent the mean ± SEM (n = 6–8).
Table 1.
A Summary of the Numbers of Biochemicals That Achieved Statistical Significance (p ≤ .05) Among the Different Comparisons
| Statistical Comparisons |
||||
|---|---|---|---|---|
| ANOVA contrasts | ||||
| Total biochemicals (p< .05) | 124 | 213 | 7 | 129 |
| Biochemicals (up | down ) | 75 | 49 | 163 | 50 | 4 | 3 | 111 | 18 |
DISCUSSION
We previously reported that TIPARP acts as part of negative feedback loop to regulate AHR activity, and that global loss of TIPARP expression increases sensitivity to dioxin-induced toxicities such as steatohepatitis and wasting syndrome (Ahmed et al., 2015; MacPherson et al., 2013). Since hepatocyte-specific deletion of AHR prevents dioxin-induced hepatotoxicity, we reasoned that hepatocyte-specific deletion of TIPARP would result in increased dioxin-induced hepatotoxicity. We therefore generated a hepatocyte-specific TIPARP deletion (Tiparpfl/flCreAlb) mouse strain. We also generated a whole-body knockout TIPARP (TiparpEx3−/−) strain in which Tiparp is deleted by the removal of exon 3, making it distinct from other TIPARP null lines (Ahmed et al., 2015; Kozaki et al., 2017; Schmahl et al., 2007). Here we show that TiparpEx3−/− and Tiparpfl/flCreAlb mice are both more sensitive than WT mice to dioxin-induced hepatotoxicity and lethality. These findings provide further support for the importance of TIPARP in AHR signaling and for its role in protecting against dioxin-induced toxicity (Ahmed et al., 2015; Matthews, 2017), as well as demonstrating that the expression of TIPARP in hepatocytes plays a key role in the manifestations of this toxicity.
Tiparpfl/flCreAlb mice treated with dioxin lost significant body weight without any reduction in food intake. Hepatic glycogen and epididymal WAT levels were also reduced, pointing to a possible deficiency in the efficiency of intestinal nutrient absorption resulting in altered metabolism. Dioxin-induced hypophagia contributes to body weight and adipose tissue loss in many species, but numerous studies using pair-feeding or total parenteral nutrition have failed to identify a single explanation for the weight loss (Linden et al., 2010; Seefeld et al., 1984). The severe hepatotoxicity and extensive hepatosteatosis in dioxin-treated Tiparpfl/flCreAlb and TiparpEx3−/− mice, would cause impaired liver function that could result in reduced intestinal nutrient absorption and impaired liver homeostasis (Kalaitzakis, 2014).
Dioxin-treated Tiparpfl/flCreAlb and TiparpEx3−/− mice exhibit many of the alterations in lipid homeostasis, increased hepatic inflammation and other toxic endpoints that have been reported in other studies (Boverhof et al., 2006; Duval et al., 2017). Tiparpfl/flCreAlb and TiparpEx3−/− mice exhibited increased hepatosteatosis due to increased expression of genes regulating lipid uptake (Cd36), but decreased expression of those involved in fatty acid β-oxidation and de novo lipogenesis (Scd1, Srebp1, Ppara) (Ahmed et al., 2015; Duval et al., 2017; Lee et al., 2010). Scd1 expression was increased in Tiparpfl/fl mice, but decreased in Tiparpfl/flCreAlb mice. One possible explanation is that the increased hepatosteatosis in Tiparpfl/flCreAlb mice results in negative regulation of Scd1 by other factors or hormones (Mauvoisin and Mounier, 2011). Srebp1 expression levels were decreased in Tiparpfl/flCreAlb and TiparpEx3−/− mice, supporting findings from a recent high-dose dioxin exposure study (Duval et al., 2017). Srebp1 expression is positively regulated by the liver X receptor (LXR) and PPARα. TIPARP is an LXR coactivator, so the loss of Tiparp expression combined with the reduced PPARα expression levels could also contribute to reduced SREBP1 levels (Bindesboll et al., 2016).
CYP7A1 plays a critical role in the control of bile acid and cholesterol homeostasis as the rate-limiting enzyme in the classic bile acid synthesis pathway (Gupta et al., 2001). Overexpression of mouse CYP7A1 protects against high-fat diet induced obesity, fatty liver and insulin resistance (Li et al., 2010), whereas in humans, genetic deficiency of CYP7A1 leads to hyperlipidemia (Pullinger et al., 2002). The decrease in CYP7A1 expression levels in treated Tiparpfl/flCreAlb and TiparpEx3−/− mice is in agreement with studies of male C57BL/6 mice treated with 0.01 to 30 μg/kg dioxin every 4 for 28 days (Fader et al., 2017), and could be a contributing factor to the increased hepatosteatosis and reduced WAT levels observed, through reduced lipid absorption resulting from altered bile acid homeostasis (Fader et al., 2017). In support of this, we observed increased hepatic levels of taurochenodeoxycholic acid and taurocholic acid in dioxin-treated Tiparpfl/flCreAlb mice but not in Tiparpfl/fl mice.
Metabolomic profiling studies identified significant changes in metabolite levels in dioxin-treated Tiparpfl/flCreAlb mice compared with Tiparpfl/fl mice. Many of the major treatment-based changes were conserved in the 2 cohorts (ie, accumulated fatty acids, changes related to nucleotides and polyamines). However, the extent of change of the affected metabolites differed between the Tiparpfl/fl and Tiparpfl/flCreAlb groups. Dioxin-treated Tiparpfl/flCreAlb mice, however, exhibited significant increases in gamma-glutamyl-epsilon lysine levels compared with their untreated counterparts or dioxin-treated Tiparpfl/fl mice. This metabolite is formed by tissue transglutaminase (TG2), which catalyzes crosslinks between glutamine and lysine residues of proteins (Iismaa et al., 2009). TG2 activity increases following acute and chronic liver injury, and aberrant TG2 activation has been implicated in the development of fibrosis and cancer (Iismaa et al., 2009). In contrast, retinoid-induced TG2 mRNA up-regulation is reduced by dioxin treatment in a human squamous cell carcinoma cell line (Krig and Rice, 2000). The increased γ-glutamyl-ε-lysine observed in Tiparpfl/flCreAlb mice suggests that TIPARP might influence TG2 activity following dioxin-induced liver damage.
We observed that the expression of most of the AHR target genes examined was increased in response to dioxin in Tiparpfl/flCreAlb and Tiparpfl/fl compared with WT mice, including Cxcl2 (macrophage inflammation protein-2) and Serpine1 genes (Son and Rozman, 2002). CXCL2 is a member of the CXC subfamily of chemokines that are crucial for neutrophil recruitment to sites of inflammation following hepatic injury (Marra and Tacke, 2014). Levels of PAI-1, the product of Serpine1 gene, were higher in Tiparpfl/flCreAlb mice than in Tiparpfl/fl mice. PAI-1 is a physiologic inhibitor of plasminogen activators that regulate fibrosis via regulation of the extracellular matrix. PAI-1 expression is increased in dioxin-induced fibrosis (Nault et al., 2016a).
The mRNA levels of AHR repressor (AHRR), a negative regulator of AHR (Mimura et al., 1999), were increased in Tiparpfl/flCreAlb compared with Tiparpfl/fl mice. AHRR is a potent inhibitor of AHR activity in vitro (Karchner et al., 2009) that exhibits gene- and tissue-specific inhibition of AHR signaling in mice (Hosoya et al., 2008). Although the effect of Ahrr loss on dioxin-induced wasting syndrome has not been reported, AHRR transgenic mice are protected from dioxin-induced lethality and hepatotoxicity (Vogel et al., 2016).
Increased TIPARP and PARP1 activity have been proposed to be important in augmenting dioxin toxicity through the depletion of NAD+. Indeed, NAD+ repletion or treatment with the pan-PARP inhibitor PJ34 can prevent dioxin-induced thymic atrophy and hepatosteatosis in a chicken embryo model (Diani-Moore et al., 2017). However, we observed decreased hepatic NAD+ levels in dioxin-treated Tiparpfl/flCreAlb mice, suggesting that PARP1 or an NAD+-consuming enzyme other than TIPARP is responsible for the reduced NAD+ levels after dioxin treatment in mice. Another possible explanation is that there are species differences in the AHR-TIPARP signaling axis, such that TIPARP protects against dioxin toxicity in mice but enhances it in avian species. Further studies using gene targeting methods to delete TIPARP in nonmurine models are needed to explain these discrepancies.
AHR is also a key regulator of gut homeostasis, inflammation, immunity and T-cell differentiation (Stockinger et al., 2014). AHR is required for the maintenance of intraepithelial lymphocytes, which are the first line of immune defense in the intestine. Loss of AHR or reduced exposure to dietary AHR ligands compromises these cells, leading to increased microbial load, immune activation and epithelial damage (Li et al., 2011). Moreover, AHR activation by dietary indoles (indole-3-carbinol) improves colitis and protects against experimental autoimmune encephalomyelitis, a murine model of multiple sclerosis (Lamas et al., 2016; Li et al., 2011; Monteleone et al., 2011; Rouse et al., 2013). An unanswered question is whether TIPARP also regulates endogenous AHR signaling and if so, how would its loss affect the protective role of the AHR signaling pathway in models of inflammatory disease? Kynurenine, an endogenous AHR ligand, was reported to repress type-I interferon responses during viral infection in an AHR- and TIPARP-dependent manner, supporting the notion that TIPARP has a broad role in AHR biology and regulates endogenous ligand-induced AHR activation (Yamada et al., 2016).
In summary, we provide evidence from 2 additional mouse models that the loss of TIPARP expression increases sensitivity to dioxin toxicity and lethality. Hepatocyte-specific loss of TIPARP is sufficient to increase sensitivity to dioxin hepatotoxicity, steatosis and lethality, highlighting the importance of liver damage in the dioxin-induced wasting syndrome. Our results provide further support for the importance of the AHR-TIPARP axis in regulating dioxin toxicity, and potentially in regulating the biological actions of AHR following its activation by endogenous or dietary ligands.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
FUNDING
This work was supported by Canadian Institutes of Health Research (CIHR) operating grants (MOP-494265 and MOP-125919), CIHR New Investigator Award, an Early Researcher Award from the Ontario Ministry of Innovation (ER10-07-028), an unrestricted research grant from the DOW Chemical Company, the Johan Throne Holst Foundation, Novo Nordic Foundation and the Norwegian Cancer Society to J.M.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank all members of the Matthews and Grant laboratories for their help with the preparation of the article.
REFERENCES
- Ahmed S., Bott D., Gomez A., Tamblyn L., Rasheed A., MacPherson L., Sugamori K. S., Cho T., Yang Y., Grant D. M., et al. (2015). Loss of the Mono-ADP-Ribosyltransferase, TIPARP, Increases Sensitivity to Dioxin-Induced Steatohepatitis and Lethality. J. Biol. Chem. 290, 16824–16840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindesboll C., Tan S., Bott D., Cho T., Tamblyn L., MacPherson L., Gronning-Wang L. M., Nebb H. I., Matthews J. (2016). TCDD-inducible poly-ADP-ribose polymerase (TIPARP/PARP7) mono-ADP-ribosylates and coactivates liver X receptors. Biochem. J. 473, 899–910. [DOI] [PubMed] [Google Scholar]
- Birnbaum L. S. (1994). Endocrine effects of prenatal exposure to PCBs, dioxins, and other xenobiotics: Implications for policy and future research. Environ. Health Perspect. 102, 676–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum L. S. (1995). Developmental effects of dioxins and related endocrine disrupting chemicals. Toxicol. Lett. 82-83, 743–750. [DOI] [PubMed] [Google Scholar]
- Boverhof D. R., Burgoon L. D., Tashiro C., Sharratt B., Chittim B., Harkema J. R., Mendrick D. L., Zacharewski T. R. (2006). Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol. Sci. 94, 398–416. [DOI] [PubMed] [Google Scholar]
- Casado F. L., Singh K. P., Gasiewicz T. A. (2011). Aryl hydrocarbon receptor activation in hematopoietic stem/progenitor cells alters cell function and pathway-specific gene modulation reflecting changes in cellular trafficking and migration. Mol Pharmacol 80, 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. S., Nagy S. R. (2003). Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43, 309–334. [DOI] [PubMed] [Google Scholar]
- Diani-Moore S., Ram P., Li X., Mondal P., Youn D. Y., Sauve A. A., Rifkind A. B. (2010). Identification of the aryl hydrocarbon receptor target gene TiPARP as a mediator of suppression of hepatic gluconeogenesis by 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin and of nicotinamide as a corrective agent for this effect. J. Biol. Chem. 285, 38801–38810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diani-Moore S., Shoots J., Singh R., Zuk J. B., Rifkind A. B. (2017). NAD(+) loss, a new player in AhR biology: Prevention of thymus atrophy and hepatosteatosis by NAD(+) repletion. Sci. Rep. 7, 2268.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval C., Teixeira-Clerc F., Leblanc A. F., Touch S., Emond C., Guerre-Millo M., Lotersztajn S., Barouki R., Aggerbeck M., Coumoul X. (2017). Chronic exposure to low doses of dioxin promotes liver fibrosis development in the C57BL/6J diet-induced obesity mouse model. Environ. Health Perspect. 125, 428–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader K. A., Nault R., Zhang C., Kumagai K., Harkema J. R., Zacharewski T. R. (2017). 2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin (TCDD)-elicited effects on bile acid homeostasis: Alterations in biosynthesis, enterohepatic circulation, and microbial metabolism. Sci. Rep. 7, 5921.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salguero P. M., Hilbert D. M., Rudikoff S., Ward J. M., Gonzalez F. J. (1996). Aryl-hydrocarbon receptor-deficient mice are resistant to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 140, 173–179. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stravitz R. T., Dent P., Hylemon P. B. (2001). Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J. Biol. Chem. 276, 15816–15822. [DOI] [PubMed] [Google Scholar]
- He J., Hu B., Shi X., Weidert E. R., Lu P., Xu M., Huang M., Kelley E. E., Xie W. (2013). Activation of the aryl hydrocarbon receptor sensitizes mice to nonalcoholic steatohepatitis by deactivating mitochondrial sirtuin deacetylase Sirt3. Mol. Cell Biol. 33, 2047–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T., Harada N., Mimura J., Motohashi H., Takahashi S., Nakajima O., Morita M., Kawauchi S., Yamamoto M., Fujii-Kuriyama Y. (2008). Inducibility of cytochrome P450 1A1 and chemical carcinogenesis by benzo[a]pyrene in AhR repressor-deficient mice. Biochem. Biophys. Res. Commun. 365, 562–567. [DOI] [PubMed] [Google Scholar]
- Hottiger M. O., Hassa P. O., Luscher B., Schuler H., Koch-Nolte F. (2010). Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 35, 208–219. [DOI] [PubMed] [Google Scholar]
- Iismaa S. E., Mearns B. M., Lorand L., Graham R. M. (2009). Transglutaminases and disease: Lessons from genetically engineered mouse models and inherited disorders. Physiol. Rev. 89, 991–1023. [DOI] [PubMed] [Google Scholar]
- Kalaitzakis E. (2014). Gastrointestinal dysfunction in liver cirrhosis. World J. Gastroenterol. 20, 14686–14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner S. I., Jenny M. J., Tarrant A. M., Evans B. R., Kang H. J., Bae I., Sherr D. H., Hahn M. E. (2009). The active form of human aryl hydrocarbon receptor (AHR) repressor lacks exon 8, and its Pro 185 and Ala 185 variants repress both AHR and hypoxia-inducible factor. Mol. Cell Biol. 29, 3465–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki T., Komano J., Kanbayashi D., Takahama M., Misawa T., Satoh T., Takeuchi O., Kawai T., Shimizu S., Matsuura Y., et al. (2017). Mitochondrial damage elicits a TCDD-inducible poly(ADP-ribose) polymerase-mediated antiviral response. Proc. Natl. Acad. Sci. U.S.A. 114, 2681–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus W. L., Hottiger M. O. (2013). PARP-1 and gene regulation: Progress and puzzles. Mol. Aspects Med. 34, 1109–1123. [DOI] [PubMed] [Google Scholar]
- Krig S. R., Rice R. H. (2000). TCDD suppression of tissue transglutaminase stimulation by retinoids in malignant human keratinocytes. Toxicol. Sci. 56, 357–364. [DOI] [PubMed] [Google Scholar]
- Lamas B., Richard M. L., Leducq V., Pham H. P., Michel M. L., Da Costa G., Bridonneau C., Jegou S., Hoffmann T. W., Natividad J. M., et al. (2016). CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Wada T., Febbraio M., He J., Matsubara T., Lee M. J., Gonzalez F. J., Xie W. (2010). A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology 139, 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Owsley E., Matozel M., Hsu P., Novak C. M., Chiang J. Y. (2010). Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology 52, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Innocentin S., Withers D. R., Roberts N. A., Gallagher A. R., Grigorieva E. F., Wilhelm C., Veldhoen M. (2011). Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147, 629–640. [DOI] [PubMed] [Google Scholar]
- Linden J., Lensu S., Tuomisto J., Pohjanvirta R. (2010). Dioxins, the aryl hydrocarbon receptor and the central regulation of energy balance. Front. Neuroendocrinol. 31, 452–478. [DOI] [PubMed] [Google Scholar]
- Lo R., Celius T., Forgacs A., Dere E., MacPherson L., Zacharewski T., Matthews J. (2011). Identification of aryl hydrocarbon receptor binding targets in mouse hepatic tissue treated with 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Toxicol. Appl. Pharmacol. doi: 10.1016/j.taap.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Lu H., Cui W., Klaassen C. D. (2011). Nrf2 protects against 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD)-induced oxidative injury and steatohepatitis. Toxicol. Appl. Pharmacol. 256, 122–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Baldwin K. T., Renzelli A. J., McDaniel A., Dong L. (2001). TCDD-inducible poly(ADP-ribose) polymerase: A novel response to 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 289, 499–506. [DOI] [PubMed] [Google Scholar]
- MacPherson L., Tamblyn L., Rajendra S., Bralha F., McPherson J. P., Matthews J. (2013). 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 41, 1604–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra F., Tacke F. (2014). Roles for chemokines in liver disease. Gastroenterology 147, 577–594 e1. [DOI] [PubMed] [Google Scholar]
- Matsubara T., Tanaka N., Krausz K. W., Manna S. K., Kang D. W., Anderson E. R., Luecke H., Patterson A. D., Shah Y. M., Gonzalez F. J. (2012). Metabolomics identifies an inflammatory cascade involved in dioxin- and diet-induced steatohepatitis. Cell Metab. 16, 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J. (2017). AHR Toxicity and Signalling: Role of TIPARP and ADP-ribosylation. Curr. Opin. Toxicol. 2, 50–57. [Google Scholar]
- Mauvoisin D., Mounier C. (2011). Hormonal and nutritional regulation of SCD1 gene expression. Biochimie 93, 78–86. [DOI] [PubMed] [Google Scholar]
- Mimura J., Ema M., Sogawa K., Fujii-Kuriyama Y. (1999). Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev. 13, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone I., Rizzo A., Sarra M., Sica G., Sileri P., Biancone L., MacDonald T. T., Pallone F., Monteleone G. (2011). Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 141, 237–248, 248 e1. [DOI] [PubMed] [Google Scholar]
- Moura-Alves P., Fae K., Houthuys E., Dorhoi A., Kreuchwig A., Furkert J., Barison N., Diehl A., Munder A., Constant P., et al. (2014). AhR sensing of bacterial pigments regulates antibacterial defence. Nature 512, 387–392. [DOI] [PubMed] [Google Scholar]
- Nault R., Fader K. A., Ammendolia D. A., Dornbos P., Potter D., Sharratt B., Kumagai K., Harkema J. R., Lunt S. Y., Matthews J., et al. (2016a). Dose-dependent metabolic reprogramming and differential gene expression in TCDD-elicited hepatic fibrosis. Toxicol. Sci. 154, 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault R., Fader K. A., Kirby M. P., Ahmed S., Matthews J., Jones A. D., Lunt S. Y., Zacharewski T. R. (2016b). Pyruvate Kinase Isoform Switching and Hepatic Metabolic Reprogramming by the Environmental Contaminant 2, 3, 7, 8-Tetrachlorodibenzo-p-Dioxin. Toxicol. Sci. 149, 358–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjanvirta R., Tuomisto J. (1994). Short-term toxicity of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin in laboratory animals: Effects, mechanisms, and animal models. Pharmacol. Rev. 46, 483–549. [PubMed] [Google Scholar]
- Poland A., Knutson J. C. (1982). 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: Examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 22, 517–554. [DOI] [PubMed] [Google Scholar]
- Poland A., Palen D., Glover E. (1994). Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol. Pharmacol. 46, 915–921. [PubMed] [Google Scholar]
- Pullinger C. R., Eng C., Salen G., Shefer S., Batta A. K., Erickson S. K., Verhagen A., Rivera C. R., Mulvihill S. J., Malloy M. J., et al. (2002). Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J. Clin. Invest. 110, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana F. J., Basso A. S., Iglesias A. H., Korn T., Farez M. F., Bettelli E., Caccamo M., Oukka M., Weiner H. L. (2008). Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71. [DOI] [PubMed] [Google Scholar]
- Rouse M., Singh N. P., Nagarkatti P. S., Nagarkatti M. (2013). Indoles mitigate the development of experimental autoimmune encephalomyelitis by induction of reciprocal differentiation of regulatory T cells and Th17 cells. Br. J. Pharmacol. 169, 1305–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahl J., Raymond C. S., Soriano P. (2007). PDGF signaling specificity is mediated through multiple immediate early genes. Nat. Genet. 39, 52–60. [DOI] [PubMed] [Google Scholar]
- Seefeld M. D., Corbett S. W., Keesey R. E., Peterson R. E. (1984). Characterization of the wasting syndrome in rats treated with 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Toxicol. Appl. Pharmacol. 73, 311–322. [DOI] [PubMed] [Google Scholar]
- Son D. S., Rozman K. K. (2002). 2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin (TCDD) induces plasminogen activator inhibitor-1 through an aryl hydrocarbon receptor-mediated pathway in mouse hepatoma cell lines. Arch. Toxicol. 76, 404–413. [DOI] [PubMed] [Google Scholar]
- Stevens E. A., Mezrich J. D., Bradfield C. A. (2009). The aryl hydrocarbon receptor: A perspective on potential roles in the immune system. Immunology 127, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B., Di Meglio P., Gialitakis M., Duarte J. H. (2014). The aryl hydrocarbon receptor: Multitasking in the immune system. Annu. Rev. Immunol. 32, 403.. [DOI] [PubMed] [Google Scholar]
- Vogel C. F., Chang W. L., Kado S., McCulloh K., Vogel H., Wu D., Haarmann-Stemmann T., Yang G., Leung P. S., Matsumura F., et al. (2016). Transgenic overexpression of aryl hydrocarbon receptor repressor (AhRR) and AhR-mediated induction of CYP1A1, cytokines, and acute toxicity. Environ. Health Perspect. 124, 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walisser J. A., Glover E., Pande K., Liss A. L., Bradfield C. A. (2005). Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc. Natl. Acad. Sci. U.S.A. 101, 16677–16682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr. (1999). Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 39, 103–125. [DOI] [PubMed] [Google Scholar]
- Xia J., Wishart D. S. (2016). Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics 55, 14.10.1.. [DOI] [PubMed] [Google Scholar]
- Yamada T., Horimoto H., Kameyama T., Hayakawa S., Yamato H., Dazai M., Takada A., Kida H., Bott D., Zhou A. C., et al. (2016). Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat. Immunol. 17, 687–694. [DOI] [PubMed] [Google Scholar]
- Zhang L., Savas U., Alexander D. L., Jefcoate C. R. (1998). Characterization of the mouse Cyp1B1 gene. Identification of an enhancer region that directs aryl hydrocarbon receptor-mediated constitutive and induced expression. J. Biol. Chem. 273, 5174–5183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.