Abstract
Background & Aims
Although nearly half of pancreatic ductal adenocarcinoma (PDAC) patients have diabetes mellitus with episodes of hyperglycemia, its tumor microenvironment is hypoglycemic. Thus, it is crucial for PDAC cells to develop adaptive mechanisms dealing with oscillating glucose levels. So far, the biological impact of such glycemic variability on PDAC biology remains unknown.
Methods
Murine PDAC cells were cultured in low- and high-glucose medium to investigate the molecular, biochemical, and metabolic influence of glycemic variability on tumor behavior. A set of in vivo functional assays including orthotopic implantation and portal and tail vein injection were used. Results were further confirmed on tissues from PDAC patients.
Results
Glycemic variability has no significant effect on PDAC cell proliferation. Hypoglycemia is associated with local invasion and angiogenesis, whereas hyperglycemia promotes metastatic colonization. Increased metastatic colonization under hyperglycemia is due to increased expression of runt related transcription factor 3 (Runx3), which further activates expression of collagen, type VI, alpha 1 (Col6a1), forming a glycemic pro-metastatic pathway. Through epigenetic machinery, retinoic acid receptor beta (Rarb) expression fluctuates according to glycemic variability, acting as a critical sensor relaying the glycemic signal to Runx3/Col6a1. Moreover, the signal axis of Rarb/Runx3/Col6a1 is pharmaceutically accessible to a widely used antidiabetic substance, metformin, and Rar modulator. Finally, PDAC tissues from patients with diabetes show an increased expression of COL6A1.
Conclusions
Glycemic variability promotes both local invasion and metastatic colonization of PDAC. A pro-metastatic signal axis Rarb/Runx3/Col6a1 whose activity is controlled by glycemic variability is identified. The therapeutic relevance of this pathway needs to be explored in PDAC patients, especially in those with diabetes.
Keywords: Pancreatic Cancer, Glucose Metabolism, Metastasis, Retinoic Acid
Abbreviations used in this paper: ADP, adenosine diphosphate; ATP, adenosine triphosphate; Caix, carbonic anhydrase IX; Col6a1, collagen; type VI, alpha 1; CT, computed tomography; 2DG, 2-deoxy-D-glucose; ECM, extracellular matrix; Egr2, early growth response 2; FBS, fetal bovine serum; IHC, immunohistochemistry; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PDAC, pancreatic ductal adenocarcinoma; PET, positron emission tomography; qRT-PCR, quantitative real-time polymerase chain reaction; RA, retinoic acid; Rarb, retinoic acid receptor beta; Runx3, runt related transcription factor 3
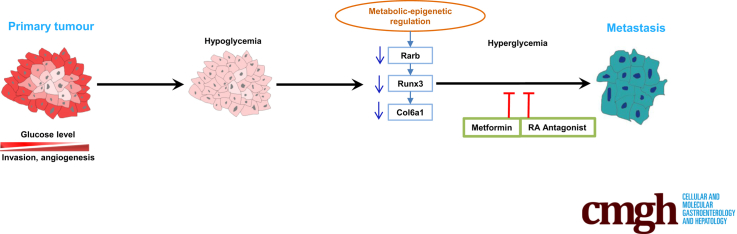
Graphical abstract
Summary.
Glycemic variability constitutes a major driving force for pancreatic ductal adenocarcinoma local invasion, angiogenesis, and metastatic colonization. Through epigenetic machinery, retinoic acid signaling acts as a critical sensor of glycemic variability that relays the glycemic signal to pro-metastatic Runx3/Col6a1 pathway.
Pancreatic ductal adenocarcinoma (PDAC) is a devastating cancer.1 Unlike other malignant entities, PDAC has a bi-directional connection with diabetes mellitus,2, 3 with diabetes being both a risk factor and an early sign of the disease; nearly 50% of PDAC patients have diabetes at the time of diagnosis.4 PDAC patients with diabetes tend to have larger tumors and worse prognosis.5, 6, 7 PDAC is poorly vascularized, and its tumor microenvironment is hypoxic and severely hypoglycemic.8 Thus, PDAC cells (particularly those in diabetic patients) are likely to experience oscillating extracellular glucose levels. So far, the biological significance of transient presumed hypoglycemia and hyperglycemia in pancreatic carcinogenesis remains undefined.
Recent sequencing results of human PDAC have uncovered a limited heterogeneity in the genomes of primary tumors and metastases; the metastatic lesion has the same driver gene mutation as its primary cancer. In particular, no metastasis-specific driver mutation was discovered.9 This raises an important question: what is the driving force triggering early metastasis. In this regard, a recent landmark study demonstrated that metastatic PDAC carried a distinctive large-scale reprogramming of chromatin modifications targeting genes related to anabolic glucose metabolism,10 underscoring the importance of the metabolic–epigenetic program for glucose metabolism in promoting PDAC metastases. These findings are in line with clinical observations that PDAC metastases are largely tropic for organs (liver, lung) with glucose replete condition relative to hypoglycemic primary tumors.11
Collagen, type VI, alpha 1 (Col6a1) is a secreted pro-metastatic mediator promoting metastatic colonization of PDAC,12, 13 whose expression is specifically controlled by runt related transcription factor 3 (Runx3), forming a distinct pro-metastatic signaling axis. In PDAC, Runx3 expression is affected by 2 major driver mutations, p53 (TP53) and Smad4.12, 13 Here, Runx3 is induced by either p53 gain-of-function mutation or complete loss of Smad4. In other biological contexts, Runx3 has been previously shown to be activated by retinoic acid (RA) signaling.14, 15 Although RA signaling and its receptors are dysregulated in PDAC,16, 17 it is unclear whether RA-mediated Runx3 activation is functionally relevant in promoting a metastatic phenotype of PDAC.
Here we investigated the biological impact of glycemic variability on PDAC cells. This analysis reveals that hypoglycemia is associated with local invasion and angiogenesis, whereas hyperglycemia promotes metastatic colonization. The phenotypic difference in metastatic colonization is due to the increased expression of Runx3/Col6a1. Further analysis identifies RA signaling as the key glucose-sensing pathway that promotes Runx3/Col6a1 expression, which is operated by epigenetically regulated retinoic acid receptor beta (Rarb) expression. Moreover, this glycemic pathway is pharmaceutically accessible by using metformin or Rar modulator. Finally, PDAC sections from patients with diabetes show increased expression of COL6a1. These data define the signal axis of Rarb/Runx3/Col6a1 as a potential target for PDAC patients with diabetes.
Results
Glycemic Variability Has No Effect on Pancreatic Ductal Adenocarcinoma Proliferation
Previously, we characterized a highly metastatic mouse model of PDAC that was driven by one allele loss of tuberous sclerosis 1 together with pancreas-specific oncogenic KrasG12D expression (p48Cre; KrasG12D/+; Tsc1-/+).18, 19, 20 A particular signature of "Mek/Erk-mTOR" was identified by using cancer cell lines from those animals. The Kyoto Encyclopedia of Genes and Genomes pathway analysis of this signature uncovered the glucose-related metabolism as the major enriched pathway.20 Thus, we chose one representative cell line, 399 cells (showing epithelial-like morphology in 2-dimensional culture system), to investigate the biological influence of glycemic variability on tumor behavior (Figure 1A).
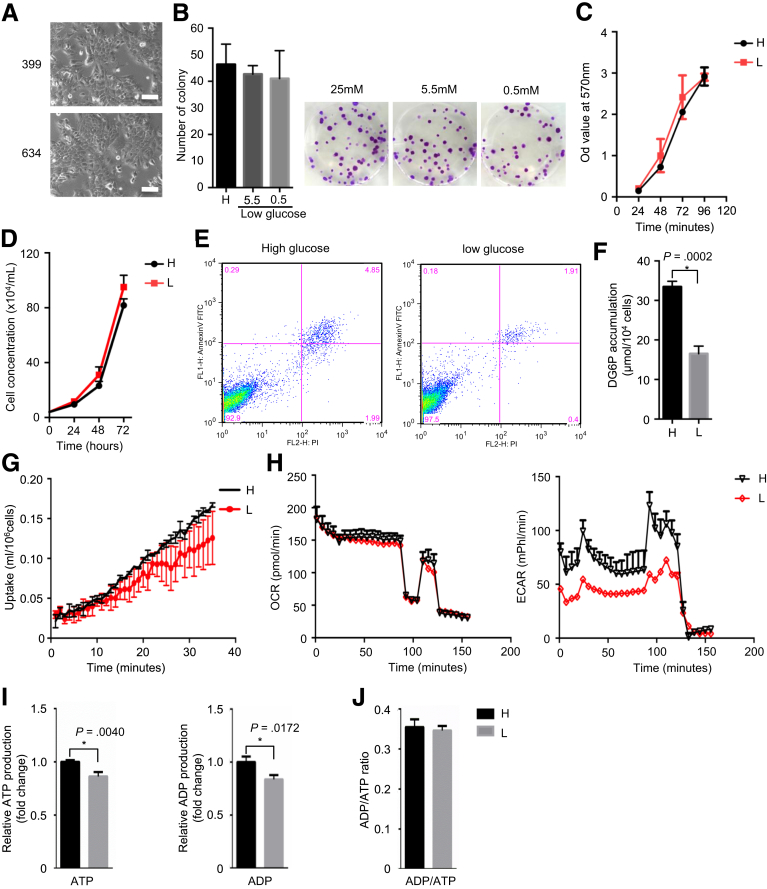
Figure 1.
Glycemic variability has no effect on PDAC proliferation. (A) Representative pictures of 399 and 634 cells in 2-dimensional culture system; scale bars, 25 μmol/L. (B) Colony formation results (left panel) and representative pictures (right panel) show glycemic variability has no effect on cell proliferation. (C) MTT assay shows comparable proliferation rate after long-term culturing in high-glucose (H) and low-glucose (L) medium. (D) Live cell counting results show similar proliferation rate of hyperglycemic (H) and hypoglycemic (L) PDAC cells. (E) FACS analysis for annexin V and propidium iodide (PI) demonstrates comparable apoptotic and necrotic rates of hyperglycemic (H) and hypoglycemic (L) PDAC cells. (F and G) 2DG and 18F-fludeoxyglucose–based glucose uptake assays determine glucose uptake rate in high-glucose cultured and low-glucose cultured PDAC cells. (H) Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) analysis show mitochondrial respiration and glycolytic activity, respectively, in high-glucose and low-glucose cultured PDAC cells. (I and J) ATP, ADP, and ADP/ATP ratio analysis determine basic cellular energy homeostasis in high-glucose and low-glucose cultured PDAC cells. All data are presented as mean ± standard deviation; data from 3 independent experiments are shown. Unpaired t test is used to examine statistical significance, *P < .05.
First, we tested the influence of glycemic variability on anchorage-dependent growth by culturing 399 cells in medium supplemented with 10% non-dialyzed fetal bovine serum (FBS) containing 2 mmol/L glutamine and a range of glucose levels. Here, neither high levels of glucose (25 mmol/L) nor previously defined low levels of glucose (0.5 mmol/L) had significant effects (Figure 1B)21; thus we chose medium containing 0.5 mmol/L medium as hypoglycemia culture medium for further experiment. Even after long-term culturing in low-glucose medium (>30 days), no difference in cell proliferation was observed (as determined by MTT and live cell counting assays; Figure 1C and D). Furthermore, no difference in the apoptotic and necrotic rates was found by fluorescence-activated cell sorting analysis (Figure 1E). However, PDAC cells maintained in low-glucose medium showed around 50% decrease in glucose uptake compared with those in high-glucose medium (Figure 1F, P = .0002). A fludeoxyglucose F 18 uptake assay based on microfluidic system confirmed these results by showing a real-time reduction in glucose uptake in PDAC cells maintained in low-glucose medium22 (Figure 1G). The reduced glucose uptake was reflected in reduced glycolytic activity as judged by extracellular acidification rates. The simultaneous assessment of cellular respiration by plate-based respirometry revealed no differences in mitochondrial energy turnover (Figure 1H). The reduced metabolic status of PDAC cells in low glucose was seen in lower adenosine triphosphate (ATP) and adenosine diphosphate (ADP) concentrations (Figure 1I) but without changing the ADP/ATP ratio (Figure 1J). Taken together, glycemic variability has no effect on cell proliferation; PDAC cells maintained in high-glucose medium (25 mmol/L) were defined as hyperglycemic PDAC cells, whereas PDAC cells maintained in low-glucose medium (0.5 mmol/L) were defined as hypoglycemic PDAC cells.
Hypoglycemia Is Associated With Local Invasion/Angiogenesis and Hyperglycemia Promotes Metastatic Colonization
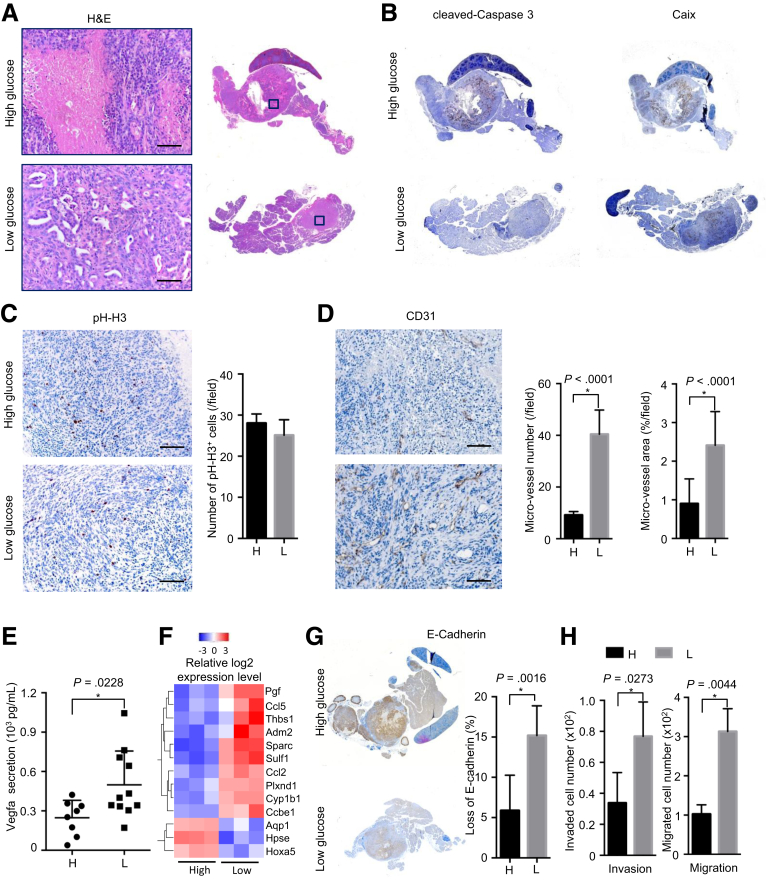
First, we performed orthotopic injection to determine the in vivo phenotype of PDAC cells maintained in hyperglycemic and hypoglycemic conditions. Although no difference in tumor size and stroma density was observed, tumors formed by hypoglycemic PDAC cells contained less tumor necrosis (Figure 2A, validated by cleaved-caspase 3 and carbonic anhydrase IX (Caix) staining, Figure 2B). The proliferation rate was similar (Figure 2C). As determined by CD31 staining, microvessel density was significantly elevated in tumors formed by hypoglycemic PDAC cells compared with those formed by hyperglycemic PDAC cells (Figure 2D). In line, hypoglycemic PDAC cells secreted a higher amount of vascular endothelial growth factor A, a potent pro-angiogenic factor, compared with hyperglycemic cells (Figure 2E). Transcriptome analysis revealed that genes related to the regulation of angiogenesis and vasculature development were mostly enriched in hypoglycemic PDAC cells (as determined by Gene Ontology; Figure 2F, Table 1). Meanwhile, hypoglycemic PDAC cells gave rise to tumors showing a locally more invasive phenotype. The pathological analysis revealed that the ratio of the solid (dedifferentiated) versus ductal component was higher in the hypoglycemic cells–derived tumors than the hyperglycemic ones. This was also reflected by the staining pattern of E-cadherin. The quantitative analysis revealed that the tumor area negative for E-cadherin staining was significantly larger in hypoglycemic cells–derived tumors (Figure 2G). In vitro invasion and migration assays confirmed that hypoglycemic PDAC cells were more invasive than hyperglycemic cells (Figure 2H).
Figure 2.
Hypoglycemia links with local invasion/angiogenesis. (A) Representative H&E staining pictures show larger necrotic area in tumors formed by hyperglycemic cells; high glucose (H), hyperglycemic cells; low glucose (L), hypoglycemic cells; scale bar: 100 μm. (B) Representative IHC pictures of cleaved-caspases 3 (left panel) and Caix (right panel) show tumor necrosis in tumors formed by hyperglycemic (H) and hypoglycemic (L) PDAC cells; H, high glucose; L, low glucose. (C) Representative IHC pictures of phospho-histone H3 (pH-H3, left panel) and quantitative analysis (right panel) demonstrate the proliferation status in vivo, scale bar: 100 μm. (D) Representative pictures of CD31 staining (left panel) and microvessel density calculation (right panel) demonstrate increased angiogenesis in tumors formed by hyperglycemic cells, scale bar: 100 μm. (E) Vascular endothelial growth factor type A (Vegfa) enzyme-linked immunosorbent assay shows vascular endothelial growth factor type A secretion in hyperglycemic and hypoglycemic PDAC cells. (F) Heatmap of genes related to angiogenesis indicated on the right; voxel color: transcriptional up-regulation (red) and down-regulation (blue) as compared with the mean. (G) Representative IHC pictures (left panel) and quantitative analysis (right panel) show higher rate of E-cadherin loss in tumors derived from hypoglycemic (L) cells compared with hyperglycemic (H) cells. (H) Invasion (left panel) and migration (right panel) assay demonstrate invasion and migration capacities of hyperglycemic and hypoglycemic PDAC cells in vitro. All data are presented as mean ± standard deviation, and data from 3 independent experiments are shown. Unpaired t test is applied, *P < .05.
Table 1.
Gene Ontology (GO) Terms Analysis
| GOBPID | P value | Count | Size | Term | |
|---|---|---|---|---|---|
| 1 | GO:0052547 | 1.05E-06 | 12 | 319 | Regulation of peptidase activity |
| 2 | GO:0010466 | 5.79E-06 | 9 | 199 | Negative regulation of peptidase activity |
| 3 | GO:0045765 | 7.83E-06 | 8 | 156 | Regulation of angiogenesis |
| 4 | GO:0001525 | 8.09E-06 | 11 | 324 | Angiogenesis |
| 5 | GO:0001944 | 1.47E-05 | 13 | 484 | Vasculature development |
| 6 | GO:1901342 | 1.53E-05 | 8 | 171 | Regulation of vasculature development |
| 7 | GO:0051346 | 2.16E-05 | 10 | 295 | Negative regulation of hydrolase activity |
| 8 | GO:0044767 | 6.75E-05 | 42 | 3722 | Single-organism developmental process |
| 9 | GO:0032502 | 7.54E-05 | 42 | 3739 | Developmental process |
| 10 | GO:0043086 | 9.28E-05 | 12 | 499 | Negative regulation of catalytic activity |
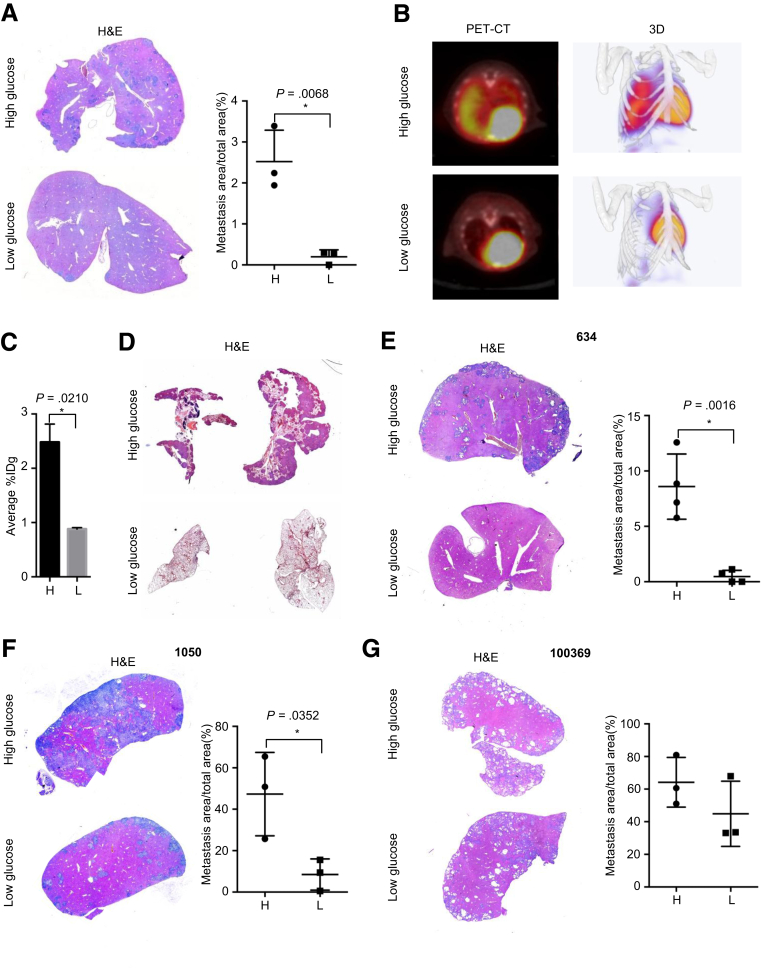
To characterize the hepatic metastatic colonization of hypoglycemic and hyperglycemic PDAC cells, we performed portal vein injections. Here, the percentage of the area of hepatic metastasis in hyperglycemic PDAC cells was 5-fold higher than that of hypoglycemic cells (Figure 3A, P = .0068). To further validate these results, we performed tail vein injections to assess the pulmonary colonization of these cells. As determined by standardized uptake values, a quantitative analysis using positron emission tomography (PET)–computed tomography (CT) scans, the pulmonary metastasis load of animals injected with hyperglycemic PDAC cells was 2.48% ± 0.24% IDg, which was higher than that of hypoglycemia cells (0.88% ± 0.02% IDg; Figure 3B and C, P = .02), which was confirmed by histologic analysis (Figure 3D). Because of significant difference in metastatic colonization, we repeated the experiments in 3 other murine PDAC cell lines: 634 cells established also from p48Cre; KrasG12D/+; Tsc1-/+ mice; less metastatic 1050 cells isolated from p48Cre; KrasG12D/+; p53-/+ mice; and 10069 cells containing a p53R172H mutation obtained from Pdx1Cre; KrasG12D/+; p53R172H/+ mice.20 Consistently, this analysis revealed that the hypoglycemia dramatically inhibited metastatic capacities of 634 and 1050 cells (Figure 3E and F). However, no effect was observed in 10069 cells (Figure 3G).
Figure 3.
Hyperglycemia promotes metastatic colonization. (A) Representative H&E staining pictures (left panel) and metastatic area calculation (right panel) show hepatic colonization of hyperglycemic and hypoglycemic cells. (B) Representative PET-CT scan and reconstructed 3-dimensional pictures show pulmonary colonization of hyperglycemic and hypoglycemic cells. (C) Quantitative analysis of average standard uptake value demonstrates pulmonary colonization of hyperglycemic and hypoglycemic cells. (D) Representative pictures of H&E staining show increased pulmonary colonization by hyperglycemic PDAC cells. (E–G) Representative H&E staining pictures (left panel) and metastatic area calculation (right panel) show hepatic colonization of hyperglycemic and hypoglycemic 634, 1050, and 100369 cells (n ≥ 3 for each group); All data are presented as mean ± standard deviation, and data from 3 independent experiments are shown. Unpaired t test is used to examine statistical significance, *P < .05.
Taken together, these data demonstrate that hypoglycemia is associated with local invasion/angiogenesis, whereas hyperglycemia promotes metastatic colonization.
Collagen, Type VI, Alpha 1 Is Regulated by Glycemic Variability to Promote Metastatic Colonization
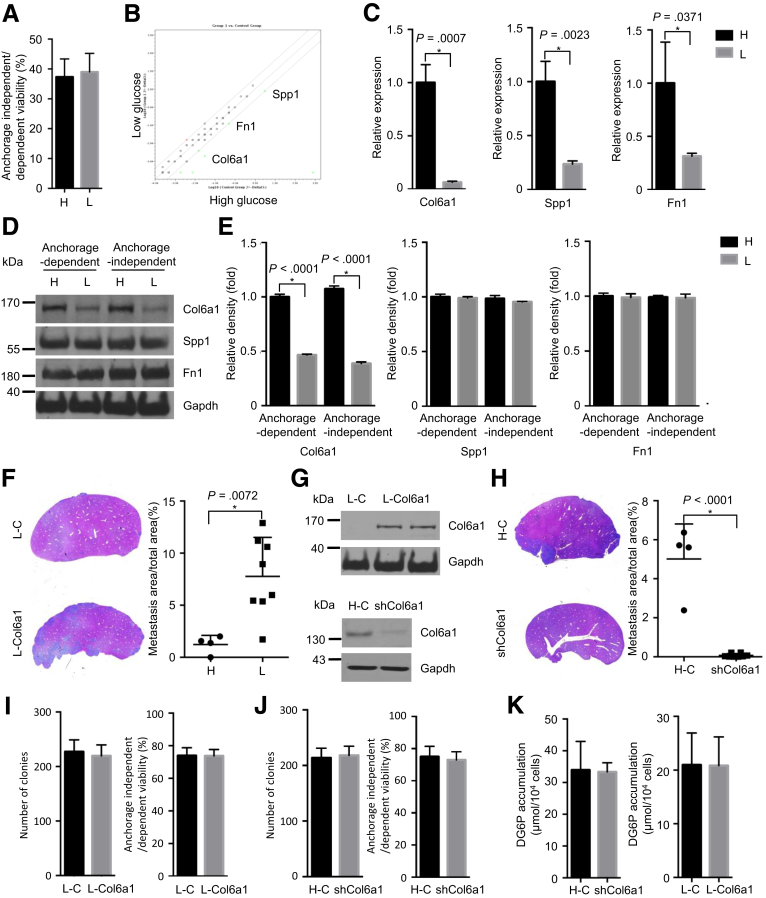
Because a pronounced difference in metastatic colonization between hypoglycemic and hyperglycemic PDAC cells was observed, we investigated the underlying molecular mechanism responsible for this difference in metastatic colonization. An anoikis assay was performed to test the ability of hypoglycemic and hyperglycemic PDAC cells to survive under anchorage-independent conditions, which is the first step after extravasation to form metastatic colonization.23 This analysis revealed no difference (Figure 4A), excluding the anoikis effect on hypoglycemic and hyperglycemic PDAC cells. Because previously published data showed extracellular matrix (ECM) plays an important role in mediating metastatic colonization,12, 24 we evaluated ECM protein expression under anchorage-independent conditions by using a polymerase chain reaction (PCR) array. Interestingly, 3 previously defined pro-metastatic ECM mediators, Col6a1, secreted phosphoprotein 1, also known as osteopontin, and fibronectin 1, were up-regulated in hyperglycemic PDAC cells at mRNA levels (confirmed by independent quantitative real-time polymerase chain reaction [qRT-PCR] assays; Figure 4B and C).12 However, only the change of Col6a1 could be confirmed on protein levels (as determined by Western blot analysis in cell lysates and densitometry analysis; Figure 4D and E). To test the functional relevance of Col6a1 in mediating metastatic colonization, we overexpressed Col6a1 in hypoglycemic PDAC cells by transfecting a Col6a1-expressing vector and down-regulated its expression in hyperglycemic PDAC cells by transfecting a Col6a1-shRNA vector. This analysis revealed that Col6a1 overexpression restored the metastatic capacity of hypoglycemic PDAC cells (Figure 4F and G, P = .0072), whereas Col6a1 down-regulation inhibited the metastatic ability of hyperglycemic PDAC cells (Figure 4G and H, P < .0001). Cell proliferation and cell viability under anchorage-independent conditions were not altered (Figure 4I and J). Furthermore, the altered expression of Col6a1 had no effect on glucose uptake in hypoglycemic and hyperglycemic PDAC cells (Figure 4K).
Figure 4.
Col6a1 is regulated by glycemic variability to promote metastatic colonization. (A) Anoikis assay shows cell viability under anchorage-independent state; H, hyperglycemic cells; L, hypoglycemic cells. (B and C) PCR array (left panel) and qRT-PCT results (right panel) demonstrate altered expression in ECM genes between hyperglycemic and hypoglycemic PDAC cells; H, high glucose; L, low glucose. (D) Western blot analyses show expressions of Col6a1, Spp1, Fn1 under anchorage-dependent and -independent states in hyperglycemic and hypoglycemic PDAC cells; Gapdh is used as a housekeeping gene. (E) Densitometry analysis reveals reduced expression of Col6a1, but not Spp1 and Fn1. (F) Representative pictures of H&E staining (left panel) and metastatic area quantification (right panel) results demonstrate that Col6a1 overexpression restores the ability of metastatic colonization of hypoglycemic PDAC cells. (G) Representative pictures of H&E staining (left panel) and metastatic area quantification (right panel) show that Col6a1 down-regulation impairs the capacity of metastatic colonization in hyperglycemic PDAC cells. (I) Western blot analysis shows Col6a1 overexpression in hypoglycemic PDAC cells and Col6a1 down-regulation in hyperglycemic PDAC cells. (H) Representative pictures of H&E staining (left panel) and metastatic area quantification (right panel) show that Col6a1 down-regulation impairs the capacity of metastatic colonization in hyperglycemic PDAC cells. (I) Colony formation (left panel) and anoikis assay (right panel) determine the cell proliferation rate under anchorage-dependent and -independent state in control and Col6a1-overexpressing hypoglycemic PDAC cells. (J) Colony formation (left panel) and anoikis assay (right panel) show cell proliferation rate under anchorage-dependent and -independent state in control and Col6a1 down-regulated hyperglycemic PDAC cells. (K) 2DG assay determines glucose uptake rate in Col6a1-overexpressing hypoglycemic cells (L-Col6a1), hypoglycemic controls (L-C), Col6a1-downregulating hyperglycemic cells (shCol6a1), and hyperglycemic control cells (H-C). All data are presented as mean ± standard deviation, and data from 3 independent experiments are shown. Unpaired t test is used to examine statistical significance, *P < .05.
Collagen, Type VI, Alpha 1 Is Controlled by the Retinoic Acid Receptor Beta/Runt Related Transcription Factor 3 Signal Axis
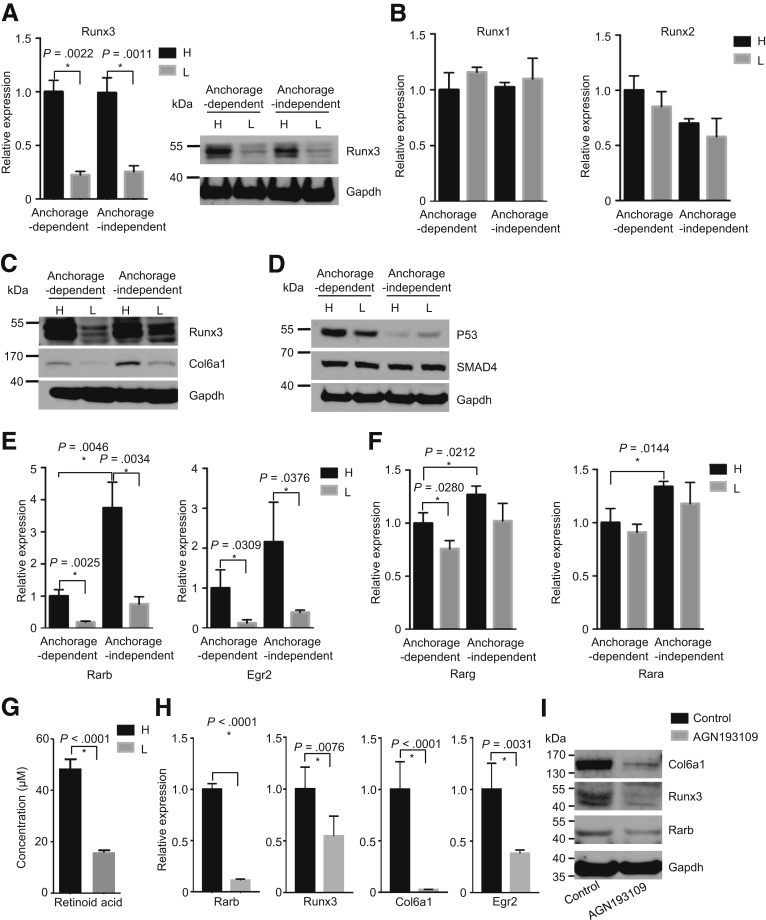
Next, we set out to investigate the molecular mechanism underlying increased Col6a1 expression in hyperglycemic cells. The transcription factor Runx3 controls the expression of Col6a1 via direct binding to its promoter, forming a distinctive pro-metastatic signal axis.12 Here we show that the expression of Runx3 (rather than Runx1 or Runx2) is increased in hyperglycemic PDAC cells (Figure 5A and B). Furthermore, this hyperglycemia-associated elevation in Runx3/Col6a1 expression was confirmed in another cell line (634 cells, Figure 5C).
Figure 5.
Col6a1 is controlled by the Rarb/Runx3 signal axis. (A) Quantitative RT-PCR (left panel) and Western blot (right panel) analysis show expression of Runx3; H, hyperglycemic cells; L, hypoglycemic cells. (B) Quantitative RT-PCR analysis shows expression of Runx1 and Runx2 in hyperglycemic and hypoglycemic PDAC cells. (C) Western blot analysis shows expression of Runx/Col6a1 in another (634 cells) hyperglycemic (H) and hypoglycemic (L) PDAC cells. (D) Western blot analysis shows expression of Smad4 and p53 in hyperglycemic (H) and hypoglycemic (L) PDAC cells. (E) Quantitative RT-PCR analysis shows expression of Rarb (left panel) and its target gene Egr2 (right panel) in hyperglycemic and hypoglycemic cells. (F) Real-time PCR analysis shows expression of Rarg and Rara in hyperglycemic (H) and hypoglycemic (L) PDAC cells. (G) Mass spectrometry analysis shows intracellular levels of RA in hyperglycemic and hypoglycemic PDAC cells. (H) Quantitative PCR analysis shows expression of Rarb, Runx3, and Col6a1 after Rar inhibition (by pan-Rar antagonist AGN193109, 50 μmol/L) for 24 hours. (I) Western blot analysis shows expression of Rarb, Runx3, and Col6a1 after Rar inhibition in hyperglycemic PDAC cells. All data are presented as mean ± standard deviation, and data from 3 independent experiments are shown. Unpaired t test is applied, *P < .05.
Because it has been previously demonstrated that Runx3 expression is affected by Smad4 and p53 status,12, 13 we compared the expression of p53 and Smad4 (SMAD Family Member 4) between hypoglycemic and hyperglycemic PDAC cells. As such, no difference was found (Figure 5D). We set out to look for an alternative mechanism. In this regard, it has been shown that Runx3 can be induced by all-trans-retinoic acids in a RA receptor–dependent manner.12, 13 Thus, we tested the expression of retinoic acid receptor alpha, Rarb, retinoic acid receptor gamma, and early growth response 2 (Egr2) (a downstream target of RA signaling) in hypoglycemic and hyperglycemic PDAC cells.17 This analysis revealed that the expression of Rarb and Egr2 was dramatically increased in hyperglycemic PDAC cells as compared with that in hypoglycemic cells (Figure 5E). In line, a slightly increased expression of retinoic acid receptor gamma was also observed (Figure 5F, left panel). However, no difference in the expression of retinoic acid receptor alpha was seen (Figure 5F, right panel). Furthermore, the intracellular uptake of RA was also higher in hyperglycemic PDAC cells (Figure 5G, as determined by mass spectrometry analysis), suggesting an activated RA signaling. To further confirm this, we treated PDAC cells with a pan-Rar antagonist (AGN193109) to inhibit the activity of Rar receptors. Indeed, inhibition of Rar receptors reduced the expression of Rarb, Runx3, and Col6a1 in PDAC cells (Figure 5H and I). In summary, Rarb/Runx3/Col6a1 signal axis constitutes an important pro-metastatic signal axis whose activity is affected by glycemic variability.
Retinoic Acid Receptor Beta Relays the Signal of Glycemic Variability to Runt Related Transcription Factor 3/Collagen, Type VI, Alpha 1 via Epigenetic Regulation
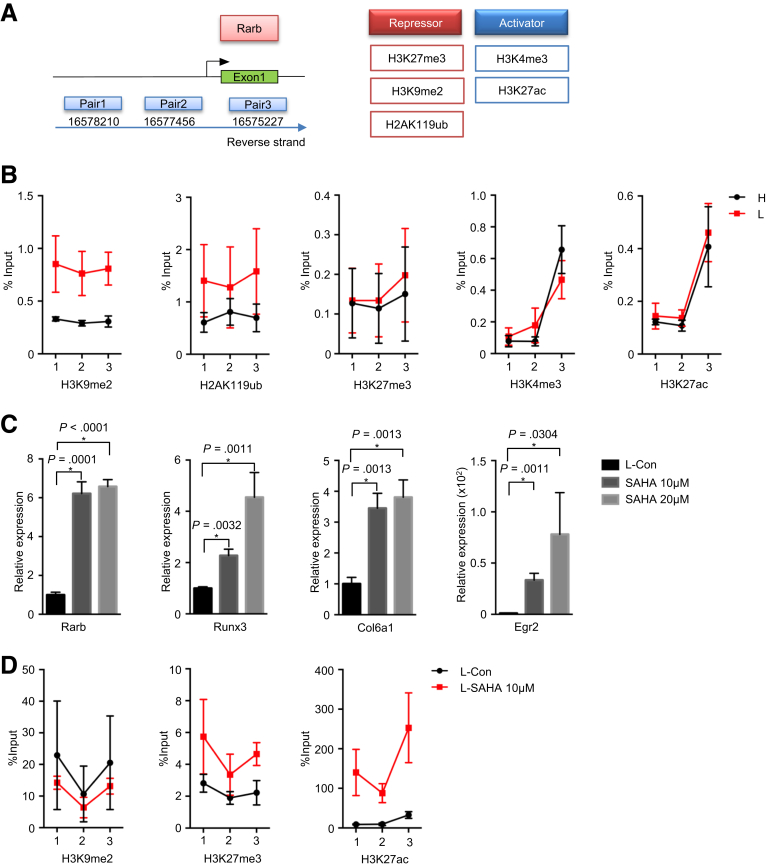
Previous studies suggested that an epigenetic-metabolic interplay played a key role in tumorigenicity.25 As for PDAC, a recent study demonstrated that the epigenetic-metabolic control of genes responsible for anabolic glucose metabolism promoted PDAC metastases.10 Using chromatin immunoprecipitation analysis, we analyzed the promoter of Rarb for occupation of a set of repressive (H3K27me3, H3K9me2, and H2AK119ub) and activating (H3K4me3 and H3K27ac) histone modification markers in hypoglycemic and hyperglycemic PDAC cells (Figure 6A). This analysis revealed that the Rarb promoter in hyperglycemic PDAC cells possessed lower levels of repressive histone markers H3K9me2 and H2AK119ub as compared with hypoglycemic cells. No difference was found for other histone modification markers (Figure 6B). In line, inhibition of histone deacetylases by Vorinostat (also known as SAHA) reconstituted the expression of Rarb, Runx3, Col6a1, and Egr2 in hypoglycemic PDAC cells (Figure 6C). In line, the histone deacetylase inhibition led to the significantly increased occupation levels of acetylated histone marker (H3K27ac) on Rarb promoter but not the methylated histone markers (Figure 6D).
Figure 6.
Rarb relays the signal of glycemic variability to Runx3/Col6a1 via histone modification. (A) Sketch view of chromatin immunoprecipitation (ChIP) assay. (B) ChIP analysis of Rarb gene shows percentage input of H3K9me2, H2AK119ub, H3K27me3, H3K4me3, and H3K27ac in hyperglycemic and hypoglycemic PDAC cells. Data are presented as mean ± standard deviation, and data from 3 independent experiments are shown; H, hyperglycemic cells; L, hypoglycemic cells. (C) Quantitative RT-PCR analysis shows expression of Rarb, Runx3, and Col6a1 after 10 μmol/L, 20 μmol/L SAHA treatment for 24 hours. Data are presented as mean ± standard deviation, and data from 3 independent experiments are shown. Unpaired t test is applied, *P < .05. (D) ChIP analysis of Rarb gene shows percentage input of H3K9me2, H3K27me3, and H3K27ac in control and SAHA treated hypoglyemic (L) PDAC cells. Data are presented as mean ± standard deviation, and data of 2 independent experiments are shown.
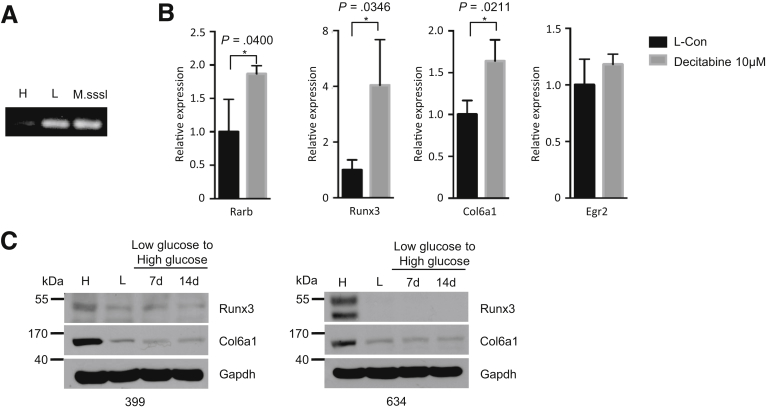
Furthermore, methylation-specific PCR analysis (targeted at the Rarb promoter CpG island) uncovered that these CpG islands were highly methylated in hypoglycemic PDAC cells (Figure 7A). After decitabine treatment (an inhibitor of DNA methylation), the expression of Rarb, Runx3, and Col6a1 was also reconstituted (Figure 7B). No difference in the expression of Egr2 was found. In addition, these histone modifications seemed to be long-lasting in that a short-term restoration (2 weeks) of hyperglycemic conditions was not able to reverse the suppressed expression of Runx3/Col6a1 in hypoglycemic cells (Figure 7C). Collectively, these data demonstrated that the epigenetic-metabolic control of Rarb was responsible for relaying signal of glycemic variability to Runx3/Col6a1 signal axis.
Figure 7.
Rarb relays the signal of glycemic variability to Runx3/Col6a1 via promoter methylation. (A) Methylation-specific PCR analysis shows the methylation status of Rarb gene in hypoglycemic PDAC cells, but not in hyperglycemic PDAC cells; M.SssI, CpG methyltransferase. (B) Quantitative RT-PCR analysis shows the expression of Rarb, Runx3, Col6a1, and Egr2 after decitabine treatment in hypoglycemic PDAC cells. All data are presented as mean ± standard deviation, and data from 3 independent experiments are shown. Unpaired t test is applied, *P < .05. (C) Western blot analysis shows expression of Runx3 and Col6a1 in hypoglycemic 399 (left panel) and 634 (right panel) cells after restoration of hyperglycemic culturing conditions.
The Activity of Retinoic Acid Receptor Beta/Runt Related Transcription Factor 3/Collagen, Type VI, Alpha 1 Signal Axis Could Be Modulated by Metformin
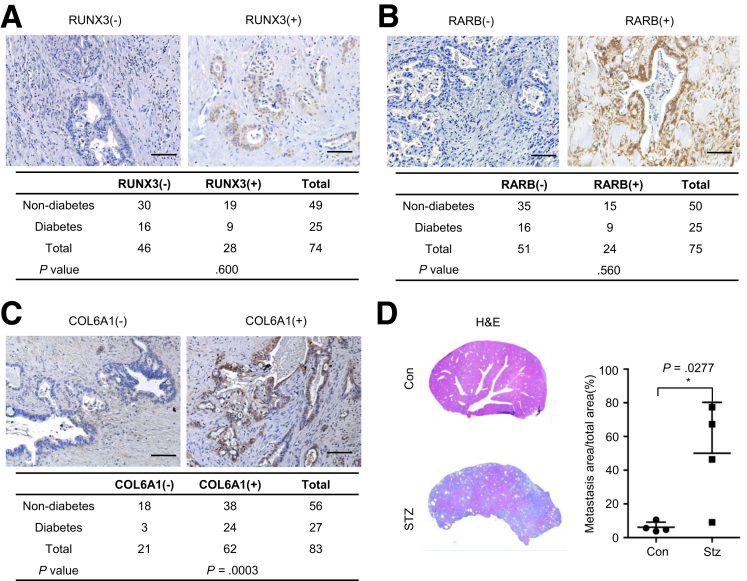
We hypothesized that the signal axis of Rarb/Runx3/Col6a1 permissive to glycemic variability was more likely to be relevant in PDAC patients who had diabetes. Thus, we stained a cohort of consecutive PDAC sections for RARB, RUNX3, and COL6A1. Although no correlation between either RARB or RUNX3 staining and diabetes was observed (Figure 8A and B), COL6A1 staining was positive in 88.9% of diabetic PDAC samples (24/27), which was significantly higher compared with 67.9% in nondiabetic PDAC samples (38/56) (Figure 8C, P = .039). Using a diabetic mouse (induced by streptozotocin treatment), we observed that diabetes indeed promoted metastatic colonization (Figure 8D, P = .0277).
Figure 8.
PDAC tissues of diabetic patients show increased expression of COL6A1. (A and B) Representative IHC pictures and statistical analysis show positive rate of RUNX3 and RARB in diabetic and non-diabetic PDAC slides; scale bar, 50 μm. (C) Representative COL6A1 IHC pictures and statistical analysis show positivity rate of COL6A1 in diabetic and non-diabetic PDAC slides, scale bar, 50 μm. (D) Representative H&E pictures (left panel) and metastatic area quantification (right panel) show metastatic colonization of PDAC cells in normal (Con) and diabetic samples (STZ). Data are presented as mean ± standard deviation, and data from 3 independent experiments are shown. Unpaired t test is applied, *P < .05.
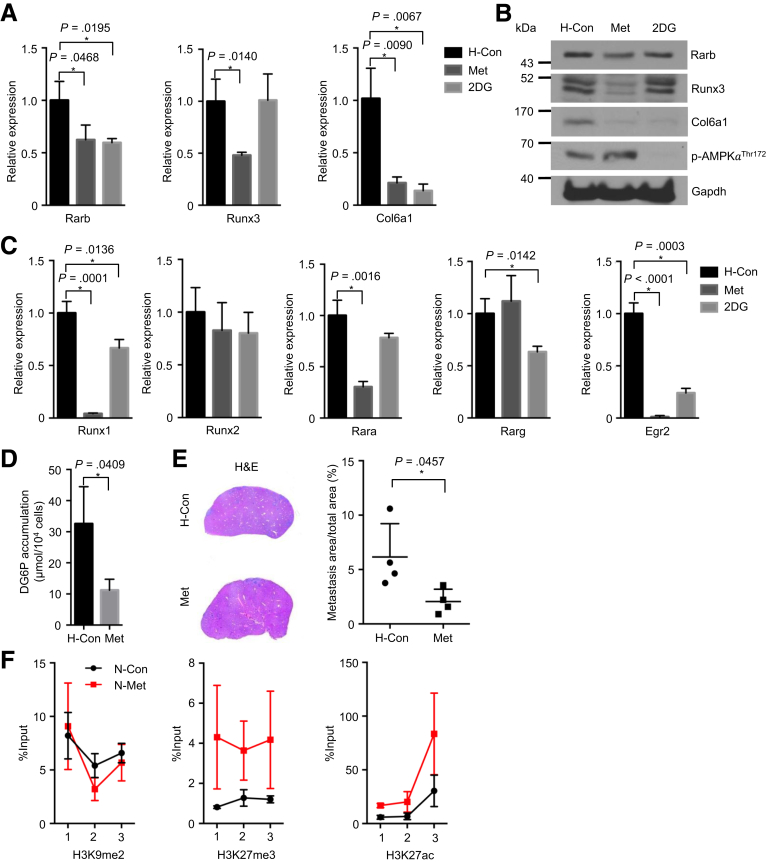
Because metformin is a widely used antidiabetic substance associated with favorable prognosis in diabetic patients with PDAC,26, 27, 28 we tested whether the glycemic Rarb/Runx3/Col6a1 pathway is affected by metformin. A glucose uptake inhibitor 2-deoxy-D-glucose (2DG) was also tested. Here, metformin consistently reduced the expression of Rarb, Runx3, and Col6a1, and it decreased the glucose uptake of PDAC cells (Figure 9A–D), showing a similar effect of hypoglycemic culturing. In addition, Runx1, retinoic acid receptor alpha, and Egr2 were also found to be reduced by metformin treatment (Figure 9C). As an internal control, protein kinase ADP-activated catalytic subunit alpha 1 was activated by metformin (Figure 9B). Except for Runx3, 2DG reduced the expression of Rarb and Col6a1 (Figure 9A and B). Finally, metformin treatment significantly impaired the hepatic metastasis of hyperglycemic PDAC cells (Figure 9E, P = .0457). Mechanistically, chromatin immunoprecipitation analysis revealed that metformin treatment increased the occupation levels of a suppressive histone marker H3K27me on the Rarb promoter; no difference on either H3K9me2 or H3K27ac was found (Figure 9F).
Figure 9.
Activity of the Rarb/Runx3/Col6a1 signal axis is inhibited by metformin. Quantitative RT-PCR results show expression of Rarb, Runx3, and Col6a1 after metformin (20 mmol/L) and 2DG (10 mmol/L) treatment for 48 hours in hyperglycemic PDAC cells. (B) Western blot analysis shows expression of Rarb, Runx3, Col6a1, and phospho-AMPKαThr172 after metformin and 2DG treatment for 48 hours in hyperglycemic PDAC cells. (C) Quantitative RT-PCR analysis shows expression of Runx1, Runx2, Rara, Rarg, and Egr2 after metformin (20 mmol/L) and 2DG (10 mmol/L) treatment for 48 hours in hyperglycemic PDAC cells. (D) Glucose uptake assay shows reduced cellular uptake after metformin treatment for 24 hours. (E) Representative H&E pictures (left panel) and metastatic area quantification (right panel) show metastatic colonization in control and metformin treated hyperglycemic PDAC cells. All data are presented as mean ± standard deviation; data of 3 independent experiments are shown. Unpaired t test is applied, *P < .05. (F) Chromatin immunoprecipitation (ChIP) analysis of Rarb gene shows percentage input of H3K9me2, H3K27me3, and H3K27ac in control and metformin treated hyperglycemic PDAC cells. All data are presented as mean ± standard deviation, and data from 2 independent experiments are shown. H, hyperglycemic cells; Met, metformin.
Discussion
In this study, we provided descriptive and functional evidence defining the glycemic variability as major driving force for PDAC local invasion, angiogenesis, and metastatic colonization. A well-defined pro-metastatic pathway, Runx3/Col6a1, was found to be affected by glycemic variability. Through epigenetic machinery, RA signaling acts as a critical sensor of glycemic variability that relays the glycemic signal to oncogenic Runx3/Col6a1 expression. This was achieved through epigenetically regulated Rarb receptor expression. Finally, the signal axis of Rarb/Runx3/Col6a1 was pharmaceutically accessible to metformin and Rar modulator.
In essence, we uncovered a novel mechanism defining Runx3/Col6a1 as the central player in coupling glucose metabolism with PDAC metastatic colonization. Interestingly, Runx2 was previously reported to favor glucose uptake and promote synthesis of type I collagen in osteoblasts.29 Although no difference was observed in Runx2 in our analysis, these data collectively highlight a particular role of the Runx family in linking glucose metabolism and synthesis of ECM proteins. As for Runx3, a context-dependent role was previously reported in PDAC; it suppresses cell proliferations by inducing expression of cyclin-dependent kinase inhibitor 1A (also known as p21) but promotes distant colonization via up-regulating a number of pro-metastatic mediators including Col6a1. Thus, patients with high RUNX3 levels are at increased risk for metastatic disease, whereas the ones with lower RUNX3 levels are prone to have a locally destructive disease.12, 13 On the basis of phenotypic analysis of different transgenic mouse models, it is proposed that p53 acts cooperatively with Smad4 in determining Runx3 levels: (1) wild-type p53 induces Runx3 degradation, whereas p53 gain-of-function mutation stabilizes Runx3; (2) a single-allele loss of Smad4 decreases Runx3 expression, whereas the double-allele loss of Smad4 promotes Runx3 expression. We reported that Runx3 levels were also subjected to glucose metabolism, adding another layer of complexity to the existing scenario.
Nevertheless, our data demonstrate that the microenvironmental hypoglycemia constitutes the initial driving force for invasive growth and angiogenesis; however, these early disseminating cells may not be able to establish metastases immediately because of the lack of ability in metastatic colonization. In line, this is also achieved by epigenetically down-regulated Rarb expression, which decreases the activity of a pro-metastatic Runx3/Col6a1 signal. However, this loss of capacity in colonization can be overcome by systemic hyperglycemia (eg, diabetes), genetic mutations (eg, p53 mutation), and other alternative metastatic pathways (eg, formation of a pre-metastatic niche).24
Metformin is a widely used antidiabetic oral medication. Long-term use of metformin not only decreases PDAC risk30, 31 but is also associated with improved patient survival.26, 27, 28 We demonstrated that the pro-metastatic pathway of Rarb/Runx3/Col6a1 axis could be directly/indirectly affected by metformin, which may contribute to its beneficiary effect. Despite these encouraging results, the addition of metformin to the conventional chemotherapy did not improve outcome in patients with metastatic PDAC in two phase II trials.32, 33 A possible explanation is that the patients involved in the clinical trials were in metastatic stage. The disseminating PDAC cells evading from their primary hypoglycemic niches have likely overcome the defect in remote colonization. Thus, it is likely that metformin needs to be offered at the early stage of disease or as an adjuvant agent.34 In addition, RAR inhibitors have been previously shown to have anti-metastatic properties in PDAC.35 Thus, it is reasonable to speculate that a combination of highly selective RAR modulators (eg, against RARB) and metformin might be more effective in inhibiting PDAC metastasis.36 This is particularly important for PDAC patients with diabetes.
Materials and Methods
Cell Culture
High-glucose medium was commercially available (glucose 25 mmol/L, D5796; Sigma-Aldrich, Munich, Germany). Low-glucose medium was prepared by using glucose-free Dulbecco modified Eagle medium (11966025; Thermo Fischer Scientific, Dreieich, Germany) supplemented with 0.5 mmol/L D(+)-Glucose (X997.1; Carl Roth, Karlsruhe, Germany). All PDAC cells were cultured with specific conditional medium supplemented with 10% FBS (F0804; Sigma-Aldrich), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37ºC, 5% CO2. The non-dialyzed FBS was used, which contained 1.1 ± 0.1 mmol/L glucose. Thus, the actual glucose concentration in medium containing 10% FBS is 0.1 mmol/L higher than the presumed ones.
Primary Tumor Cell Isolation
Primary tumor cells were isolated from genetically engineered mice with the genotypes of p48Cre; LSL-KrasG12D/+; Tsc1fl/+ (399 and 634 cells), p48Cre; LSL-KrasG12D/+; p53-/+ (1050 cells) and Pdx1Cre; LSL-KrasG12D/+; p53R172H/+ (10069 cells), as previously described.20 In brief, dissected tumor tissues were cut into small pieces and incubated with culture medium supplemented with 1.2 mg/mL collagenase (C6885-1G; Sigma-Aldrich) at 37°C for 30–40 minutes. Afterwards, the collagenase was washed out by using collagenase-free culture medium with centrifugation at 300 rpm for 5 minutes. After an additional collagenase incubation and washout, the achieved cell suspensions were seeded into a 10-cm2 dish with complete culture medium.
Inhibitor Treatment Experiment
PDAC cells were treated with either 20 mmol/L metformin hydrochloride (PHR 1084; Sigma-Aldrich) or 2 mmol/L 2DG (D8375; Sigma-Aldrich) for 48 or 72 hours. Hypoglycemic PDAC cells were treated with 10 or 20 μmol/L Vorinostat (also known as SAHA, SML0061; Sigma-Aldrich), 50 μmol/L AGN193109 for 48 hours (5758; Tocris, Wiesbaden-Nordenstadt, Germany), or with 10 μmol/L 5-aza-2′-deoxycytidine (Decitabine, A3656; Sigma-Aldrich) for 120 hours.
Mouse Extracellular Matrix and Adhesion Molecules Polymerase Chain Reaction Array
The assay was performed according to the manufacturer’s instructions of the mouse ECM and the adhesion molecule array kit (PAMM-013Z; Qiagen, Hilden, Germany). Data analysis was performed according to manual instructions on website http://www.qiagen.com/de/shop/genes-and-pathways/data-analysis-center-overview-page.
Chromatin Immunoprecipitation
The chromatin immunoprecipitation assay was performed as previously described.37 In brief, cells were cross-linked with 1% (vol/vol) formaldehyde in phosphate-buffered saline (PBS) for 10 minutes, lysed in 1% (wt/vol) sodium dodecyl sulfate, and sonicated. Precleared cell lysates were incubated with the antibodies at 4°C overnight. Immune complexes were recovered with salmon sperm DNA/protein A agarose slurry. After washing and elution, genomic DNA was extracted with phenol/chloroform for PCR analysis. Anti-H2AK119ub, Anti-H3K9me2, anti-H3K27me3, anti-H3K27ac, and anti-H3K4me3 antibodies were used for chromatin immunoprecipitation; the detailed antibody information is listed in Table 2. QIAquick PCR purification kit (28104; Qiagen) was used for chromatin purification before amplification of promoter regions of Rarb with qPCR. A detailed primer sequence is listed in Table 3.
Table 2.
Antibody List
| Antibody name | Catalogue number | Application | Producer |
|---|---|---|---|
| Rabbit anti-p-histone H3 (Ser10) Ab | 9701 | IHC | Cell Signaling Technology |
| Rabbit anti-cleaved Casp3 mAb | 9664 | IHC | Cell Signaling Technology |
| Rat anti-Cd31 Ab | DIA-310 | IHC | Dianova (Hamburg, Germany) |
| Rabbit anti-GAPDH Ab | sc-25778 | WB | Santa Cruz |
| Rabbit anti-carbonic anhydrase IX Ab | ab15086 | IHC | Abcam |
| Rabbit anti-COL6A1 Ab (H-200) | sc-20649 | IHC WB | Santa Cruz |
| Rabbit anti-fibronectin 1 C-terminal Ab | Sab4500974 | WB | Sigma-Aldrich |
| Mouse anti-OPN Ab (LFMb-14) | SC-73631 | WB | Santz Cruz |
| Mouse anti-RUNX3/AML2 (D9K6L) mAb | 13089s | WB IHC | Cell Signaling Technology |
| Mouse anti-RARβ2 Ab (B-12) | SC-514585 | WB | Santa Cruz |
| Mouse anti-RARB Ab clone 11E8.1 | MABC302 | IHC | Merck (Darmstadt, Germany) |
| Rabbit anti-p-AMPKα (Thr172) mAb | 2535 | WB | Cell Signaling Technology |
| Rabbit anti-ubiquityl-histone H2A (Lys119) (D27C4) mAb | 8240 | ChIP | Cell Signaling Technology |
| Mouse anti-histone H3 (di-methyl K9) Ab [mAbcam 1220] - ChIP grade | ab1220 | ChIP | Abcam (Cambridge, UK) |
| Mouse anti-histone H3 (tri-methyl K27) Ab [mAbcam 6002] - ChIP grade | ab6002 | ChIP | Abcam |
| Rabbit anti-histone H3 (acetyl K27) Ab - ChIP grade | ab4729 | ChIP | Abcam |
| Rabbit anti-tri-methyl-histone H3 (Lys4) (C42D8) mAb | 9751 | ChIP | Cell Signaling Technology |
ChIP, chromatin immunoprecipitation; WB, Western blot.
Table 3.
Sequences of Primers for Rarb Chromatin Immunoprecipitation Analysis
| Name | Sense (5′→3′) | Antisense (5′→3′) |
|---|---|---|
| Pair1 | ATACTGCTCCACCCCAGGAA | GCCATGGGCAATTAAGGCAG |
| Pair2 | CACGCTGAGCTTCCTATGCT | GTACACCCACCCTTGCTGAA |
| Pair3 | AGCGAGCCTGGAAAATGGTAA | CTCCATCAAACTCTGCCCCT |
Cell Transplantation Experiment
For the orthotopic xenograft models, 1 × 106 murine PDAC cells in 100 μL PBS buffer were resuspended and injected into the pancreatic tails of 8-week-old C57BL/6J mice. The pancreatic tumors were collected after 4 weeks of injection. For the portal vein injection models, either 5 × 105 cells (399 and 634 cells) or 2 × 106 (1050 and 10069 cells) in 50 μL PBS buffer were inoculated into the portal vein of 8-week-old C57BL/6J mice. The entire liver lobes were collected after 2 weeks of injection. In the tail vein injection models, 1 × 106 PDAC cells were injected slowly into the tail vein. The mice were kept for 3 weeks before PET-CT scanning. The entire lung tissue was collected. All studies were performed under the agreement of a protocol approved by the Animal Care and Use Committee of the Technical University of Munich (55.2.1.54-2532-42-13).
Streptozotocin and Metformin Treatment Experiment
For streptozotocin treatment, 8-week-old C57BL/6J mice were injected with 50 mg/kg streptozotocin (S0130; Sigma-Aldrich) for 5 days consecutively as previously described in http://www.mmpc.org/shared/protocols.aspx when fasting blood glucose level is higher than 200 mg/dL (defined as hyperglycemia). These studies were performed within frame of collaboration with the Department of Gastroenterology, Nanjing Drum Tower Hospital, Nanjing University and approved by local authorities.
For metformin treatment, 8-week-old C57BL/6J mice received portal vein injection with murine PDAC cells. Then all animals were daily applied with either saline for the control group or 250 mg/kg metformin hydrochloride (PHR1084; Sigma-Aldrich) for 14 days before tissue collection. All studies were performed under the agreement of a protocol approved by the Animal Care and Use Committee of the Technical University of Munich (55.2.1.54-2532-42-13).
Colony Formation Assay
Three × 102 PDAC cells were counted and seeded in a single well of the 6-well plate. After cell adherence, cells were washed with PBS and incubated with the respective conditioned medium containing 25 mmol/L or 0.5 mmol/L glucose for 7 days. A mixture of 75% methanol and 25% acetone was used for colony fixation. Ten percent crystal violet was applied for colony staining. The colony number containing more than 50 cells in every well was counted by 2 independent researchers (Z.J., T.C.).
MTT Assay
Two × 103 PDAC cells were seeded with the 100 μL respective conditioned medium in 96-well plates. Fifty μg thiazolyl blue tetrazolium bromide (MTT, M2128; Sigma-Aldrich) was added to every well after 24, 48, 72, and 96 hours. Proliferation rate was determined and calculated by measuring absorbance values at 570 nm.
Live Cell Counting
Two mL PDAC cells in logarithmic phase were trypsinized and seeded in 6-well plate at the concentration of 4 × 104/mL. After 24, 48, and 72 hours, cells were trypsinized. Afterwards, 10 μL cell suspension was mixed with 10 μL 0.4% trypan blue (T10282; Thermo Fischer Scientific), and unstained cells (live cells) were counted with a hemocytometer under microscope. The cell concentration and doubling time were calculated accordingly.
Annexin V and Propidium Iodide Staining
Cells seeded at 100-mm dish were cultured under hypoglycemic and hyperglycemic conditions. For quantification of cell apoptosis/necrosis, cells were trypsinized, washed in PBS, and then resuspended in 1× binding buffer. Cells were incubated with annexin (A13203; Thermo Fischer Scientific) and propidium iodide (P4864; Sigma-Aldrich, Taufkirchen, Germany) for 15 minutes and analyzed with a FACScan flow cytometer (BD Biosciences, San Jose, CA).
Invasion and Migration Assay
Cell invasion migration assay was performed by using a Transwell chamber (354480; Thermo Fischer Scientific). Briefly, 2 × 104 cells were resuspended in 200 μL FBS-free conditioned medium in the upper chamber; 500 μL normal conditioned medium (10% FBS) was added to the lower chamber. After 24 hours, migrated and invaded cells were fixed with 100% methanol and stained with 10% crystal violet.
Vascular Endothelial Growth Factor A Enzyme-linked Immunosorbent Assay Experiment
The same amount of PDAC cells were seeded in a 6-well plate. After adherence, 1 mL FBS-free conditioned culture medium was applied on each group for 24 hours. Afterwards, the supernatant was collected for determining vascular endothelial growth factor A secretion with mouse vascular endothelial growth factor quantikine enzyme-linked immunosorbent assay kit (MMV00; R&D Systems, Wiesbaden, Germany).
Microarray Experiment and Data Analysis
Microarray experiments were performed as previously described.38 In brief, high- and low-glucose cell samples were analyzed by Affymetrix GeneChip Mouse Gene 1.0 ST microarrays. Normalization was performed according to the robust multichip average method. A linear model and one-way analysis of variance were used for identifying differentially expressed genes. Those differentially expressed genes involved in angiogenesis and the regulation of angiogenesis were determined through gene ontology analysis and KEGG analysis.
Glucose Uptake Assay
The same amount of hypoglycemic or hyperglycemic PDAC cells were seeded and maintained overnight in their specific conditioned mediums. Before measurement, cells were washed 3 times with either KRH buffer (non-radioactive 2DG-based assay) or PBS buffer (microfluidic radioassay-based assay) and incubated with same amount of 2DG or 18F-fludeoxyglucose. Afterwards, the glucose uptake was determined either by a previously published protocol39 or by a microfluidic radioassay system.22 ATP, ADP, and ADP/ATP ratio were determined by a commercial EnzyLight ADP/ATP Ratio Assay Kit (ELDT-100; Bioassay Systems, Hayward, CA).
Anoikis Assay
One × 104 cells were seeded and cultured in normal and ultra-low adherent plate (CLS3473; Sigma-Aldrich) for 48 hours. Afterwards, 50 μL of 250 μg MTT was supplemented and incubated for 4 hours at 37ºC. The precipitate was dissolved in cell lysis buffer (10% sodium dodecyl sulfate, 0.01 mol/L HCl), and cell viability was determined by the absorbance value at 570 nm.
Western Blotting
Twenty μg total proteins was loaded in 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Then, proteins were transferred to a polyvinylidene difluoride membrane (10600002; GE Healthcare, Freiburg, Germany) and incubated with blocking buffer, first antibodies, and second antibodies subsequently incubation; specific bands were detected with the enhanced chemiluminescence system (Amersham Life Science Ltd, Bucks, United Kingdom). The antibodies used are listed in Table 2.
Patient Tissue Collection and Evaluation
The PDAC patients’ tissue were collected, fixed in paraformaldehyde, and embedded in paraffin. The use of the patient material was approved by the patients before surgery as well as by the local ethics committee (Department of Surgery, Klinikum rechts der Isar, Technical University of Munich). Stainings of patients’ tissue were evaluated by 2 independent researchers; the χ2 test was performed.
Immunohistochemistry Staining and Positive Cell Calculation
The paraffin-embedded tissue sections were deparaffinized and rehydrated. Antigen retrieval, endogenous peroxidase, and non-specific blocking were performed before sections were incubated with anti-cleaved caspase-3, anti-phospho-histone H3, anti-CAIX, anti-CD31, anti-E-cadherin, anti-COL6A1, and anti-RUNX3 antibodies, respectively. After application of secondary antibodies, sections were applied with a Liquid DAB+ Substrate Chromogen System (K3468; Dako, Hamburg, Germany) for the color reaction and with Mayer’s hematoxylin for counterstaining. Then the sections were dehydrated and mounted, and pictures were taken by a Carl Zeiss microscope (Carl Zeiss, Munich, Germany) at 5 random views for positive cell calculation. The number of positively stained cells was counted by 2 independent researchers with Image J software (National Institutes of Health, https://imagej.nih.gov/ij/). The antibody used is listed in Table 2.
Stable Transfection Experiment
For Col6a1 overexpression transfection, hypoglycemic PDAC cells were transfected with 1 μg of either mouse cDNA ORF clone (MG223027; OriGene, Herford, Germany) or control ORF clone (PS100010; OriGene). Then, 0.5 mg/mL geneticin (10131027; Thermo Fisher Scientific) was supplemented in culture medium for selection. For Col6a1 stable knock-down transfection, hyperglycemic PDAC cells were transfected with 1 μg of either COL6A1 shRNA plasmid (sc-35086-SH; Santa Cruz Biotechnology, Heidelberg, Germany) or control shRNA plasmid (sc-108060; Santa Cruz Biotechnology). Then, 4 μg/mL puromycin (sc-108071; Santa Cruz Biotechnology) was supplemented in culture medium for selection.
Mitochondrial Respiration and Glycolysis Evaluation
Mitochondrial respiration was determined by oxygen consumption rate, and glycolysis rate was determined by extracellular acidification rate as previously described.40 Briefly, 2 × 104 hypoglycemic or hyperglycemic cells were seeded in XF96 culture microplates (101085-004; Agilent Technologies Deutschland GmbH, Waldbronn, Germany) overnight at cultivation condition. For basal cellular respiration measurement, cells were treated sequentially with 2,4-DNP (0.1 mmol/L), 2DG (100 mmol/L), oligomycin (1.5 μg/mL), and eventually rotenone/antimycin A (2.5 μmol/L and 2.5 μmol/L, respectively). Data were acquired by the Seahorse XF96 analyzer. The oxygen consumption rate and extracellular acidification rate data were analyzed and interpreted by using Agilent Seahorse Wave 2.4 Software. The experiments were performed within frame of collaboration with Dr Jastroch (German Center for Diabetes Research, Helmholtz Zentrum München).
Quantitative Real-Time Polymerase Chain Reaction
RNA extraction (74104; Qiagen) and cDNA synthesis (K1632; Thermo Fisher Scientific) were conducted according to the manufacturer’s instructions. Quantitative real-time PCR was performed by using the KAPA SYBR FAST Kit (KK4610; Sigma-Aldrich) in a Light-CyclerTM480 system. Peptidylprolyl isomerase B was used as housekeeping gene. Detailed primer sequences are listed in Table 4.
Table 4.
Sequences of Primers for qRT-PCR Analysis
| Name | Sense (5′→3′) | Antisense (5′→3′) |
|---|---|---|
| Spp1 | AAGAAGCATCCTTGCTTGGGT | ATGGTCGTAGTTAGTCCTTGGC |
| Col6a1 | CAGCCAGACCATTGACACCA | TTCCTCGCTCCCCCTCATAC |
| Fn1 | ATGTGGACCCCTCCTGATAGT | GCCCAGTGATTTCAGCAAAGG |
| Ppib | GGAGCGCAATATGAAGGTGC | CTTATCGTTGGCCACGGAGG |
| Rara | ATCTGTGGAGACCGACAGGA | CCGTTTCCGGACGTAGACTT |
| Rarb | CAAGACACCGAGGTTGTGGA | ACAGAGGCCCTGGTTCCTTA |
| Rarg | TGCTGCGGATCTGTACAAGG | TTGTGCATCTGGGTTCGGTT |
| Runx3 | AGAGTTTCACGCTCACAATC | GGAGAAGGGGTTCAGGTC |
| Egr2 | GCCAAGGCCGTAGACAAAATC | CCACTCCGTTCATCTGGTCA |
| Runx1 | TTTGGGGTGCATCAGTGTGA | AACCAGCGGTTAGGCTTCAT |
| Runx2 | GGGAACCAAGAAGGCACAGA | GGATGAGGAATGCGCCCTAA |
Bisulfite Modification and Methylation Specific Polymerase Chain Reaction
Genomic DNA was isolated from murine PDAC cells with DNeasy Blood & Tissue Kit (69504; Qiagen). Genomic DNA was modified with EpiTect Bisulfite Kit (59104; Qiagen) according to manual instructions. Genomic DNA modified with CpG methyltransferase (M0226S; Biolabs, Frankfurt am Main, Germany) was used as positive control. Methylation specific PCR analyses and the according primers were used as described before.41 Afterwards, methylation specific PCR analyses were described as follows: 95°C 5 minutes, 40 cycles of thermocycling: 95°C 30 seconds, 56.5°C 30 seconds, 72°C 45 seconds, and ended with 72°C for 5 minutes.
Measurement of Intracellular Uptake of Retinoid Acid by Mass Spectrometry
Two × 105 399 cells were plated either in low-glucose medium or in high-glucose medium. After 24 hours, 100 μmol/L RA (Sigma-Aldrich; R2625) was added in the dark. After additional 48 hours, the remaining cells were washed with 0.5 mL PBS and harvested with 300 μL ice-cold isopropanol. Afterwards, the cells were sonicated, vortexed, and stored in dark tubes at –80°C. To prevent isomerization of the RA, amber glassware was used. Before measurement, the samples were centrifuged for 5 minutes at 4°C with 13,000 rpm to remove cell debris. The supernatant was directly used for liquid chromatography–mass spectrometry analysis.
The samples were subjected to high-performance chromatography–mass spectrometry analysis by using a Prominence HPLC system (Shimadzu, Hannover, Germany) and a 4000 QTRAP (Applied Biosystems, Foster City, CA). The separation was performed by using a 100 × 2.1 mm inner diameter, 5 μm Kinetex 5u XB-C18 100 A column (Phenomenex, Aschaffenburg, Germany) with a 300 μL/min flowrate and a linear gradient from 50% A (A: acetonitrile/isopropanol/water with 5 mmol/L ammonia acetate [55/40/5, v/v/v, pH 5], B: 5 mmol/L ammonia acetate [pH 5]) for 1 minute to 100% A in 8 minutes, which was maintained for 4 minutes. Afterwards, the column was equilibrated to starting conditions. Ions were analyzed by mass spectrometry in the positive mode using a Turbo V Ion Source (Applied Biosystems) in the APCI mode. The nebulizer current was set to 5 μA at a source temperature of 300°C and using nitrogen as collision gas. The parameters for the collision activated dissociation were medium, curtain gas: 20 psi, ion source gas 1: 90 psi, declustering potential 31 V, entrance potential 10 V. The multiple reaction monitoring transition 301.23 → 123.10 (collision energy 7 V, cell exit potential 14 V) was used as quantifier, and 301.23 → 80.90 (collision energy 23 V, cell exit potential 10 V) was used as qualifier. The quantification was performed by using an external calibration from 0.1 to 500 μmol/L RA.
Liver Metastasis Area Analysis
The tissue sections were scanned with a 1.25× object of a Zeiss microscope. The tumor’s metastasis area and total liver area were counted by Image J (1.48v). The metastasis area ratio was calculated as follows:
Positron Emission Tomography/Computed Tomography Scan
The animals were anesthetized with isoflurane and imaged by using the Siemens Inveon microPET/CT (Siemens Healthcare, Erlangen, Germany). Fifty minutes before the PET scan, 10–15 MBq 18F-fludeoxyglucose was injected intravenously. All measurements were corrected for physical decay, dead time, and non-uniformity of microPET response, and the images were reconstructed by using the 3-dimensional ordered subset expectation maximization algorithm. The standard uptake values were calculated by normalizing the injection dose and the body weight. Regions of interest were defined manually on the basis of the fusion of CT and PET images by using PMOD software (version 3.1; PMOD Technologies, Zurich, Switzerland).
Cellular Microfluidic Radioassay
A continuously infused microfluidic radioassay system was used to measure real-time cellular uptake.22 Two days before the continuously infused microfluidic radioassay measurements, approximately 1.5 × 104 cells in 30 μL suspension were inoculated into the cell culture chamber of a microfluidic chip (μ-Slide VI0.4; ibidi GmbH, Munich, Germany) and formed a homogeneity single layer on the bottom surface of the chamber. The cells were then cultured in 25 mmol/L glucose Dulbecco modified Eagle medium (Biochrom GmbH, Berlin, Germany), supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at controlled condition of 37ºC and 5% CO2. One day before the measurements, the cell chamber for low-glucose medium was changed with 0.5 mmol/L glucose concentration separately. The corresponding medium was exchanged every 8 hours until the start of the measurements to sustain glucose concentration. At the time of the continuously infused microfluidic radioassay measurements, the cells reached approximately 60%–90% confluence inside the cell chamber. Before the infusion, the flow control unit (Cavro XLP 6000; Tecan Group Ltd, Männedorf, Switzerland) was sterilized and cleaned by using 70% ethanol and ultrapure water. [18F]Fludeoxyglucose was diluted into the cell culture medium to generate a radioactive solution of 0.2–5 MBq/mL with the culturing glucose concentration. The tracer medium solution was pumped through the medium chamber and cell chamber at a constant flow of 1.25 μl/s. Positrons emitted both from the medium chamber and the cell chamber were measured by the positron camera (Crytur Ltd, Turnov, Czech Republic) for 75 minutes. Measurements were binned into frames of 1 minute each. Each measurement was repeated 6 times. After the continuously infused microfluidic radioassay measurements, the cell chamber was imaged by using a microscope (BZ-9000; Keyence Co Ltd, Osaka, Japan), and 5 distributed fields of view were selected for counting the number of cells. The average number of cells per detector pixel was calculated.
Statistical Analysis
Statistical analysis was performed by GraphPad Prism 6 (GraphPad, La Jolla, CA) or IBM SPSS 22.0 Software (Statistical Package for the Social Sciences; IBM, New York, NY). All experiments were performed independently at least 3 times. Unless otherwise stated, an unpaired Student t test was used for group-wise comparisons of 2 groups. The data significance level was set at P < .05. Results are expressed as mean ± standard deviation unless indicated otherwise.
Footnotes
Author contributions B.K. and J.K. designed the study. Z.J., T.C., Z.Z., N.M., S.N., K.K., P.B., J.A., and C.J. performed experiments and acquired data. K.S. provided expertise in histologic analysis. B.K., Z.J., and J.K. drafted the manuscript. S.R., K.S., T.H., S.B., D.L., M.J., P.H., S.S., X.Z., G.O.C., H.F., and C.W.M. revised the manuscript critically for important intellectual content. All authors approved the final version of the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding Supported in part by the AiF Projekt (ZIM-Kooperationsprojekte, Central Innovation Program Cooperation Projects, ZF4353701SK6, to B.K.), the German Research Foundation (DFG, MI 1173/5-1, to C.W.M., B.K., and JK), DFG Collaborative Research Centre 824 (SFB824) and the China Scholarship Council (CSC), Nanjing International Joint Research Project (201605082, to B.K.), the Youngth National Natural Science Foundation of China (81602147, to B.K.), the Youngth Natural Science Foundation of Jiangsu Province (BK20160110, to B.K.), and the Outstanding Youth Project of Nanjing City (JQX16026, to S.S., Z.X.P., and B.K.).
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Bosetti C., Rosato V., Li D., Silverman D., Petersen G.M., Bracci P.M., Neale R.E., Muscat J., Anderson K., Gallinger S., Olson S.H., Miller A.B., Bas Bueno-de-Mesquita H., Scelo G., Janout V., Holcatova I., Lagiou P., Serraino D., Lucenteforte E., Fabianova E., Ghadirian P., Baghurst P.A., Zatonski W., Foretova L., Fontham E., Bamlet W.R., Holly E.A., Negri E., Hassan M., Prizment A., Cotterchio M., Cleary S., Kurtz R.C., Maisonneuve P., Trichopoulos D., Polesel J., Duell E.J., Boffetta P., La Vecchia C. Diabetes, antidiabetic medications, and pancreatic cancer risk: an analysis from the International Pancreatic Cancer Case-Control Consortium. Ann Oncol. 2014;25:2065–2072. doi: 10.1093/annonc/mdu276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boursi B., Finkelman B., Giantonio B.J., Haynes K., Rustgi A.K., Rhim A.D., Mamtani R., Yang Y.X. A clinical prediction model to assess risk for pancreatic cancer among patients with new-onset diabetes. Gastroenterology. 2017;152:840–850 e3. doi: 10.1053/j.gastro.2016.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pannala R., Leirness J.B., Bamlet W.R., Basu A., Petersen G.M., Chari S.T. Prevalence and clinical profile of pancreatic cancer-associated diabetes mellitus. Gastroenterology. 2008;134:981–987. doi: 10.1053/j.gastro.2008.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hart P.A., Law R.J., Frank R.D., Bamlet W.R., Burch P.A., Petersen G.M., Rabe K.G., Chari S.T. Impact of diabetes mellitus on clinical outcomes in patients undergoing surgical resection for pancreatic cancer: a retrospective, cohort study. Am J Gastroenterol. 2014;109:1484–1492. doi: 10.1038/ajg.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleeff J., Costello E., Jackson R., Halloran C., Greenhalf W., Ghaneh P., Lamb R.F., Lerch M.M., Mayerle J., Palmer D., Cox T., Rawcliffe C.L., Strobel O., Buchler M.W., Neoptolemos J.P. The impact of diabetes mellitus on survival following resection and adjuvant chemotherapy for pancreatic cancer. Br J Cancer. 2016;115:887–894. doi: 10.1038/bjc.2016.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter U., Kohlert T., Rahbari N.N., Weitz J., Welsch T. Impact of preoperative diabetes on long-term survival after curative resection of pancreatic adenocarcinoma: a systematic review and meta-analysis. Ann Surg Oncol. 2014;21:1082–1089. doi: 10.1245/s10434-013-3415-6. [DOI] [PubMed] [Google Scholar]
- 8.Kamphorst J.J., Nofal M., Commisso C., Hackett S.R., Lu W., Grabocka E., Vander Heiden M.G., Miller G., Drebin J.A., Bar-Sagi D., Thompson C.B., Rabinowitz J.D. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makohon-Moore A.P., Zhang M., Reiter J.G., Bozic I., Allen B., Kundu D., Chatterjee K., Wong F., Jiao Y., Kohutek Z.A., Hong J., Attiyeh M., Javier B., Wood L.D., Hruban R.H., Nowak M.A., Papadopoulos N., Kinzler K.W., Vogelstein B., Iacobuzio-Donahue C.A. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet. 2017;49:358–366. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald O.G., Li X., Saunders T., Tryggvadottir R., Mentch S.J., Warmoes M.O., Word A.E., Carrer A., Salz T.H., Natsume S., Stauffer K.M., Makohon-Moore A., Zhong Y., Wu H., Wellen K.E., Locasale J.W., Iacobuzio-Donahue C.A., Feinberg A.P. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet. 2017;49:367–376. doi: 10.1038/ng.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iacobuzio-Donahue C.A., Fu B., Yachida S., Luo M., Abe H., Henderson C.M., Vilardell F., Wang Z., Keller J.W., Banerjee P., Herman J.M., Cameron J.L., Yeo C.J., Halushka M.K., Eshleman J.R., Raben M., Klein A.P., Hruban R.H., Hidalgo M., Laheru D. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittle M.C., Izeradjene K., Rani P.G., Feng L., Carlson M.A., DelGiorno K.E., Wood L.D., Goggins M., Hruban R.H., Chang A.E., Calses P., Thorsen S.M., Hingorani S.R. RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell. 2015;161:1345–1360. doi: 10.1016/j.cell.2015.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong Y., Macgregor-Das A., Saunders T., Whittle M.C., Makohon-Moore A., Kohutek Z.A., Poling J., Herbst B.T., Javier B.M., Cope L., Leach S.D., Hingorani S.R., Iacobuzio-Donahue C.A. Mutant p53 together with TGFbeta signaling influence organ-specific hematogenous colonization patterns of pancreatic cancer. Clin Cancer Res. 2017;23:1607–1620. doi: 10.1158/1078-0432.CCR-15-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le X.F., Groner Y., Kornblau S.M., Gu Y., Hittelman W.N., Levanon D., Mehta K., Arlinghaus R.B., Chang K.S. Regulation of AML2/CBFA3 in hematopoietic cells through the retinoic acid receptor alpha-dependent signaling pathway. J Biol Chem. 1999;274:21651–21658. doi: 10.1074/jbc.274.31.21651. [DOI] [PubMed] [Google Scholar]
- 15.Otto F., Lubbert M., Stock M. Upstream and downstream targets of RUNX proteins. J Cell Biochem. 2003;89:9–18. doi: 10.1002/jcb.10491. [DOI] [PubMed] [Google Scholar]
- 16.Bleul T., Ruhl R., Bulashevska S., Karakhanova S., Werner J., Bazhin A.V. Reduced retinoids and retinoid receptors' expression in pancreatic cancer: a link to patient survival. Mol Carcinog. 2015;54:870–879. doi: 10.1002/mc.22158. [DOI] [PubMed] [Google Scholar]
- 17.Segara D., Biankin A.V., Kench J.G., Langusch C.C., Dawson A.C., Skalicky D.A., Gotley D.C., Coleman M.J., Sutherland R.L., Henshall S.M. Expression of HOXB2, a retinoic acid signaling target in pancreatic cancer and pancreatic intraepithelial neoplasia. Clin Cancer Res. 2005;11:3587–3596. doi: 10.1158/1078-0432.CCR-04-1813. [DOI] [PubMed] [Google Scholar]
- 18.Cheng T., Jian Z., Li K., Raulefs S., Regel I., Shen S., Zou X., Ruland J., Ceyhan G.O., Friess H., Michalski C.W., Kleeff J., Kong B. In vivo functional dissection of a context-dependent role for Hif1alpha in pancreatic tumorigenesis. Oncogenesis. 2016;5:e278. doi: 10.1038/oncsis.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong B., Cheng T., Wu W., Regel I., Raulefs S., Friess H., Erkan M., Esposito I., Kleeff J., Michalski C.W. Hypoxia-induced endoplasmic reticulum stress characterizes a necrotic phenotype of pancreatic cancer. Oncotarget. 2015;6:32154–32160. doi: 10.18632/oncotarget.5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong B., Wu W., Cheng T., Schlitter A.M., Qian C., Bruns P., Jian Z., Jager C., Regel I., Raulefs S., Behler N., Irmler M., Beckers J., Friess H., Erkan M., Siveke J.T., Tannapfel A., Hahn S.A., Theis F.J., Esposito I., Kleeff J., Michalski C.W. A subset of metastatic pancreatic ductal adenocarcinomas depends quantitatively on oncogenic Kras/Mek/Erk-induced hyperactive mTOR signalling. Gut. 2016;65:647–657. doi: 10.1136/gutjnl-2014-307616. [DOI] [PubMed] [Google Scholar]
- 21.Yun J., Rago C., Cheong I., Pagliarini R., Angenendt P., Rajagopalan H., Schmidt K., Willson J.K., Markowitz S., Zhou S., Diaz L.A., Jr., Velculescu V.E., Lengauer C., Kinzler K.W., Vogelstein B., Papadopoulos N. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z., Jian Z., Wang Q., Cheng T., Feuerecker B., Schwaiger M., Huang S.C., Ziegler S.I., Shi K. A continuously infused microfluidic radioassay system for the characterization of cellular pharmacokinetics. J Nucl Med. 2016;57:1548–1555. doi: 10.2967/jnumed.115.169151. [DOI] [PubMed] [Google Scholar]
- 23.Buchheit C.L., Weigel K.J., Schafer Z.T. Cancer cell survival during detachment from the ECM: multiple barriers to tumour progression. Nat Rev Cancer. 2014;14:632–641. doi: 10.1038/nrc3789. [DOI] [PubMed] [Google Scholar]
- 24.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., Xiang J., Zhang T., Theilen T.M., Garcia-Santos G., Williams C., Ararso Y., Huang Y., Rodrigues G., Shen T.L., Labori K.J., Lothe I.M., Kure E.H., Hernandez J., Doussot A., Ebbesen S.H., Grandgenett P.M., Hollingsworth M.A., Jain M., Mallya K., Batra S.K., Jarnagin W.R., Schwartz R.E., Matei I., Peinado H., Stanger B.Z., Bromberg J., Lyden D. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong C.C., Qian Y., Yu J. Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene. 2017;36:3359–3374. doi: 10.1038/onc.2016.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amin S., Mhango G., Lin J., Aronson A., Wisnivesky J., Boffetta P., Lucas A.L. Metformin improves survival in patients with pancreatic ductal adenocarcinoma and pre-existing diabetes: a propensity score analysis. Am J Gastroenterol. 2016;111:1350–1357. doi: 10.1038/ajg.2016.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerullo M., Gani F., Chen S.Y., Canner J., Pawlik T.M. Metformin use is associated with improved survival in patients undergoing resection for pancreatic cancer. J Gastrointest Surg. 2016;20:1572–1580. doi: 10.1007/s11605-016-3173-4. [DOI] [PubMed] [Google Scholar]
- 28.Sadeghi N., Abbruzzese J.L., Yeung S.C., Hassan M., Li D. Metformin use is associated with better survival of diabetic patients with pancreatic cancer. Clin Cancer Res. 2012;18:2905–2912. doi: 10.1158/1078-0432.CCR-11-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei J., Shimazu J., Makinistoglu M.P., Maurizi A., Kajimura D., Zong H., Takarada T., Lezaki T., Pessin J.E., Hinoi E., Karsenty G. Glucose uptake and Runx2 synergize to orchestrate osteoblast differentiation and bone formation. Cell. 2015;161:1576–1591. doi: 10.1016/j.cell.2015.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bodmer M., Becker C., Meier C., Jick S.S., Meier C.R. Use of antidiabetic agents and the risk of pancreatic cancer: a case-control analysis. Am J Gastroenterol. 2012;107:620–626. doi: 10.1038/ajg.2011.483. [DOI] [PubMed] [Google Scholar]
- 31.Li D., Yeung S.C., Hassan M.M., Konopleva M., Abbruzzese J.L. Antidiabetic therapies affect risk of pancreatic cancer. Gastroenterology. 2009;137:482–488. doi: 10.1053/j.gastro.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kordes S., Pollak M.N., Zwinderman A.H., Mathot R.A., Weterman M.J., Beeker A., Punt C.J., Richel D.J., Wilmink J.W. Metformin in patients with advanced pancreatic cancer: a double-blind, randomised, placebo-controlled phase 2 trial. Lancet Oncol. 2015;16:839–847. doi: 10.1016/S1470-2045(15)00027-3. [DOI] [PubMed] [Google Scholar]
- 33.Reni M., Dugnani E., Cereda S., Belli C., Balzano G., Nicoletti R., Liberati D., Pasquale V., Scavini M., Maggiora P., Sordi V., Lampasona V., Ceraulo D., Di Terlizzi G., Doglioni C., Falconi M., Piemonti L. (Ir)relevance of metformin treatment in patients with metastatic pancreatic cancer: an open-label, randomized phase II trial. Clin Cancer Res. 2016;22:1076–1085. doi: 10.1158/1078-0432.CCR-15-1722. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y.X., Rustgi A.K. Impact of metformin on advanced pancreatic cancer survival: too little, too late? Clin Cancer Res. 2016;22:1031–1033. doi: 10.1158/1078-0432.CCR-15-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss F.U., Marques I.J., Woltering J.M., Vlecken D.H., Aghdassi A., Partecke L.I., Heidecke C.D., Lerch M.M., Bagowski C.P. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137 doi: 10.1053/j.gastro.2009.08.065. 2136–2145 e1-7. [DOI] [PubMed] [Google Scholar]
- 36.Altucci L., Leibowitz M.D., Ogilvie K.M., de Lera A.R., Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- 37.Benitz S., Regel I., Reinhard T., Popp A., Schaffer I., Raulefs S., Kong B., Esposito I., Michalski C.W., Kleeff J. Polycomb repressor complex 1 promotes gene silencing through H2AK119 mono-ubiquitination in acinar-to-ductal metaplasia and pancreatic cancer cells. Oncotarget. 2016;7:11424–11433. doi: 10.18632/oncotarget.6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong B., Bruns P., Behler N.A., Chang L., Schlitter A.M., Cao J., Gewies A., Ruland J., Fritzsche S., Valkovskaya N., Jian Z., Regel I., Raulefs S., Irmler M., Beckers J., Friess H., Erkan M., Mueller N.S., Roth S., Hackert T., Esposito I., Theis F.J., Kleeff J., Michalski C.W. Dynamic landscape of pancreatic carcinogenesis reveals early molecular networks of malignancy. Gut. 2018;67:146–156. doi: 10.1136/gutjnl-2015-310913. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto N., Ueda-Wakagi M., Sato T., Kawasaki K., Sawada K., Kawabata K., Akagawa M., Ashida H. Measurement of glucose uptake in cultured cells. Curr Protoc Pharmacol. 2015;71 doi: 10.1002/0471141755.ph1214s71. 12.14.1–26. [DOI] [PubMed] [Google Scholar]
- 40.Kabra U.D., Pfuhlmann K., Migliorini A., Keipert S., Lamp D., Korsgren O., Gegg M., Woods S.C., Pfluger P.T., Lickert H., Affourtit C., Tschop M.H., Jastroch M. Direct substrate delivery into mitochondrial fission-deficient pancreatic islets rescues insulin secretion. Diabetes. 2017;66:1247–1257. doi: 10.2337/db16-1088. [DOI] [PubMed] [Google Scholar]
- 41.Demircan B., Dyer L.M., Gerace M., Lobenhofer E.K., Robertson K.D., Brown K.D. Comparative epigenomics of human and mouse mammary tumors. Genes Chromosomes Cancer. 2009;48:83–97. doi: 10.1002/gcc.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]