Abstract
Globally, the microbe Neisseria gonorrhoeae (NG) causes 106 million newly documented sexually transmitted infections each year. Once appropriately diagnosed, NG infections can be readily treated with antibiotics, but high-risk patients often do not return to the clinic for treatment if results are not provided at the point of care. A rapid, sensitive molecular diagnostic would help increase NG treatment and reduce the prevalence of this sexually transmitted disease. Here, we report on the design and development of a rapid, highly sensitive, paperfluidic device for point-of-care diagnosis of NG. The device integrates patient swab sample lysis, nucleic acid extraction, thermophilic helicase-dependent amplification (tHDA), an internal amplification control (NGIC), and visual lateral flow detection within an 80 min run time. Limits of NG detection for the NG/NGIC multiplex tHDA assay were determined within the device, and clinical performance was validated retroactively against qPCR-quantified patient samples in a proof-of-concept study. This paperfluidic diagnostic has a clinically relevant limit of detection of 500 NG cells per device with analytical sensitivity down to 10 NG cells per device. In triplicate testing of 40 total urethral and vaginal swab samples, the device had 95% overall sensitivity and 100% specificity, approaching current laboratory-based molecular NG diagnostics. This diagnostic platform could increase access to accurate NG diagnoses to those most in need.
Keywords: Neisseria gonorrhoeae, point of care, molecular diagnostic, thermophilic helicase dependent amplification, isothermal amplification, internal amplification control, vaginal swab, urethral swab, paperfluidic
1. Introduction
Sexually transmitted infections (STIs) caused by Neisseria gonorrhoeae (NG) account for 106 million newly documented STI cases each year (World Health Organization 2012). In low-resource settings, many NG infections remain untreated, both because they can remain asymptomatic for years and because rapid and sensitive diagnostics are still lacking (Aledort et al. 2006; Bourgeois et al. 1998; World Health Organization 2007). Untreated gonorrhea infections can cause pelvic inflammatory disease, infertility, and ectopic pregnancy among women, and additional complications via neonatal transmission. NG infections also facilitate HIV transmission (Aledort et al. 2006; Fleming and Wasserheit 1999); people with NG infections are up to five times more likely to contract HIV (Wasserheit 1992), and are three times more likely to transmit HIV to someone else (Fleming and Wasserheit 1999). Once diagnosed, this serious but treatable infection is often cured with a one-time dose of appropriate antibiotics. However, diagnosis through current state-of-the-art methods, such as swab sample culturing or PCR, require an equipped laboratory, trained staff, and several days or even weeks to complete.
Due to long turnaround times, patients often fail to return for laboratory diagnostic results and follow up treatment. As a result, the United States Centers for Disease Control urges clinics to use point-of-care (POC) tests to ensure that high-risk patients receive treatment in the same visit (Johnson et al. 2002). Despite some progress in developing immunoassay based rapid diagnostic tests (RDTs) and near-patient qPCR systems, a POC NG diagnostic that combines high specificity and sensitivity with low-cost is not yet available (Gaydos et al. 2013; Greer and Wendel 2008; Guy et al. 2017). While manufacturer reported sensitivities are as high as 98.3% for both urethral and vaginal swab samples (Huppert et al. 2010; Cortez 2006), clinical studies comparing multiple RDTs to qRT-PCR have shown RDT sensitivities of only 54% for vaginal swab samples (Alary et al. 2006). If timely results are available, much more sensitive nucleic acid amplification tests (NAATs) are still recommended over immunologic RDTs (Johnson et al. 2002; Papp et al. 2014).
In this work, we present an integrated system that bridges that gap between RDTs and laboratory-based NAATs. Our POC NG diagnostic device features integrated sample preparation, DNA amplification, and lateral flow strip (LFS) detection. We have previously developed a portable and inexpensive paperfluidic Human Papilloma Virus (HPV) diagnostic device composed of only of porous paper membranes and adhesive plastic (Rodriguez et al. 2016). Here, we expand the platform to NG and enhance clinical reliability by incorporating an internal amplification control to differentiate between negative test results and invalid tests, a feature that will inevitably be required for regulatory approval of POC NAATs (Johnson et al. 2002; U.S. Food and Drug Administration 2011; Lafleur et al. 2016). In our NG diagnostic, cells in patient urethral and vaginal samples are added to lysis buffer which is then washed through a porous polyethersulfone (PES) substrate to precipitate and concentrate sample DNA. Excess fluid is wicked away to a removable waste pad. NG DNA and internal control DNA (NGIC) are then amplified in a multiplexed isothermal, thermophilic helicase-dependent amplification (tHDA) reaction. The resulting molecular probe-labeled amplicons flow to an integrated LFS for simple visual detection. We evaluated the analytical sensitivity by determining the limit of detection, the selectivity against 29 bacterial and viral strains that can also colonize the genital tract, and performed a small pilot study using 40 retrospective clinical samples to establish repeatability and clinical diagnostic metrics of the test.
2. Methods
2.1. DNA Stocks, Cell Stocks
NG strain NCTC 8375 (ATCC, Manassas, VA) was grown on chocolate agar in 5% CO2 at 37 °C for 24–36 h. Genomic NG DNAwas harvested from cultured cells by incubating colonies in 10% sodium dodecyl sulfate in 5 M guanidinum thiocya-nate followed by phenol:chloroform DNA extraction and ethanol precipitation (Dillard 2011). DNA was then quantified via Nanodrop. Nucleic acids isolated from 29 co-inhabitant commensal and pathogenic microbes of the genital tract were supplied from the Johns Hopkins Center for the Development of Point of Care Tests for Sexually Transmitted Diseases at Johns Hopkins University (JHU).
2.2. Clinical Samples
Previously de-identified, discard clinical samples collected at JHU were used in the retrospective study containing a total of 40 vaginal and urethral patient swab samples with 10 NG positive and 10 NG negative specimens of each sample type. These were collected and used in accordance with Institutional Review Board (IRB) protocols approved by Boston University and JHU. The diagnostic status of the samples was previously determined at JHU by using a laboratory NAAT, the MagNA-pure extraction and qPCR system (Roche Molecular Systems, Pleasanton, CA). Vaginal swabs were qPCR tested with a duplicate swab from the same de-identified patient sample, while urethral swabs were initially tested and then reused in our study. Swabs were stored dry at −80 °C until sample processing. Samples were processed by thawing the swabs for 2 min at room temperature and then placing them into 150 μL of sterile water in a 1.7 mL tube. The tubes were vortexed for 30 s, and the reconstituted sample was aliquoted and refrozen at −80 °C for later device application and DNA extraction.
2.3. qPCR Confirmation of Human and NG DNA in Clinical Samples
DNAwas extracted from reconstituted patient samples using a column-based Qiagen Blood and Tissue kit (Qiagen Corporation, Hilden, Germany). Following DNA extraction, DNA integrity was confirmed by detection of the human RNAseP housekeeping gene and NG genomic loads were quantified via SureStart Taq polymerase (Agilent, Santa Clara, CA) qPCR per manufacturer’s instructions on a QuantStudio5 Real-Time PCR system (Thermo Scientific, Waltham, MA). Primers and probes listed in Table S1 were purchased from Integrated DNA Technologies (Coralville, IA). Samples were heated to 95 °C for 10 min followed by 45 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 15 s.
2.4. tHDA Assay
Previously confirmed as PES compatible (Linnes et al. 2016), our tHDA assay amplifies the highly conserved and single-copy porA pseudogene, which is specific to the NG genome (Unemo et al. 2005). Like any molecular target, mutations of the porA gene could result in false-negative results test results. However, recent antimicrobial resistance studies have shown that porA mutants represented less than 0.4% of clinical NG isolates (Toby et al. 2017). Primer sequences and assay components, originally designed by Biohelix Inc. (Quidel Corporation, San Diego, CA), are listed in Table S1. In tube tHDA assays were performed for 30 min at 65 °C via the QuantStudio5 thermocycler. The internal amplification control, NGIC, is an engineered plasmid that uses the same amplification primers as the target NG porA pseudogene but has a different molecular probe sequence, enabling differential detection. NG/NGIC tHDA multiplexing was optimized through titration (data not shown), and all subsequent tHDA reactions contained 104 spiked in copies of NGIC plasmid. Successful amplification of NG and NGIC targets was confirmed via gel electrophoresis as well as detection of amplicon-probe complexes on the LFS. Selective amplification of the NG porA pseudogene and NGIC was confirmed by sequencing gel extraction products.
2.5. Lateral Flow Strip Detection
Commercial LFS were purchased from Ustar Biotechnologies (Hangzhou, China). NG probes were labeled with fluoresce-in isothiocyanate (FITC) while NGIC probes were tagged with digoxigenin (DIG) for detection on LFS via anti-DIG and anti-FITC stripes, respectively. LFS band intensities were imaged and quantified using ImageJ (NIH, Bethesda, MD) and objectively assessed via threshold-based densitometry, where NG and NGIC band intensities were compared to the LFS background.
2.6. Paperfluidic Device Fabrication
The 30 mm × 30 mm reaction housing contained an 8.5 mm circular hole as the center reaction chamber and was fabricated from 6 layers of 188 μm thick cyclic olefin polymer (Zeon Chemicals, L.P., Louisville KY) with a GraphTec CE6000–40 cutting plotter. A 9.5 mm diameter punch of a 0.22 μm pore size PES membrane (EMD Millipore, Billerica, MA) was placed between the two bottom layers. The chip was bonded together with a heat press as previously described (Roy et al. 2012). A waste pad laser-cut from Whatman #3 paper (Sigma Aldrich, St Louis, MO), and a commercial LFS with fluorescein isothiocyanate (FITC), digigoxin (DIG), and streptavidin detection lines (#D005–05, Ustar Biotechnologies, Hangzhou China) were attached using 3 mm thick self-adhesive plastic (Fellowes, Itasca, IL). Adhesive reaction chamber lids were aligned using pre-marked ‘X’s on the device housing tabs and re-covered to prevent contamination. Completed chips were stored in a sterile container until use.
2.7. Integrated On-Chip Assay
On-chip DNA precipitation, washing, amplification, and LFS detection were modified from the protocol of Rodriguez et al. (2016) and performed using 5 μL of DNA-containing samples added to 95 μL of guanidinium thiocyanate lysis/DNA precipitation buffer with NGIC plasmid (Fig. 1). In addition to using COP reaction housing instead of adhesive, modifications included adding 15 μL of mineral oil to increase heat transfer efficiency, as well as incubating the tHDA reaction on a heat block at 65 °C for 45 min under a Styrofoam insulator. After adding 60 μL of commercially supplied LFS running buffer, the detection and control lines on the attached LFS were read within 5 min. Acrylamide gel analysis of heated reactions additionally confirmed amplification.
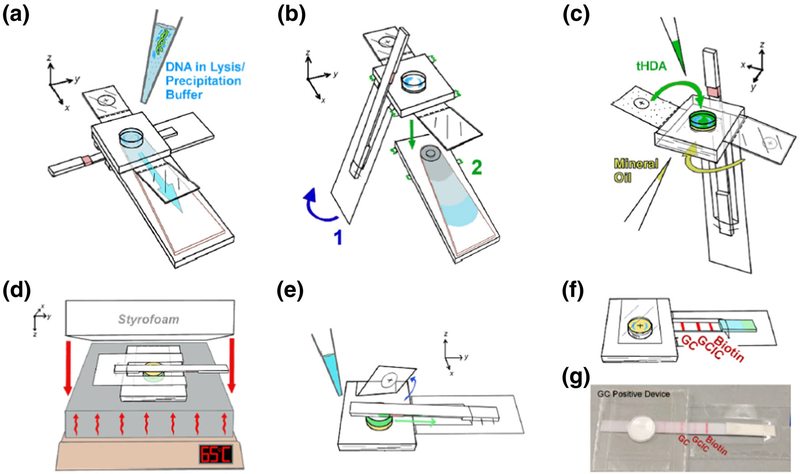
Fig. 1.
Workflow for NG precipitation, amplification, and detection within a low-cost paperfluidic device. Before device application, 5 μL of patient sample is pipetted and mixed with 95 μL of lysis/precipitation buffer. The precipitant mix is then applied to the reaction chamber of the device via pipette (a). After washing the reaction chamber with 70% and 95% ethanol, the lateral flow strip (LFS) is folded over and the waste pad is torn off and discarded (b). After drying, mineral oil is added to the smaller reaction chamber cavity and sealed with a tab (c, yellow) and tHDA amplification mix including NGIC is added to the larger cavity and sealed (c, green). The LFS is folded over top of the mineral oil side of the device before the device is heated on a standard hot plate underneath a Styrofoam insulator (d). Following heating, the tab sealing the tHDA mix is removed, allowing the LFS sample pad to touch the reaction chamber and LFS running buffer is applied to the end of the LFS (e) to enable visual detection of amplification results (f, photo g)
2.8. Statistical Analysis
Dunnet’s multiple comparison test was used to calculate the limit of detection using GraphPad Prism software. The limit of detection was determined to be the lowest NG DNA concentration input into the assay that resulted in a statistically significant difference (p < 0.05) from the no DNA control as detected by LFS. In-tube assays were run with nine replicates at each NG concentration while paperfluidic device assays were run with six replicates per NG concentration tested. Device and benchtop reactions with neither NG nor NGIC LFS lines were discarded from analysis due to failed tHDA, and reactions with no LFS lines were discarded for faulty flow conditions. Chips with weak NGIC or flow control lines but strong NG lines were included in analysis, as positive NG detection was still observed (Gervais and Delamarche 2009).
Using the positive and negative patient samples as a comparative test set, we determined the paperfludic device’s preliminary diagnostic sensitivity and specificity. Three device trials per patient sample were used to assess repeatability and results were binned together. We used stringent requirements that all three-device results be concordant with the JHU qPCR reference standard in order to be considered a true positive or true negative as detailed in the results.
3. Results
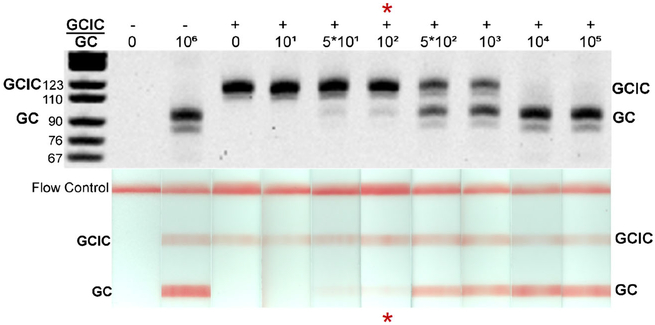
We optimized the tHDA assay to detect NG and NGIC in both a tube and integrated device format. Using a NGIC concentration of 104 copies per reaction, the multiplex tube assay detected 100 genomic copies per reaction (Table 1) with statistical significance compared to the negative, no template control (p < 0.05). Further, in some replicates, the assay was capable of detecting as few as 10 genomic copies per reaction (Fig. 2).
Table 1.
Comparison of tHDA multiplex amplification in optimal heating conditions via thermocycler and in the paperfluidic device
| tHDA Heating Conditions | Genomic NG copies per reaction | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 101 | 5*101 | 102 | 5*102 | 103 | 5*103 | 104 | 105 | |
| Positive NG amplification in thermocycler (n = 9) | 0% | 33% | 56% | 78%* | 100%* | 100%* | 100%* | 100%* | 100%* |
| Positive NG amplification in device (n = 6) | 0% | 17% | 17% | 33% | 83%* | 100%* | 100%* | 100%* | 100%* |
All reactions contained 104 copies of NGIC plasmid per reaction. Bold underlined values designate the limit of detection and
indicates p < 0.05 versus the no NG (0 copy) control using Dunnett’s multiple comparison test.
Fig. 2.
To determine the multiplex tHDA assay’s limit of NG detection, serially diluted genomic NG DNA was amplified in a thermocycler and analyzed via gel electrophoresis (top) and LFS threshold densitometry (bottom). Representative reactions for each concentration are illustrated here. The lowest concentration of NG DNA that yielded statistically significant amplification versus the negative control (p < 0.05, N = 9 for each condition) is considered the assay’s limit of NG detection (red asterisk)
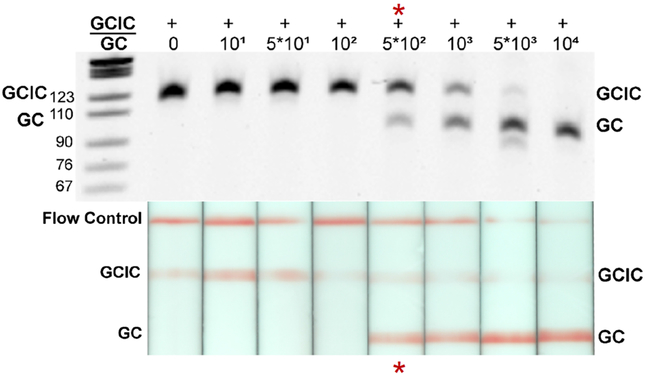
When transferred to the device, the limit of NG detection was 500 genomic NG copies (p < 0.05 compared to negative control), with digital amplification down to 10 genomic copies per device (Table 1, Fig. 3). The slightly poorer detection may have been caused by partial sample recovery during DNA precipitation or by tHDA amplification inhibition. Amplification inhibition can be the result of residual lysis and precipitation buffer components, device materials, or a reduction in reaction heating efficiency. However, the device detection limit was still well above expected patient sample bacterial loads (Priest et al. 2017; Lowe and Kraus 1976). The frequency of device failure in our limit of detection study was 4%; only 2 out of 50 chips tested failed to amplify either NG or NGIC targets.
Fig. 3.
Limit of NG DNA detection with the NG/NGIC tHDA multiplex assay in the paperfluidic device. To determine the paperfluidic device’s limit of NG detection, serially diluted genomic NG DNA was amplified under the device protocol (Fig. 1). Reactions extracted from the devices were analyzed via gel electrophoresis (top) and LFS threshold densitometry (bottom). Representative device reactions for each concentration are illustrated here. The lowest concentration of DNA that yielded statistically significant amplification success compared to the no NG DNA controls (N = 6 for each condition) is considered the limit of NG detection for the device (red asterisk). In these studies, the frequency of device success was 96%
To confirm assay selectivity for the NG-specific porA pseudogene, we tested our multiplexed tHDA assay against range of potential confounding microbes. These selectivity tests found no cross-species amplification within any of the 29 bacterial and viral genomes tested in our NG and NG/NGIC multiplex tHDA assays (Table S2). These results confirm our assay’s selectivity towards NG DNA amplification. Selectivity for NG and NGIC was further confirmed by sequencing the amplification products generated from genomic NG and NGIC plasmid tHDA reactions (data not shown).
Having demonstrated assay selectivity and a clinically relevant device limit of detection for NG in our integrated device, we next determined the expected NG bacterial load in clinical urethral and vaginal swab samples (Table S3). No NG DNA was detected in any of the NG-negative swabs. In the NG-positive patient samples, qPCR quantification of patient sample DNA extracts showed an average of 6.59*104 porA pseudogene copies per swab (range: 1.56*102 to 2.41*105) across the NG-positive urethral swab isolates and an estimated average of 2.86*106 copies per swab (range: 3.56*102 to 1.87*107) over the NG-positive vaginal swab isolates. Using these qPCR values, the approximate average NG DNA concentrations input into the device were 2.53*103 organisms per urethral swab and 1.81*105 organisms per vaginal swab.
To maximize the evaluation of device reproducibility, we tested each clinical sample in three separate devices. Overall conditions for successful amplification were present in 88% (53 out of 60) of the tests performed on vaginal samples and in 92% (55 out of 60) of the tests performed on urethral samples (Table S3). Device trials were only considered correlative with the JHU standard if all three of the trials concurred with the reference standard. Our paperfluidic device repeatedly detected NG DNA from reconstituted swabs across all three device trials in 9 out of 10 NG-positive urethral swab samples and in 10 out of 10 NG-positive vaginal swabs (Table S3). Additionally, we saw no false positive NG detection in replicate tests of the 10 negative vaginal and 10 negative urethral swab samples.
4. DISCUSSION
In this proof-of-concept study, we developed a low-cost paperfluidic device for detection of NG DNA from urethral and vaginal samples. Device run time approaches suitability for the clinical point of care at 80 min turnaround time from sample-to-result. In the tHDA limit of detection studies, there is a slight reduction in the device detection limit compared to in-tube experiments, however, both methods were able to detect as few as 10 copies of DNA in a subset of reactions. Further development of this platform will focus on reducing the turnaround time from 80 min to less than 60 min and decreasing the required user interaction to fewer than three steps in order to align it with the recently published target produce profile for point-of-care NG diagnostic tests (Toskin et al. 2017). Presently, patient sample processing and reaction chamber drying require 30 min, amplification requires 45 min, and LFS interpretation another 5 min. To reduce device run time and amplification success, we are optimizing reaction chamber drying and tHDA efficiency.
The lower detection limit of 500 genomic NG copies per reaction in our device is sensitive enough to detect infections in both urethral swabs (Priest et al. 2017) and vaginal samples (Lowe and Kraus 1976). Of note, some apparent qPCR estimates of bacterial loads fell below our projected limit of detection (e.g., NG positive urethral samples #2, 3, 6, and 9 (Table S3)). We hypothesize that the extraction efficiency of the membrane-based precipitation used in the device may have actually been better than the silica-based commercial kit used to prepare our qPCR swab samples. Membrane-based precipitation has been shown to be effective at even low concentrations of DNA, while previous reports have shown significant genomic DNA losses from silica-based kits at DNA concentrations below 104 CFU/mL (Kulinski et al. 2009). We saw a reduced bacterial load in our qPCR measurements when comparing our urethral swab NG bacterial load quantifications to a previous study (Priest et al. 2017). While extraction efficiency may play a role in this reduction, the urethral swabs we received had already been used for initial diagnostic testing and some material was certainly removed during this process.
By testing 29 bacterial and viral species that commonly inhabit human genitalia (Brotman 2011; Price et al. 2010; Linnes et al. 2014) with the tHDA multiplex assay, we screened for non-specific amplification even with high DNA genomic loads under optimal amplification conditions. Assay selectivity was confirmed, as none of the potentially confounding targets amplified.
An important characteristic of all diagnostic test development is the evaluation of replicate trials of the same sample in different devices. We performed critical experiments to ensure clinical device reliability by evaluating multiple device trials in our limit of detection as well as patient sample validation studies. In patient sample trials, we performed 120 total device trials with the 40 clinical samples (triplicate tests for each sample). Conditions for successful amplification were present in 88% of the vaginal samples and in 92% of the urethral samples. All invalid tests arose in the NG negative samples due to assay conditions that were unfavorable for NGIC amplification. Inclusion of NGIC enabled this important differentiation between false negative results, and failed amplification conditions. When comparing to the 96% success rate of the pre-isolated NG DNA trials, the lowered success of NGIC amplification in NG-negative swab device trials suggests that both urethral and vaginal patient sample matrices introduce some inhibition to amplification. Indeed, this has been seen in other recent paperfluidic NAATs, which had a 62% success rate of Staphylococcus aureus detection using clinical nasal samples (Lafleur et al. 2016). To improve our device success rate in future trials with NG negative patient samples, NGIC amplification will require further optimization, such as adjusting assay salt balance and ensuring complete washing for removal of the patient sample matrices.
The United States Food and Drug Administration currently advises that rapid NG NAAT diagnostics in development should reach 95% sensitivity and 95% specificity (U.S. Food and Drug Administration 2011). A more recent target product profile developed by international STI experts indicated that 90% sensitivity and 90% specificity would be acceptable for surveillance and screening in a test that required less than 60 min (Toskin et al. 2017). A limitation to our pilot study is that the sample size is underpowered to determine true diagnostic sensitivity and specificity of the paperfluidic device. We would have required 146 clinical samples for 95% sensitivity and 95% specificity measurements (or 279 samples for 90% sensitivity and 90% specificity) using a prevalence of 50%, confidence level of 95%, and accuracy of 0.05. Nevertheless, in our 40-sample study, 19 of the 20 NG positive samples amplified and no false positives were detected in the 20 NG negative samples. These promising results indicate the targeted clinical diagnostic sensitivity and specificity are feasible with a larger sample size. As our device reliability trials illustrated diagnostic disparity in only 1 out of the 40 patient samples, likely due to NG concentrations near the assay limit of detection, future validation studies could be performed with a single device per sample with minimal adverse impact on sensitivity and specificity. These future validation trials will include a larger sample size using blinded samples of fresh, prospectively collected swabs from patients with both asymptomatic and symptomatic NG infections.
5. CONCLUSIONS
Our device yielded a statistically significant lower detection limit of 500 genomic NG copies per reaction, digital detection from as few as 10 NG copies, which resulted in an assay that is sensitive enough to detect infections in clinical urethral and vaginal swabs. We validated the device in a rigorous proof-of-concept study using forty discarded de-identified urethral and vaginal swabs, each tested in three separate device trials. Our device had an overall sensitivity of 95% (detecting 19 of 20 positive samples) and specificity of 100% for all sample types. These promising results are an exciting proof-of-principle demonstration for this POC NG NAAT platform. The further development of this diagnostic platform could overcome many NG testing barriers currently faced in limited-resource settings and increase access to accurate NG diagnoses for those most in need.
Supplementary Material
Acknowledgements
This work was funded by the National Institute of Health National Institute of Allergy and Infectious Diseases award number R01 AI113927 to Boston University and the NIH National Institute of Biomedical and Bioengineering award number U54 EB007958 to Johns Hopkins University.
Footnotes
Electronic supplementary material
The online version of this article (https://doi.org/10.1007/s10544018-0280-x) contains supplementary material, which is available to authorized users.
References
- Alary M, Gbenafa-Agossa C, Aina G, et al. , Evaluation of a rapid point-of-care test for the detection of gonococcal infection among female sex workers in Benin. Sex. Transm. Infect 82, 29–32 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aledort JE, Ronald A, Rafael ME, et al. , Reducing the burden of sexually transmitted infections in resource-limited settings: the role of improved diagnostics. Nature 444, 59–72 (2006) [DOI] [PubMed] [Google Scholar]
- Bourgeois A, Henzel D, Malonga-Mouelet G, et al. , Clinical algorithms for the screening of pregnant women for STDs in Libreville, Gabon: which alternatives? Sex. Transm. Infect 74, 35–39 (1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotman RM, Vaginal microbiome and sexually transmitted infections: an epidemiologic perspective. J. Clin. Investig 121, 4610–4617 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez Diagnostics, Inc. OneStep RapiDip Gonorrhea InstaTest Package Insert (Cortez Diagnostics, Inc., 2006). http://www.rapidtest.com/Gonorrhea_176512.pdf. Accessed 2 Dec 2016
- Dillard JP, Genetic Manipulation of Neisseria gonorrhoeae. Curr.Protoc. Microbiol 23, 4A.1.1–4A.1.24 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming DT, Wasserheit JN, From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex. Transm. Infect 75, 3–17 (1999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos CA, Pol BVD, Jett-Goheen M, et al. , Performance of the Cepheid CT/NG Xpert Rapid PCR Test for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol 51, 1666–1672 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais L, Delamarche E, Toward one-step point-of-care immunodiagnostics using capillary-driven microfluidics and PDMS substrates. Lab. Chip 9, 3330 (2009) [DOI] [PubMed] [Google Scholar]
- Greer L, Wendel GD, Rapid diagnostic methods in sexually transmitted infections. Infect. Dis. Clin. N. Am 22, 601–617 (2008) [DOI] [PubMed] [Google Scholar]
- Guy RJ, Cuaser LM, Klausner JD, et al. , Performance and operational characteristics of point-of-care tests for the diagnosis of urogenital gonococcal infections. Sex. Transm. Dis 93, S16–S21 (2017) [DOI] [PubMed] [Google Scholar]
- Huppert J, Hesse E, Gaydos CA, What is the point? How point-of-care sexually transmitted infection tests can impact infected patients. Point Care 9, 36–46 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Newhall WJ, Papp JR, et al. , Screening tests to detect Chlamydia trachomatis and Neisseria gonorrhoeae infections–2002. MMWR Recomm. Rep 51, 1–38 (2002) [PubMed] [Google Scholar]
- Kulinski MD, Mahalanabis M, Gillers S, et al. , Sample preparation module for bacterial lysis and isolation of DNA from human urine. Biomed. Microdevices 11, 671–678 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafleur LK, Bishop JD, Heiniger EK, et al. , A rapid, instrument-free, sample-to-result nucleic acid amplification test. Lab Chip 16, 3777–3787 (2016) [DOI] [PubMed] [Google Scholar]
- Linnes JC, Fan A, Rodriguez NM, et al. , Paper-based molecular diagnostic for Chlamydia trachomatis. RSC Adv 4, 42245–42251 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnes JC, Rodriguez NM, Liu L, et al. , Polyethersulfone improves isothermal nucleic acid amplification compared to current paper-based diagnostics. Biomed. Microdevices 18, 30 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TL, Kraus SJ, Quantitation of Neisseria gonorrhoeae from women with gonorrhea. J. Infect. Dis 133, 621–626 (1976) [DOI] [PubMed] [Google Scholar]
- Papp JR, Schachter J, CA Gaydos, et al. , Recommendations for the laboratory-based detection of Chlamydia trachomatis and Neisseria gonorrhoeae–2014. MMWR Recomm. Rep 63, 1–19 (2014) [PMC free article] [PubMed] [Google Scholar]
- Price LB, Liu CM, Johnson KE, et al. , The effects of circumcision on the penis microbiome. PLoS One 5, 1–12 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest D, Ong JJ, Chow EPF, et al. , Neisseria gonorrhoeae DNA bacterial load in men with symptomatic and asymptomatic gonococcal urethritis. Sex. Transm. Infect 93(7), 478–481 (2017) [DOI] [PubMed] [Google Scholar]
- Rodriguez NM, Wong WS, Liu L, et al. , A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip 16, 753–763 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Yue C, Wang Z, Anand L, Thermal bonding of microfluidic devices: Factors that affect interfacial strength of similar and dissimilar cyclic olefin copolymers. Sensors Actuators B Chem 161, 1067–1073 (2012) [Google Scholar]
- Toby M, Saunders P, Cole M, et al. , Prevalence of porA pseudogene deletion among Neisseria gonorrhoeae isolates referred to the UK’s Gonococcal Resistance to Antimicrobials Surveillance Program. Sex. Health 14(4), 392–393 (2017) [DOI] [PubMed] [Google Scholar]
- Toskin I, Murtagh M, Peeling RW, et al. , Advancing prevention of sexually transmitted infections through point-of-care testing: target product profiles and landscape analysis. Sex. Transm. Dis 93, S69–S80 (2017) [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration. Draft Guidance for Industry and Food and Drug Administration Staff Establishing the Performance Characteristics of In Vitro Diagnostic Devices for Chlamydia trachomatis and/or Neisseria gonorrhoeae: Screening and Diagnostic Testing. U.S. Federal Register 76(91), 19 (2011) [Google Scholar]
- Unemo M, Norlen O, Fredlund H, The porA pseudogene of Neisseria gonorrhoeae - low level of genetic polymorphism and a few, mainly identical, inactivating mutations. APMIS 113, 410–419 (2005) [DOI] [PubMed] [Google Scholar]
- Wasserheit JN, Epidemiological synergy. Interrelationships between human immunodeficiency virus infection and other sexually transmitted diseases. Sex. Transm. Dis 19, 61–77 (1992) [PubMed] [Google Scholar]
- World Health Organization, Global strategy for the prevention and control of sexually transmitted infections : 2006 – 2015 : breaking the chain of transmission (WHO Press, Geneva, Switzerland: 2007) p. 26 [Google Scholar]
- World Health Organization, Global incidence and prevalence of selected curable sexually transmitted infections – 2008 (WHO Press, Geneva, Switzerland, 2012) p. 2 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.