Abstract
Circular RNAs (circRNAs) are long-lived, covalently closed RNAs that are abundantly expressed and evolutionarily conserved across eukaryotes. Possible functions ranging from microRNA (miRNA) and RNA binding protein sponges to regulators of transcription and translation have been proposed. Here we describe the design and characterization of recombinant adeno-associated viral (AAV) vectors packaging transgene cassettes containing intronic sequences that promote backsplicing to generate circularized RNA transcripts. Using a split GFP transgene, we demonstrate the capacity of vectors containing different flanking intronic sequences to efficiently drive persistent circRNA formation in vitro. Further, translation from circRNA is efficiently driven by an internal ribosomal entry site (IRES). Upon injecting AAV vectors encoding circRNA in mice, we observed robust transgene expression in the heart, but low transduction in the liver for the intronic elements tested. Expression in the murine brain was restricted to astrocytes following systemic or intracranial administration, while intravitreal injection in the eye yielded robust transgene expression across multiple retinal cell layers. These results highlight the potential for exploiting AAV-based circRNA expression to study circRNA function and tissue-specific regulation in animal models, as well as development of therapeutic platforms using this approach.
Keywords: adeno-associated virus, circular RNA, gene therapy
Introduction
Circular RNAs (circRNAs) are a class of regulatory RNAs in which the RNA is covalently closed to form a circle. They are found in a range of organisms spanning from Archaea to humans.1, 2 In metazoans, they are predominantly formed from pre-mRNAs through a process known as backsplicing, in which a splice donor site is spliced to an upstream acceptor site, thus resulting in a mature circRNA that consists of an exon (or multiple exons).3, 4, 5 There is thus competition between production of the circRNA product and the linear mRNA product, and the ratio of circular and linear products varies for each gene; for some genes, the circRNA is the predominant RNA product.6, 7 Signals that affect the circularization of a given exon can be encoded in the surrounding intron sequences. In humans, the presence of inverted repeats (such as Alu elements) has been shown to promote exon circularization.8, 9, 10 Additionally, there are examples of circRNAs whose formation is stimulated by RNA binding proteins such as Quaking or Muscleblind, which can bind to intronic sequences and dimerize, and hence may be involved in regulation of circularization of an intervening exon(s);11, 12 spliceosomal factors are also known to promote the process.13
Recent studies have shown that circRNAs are highly expressed in multiple cell and tissue types.8, 14, 15, 16 However, the function, if any, of the majority of circRNAs is unknown. There are exceptions, such as ciRS-7/Cdr1as and circMbl, which are well characterized as microRNA 7 (miRNA-7) and MBL protein sponges, respectively.12, 17, 18 Recent studies have indicated that some endogenous circRNAs may have coding potential; however, the translation efficiency may be low, because other studies have failed to see association of endogenous circRNAs with ribosomes at appreciable levels.19, 20, 21, 22 circRNAs that are engineered to contain an IRES (internal ribosomal entry site) drive translation of an open reading frame (ORF) contained in the circRNA.23 Our lab recently created a system of inducing the expression of circRNA encoding an IRES-driven GFP ORF, in which we demonstrated association of the circRNA with actively translating polyribosomes.24 An important property of circRNAs is their enhanced stability compared with linear RNAs, which is due to their lack of free ends, and therefore resistance to cellular exonucleases. Studies have shown that circRNAs are more stable compared with their linear counterparts in cultured cells.25 Thus, it is possible to design circRNAs that will allow for expression of proteins from a very stable RNA molecule. This property makes circRNAs an intriguing possibility for use in gene delivery applications, where long-term gene expression is a key concern.
We designed circRNA expression cassettes that could be delivered using recombinant adeno-associated viral (AAV) vectors, commonly utilized in gene transfer applications in vivo. AAV is a small, non-pathogenic member of the Parvovirus family that requires a helper virus for replication.26, 27, 28 AAV packages a single-stranded DNA genome of approximately 4.7 kb with 145-bp inverted terminal repeats (ITRs) that are the sole packaging signal.29, 30 Although wild-type AAV can integrate into the host genome, recombinant vectors will not integrate in the absence of the viral Rep proteins, but persist instead as episomes.31, 32 There are many AAV capsid variants, both natural and engineered, with a broad spectrum of host tissue tropisms and cell-type specificities, allowing for transgene expression in different organ systems in different animal models and humans.33, 34, 35 In this study, we present proof-of-concept studies describing the use of recombinant AAV (rAAV) vectors to deliver circRNA producing transgenes. We created reporter transgenes with two different intron pairs that drive exon circularization and demonstrate expression in multiple cell and tissue types. Our studies help establish new AAV-circRNA vectors as tools that could prove useful for exerting spatio-temporal control over transgene expression.
Results
Design and Construction of AAV-circRNA Vectors
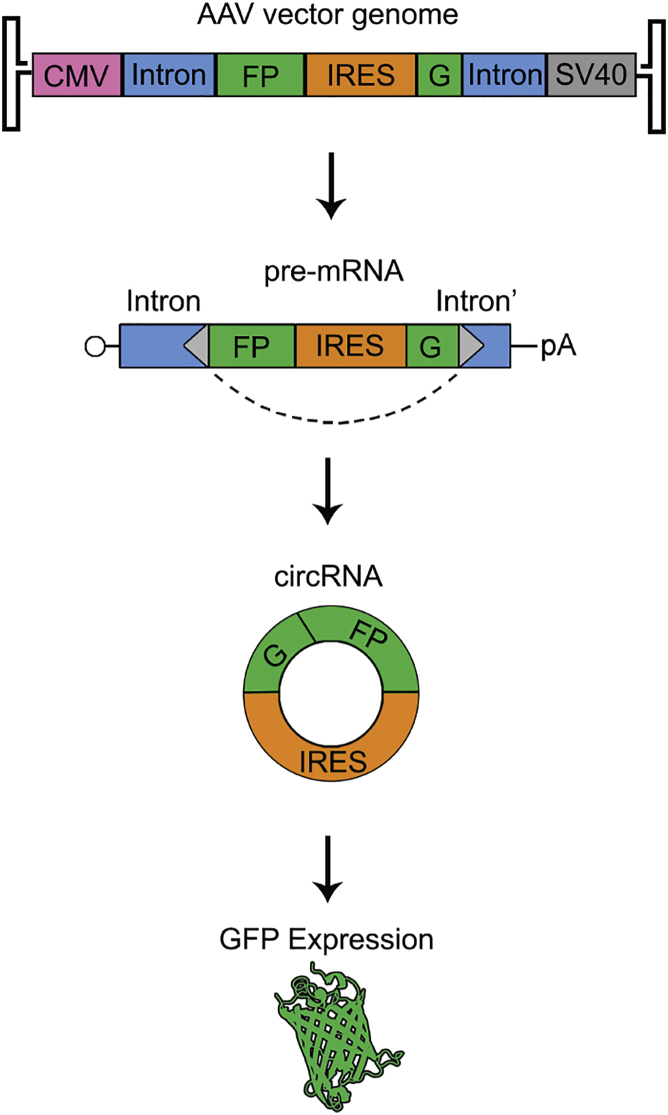
To show that rAAV vectors can be used to deliver transgene cassettes that express circRNAs, we designed reporters based on intronic sequences from the human ZKSCAN1 and HIPK3 genes that are known to drive production of endogenous circRNAs.9 Portions of these intronic sequences that flank the endogenously circularized exons were placed into AAV vector genome packaging cassettes. Specifically, a split GFP ORF was placed between the intronic sequences such that full-length GFP would be reconstituted only after backsplicing to make the circRNA.36 The EMCV IRES was additionally positioned upstream of the N terminus of GFP so that these constructs should only be translated into GFP upon backsplicing to form a circRNA (Figure 1). rAAV vectors were then generated using both sets of intron sequences, creating the ZKSCAN1 split GFP and HIPK3 split GFP vectors. As a control, we created a vector (linIRES-GFP) lacking intronic elements, but containing the EMCV IRES followed by a contiguous GFP ORF; this creates a linear mRNA containing the same sequences as the circRNA generated from the above cassettes.
Figure 1.
Schematic of AAV Vector Design Encoding circRNA Constructs
The RNAs derived from either ZKSCAN1 split GFP or HIPK3 split GFP constructs are expressed from the CMV promoter and are capped and poly-adenylated in their linear isoforms. The precursor split GFP transcript contains a split GFP cassette flanked by intronic sequences derived from the human ZKSCAN1 or HIPK3 genes and will not express GFP. Pre-mRNAs are predicted to be ∼2.8 and ∼2.7 kb in length, respectively. Donor and acceptor splice sites are represented by gray triangles, and the dotted lines indicate the backsplicing pattern. The GFP fragments are separated by an EMCV IRES such that upon RNA circularization (forming a 1.3-kb circRNA), full-length GFP can be expressed.
AAV-Mediated circRNA Expression in Cell Culture
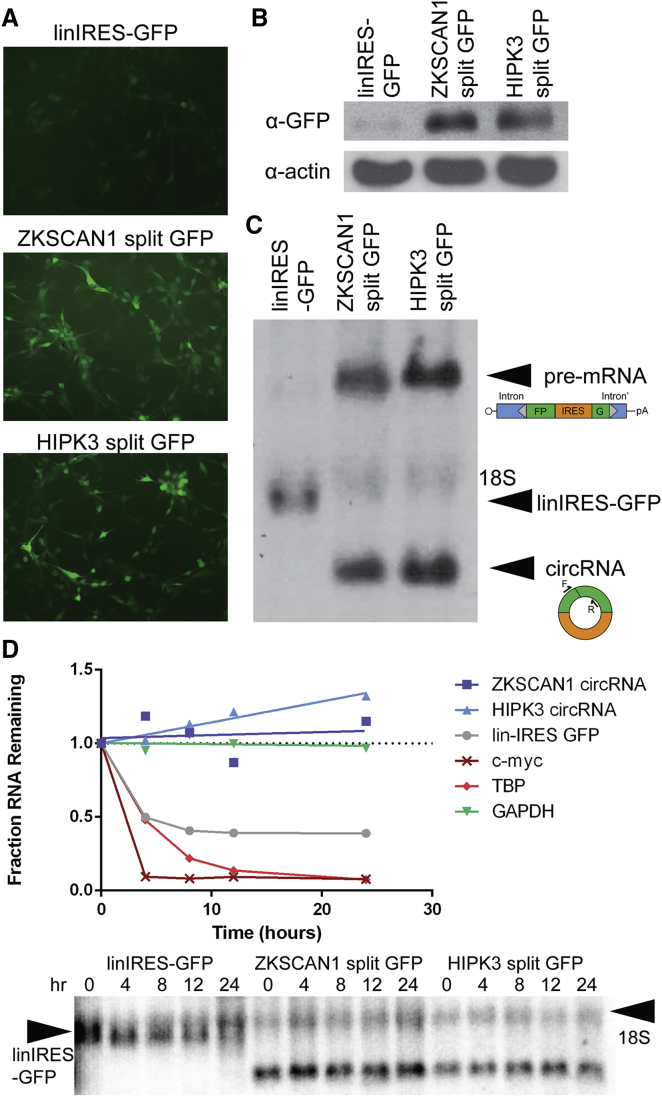
The different constructs were packaged into rAAV2 vectors to efficiently transduce human glioma (U87) cells in culture. The two circular cassettes showed roughly equal expression based on assessment of fluorescent images, whereas the control expressed low levels (Figure 2A). This was verified by western blot analysis of GFP expression (Figure 2B). To further characterize circRNA expression, we extracted RNA from cells and assessed by northern blot. When probed for GFP-specific sequences, the two split GFP vectors showed two bands: one corresponding to the unspliced polyadenylated RNA and the other corresponding to the spliced circRNA as described previously by our lab using plasmid vectors.24 linIRES-GFP also produced an mRNA at the expected size (Figure 2C). Northern blots demonstrated that the levels of circRNA produced from ZKSCAN1 split GFP and HIPK3 split GFP were not significantly different from each other, consistent with GFP protein levels observed. Approximately 50% of the transcripts were linear, polyadenylated RNA, and 50% were circularized at 4 days post-transduction. These cell culture experiments validated that our vectors could be successfully packaged into AAV for delivery and expression of translatable circRNAs.
Figure 2.
AAV-Mediated Expression of circRNAs In Vitro
(A) GFP fluorescence at 4 days post-transduction in U87 cells transduced with 100,000 vector genomes per cell of recombinant AAV2 vectors packaging linIRES-GFP, ZKSCAN1 split GFP, or HIPK3 split GFP (n = 3). (B) GFP expression verified by western blot; Actin loading control is also shown. (C) Northern blotting of total cellular RNA from the above cells, probed against GFP, revealed the following species: the linIRES-GFP mRNA, the unspliced pre-mRNA, and the spliced circRNA. Schematic of RNA species is depicted alongside the bands. The position of 18S rRNA is noted as a marker of size. (D) U87 cells were transduced with 100,000 vector genomes per cell of recombinant AAV2 vectors packaging linIRES-GFP, ZKSCAN1 split GFP, or HIPK3 split GFP (n = 2 biological replicates). At 3 days after transduction, cells were treated with actinomycin D, then harvested for RNA at 0-, 4-, 8-, 12-, and 24-hr time points. qRT-PCR was performed for GAPDH, c-myc, and TBP RNA quantification. Northern blotting was performed for the GFP-containing species and is shown in the panel below.
We next utilized this in vitro system to probe the stability of the RNAs produced from our vectors. U87 cells were transduced as above; after 3 days, cells were treated with the transcription inhibitor actinomycin D, and RNA was harvested at time points ranging from 0 to 24 hr. To verify our assay, we performed qRT-PCR for GAPDH, TBP, and c-myc mRNAs across the time course. Our results agreed with previous reports,8 showing GAPDH to be highly stable, c-myc to completely degrade by the 4-hr time point, and TBP to have an intermediate phenotype (Figure 2D). Next, we performed northern blots to investigate circRNA stability; we compared the circRNA with the RNA from linIRES-GFP, because it is a capped and poly-adenylated RNA containing the same sequences as the circRNA. The circRNAs generated by either ZKSCAN1 split GFP or HIPK3 split GFP were extremely stable and showed no evidence of degradation after actinomycin D treatment (Figure 2D). In contrast, the linIRES-GFP RNA showed degradation kinetics similar to that of TBP. Quantification of this RNA was difficult due to the slightly overlapping 18S RNA background band (see image of northern blot below the plotted quantifications). However, it can clearly be seen that the linIRES-GFP RNA levels decrease, whereas the circRNA levels do not. These experiments convincingly demonstrate the inherent stability of the circRNAs generated by our system compared with otherwise linear mRNAs.
AAV-Mediated circRNA Expression after Intravenous Delivery in Mice
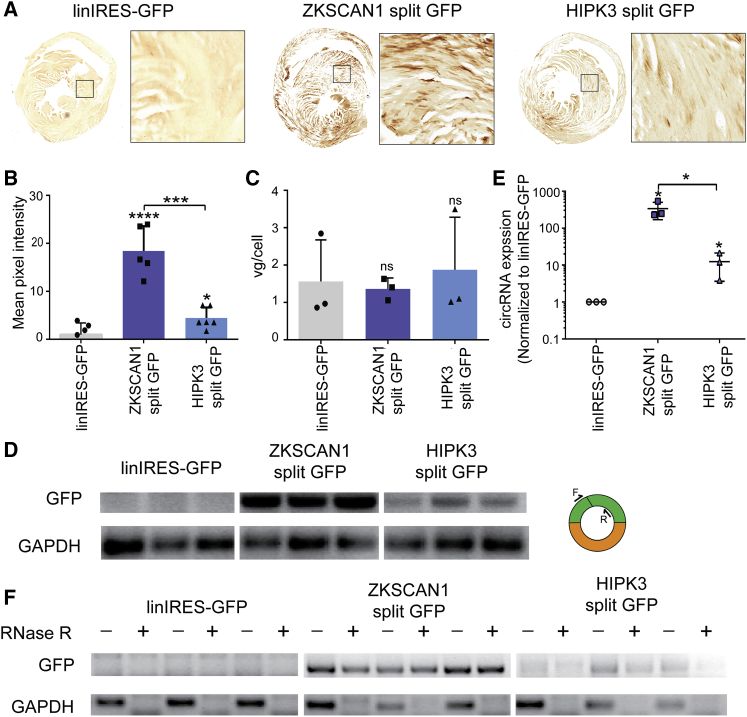
To determine whether these vectors were functional in vivo, we packaged ZKSCAN1 split GFP and HIPK3 split GFP cassettes into recombinant AAV9 vectors, a serotype known to transduce several tissues in vivo.37 Mice were injected intravenously at a dose of 5.5 × 1011 vector genomes (vg)/mouse, and immunohistochemical staining against GFP was carried out in cardiac tissue at 4 weeks post-administration. The two circRNA vectors showed largely uniform expression throughout the heart with ZKSCAN1 split GFP showing more robust expression than HIPK3 split GFP (Figure 3A). In contrast, linIRES-GFP showed little to no GFP staining. This was confirmed by quantifying the mean pixel intensities of stained sections (Figure 3B). We further verified that differences in expression were not due to dosing, because all animals had similar numbers of vector genomes per cell in the heart (Figure 3C).
Figure 3.
Systemic Expression of circRNAs in Murine Cardiac Tissue
The indicated constructs were packaged into recombinant AAV9 vectors and injected intravenously into C57/BL6 mice (n = 3) at a dose of 5.5 × 1011 vector genomes per animal, then harvested 4 weeks post-injection. (A) Immunohistochemical staining of sectioned cardiac tissue showed GFP expression for ZKSCAN1 split GFP and HIPK3 split GFP. (B) The level of GFP expression in stained cardiac sections was analyzed semiquantitatively by mean pixel intensity. (C) AAV vector genomes per cell were quantified by qPCR for the CMV promoter, normalized to the mouse lamin B2 locus. (D) RT-PCR performed with primers amplifying GFP across the backsplice junction (see schematic) showed expression of circRNA from both ZKSCAN1 split GFP and HIPK3 split GFP. (E) qRT-PCR was performed with the same samples and primer set as in (D). (F) RT-PCR bands of the circRNA are resistant to RNaseR treatment. Where indicated, *p < 0.05; ***p < 0.0005; ****p < 0.00005. Error bars represent 1 SD.
To assess expression of the vector-derived circRNA, we performed RT-PCR with primers amplifying the GFP coding region across the circRNA splice junction. Using these primers, we detected circRNA expression from both constructs, although not from linIRES-GFP, which this primer pair also detects (Figure 3D). qRT-PCR revealed detectable levels of linear RNA from the linIRES-GFP vector, although at much lower amounts compared with the circRNA. Specifically, ZKSCAN1 split GFP showed 27-fold higher expression than the HIPK3-driven construct (Figure 3E). Because both vectors have the same promoter and ORF, they should be transcribed and translated with equal efficiencies. Thus, the ZKSCAN1 intron sequences appear to drive circularization with higher efficiency than the HIPK3 introns in cardiac tissue. Finally, we verified that the vector-derived transcripts are resistant to RNase R, a 3′ to 5′ exonuclease. RT-PCR was performed on RNA samples treated with or without RNase R. Bands were observed for both split GFP vectors after treatment, providing further evidence of resistance to RNase R and circularity (Figure 3F). PCR was also performed for the linear GAPDH mRNA; as expected, this band disappeared after RNase R treatment.
AAV-Mediated circRNA Expression in Murine Liver
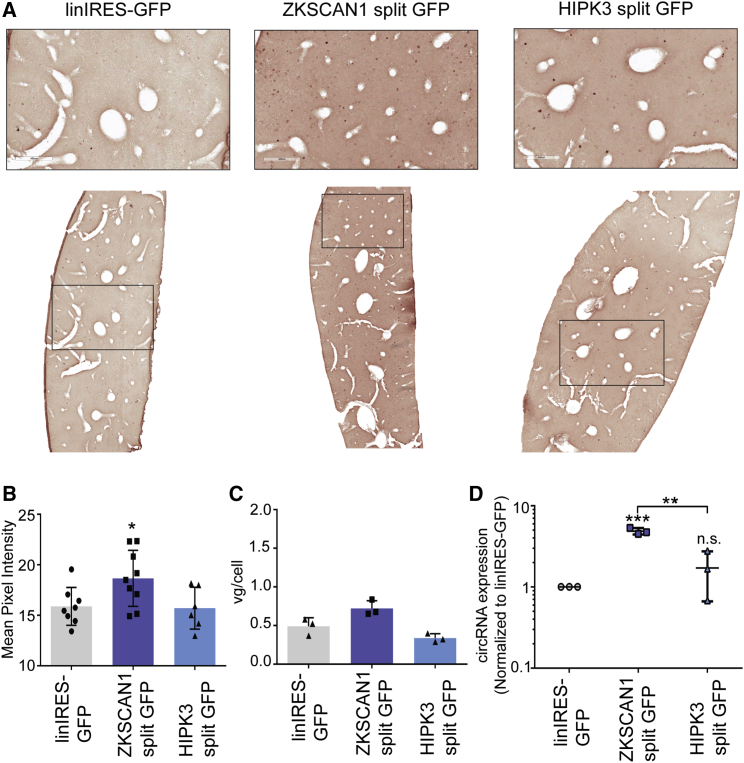
We next determined whether these circRNA expression vectors were expressed and translated in other tissue types. From the same cohort analyzed above, liver tissue was harvested and processed. Immunohistochemical staining for GFP was performed, revealing that both constructs displayed extremely low GFP expression in the liver (although visible compared with the control); ZKSCAN1 split GFP expression was higher than HIPK3 split GFP (Figure 4A). This was confirmed by quantifying the mean pixel intensities of stained sections (Figure 4B). Vector genome copy numbers determined by qPCR ranged from 0.7 to 1.4 vg/cell between constructs and were similar to those seen in the heart (Figure 4C). We directly measured the levels of circRNA in the harvested livers by performing qRT-PCR with the primers described above. This analysis showed expression of the circRNA from ZKSCAN1 and HIPK3 split GFP vectors, with ZKSCAN1 split GFP yielding greater circRNA levels (Figure 4D). The linIRES-GFP RNA was also detected, albeit at much lower levels than circRNAs (statistically significant for ZKSCAN1 split GFP). We note that consistent with the RT-PCR results, endogenous ZKSCAN1 circRNA expression has been reported in the liver.9 Because our results show RNA production but little protein expression, it is possible that the EMCV IRES does not efficiently drive translation of the circRNA in the liver. Previous work corroborates that the EMCV IRES has low activity in the liver compared with other IRES elements.38
Figure 4.
AAV-Mediated circRNA Expression in the Liver
The indicated constructs were packaged into recombinant AAV9 vectors and injected intravenously into C57/BL6 mice (n = 3) at a dose of 5.5 × 1011 vector genomes per animal, then harvested 4 weeks post-injection. (A) Immunohistochemical staining of sectioned liver tissue displayed low GFP expression for ZKSCAN1 split GFP and HIPK3 split GFP. Scale bars, 400 μm. (B) The level of GFP expression in stained cardiac sections was analyzed semi-quantitatively by mean pixel intensity. (C) AAV vector genomes per cell were quantified by qPCR for the CMV promoter, normalized to the mouse lamin B2 locus. (D) qRT-PCR performed with primers amplifying GFP across the backsplice junction revealed expression of the circRNA in the liver. Where indicated, *p < 0.05; **p < 0.005; ***p < 0.0005. Error bars represent 1 SD.
AAV-Mediated circRNA Expression in the Murine Brain and Eye
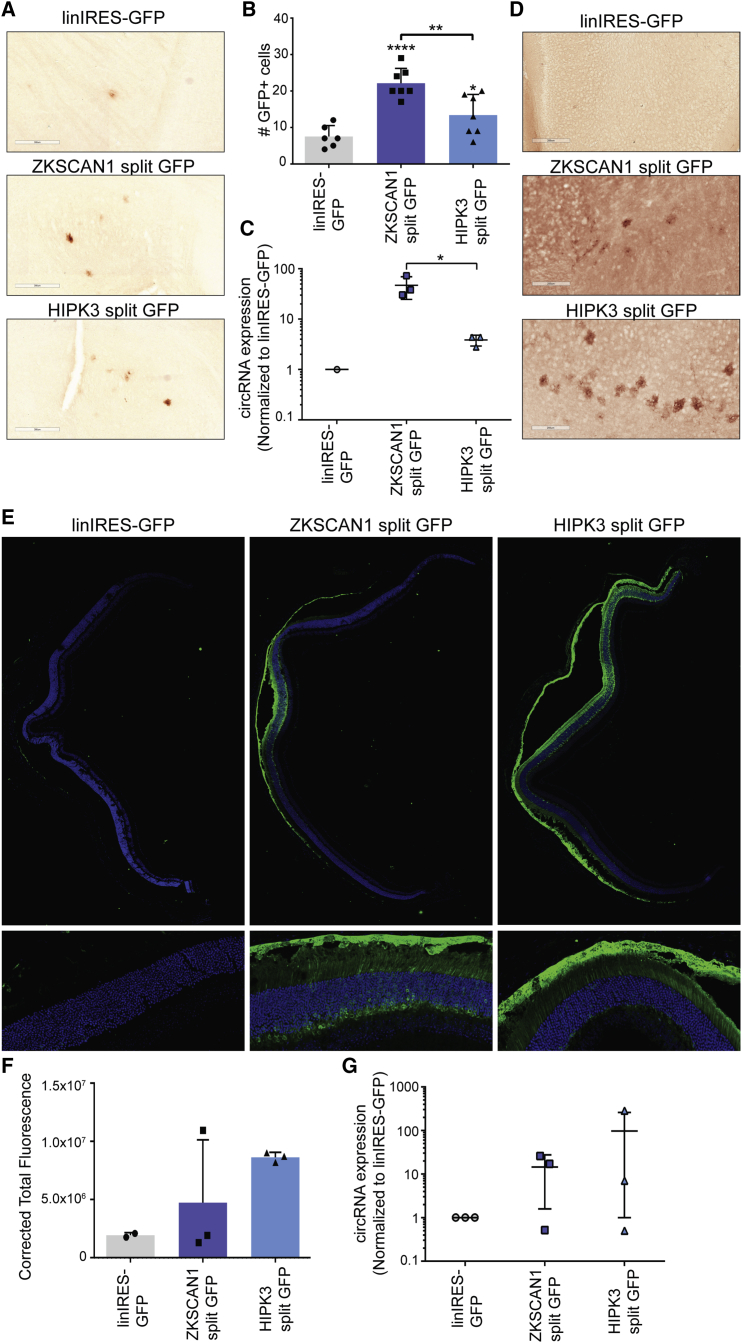
To evaluate the AAV-circRNA vectors in the nervous system, we first analyzed brain tissue from the same intravenous injection cohort. Immunohistochemical staining was performed on brain sections and showed limited expression from both split GFP vectors, mainly within the cerebral cortex and cerebellum (representative images shown in Figure 5A). This region-specific expression may be a function of vector dose or circRNA related (attributable to backsplicing due to intronic elements, IRES-mediated translation, or both). However, although AAV9 vectors are known to express broadly within the CNS,37 we observed GFP expression predominantly in astrocytes, with little neuronal expression. Quantification of GFP-positive cells showed higher expression from ZKSCAN1 split GFP, followed by HIPK3 split GFP. However, it can be seen that the number of GFP-positive cells per whole section was very low (Figure 5B). qRT-PCR to detect circRNA revealed that ZKSCAN1 split GFP had the highest RNA expression, followed by HIPK3 split GFP. However, expression of the control linIRES-GFP was below the limit of detection in all but one animal (Figure 5C).
Figure 5.
AAV-Mediated Expression of circRNAs in the Brain and the Eye
(A) The indicated constructs were packaged into recombinant AAV9 vectors and injected intravenously into C57/BL6 mice (n = 3) at a dose of 5.5 × 1011 vector genomes per animal, then harvested 4 weeks post-injection. Immunohistochemical staining of brain sections revealed astrocytic expression (representative images are shown). Scale bars, 300 μm. (B) Quantitation of the number of GFP-positive cells per brain section in the HIPK3 and ZKSCAN1 split GFP-injected animals. (C) qRT-PCR performed with primers amplifying GFP across the backsplice junction revealed expression of the circRNA in the brain. (D) The indicated constructs were packaged into recombinant AAV9 vectors and injected into the left cerebral ventricle of C57/BL6 at a dose of 4 × 109 vg per animal, then harvested 6 weeks post-injection (n = 3). Immunohistochemical staining of brain sections revealed astrocytic expression (representative images are shown). Scale bars, 300 μm. (E) The indicated constructs were packaged into recombinant AAV9 vectors and injected through the intravitreous route in C57/BL6 (n = 3) at a dose of 1 × 1010 vg per animal, then harvested 4 weeks post-injection. Immunofluorescent staining of sectioned retina revealed GFP expression in the retinal cup, in photoreceptors, and retinoid pigment epithelial cells (see zoom images). Green: immunofluorescent staining for GFP; blue: DAPI staining of cell nuclei. (F) Quantification of GFP expression by mean corrected fluorescence. (G) A separate cohort was injected as described above. RNA was extracted from retinas, and qRT-PCR was performed with a primer set amplifying GFP across the backsplice junction, revealing circRNA expression. Where indicated, *p < 0.05; **p < 0.005; ****p < 0.00005. Error bars represent 1 SD.
We also carried out direct intracerebroventricular (i.c.v.) injections of the AAV-circRNA vectors into the CSF of P0 neonatal mice at a dose of 4 × 109 vg per animal. As before, brains were harvested, and immunohistochemical staining was performed to visualize GFP expression. As with intravenous delivery, we observed GFP expression mainly in cortical astrocytes (Figure 5D). These results suggest that the astrocyte-specific expression profile is likely attributable to the biology of the circRNA constructs. Although further studies evaluating different intronic and IRES elements in combination with different AAV capsids will be required to understand the CNS expression profile of circRNAs, these proof-of-principle studies show that circRNAs can be expressed in the brain.
Further, we assessed AAV-mediated circRNA expression in the eye. In brief, rAAV9 vectors were injected into the vitreous of the mouse eye at a dose of 1 × 1010 vg per eye; eyecups were removed and the retinas sectioned, followed by immunofluorescence to visualize the GFP expression. Both ZKSCAN1 split GFP and HIPK3 split GFP showed extensive expression throughout the retinal cross section, whereas the control linIRES-GFP showed no expression (Figure 5E, quantified in Figure 5F). In this case, HIPK3 split GFP expression was higher than the ZKSCAN1 construct with predominant and widespread expression in photoreceptor cells and the retinal pigment epithelium (RPE). RNA was extracted from the retinas from a second cohort injected with rAAV-circRNA vectors for qRT-PCR analysis (Figure 5G). Consistent with GFP expression levels, ZKSCAN1 split GFP showed lower circRNA levels compared with HIPK3 split GFP, although these results were not statistically significant. These observations are particularly exciting because they provide the foundation for studying circRNA biogenesis and expression profiles in the eye for the first time.
Discussion
Our studies demonstrate that circRNAs can be overexpressed and translated in a variety of tissue types in vivo. We observed robust expression and translation in cardiac tissue, which is interesting because neither the endogenous circZKSCAN1 nor circHIPK3 are highly expressed in the heart.9 In the liver, circRNA expression was detectable, although protein translation was low, likely due to the EMCV IRES used in this study;38 ZKSCAN1 split GFP showed higher circRNA expression consistent with the endogenous profile reported for circZKSCAN1.9 In the brain, AAV-circRNA vectors showed expression in astrocytes, but not neurons. Further evaluation of circRNA constructs with neuron-specific promoters might provide additional insight into this observation, given a number of studies have shown neuronal circRNA expression, and many circRNAs are thought to function in neuronal processes.16, 18, 39, 40 The expression of endogenous circRNAs in the eye has not been reported to date. Excitingly, our vectors showed broad and robust expression in photoreceptors and the retinal pigment epithelium.
One of the distinguishing features of circRNAs is their increased stability compared with most linear mRNAs.8, 25 Indeed, we were able to demonstrate this in vitro with our vectors (Figure 2D). The current study was primarily focused on evaluating the feasibility of circRNA expression efficiency and the potential for translatability at a single time interval in vivo. However, with increased RNA stability, it is possible that the level of expression would increase over time in comparison with a linear message. Evaluating the durability of expression, persistence, and turnover rate of circRNAs expressed using AAV vectors in different tissue types would be of significant interest in future studies. In addition, it would be critical to understand whether increased stability of AAV-circRNA transcripts could help reduce the effective vector dose for gene therapy applications. This is especially important because high doses of rAAV vectors can induce inflammatory responses that result in ablation of transgene expression.41
The intronic elements characterized in the current study are derived from ZKSCAN1 and HIPK3, which are human genes containing an exon or exons that are circularized. The circularized regions are flanked by inverted Alu elements of the same family, which help drive the circularization event. Endogenous circZKSCAN1 and circHIPK3 RNAs are expressed at high levels within the brain, especially within the cerebellum;9 however, our vectors did not display a similar expression profile. One possible explanation for the latter observations is that the intronic elements utilized in our study are substantially truncated compared with the endogenous introns and might lack elements essential to maintain CNS expression. Additional studies focused on evaluating the impact of intron length and regulatory elements essential for broad CNS expression with such vector constructs are warranted. Nevertheless, the current studies provide the first proof-of-principle that artificial or naturally occurring circRNAs can be overexpressed using AAV vectors in vivo.
From a design perspective, it is important to note that the circRNA constructs described here could be modified to further adjust tissue specificity. Although the constructs used in the current study showed tissue-specific differences, we note that only two different intron pairs were evaluated. Given the large numbers of known circRNAs identified in a variety of studies,17, 39, 42 intron pairs derived from any of these could be tested for their expression level and tissue specificity. Thus, it may be possible to create a toolkit of intron pairs with targeted circRNA expression in a desired tissue and/or cell type. Similarly, we show that the EMCV IRES element used here can affect protein translation efficiency from circRNA. Consequently, evaluation of different viral or cellular IRES elements could provide another degree of control over circRNA expression. Taken together, we postulate that the current approach could afford multiple layers of regulation by modular incorporation of capsid, promoter, intronic, and IRES elements for spatio-temporal control of circRNAs.
We also note that circRNA vectors can potentially be used for expression of a wide variety of functional RNAs. For instance, noncoding RNAs could be expressed using our system, including various long non-coding RNAs (lncRNAs). In addition, circRNAs have the potential as a platform to express designer RNAs such as RNA binding proteins sponges or miRNA sponges (especially as endogenous circRNAs have been well characterized as miRNA sponges).17, 18, 43 Importantly, our platform could also serve as a powerful tool to study basic circRNA biology by enabling expression of circRNAs in vivo. Because the components of the circRNA, introns, IRES, and exon can each be readily altered in a modular fashion, it should be possible to create a variety of constructs for evaluation in a variety of tissues and animal models in vivo.
Materials and Methods
Plasmids
Plasmids containing a portion of the human ZKSCAN1 and HIPK3 genes were previously described.9 New vector cassettes were constructed using these naturally occurring intron sequences and by cloning into a plasmid backbone separated by a multiple cloning site. A cassette containing the EMCV IRES and split GFP was cloned between these intron sequences. The split GFP cassette used was derived from the circGFP plasmid (gift from Dr. Zefeng Wang).36 All plasmids were constructed in a backbone containing a CMV promoter, SV40 polyadenylation signal, and ITRs derived from the AAV2 genome.44 The linIRES-GFP construct consists of the EMCV IRES cloned directly in front of GFP, in the same backbone as the above constructs.
rAAV Vector Production
An updated triple-plasmid transfection protocol was used to generate rAAV vectors,45 with modifications. In brief, the transfection mixture contained: (1) the pXR helper plasmid; (2) the adenoviral helper plasmid pXX6-80; and (3) the indicated transgene, driven by a CMV promoter, flanked by AAV2 ITRs. Vector purification was carried out using iodixanol gradient ultracentrifugation followed by desalting with ZebaSpin desalting columns (40K MWCO; Thermo Scientific, Waltham, MA, USA). vg titers were obtained by qPCR (LightCycler 480; Roche Applied Sciences, Pleasanton, CA, USA) using primers designed to selectively bind AAV2 ITRs (forward, 5′- AACATGCTACGCAGAGAGGGAGTGG-3′; reverse, 5′- CATGAGACAAGGAACCCCTAGTGATGGAG-3′).
Cell Culture
U87 cells were cultured in DMEM (GIBCO/Life Technologies, Waltham, MA, USA) supplemented with 5% fetal bovine serum (FBS; Millipore-Sigma, St. Louis, MO, USA) and 1% gentamycin and kanamycin, and maintained at 37°C and 5% CO2. 300,000 cells were seeded into six-well plates, followed by transduction with the indicated rAAV vectors. Cells were imaged after transduction using an EVOS FL epifluorescence cell imaging system (AMC/Life Technologies, Waltham, MA, USA) equipped with the GFP light cube (excitation 470 nm, emission 510 nm).
RNA Extraction
RNA was extracted from frozen tissue stored in RNAlater or from adherent cells using TRIzol reagent (Invitrogen, Waltham, MA, USA) following the manufacturer’s protocol. For tissue samples, the tissues were first homogenized in TRIzol using a Tissue Lyser II (QIAGEN, Germantown, MD, USA).
Western Blotting
Lysates were recovered using 1× passive lysis buffer (Promega, Madison, WI, USA) and stored at −80°C. Samples were denatured in lithium dodecyl sulfate (LDS) and 100 mM DTT, and heated to 95°C before separation on a 10% Tris-glycine gel and transfer to a nitrocellulose membrane. Membranes were blocked overnight at 4°C in 5% non-fat milk in Tris-buffered saline with tween (TBST). Membranes were blotted with primary antibody against either GFP (1:1,000; SC9996; Santa Cruz Biotechnology, Dallas, TX, USA) or Actin (1:10,000; GeneTex GT5512; Irvine, CA, USA). Stabilized peroxidase-conjugated sheep anti-mouse antibody was used as secondary antibody (1:3,000; NA931V; GE Healthcare, Chicago, IL, USA). Blots were developed using SuperSignal West Femto substrate (Thermo Scientific/Life Technologies, Waltham, MA, USA) and visualized by autoradiography or by the Chemiluminescence High Sensitivity protocol on a ChemiDoc XRS+ (Bio-Rad, Hercules, CA, USA) for quantification purposes.
Northern Blotting
10 μg of total RNA was resuspended in denaturing buffer (67% deionized formamide, 6.7% formaldehyde, 1× 3-morpholinopropane-1-sulfonic acid [MOPS] running buffer), incubated at 60°C for 10 min, and cooled on ice. Samples were separated on a 1.2% denaturing agarose gel and subsequently transferred to Hybond N+ membrane (GE Healthcare, Chicago, IL, USA). A radiolabeled probe was generated using the Prime-It II Random Primer Labeling Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. DNA templates for probe labeling were generated by PCR with the following primers amplifying split GFP (5′-GCATGCTCTTCTCAGGAGCGCACCATCTTCTTCAAGGACGACGG-3′, 5′-GCATGCTCTTCTTACCTGGACGTAGCCTTCGGGCATGGC-3′). Radiolabeled probe was purified using illustra MicroSpin G-50 columns (GE Healthcare, Chicago, IL, USA) following the manufacturer’s protocol. Probe was then hybridized to the membrane in Rapid-Hyb buffer (GE Healthcare, Chicago, IL, USA). Blots were visualized by exposure to film, and radiolabel signal was quantified by exposure to a PhosphorImager screen.
Actinomycin Treatment
300,000 U87 cells were seeded into 35-mm plates and transduced with 100,000 vg/cell of the indicated rAAV2 vectors. At 3 days after transduction, media were removed and replaced with fresh media for 30 min. These media were removed and replaced with pre-warmed and equilibrated media containing 5 μg/mL actinomycin D (Sigma-Aldrich, St. Louis, MO, USA). Cells were treated for 30 min; then media were removed and replaced with fresh media. Cells were harvested in TRIzol reagent (Invitrogen, Waltham, MA, USA) at 0-, 4-, 8-, 12-, and 24-hr time points. RNA was extracted and analyzed by qRT-PCR or northern blotting as described above.
Intravenous Administration
Animal experiments reported in this study were conducted with C57/Bl6 mice bred and maintained in accordance to NIH guidelines as approved by the The University of North Carolina at Chapel Hill (UNC) Institutional Animal Care and Use Committee (IACUC). Animals were injected intravenously through the tail vein with 5.5 × 1011 vg/animal. At 4 weeks after injection, mice were overdosed with tribromoethanol (Avertin) (0.2 mL/10 g of 1.25% solution) via the intraperitoneal route. This was followed by transcardial perfusions of PBS. Portions of the harvested organs were cut and stored in RNAlater solution (Invitrogen, Waltham, MA, USA); the remainder was postfixed in 4% paraformaldehyde. For this study, n = 3 mice were injected and processed for each construct.
i.c.v. Administration
Animal experiments reported in this study were conducted with C57/Bl6 mice bred and maintained in accordance to NIH guidelines as approved by the UNC IACUC. Pups at post-natal days 1–2 were rapidly anesthetized on ice for 2 min followed by stereotaxic i.c.v. injections. Specifically, vectors packaging different transgenes were injected at a dose of 4 × 109 vector genomes into the left lateral ventricle (total volume < 3 μL) using a Hamilton 700 series syringe with a 26s gauge needle (Sigma-Aldrich, St. Louis, MO, USA), attached to a KOPF-900 small-animal stereotaxic instrument (KOPF Instruments, Tujunga, CA, USA). All neonatal injections were performed 0.5 mm relative to the sagittal sinus, 2 mm rostral to transverse sinus, and 1.5 mm deep. Following vector administration, mice were revived under a heat lamp and rubbed in the bedding before being placed back with the dam. At 6 weeks after injection, mice were overdosed with tribromoethanol (Avertin) (0.2 mL/10 g of 1.25% solution) via the intraperitoneal route. This was followed by transcardial perfusions of 4% paraformaldehyde in PBS. The brain was harvested and postfixed in 4% paraformaldehyde for 24 hr. For this study, n = 3 mice were injected and processed for each construct.
Tissue Processing and Immunohistochemistry
Using fixed tissues, we obtained 50-μm-thick sections using a Leica VT 1,200S vibrating blade microtome (Leica Biosystems, Buffalo Grove, IL, USA). Immunohistochemical analyses of GFP expression was conducted using an antibody against GFP (sc-9996; Santa Cruz Biotechnology, Dallas, TX, USA) and the Vectastain ABC-HRP kit (Rabbit IgG PK-4001 kit; Vector Biolabs, Burlingame, CA, USA) according to the manufacturer’s protocol. Zeiss CLSM 700 confocal laser scanning microscope was used for imaging sections of different organs after immunostaining (Microscopy Services Laboratory, UNC).
Quantification of GFP Expression in Tissues
Quantification of cardiac sections was performed in ImageJ. Sections were converted to 16-bit images and inverted, followed by background removal by the rolling ball method. The integrated density and area of each section was measured, as well as for a no-staining control section. Density was normalized to area, and background values (from the no-stain control) subtracted. Quantification of brain sections was performed by counting of GFP+ cells. The number of GFP+ cells in an entire brain section was counted, and at least two sections were counted for each animal.
RT-PCR
5 μg of RNA was DNase treated using the Turbo DNA-free kit (Ambion/Life Technologies, Waltham, MA, USA). Equal nanogram amounts of DNase-treated RNA were converted to cDNA using the High Capacity RNA-to-cDNA kit (Applied Biosystems/Life Technologies, Waltham, MA, USA). Products of this reverse transcription reaction were utilized as template for PCR (or qPCR) using gene-specific primers for GFP (5′-CTGCTTGTCGGCCATGATATAGACGTTGTGGC-3′, 5′-CAAGCTGACCCTGAAGTTCATCTGCACCACC-3′), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (5′-CCACTCCTCCACCTTTGAC-3′, 5′-ACCCTGTTGCTGTAGCC-3′), c-myc (5′- CGTCTCCACACATCAGCACAA-3′, 5′-CACTGTCCAACTTGACCCTCTTG-3′), and TBP (5′- TGCACAGGAGCCAAGAGTGAA-3′, 5′-CACATCACAGCTCCCCACCA-3′). Non-qPCR products were visualized on an agarose gel. For RNase R experiments, 5 μg of RNA was treated with 5 units of RNase R (Epicenter, Madison, WI, USA) at 37°C for 10 min. Enzyme was inactivated at 95°C for 5 min. The resulting RNA was used for RT-PCR as described above.
Quantification of Vector Genome Copy Number in Tissues
Genomic DNA was extracted from sections of fixed tissue using the QIAamp DNA FFPE Tissue Kit (QIAGEN, Germantown, MD, USA). To calculate viral genome copy numbers, we performed qPCR with primers specific to the CMV promoter (5′-CAAGTACGCCCCCTATTGAC-3′ and 5′-AAGTCCCGTTGATTTTGGTG-3′). The vector genome copy numbers were normalized to the mouse lamin B2 locus as the housekeeping gene (primers 5′-GGACCCAAGGACTACCTCAAGGG-3′ and 5′- AGGGCACCTCCATCTCGGAAAC-3′).
Intravitreal Administration
Animal experiments reported in this study were conducted with C57/Bl6 mice bred and maintained in accordance to NIH guidelines as approved by the UNC IACUC. Prior to injection, eyes were dilated with 1% atropine and 2.5% phenylephrine HCl (Akorn, Lake Forest, IL, USA) ophthalmic solution. Mice were anesthetized using 200 mg/kg tribromoethanol (Avertin). A small hole was made at the limbus of the eye with a 30G needle; then 1 μL of virus (at a dose of 1 × 1010 vector genomes) was slowly delivered with a 34G needle on a Hamilton gastight syringe. Four weeks post-injection, mice were sacrificed via CO2 inhalation. Limbi were marked at the top of each eye to facilitate orientation. Following this, eyes were enucleated in 4% paraformaldehyde and incubated at 4°C overnight. Cornea was dissected away from the eyecup; then the eyecup was immersed in 30% sucrose for 3 hr, cryoprotected in Optimal Cutting Temperature compound (Sakura Finetek, Torrance, CA, USA), and stored at −20°C. Where noted, the eyecup was stored in RNAlater (Invitrogen, Waltham, MA, USA) for later RNA extraction. For this study, n = 6 eyes were injected for each construct; half were processed for immunofluorescence and half for RNA.
Ocular Tissue Processing and Immunofluorescence
12-micron retinal sections were cut on a Leica CM3050 cryostat (Leica Biosystems, Buffalo Grove, IL, USA). Slides were then rinsed with 1× PBS (GIBCO, Germantown, MD, USA) three times. Retinal sections on the slides were covered with 0.5% Triton X-100 and 1% BSA (Fisher Scientific, Waltham, MA, USA), each for 1 hr. Rabbit anti-GFP (G10362; Invitrogen, Waltham, MA, USA) was diluted 1:750 in 1% BSA + 0.3% Triton X-100 and incubated with the sections at 4°C overnight. Slides were rinsed with 1× PBS and incubated with Alexa Fluor goat anti-rabbit 488 (1:500; A-11008; Invitrogen, Waltham, MA, USA) at room temperature. Following rinsing, ProLong Gold DAPI containing mounting media (Life Technologies, Waltham, MA, USA) was applied and coverslips placed. Fluorescence images were taken by Zeiss LSM 710 Spectral Confocal Laser Scanning Microscope at the UNC Microscopy Service Laboratory (MSL). Fluorescence was quantified in ImageJ by the corrected total fluorescence method.
Statistical Analysis
Statistical analysis was carried out using an unpaired, one-tailed Student t test. Where indicated, n.s. represents not significant; *p < 0.05; **p < 0.005; ***p < 0.0005; ****p < 0.00005. Error bars represent 1 SD.
Author Contributions
R.M.M., E.K.B., and A.A. designed research. R.M.M., R.M.C.R., and M.L.S. performed research. J.E.W. contributed new reagents/analytic tools. R.M.M., M.L.S., W.F.M., and A.A. analyzed data. R.M.M., W.F.M., and A.A. wrote the manuscript.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
We would like to thank Dr. Zefeng Wang for providing the split GFP plasmid. We would also like to thank Dr. Silvia Ramos for helpful advice regarding actinomycin D treatments. This work was supported by the NIH (grant R01NS099371 to A.A., W.F.M., and J.E.W. and grant 5T32GM007092 to R.M.M. and E.K.B.). J.E.W. is a Rita Allen Foundation Scholar. Funding for the open access charge was provided by the NIH.
References
- 1.Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danan M., Schwartz S., Edelheit S., Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 4.Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 5.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 6.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 7.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Liang D., Tatomer D.C., Luo Z., Wu H., Yang L., Chen L.L., Cherry S., Wilusz J.E. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell. 2017;68:940–954.e3. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glažar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 17.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebbesen K.K., Kjems J., Hansen T.B. Circular RNAs: identification, biogenesis and function. Biochim. Biophys. Acta. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. Translation of circRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C.Y., Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 24.Borchardt E.K., Meganck R.M., Vincent H.A., Ball C.B., Ramos S.B.V., Moorman N.J., Marzluff W.F., Asokan A. Inducing circular RNA formation using the CRISPR endoribonuclease Csy4. RNA. 2017;23:619–627. doi: 10.1261/rna.056838.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samulski R.J., Muzyczka N. AAV-mediated gene therapy for research and therapeutic purposes. Annu. Rev. Virol. 2014;1:427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]
- 27.Buller R.M., Janik J.E., Sebring E.D., Rose J.A. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 1981;40:241–247. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atchison R.W., Casto B.C., Hammon W.M. Adenovirus-associated defective virus particles. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 29.Srivastava A., Lusby E.W., Berns K.I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hauswirth W.W., Berns K.I. Origin and termination of adeno-associated virus DNA replication. Virology. 1977;78:488–499. doi: 10.1016/0042-6822(77)90125-8. [DOI] [PubMed] [Google Scholar]
- 31.Kotin R.M., Siniscalco M., Samulski R.J., Zhu X.D., Hunter L., Laughlin C.A., McLaughlin S., Muzyczka N., Rocchi M., Berns K.I. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakai H., Yant S.R., Storm T.A., Fuess S., Meuse L., Kay M.A. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao G., Vandenberghe L.H., Alvira M.R., Lu Y., Calcedo R., Zhou X., Wilson J.M. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao G.-P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartel M.A., Weinstein J.R., Schaffer D.V. Directed evolution of novel adeno-associated viruses for therapeutic gene delivery. Gene Ther. 2012;19:694–700. doi: 10.1038/gt.2012.20. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 38.Licursi M., Christian S.L., Pongnopparat T., Hirasawa K. In vitro and in vivo comparison of viral and cellular internal ribosome entry sites for bicistronic vector expression. Gene Ther. 2011;18:631–636. doi: 10.1038/gt.2011.11. [DOI] [PubMed] [Google Scholar]
- 39.You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mingozzi F., High K.A. Immune responses to AAV vectors: overcoming barriers to successful gene. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y., Wang J., Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jost I., Shalamova L.A., Gerresheim G.K., Niepmann M., Bindereif A., Rossbach O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018 doi: 10.1080/15476286.2018.1435248. Published online February 28, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao X., Xiao W., Li J., Samulski R.J. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J. Virol. 1997;71:941–948. doi: 10.1128/jvi.71.2.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen S., Horowitz E.D., Troupes A.N., Brown S.M., Pulicherla N., Samulski R.J., Agbandje-McKenna M., Asokan A. Engraftment of a galactose receptor footprint onto adeno-associated viral capsids improves transduction efficiency. J. Biol. Chem. 2013;288:28814–28823. doi: 10.1074/jbc.M113.482380. [DOI] [PMC free article] [PubMed] [Google Scholar]