Abstract
As next-generation sequencing technology advances and the cost decreases, whole genome sequencing (WGS) has become the preferred platform for the identification of somatic copy number alteration (CNA) events in cancer genomes. To more effectively decipher these massive sequencing data, we developed a software program named SEG, shortened from the word “segment”. SEG utilizes mapped read or fragment density for CNA discovery. To reduce CNA artifacts arisen from sequencing and mapping biases, SEG first normalizes the data by taking the log2-ratio of each tumor density against its matching normal density. SEG then uses dynamic programming to find change-points among a contiguous log2-ratio data series along a chromosome, dividing the chromosome into different segments. SEG finally identifies those segments having CNA. Our analyses with both simulated and real sequencing data indicate that SEG finds more small CNAs than other published software tools.
Keywords: SEG, Somatic Copy Number Alteration, Whole Genome Sequencing, Cancer
1. Introduction
Copy number alteration (CNA) is one of the most prominent changes found in cancer genomes [[1], [2], [3], [4], [5], [6], [7], [8], [9]], some of which contribute to cancer development and progression, e.g., deletion of tumor suppressors such as PTEN and amplification of oncogenes such as MYC.Genome wide CNA discovery is achieved via array-based technology traditionally [5,10,11] and next-generation sequencing (NGS) strategies recently [1,[12], [13], [14], [15]].Because of the high resolution and decreasing cost, NGS becomes the increasingly preferred platform for CNA-discovery [[16], [17], [18]].For example, the cost of whole genome sequencing (WGS) of a 30× coverage has already decreased to below $1000 per genome, which is actually cheaper than high density arrays considering its comprehensiveness (finding CNAs, structural rearrangements and sequence mutations) and high resolution (covering >90% of the genome).
For effective CNA-discovery, WGS of a ≥ 10× coverage is typically performed (WGS depth can be approximated by the Poisson distribution, and a ≥ 10× coverage yields a Poisson distribution that is increasingly more normal-appearing).Such sequencing generates substantially more data than even the highest density arrays currently available, such as the Affymetrix genome-wide human SNP array 6.0 that have approximately 2 million probes and have been used for CNA-finding in many projects of the cancer genome atlas (TCGA) [5,19,20].Importantly, while WGS can cover every base of the genome and could potentially identify every CNA in a cancer genome, it also presents new data analysis challenges.For example, because of the vast heterogeneity of a mammalian genome [21,22], some genomic regions (e.g.,GC-rich) are sequenced better than others, creating artificial CNAs.Moreover, mammalian genomes are very repeats-rich (e.g.,a substantial portion of the genome consists of repetitive sequences with ≥90% identities) [21,22], resulting in at least 10% of sequence reads that are unable to be mapped onto the genome unambiguously and are essentially unusable.This also leads to CNA artifacts.
A number of software tools have been developed in recent years for CNA-discovery from WGS data [13,15,[23], [24], [25], [26], [27]].However, substantial issues still exist.For example, a study has compared a total of 10 such tools with simulated and real cancer sequencing data, and has concluded that the software BICseq15 outperforms the others18. However, for detecting small CNAs of <1 kb, the sensitivity is 0.33 even with BICseq and ranges 0.0—0.35 for the other algorithms.Hence, these tools have not fully realized the great potential of WGS identifying CNA events18.To address the challenges, we have developed a software tool called SEG and evaluated its performance as described below.
2. Materials and methods
2.1.1. The algorithm of SEG
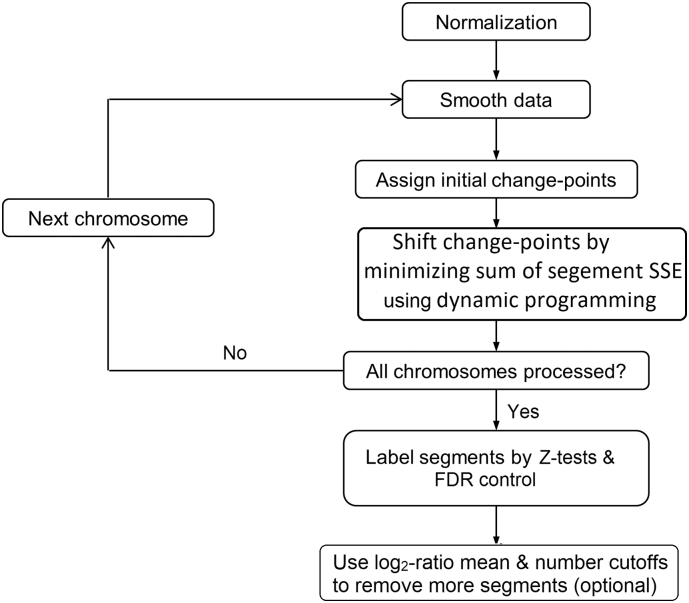
SEG consists of three major steps: 1)data normalization; 2) change-point finding; and 3)CNA identification, as illustrated in Fig. 1 and detailed below.
Fig. 1.
The algorithm of SEG. SEG will: 1)normalize the data and exclude the log2-ratio outliers (smooth data); 2)identify change-points; and 3)find CNAs (label segments).For change-point detection, SEG first depends upon the user's input to assign initial change-points, and then loops through the SSE (sum of squared error) to remove insignificant change-points using dynamic programming (see text).The program is implemented in C and can be downloaded from GitHub at https://github.com/ZhaoS-Lab/SEG.
2.1.2. Data Normalization
To identify CNAs, SEG analyzes mapped read, for single-endsequencing, or fragment, for paired-end sequencing, density calculated based on continuous and non-overlapping tiling windows along a chromosome.The window size varies with the sequencing coverage, e.g.,100 bp for 20-30× coverage based on a previous publication13. To reduce CNA artifacts arisen from sequencing and mapping biases, we first normalize the density data by , where diis the mapped read or fragment density of the ithwindow of either the tumor genome or the matching normal genome, and is the corresponding genome-wide average density.
2.1.3. Change-Point Finding
We have used the same change-point concept defined previously by the popular software tool CBS23 for change-point finding.Briefly, let x1, x2, …, xn be the log2-ratiosof a chromosome, as defined in the section above, which are also assumed to be random variables.An index sequence of A = (a1, a2, ⋯, av), where 1 ≤ v < n, would be called achange-point sequence if meeting the following requirements.A change-pointai(1 ≤ i ≤ v) divides variables xai−1+1, xai−1+2, ⋯, xai, xai+1, xai+2, ⋯, xai+1into two neighboring the ith and (i + 1)thsegments.Importantly, the variables xai−1+1, xai−1+2, ⋯, xaiof the ith segment have a common distribution function Fi.Similarly, the variables xai+1, xai+2, ⋯, xai+1 of the (i + 1)thsegment also share a common distribution function Fi+1.However, Fi differs from Fi+1.
Based on this definition, SEG finds change-points by: 1)minimizing variations of the log2-ratios within the same segment (such that these variables share a common distribution function); and 2)ensuring that the log2-ratio means between any two neighboring segments are significantly different (such that their variable distribution functions differ).To implement this algorithm, SEG adapts a bottom-up approach via dynamic programming for change-point identification, which differs from CBS where a top-down strategy is used23, as illustrated below.
2.1.4. Assign the Average Segment Size
First, SEG requires the user to input an estimated initial average segment size, w, which is the total number of log2-ratios within a segment and must be ≥2.Because w determines the upper-limit of the total change-points for which SEG can identify, it is important to have an appropriate value for w.We recommend setting w = s + 1, where s is the minimal number of continuous log2-ratios that needs to be considered collectively for CNA identification.
2.1.5. Shift change-points via minimizing the sum of squared error (SSE) using dynamic programming
The user-inputtedw divides the log2-ratiosx1, x2, x3…, xn of a chromosome into segments with a preassigned change-pointsequence of A = a1, a2, …, at−1.To find the true values of A, we first define the SSE as: let be the mean of the ith segment containing variables xai, xai+1, xai+2, …, xai+1−1, .Then, SEG scans through the chromosome via a one-segment-overlapping sliding window of a total k (2 ≤ k ≤ t), a user-defined value, consecutive segments at a time to identify the correct positions for the subset of change-point sequence of Au = (au, au+1…, au+k−1).To do this, SEGutilizes dynamic programing to shift each change-point rightward orleftward until the sum of SSE of the k segments, given by is minimized, whereSSE(au+j−1, au+j) represents SSE of the segment flanked by change-pointsau+j−1and au+j.SEG begins with au = 1 and determines the first k − 1 change-points; then repeats the process by resetting au = k − 1 and so on until the entire chromosome is examined.Note that if w × k ≥ n or k = t, dynamic programming will be applied to the whole chromosome and the entire change-point set A = a1, a2, …, at−1 will be determined at one time.
2.1.6. CNA Finding (Segment-Labeling)
The change-points identified through the procedure described above divide a chromosome into different segments.To determine which segments are significantly amplified or deleted, we use a false discovery rate (FDR) controlling procedure as follows.Let and l be the mean and total number of log2-ratios of a segment, SEG first calculates the p-value of each segment of the genome by using z-test given by , where μ and σare the genome-wide mean and standard deviation.Then, the Benjamini and Hochberg step-up method [28] is used for CNA identification by controlling the FDR at a certain desired value.We call this step as “segment-labeling” (Fig. 1), because amplified, deleted, and unchanged segments are respectively labeled with +1, −1, and 0 in the final output file.
In current implementation of SEG, two additional cutoffs can be used to make the selected segments biologically significant.First, to avoid segments with a very small but a very large l (which are unlikely to be CNA) being selected, a cutoff value m is used to select only those segments with their log2-ratio mean satisfying .Similarly, another cutoff s is used to select those segments having a total log2-ratio number l meeting l ≥ s.
2.1.7. Log2-Ratio Data Smoothing
We followed the same data smoothing procedure described by Olshen etal. [23] to exclude the log2-ratio outliers.Briefly, let x1, x2, …, xn be the log2-ratios of a chromosome, and xi and xj (j ≠ i) be the maximum (or minimum) and the next maximum (or minimum) log2-ratios in the region of xi−R, …, xi,…, xi+R where R was a small integer (we set R = 2 as suggested23), respectively.If |xi − xj| ≥ Lσ, we replaced xi by m + Mσ (if xi is the maximum) or m − Mσ(if xi is the minimum), where σ is the genome-wide log2-ratio standard deviation and m is the median of xi−R, …, xi,…, xi+R.M and L are constants, and we set L = 4, M= 2, as described23.This process modified ≤0.1% of the log2-ratios of a genome for those analyzed.
2.1.8. Simulated Data and Real Cancer Data Used to Evaluate the Performance of SEG
Both simulated and real data were used to evaluate SEG.For simulated data, we followed the same procedures as described18 to generate 10 samples of human chromosome 22.Briefly, for each sample, a total of 5 heterozygous deletions, 5 homozygous deletions, and 10 amplifications with copy number randomly choosing between three and eight were introduced to human chromosome 22.The size of these CNA events were sampled from a uniform distribution ranged between 100 bp to 10 Mb as described18.
For the real genomic sequencing data, we chose to use three canine mammary cancer cases, of which both the tumor and matching genomes were sequenced to 12-17× coverage4.These cancers were also subjected to 385 K array comparative genome hybridization (aCGH) analyses, which indicate that they represent CNA-extensive, −moderate, and -sparse genomes4.aCGH studies were conducted as previously described4 using the 385 K canine CGH array chips from Roche NimbleGen Systems, Inc.The log2-ratio value of each probe was collected and normalized following manufacturer's instruction.
2.1.9. Other Software Tools
BICseq and FREEC were run as described by Alkodsi etal. [18]. CBS was run with default parameters via DNAcopy from bioconductor.org/packages/release/bioc/html/DNAcopy.html, and CNAs were identified with the same log2-ratio cutoff as SEG.
3. Results
3.1.1. SEG Identifies more Small CNAs of <1 Kb than BICseq in Simulated Data
Alkodsi etal.18 compared a total of 10 published software tools, and concluded that BICseq15 is the best-performed among them in both sensitivity and specificity for detecting somatic CNAs from cancer genome sequencing data.We hence focused on comparing SEG to BICseq to evaluate the performance of SEG, using simulated data of ten test samples of chromosome 22 harboring twenty CNAs generated as described by Alkodsi etal.18 (see Materials and Methods).To run SEG, we first divided the chromosome into tiling windows of 100 bp, because of the 30× sequence coverage, and calculated the average mapped fragment density for each window.Then, we computed the log2-ratio of the density of a test chromosome 22 (with CNAs) against the reference chromosome 22 (without CNAs) for each window.Windows with no reads mapped to them and hence with zero density in either the test or reference chromosome are excluded from further analysis.Among these windows, those with zero density in the test chromosome and with density in the reference chromosome reaching the top 2.5% of its density distribution are considered as homozygous deletions, the reverse of which are considered as high level amplifications (in real cancer data, these windows should be rarer due to reasons such as contaminating non-tumor or tumor cells in the tumor or normal sample respectively).
For change-point identification, we tested SEG by setting w (the initial segment size, i.e.,the number of log2-ratio) and k (the number of segments for dynamic programming) to various values, and found the results are largely consistent.The analysis described below was performed by setting w = 5 and k = 1001.For CNA-finding, we set FDR ≤ 0.05, s = 1 and m = σ, where s and m represent the minimum cutoffs of the log2-ratio number and mean respectively of a segment with CNA, while σ is the genome-wide standard deviation of log2-ratios.These parameters and cutoffs are mostly the default setting ofSEG.
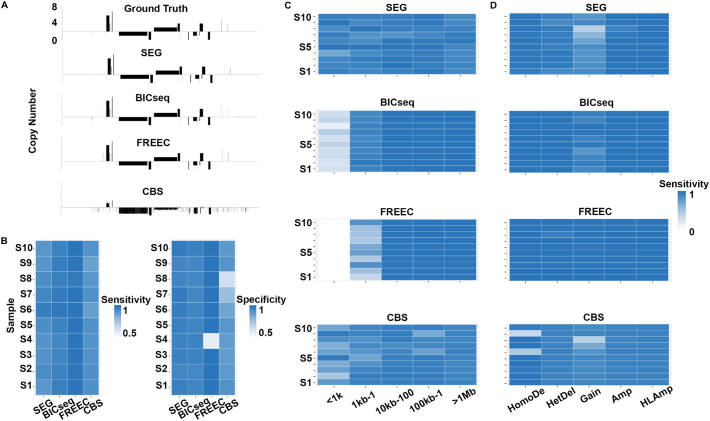
Each of the 10 simulated human chromosome 22 samples harbors 10 deletions and 10 amplifications with size ranged from 100 bp to10 Mb, with 2 amplifications and 2 deletions falling in each bin of 100bp-1 kb, 1 kb–10 kb, 10 kb–100 kb, 100 kb-1 Mb, and 1 Mb–10Mb.Overall, SEG detects these CNA events with approximately the same sensitivities, ranging from 0.90 to 0.97, and specificities, ranging from 0.95 to 0.98, as BICseq in these samples (Fig. 2A and B).However, for detecting small CNAs of 100 bp-1 kb, our analyses indicate that SEG significantly outperformed BICseq, with the sensitivity ranging from 0.72 to 1.00 with an average of 0.91, compared to a 0.28–0.44 range and a 0.34 average for BICseq18(Fig. 2C).
Fig. 2.
CNAs identified by SEG, BICseq, FREEC and CBS in 10 simulated samples of chromosome 22.A.Amplifications and deletions of ground truth, and those identified by SEG or other software tools drew as described18 for one simulated sample. B. Heatmaps showing the overall sensitivity and specificity of CNA detection in each of the 10 simulated samples by SEG or other software tools. C. Heatmaps showing the overall sensitivity of CNA detection based on the size by SEG or other software tools. D. Heatmaps showing the overall sensitivity of CNA detection for each category indicated by SEG or other software tools.
For large CNAs of >1 Mb, BICseq performed better than SEG, with an average sensitivity of 1.00 for BICseq versus 0.91 for SEG(Fig. 2C).This is especially so for detecting 1-copy gain event of >1 Mb (Fig. 2D), with an average sensitivity of 0.90 for BICseq and 0.74 for SEG.
To further evaluate SEG, we compared SEG to two additional software tools that use different segmentation strategies. One is FREEC [26], a well-cited tool for copy number and allele content determination and ranked the 2nd best performed (after BICseq) by Alkodsi etal. [18]. The other is CBS [23], the most cited CNA tool as of today to our knowledge and used by TCGA [5,19,20] and numerous others (although originally designed for the microarray platform, CBS can be applied on WGS data, e.g.,it has been used to segment the WGS data of TCGA). Moreover, as described previously, SEG utilizes the same change-point concept as CBS. Our comparison reached the same conclusion as described above–SEG is more sensitive in discovering small CNAs than either FREEC or CBS(Fig. 2C). Consistent with the evaluation by Alkodsi etal. [18], our analysis also indicates that the sensitivity of FREEC is very high for large (>10Kb) CNA discovery but very low for small CNA detection (Fig. 2). CBS is underperformed than SEG in nearly every aspect examined (Fig. 2).
3.1.2. SEG Identifies both Large and Small CNAs from Real Cancer WGSData
We applied SEG on three canine mammary cancer cases (IDed 32,510, 76 and 406,434), each with its tumor and matching normal genomes undergone paired-endWGS of 12-17× sequence coverage and 20-32× fragment coverage4.In addition, aCGH analyses find very different CNA landscapes among the three cancer genomes, with tumor 32,510 having hardly any CNAs detected, tumor 76 harboring two large amplicons of >4 Mb, and tumor 406,434 having more extensive CNAs and with whole chromosome gain4.Hence, the three tumors provide a nice dataset to test the performance of SEG.
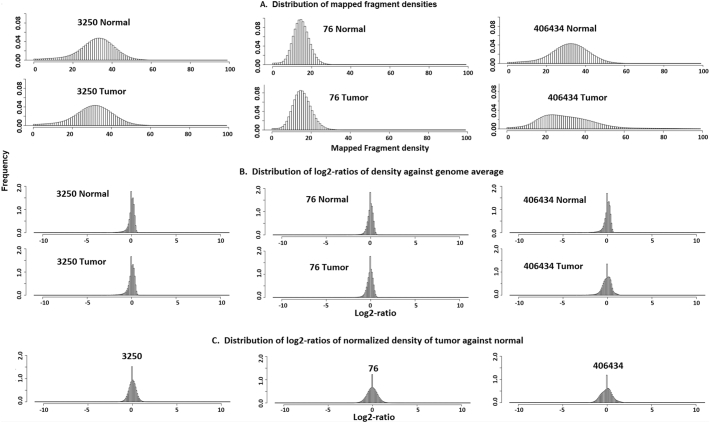
We first divided each of the 39 canine chromosomes into 100 bp window, because of >20× fragment coverage, and calculated the fragment density in each window (Fig. 3A).We then normalized each density against its genome-wide average to correct for the difference in sequencing/fragment coverage among the genomes (Fig. 3B).Afterwards, we further normalized each corrected tumor density against its counterpart from the matching normal genome (Fig. 3C), as described in Materials and Methods.As shown in Fig. 3, the distribution of final tumor against normal density log2-ratios is significantly more normal-looking than the original density distribution for each tumor, indicating that this approach is valid.
Fig. 3.
Data normalization in the three canine mammary cancer genomes. A.The distribution of average mapped fragment density, di, of 100 bp tilting window of the tumor and normal genome of the cancer cases with ID indicated. B. The distribution of the normalized density against its genome wide average by. C. The distribution of the final normalized density of the tumor against the matching normal data by (equation).
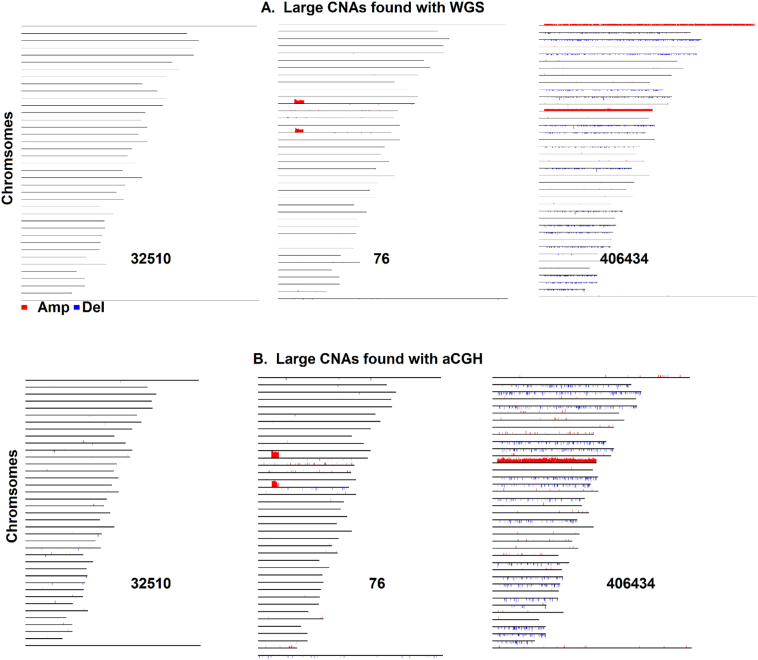
We then ran SEG on these normalized data for the three tumors and examined the identified CNA events to evaluate the SEG performance.First, SEG identified many CNAs from WGS among those found by aCGH.These include the two large amplicons of >4 Mb on chromosomes 12 and 16 of tumor 76, as well as the whole chromosome amplification of chromosome 13 and numerous deletions in tumor 406,434 (Fig. 4).
Fig. 4.
Large CNAs identified with WGS (A)and aCGH (B)by SEG.Each line represents a dog chromosome with its chromosome number indicated on the left.Red (amplifications) and blue (deletions) vertical lines shown above the chromosomes are drew as previously described4.Only CNAs of >8.5 kb were plotted, as 8.5 kb is the minimal size of CNAs found by aCGH.
SEG also identified many additional small CNAs (Table 1).In tumor 32,510 (of which aCCH found very few CNAs), these CNAs are allbelow 3 kb, averaged 418 bp and 443 bp and totaling to 9 Mb and 13 Mb for amplifications and deletions respectively (Table1).These small CNAs are significantly increased in tumors 76 and 406,434 (Table1), which also harbor large CNAs averaged >10 kb in size (Table 2).
Table 1.
Small CNAs of ≤3 kb identified by SEG from WGS data.
| Tumor ID | Amplification |
Deletion |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total Amount | Average size | Exon contenta | GC contenta | Repeats contenta | Total Amount | Average size | Exon content | GC content | Repeats content | |
| 32,510 | 8.7 Mb | 418 bp | 1/4.9kbb | 47.0% | 28.7% | 12.7 Mb | 443 bp | 1/8.5 kb | 40.8% | 36.40% |
| 76 | 36.2 Mb | 308 bp | 1/6.9 kb | 42.4% | 33.0% | 44.5 Mb | 318 bp | 1/5.5 kb | 44.6% | 27.5% |
| 406,434 | 32.1 Mb | 621 bp | 1/7.3 kb | 40.0% | 33.9% | 56.6 Mb | 673 bp | 1/10.5 kb | 40.0% | 32.7% |
The calculations are based on the canFam2 genome assembly, Ensembl gene annotation release-65 (exon content), and RepeatMasker 4.0.5 with repeats database Dfam_2.0.
One exon every 4.5 kb on average. Genome wide: 1/11.7 kb.
Table 2.
Large CNAs of >3 kb identified by SEG from WGS data.
| Tumor ID | Amplification |
Deletion |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total Amount | Average size | Exon content | GC content | Repeatscontent | Total Amount | Average size | Exon content | GC content | Repeats content | |
| 32,510 | None | None | ||||||||
| 76 | 9.2 Mb | 74,656 bp | 1/8.0 kb | 43.5% | 35.6% | None | ||||
| 406,434 | 67.5 Mb | 13,575 | 1/13.2 kb | 40.3% | 35.3% | 1.7 Mb | 4017 bp | 1/17.7 kb | 37.9% | 36.1% |
To better understand these small CNAs identified by SEG, we performed several analyses. First, to evaluate whether they are false results created by SEG, we examined the distributions of their mapped fragment densities. We found that significantly more/fewer fragments were mapped to those amplified/deleted regions in the tumor samples than in the normal samples (Fig. S1). Hence, these small CNAs are indeed amplification/deletion events, not false results created by SEG. Second, to evaluate whether these small CNAs are sequencing/mapping artifacts (i.e.,better or worse sequenced/mapped than an average genomic region) or play a role in cancer, we examined their GC, repetitive sequence and gene contents. For GC and repeat contents, we found no clear and consistent differences between small and big CNAs (Table 1, Table 2). Our analysis revealed, however, that these small CNAs harbor more genes, compared to large CNAs or an average genomic region. Specifically, the average exon density is one per 5-10 kb for small CNAs, compared to one per 8-18 kb for big CNAs and one per 12 kb genome-wide(Table 1, Table 2). Furthermore, the genes harbored by small CNAs are more enriched in cell cycle and other cancer-related functions, compared to those of large CNAs. These analyses indicate that these small CNAs may have a role in cancer development and progression.
3.1.3. SEG Performance
Because of dynamic programming, SEG runs fast.Using a PC with 2GB RAM,SEG takes a few minutes to process a sample of canine 384 K aCGH [7] or human 2 M SNP array19 studies.WGS has significantly more log2-ratios, and the speed depends on the user input for k, the number of segments on which dynamic programming is applied at a time.If setting k = 101, this will take less than half an hour to finish a 30×WGS genome using a PC with 2GB RAM.We have compared the results of having small k (101) and large k (covering the entire chromosome), the results agree >90%.SEG can be obtained from the GitHub at https://github.com/ZhaoS-Lab/SEG.
4. Discussion
Unlike microarrays that are restricted by the probes, deep WGS can cover every single base of the genome and has the potential to identify somatic CNAs of all size in a cancer genome.However, current published software tools examined have a low sensitivity (<0.35) detecting small CNAs of <1kb18, unable to realize the full potential of deep WGS in finding smaller CNAs.To address this issue, we have developed a software tool, SEG.Based on simulated data, SEG is able to detect CNAs of <1 kb with >0.9 sensitivities, outperforming other software tools compared18.
The core algorithm of SEG is change-point detection among the data series along a chromosome.We have used the same change-point definition as the popular software CBS23.However, unlike CBS23 which uses a top-down approach for change-point detection, SEG uses a bottom-up approach, with the upper limit of the total change-pointsdetermined by the user and utilizing dynamic programming for change-point discovery.These differences allow SEG to more accurately determine small CNAs.
SEG identifies substantial amount of small CNAs of <3 kb in WGS data of the three cancer genomes which are not found by aCGH.Our analysis indicates that these small CNAs are not false events created by SEG. Instead, these small CNAs could be cancer drivers (because of their higher gene content and enrichment in cancer-related functions) or passengers (e.g.,arising from increased cancer genomic instability and defective DNA repair), or simply artifacts due to sequencing or mapping biases (e.g.,GC-rich regions or repetitive sequences such as Alu, LINEs, etc.).
Sequencing/mapping originated artifact CNAs vary with the sequencing depth, as well as the window size chosen to calculate the log2-ratios (see Materials and Methods). Except for a publication that suggests using 100 bp windows for 20-30× sequence coverage for germline copy number variation discovery13, we have not yet found a study that discusses the appropriate window size for cancer CNA finding. We will try to develop a statistical model that determines the window size based on sequencing depth to minimize artifact CNAs. Second, even though SEG normalizes the tumor data against the matching normal data to reduce artificial CNAs arising from sequencing and mapping biases, substantial issues remain, especially for low coverage WGS.Data normalization remains a significant challenge and better normalization strategies need to be developed.Third, the results of SEG vary with several user-input values, including initial segment size as well as cutoffs on minimal log2-ratio number and mean. Choosing appropriate values will also reduce artifact CNAs.
To narrow down small CNAs that are more likely cancer-associated, we first plan to add a new function to SEG to identify small CNAs that are clustered in the genome.These CNA clusters should be more cancer-relevant, compared to random small CNAs.Second, we will modify SEG to give users the option to exclude copy number variations identified among normal individuals.Third, many genomic sites are already known to be recurrently amplified/deleted in human cancers (e.g.,from TCGA studies [5,19,20]). Small CNAs that locate within those genomic regions have a higher probability to be cancer-associated event. Moreover, small CNAs that harbor known cancer genes or genes with cancer-related functions (e.g.,cell proliferation, apoptosis, invasion, etc.) are more likely to be cancer drivers. Finally, we note again that small CNAs identified by SEG contain more genes, especially those with cancer-related functions. More studies are required to understand the significance of these small CNAs in cancer development and progression.
For detection of >1 Mb large gains and losses, SEG has a lower sensitivity compared to BICseq18 and FREEC [26].Hence, SEG needs further improvement in this aspect.For current CNA discovery, we recommend using SEG for more sensitive detection of small CNAs, and in combination with another program (e.g.,BICseq, FREEC, etc.) for large CNA discovery. Finally, we emphasize once again that SEG requires several user inputs, the values of which will influence the outcome of SEG.Hence, for new datasets, users may need to try different input values and choose the most appropriate ones.
The following are the supplementary data related to this article.
Distribution of normalized fragment densities within SEG-identified-CNA genomic regions in the tumor and normal genomes of the three canine cancer cases.
Authors'Contributions
MZ developed and implemented the SEG algorithm.DL performed the data analyses.YF and TW contributed to the data analyses in the manuscript revision. JT performed some of the initial analyses.KD and PS advised and helped the statistic analyses.SZ helped the analyses and wrote the manuscript.All authors contributed to the manuscript editing.
Acknowledgments
Acknowledgements
We are grateful for the data and help by Dr. Amjad Alkodsi for the data simulation and analyses, and Mr. Sheng Tao for his work.The work is supported by the NCIR01 CA182093, American Cancer Society, Georgia Cancer Coalition, and the AKC Canine Health Foundation.
Conflict of Interest
The authors declare no conflict of interests.
B.Heatmaps showing the overall sensitivity and specificity of CNA detection in each of the 10 simulated samples by SEG or other software tools.
C.Heatmaps showing the overall sensitivity of CNA detection based on the size by SEG or other software tools.
D.Heatmaps showing the overall sensitivity of CNA detection for each category indicated by SEG or other software tools.
B.The distribution of the normalized densitydi against its genome wide average by .
C.The distribution of the final normalized density of the tumor against the matching normal data by .
References
- 1.Stephens P.J., DJ McBride, Lin M.L. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462(7276):1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang J., Le S., Sun L. Copy number abnormalities in sporadic canine colorectal cancers. Genome Res. 2010;20(3):341–350. doi: 10.1101/gr.092726.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang J., Li Y., Lyon K. Cancer driver-passenger distinction via sporadic human and dog cancer comparison: a proof-of-principle study with colorectal cancer. Oncogene. 2014;33(7):814–822. doi: 10.1038/onc.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu D., Xiong H., Ellis A.E. Molecular homology and difference between spontaneous canine mammary cancer and human breast cancer. Cancer Res. 2014;74(18):5045–5056. doi: 10.1158/0008-5472.CAN-14-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zack T.I., Schumacher S.E., Carter S.L. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beroukhim R., Mermel C.H., Porter D. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D., Xiong H., Ellis A.E. Canine spontaneous head and neck squamous cell carcinomas represent their human counterparts at the molecular level. PLoS Genet. 2015;11(6) doi: 10.1371/journal.pgen.1005277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Xu J., Xiong H. Cancer driver candidate genes AVL9, DENND5A and NUPL1 contribute to MDCK cystogenesis. Oncoscience. 2014;1(12):854–865. doi: 10.18632/oncoscience.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui J., Yin Y., Ma Q. Comprehensive characterization of the genomic alterations in human gastric cancer. Int J Cancer. 2015;137(1):86–95. doi: 10.1002/ijc.29352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallioniemi A. CGH microarrays and cancer. Curr Opin Biotechnol. 2008;19(1):36–40. doi: 10.1016/j.copbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 11.McCormick M.R., Selzer R.R., Richmond T.A. Methods in high-resolution, array-based comparative genomic hybridization. Methods Mol Biol. 2007;381:189–211. doi: 10.1007/978-1-59745-303-5_9. [DOI] [PubMed] [Google Scholar]
- 12.Navin N., Kendall J., Troge J. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21(6):974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abel H.J., Duncavage E.J. Detection of structural DNA variation from next generation sequencing data: a review of informatic approaches. Cancer Genet. 2013;206(12):432–440. doi: 10.1016/j.cancergen.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xi R.B., Hadjipanayis A.G., Luquette L.J. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc Natl Acad Sci U S A. 2011;108(46):E1128–E1136. doi: 10.1073/pnas.1110574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L., Wendl M.C., McMichael J.F., Raphael B.J. Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet. 2014;15(8):556–570. doi: 10.1038/nrg3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Waters J., Leung M.L. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alkodsi A., Louhimo R., Hautaniemi S. Comparative analysis of methods for identifying somatic copy number alterations from deep sequencing data. Brief Bioinform. 2015;16(2):242–254. doi: 10.1093/bib/bbu004. [DOI] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindblad-Toh K., Wade C.M., Mikkelsen T.S. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 22.Venter J.C., Adams M.D., Myers E.W. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 23.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-basedDNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 24.Afyounian E., Annala M., Nykter M. Segmentum: a tool for copy number analysis of cancer genomes. BMC Bioinformatics. 2017;18(1):215. doi: 10.1186/s12859-017-1626-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Araujo Lima L., Wang K. PennCNV in whole-genome sequencing data. BMC Bioinformatics. 2017;18(Suppl. 11):383. doi: 10.1186/s12859-017-1802-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeva V., Popova T., Bleakley K. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28(3):423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., Gupta P., Wang J. CONSERTING: integrating copy-number analysis with structural-variation detection. Nat Methods. 2015;12(6):527–530. doi: 10.1038/nmeth.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of normalized fragment densities within SEG-identified-CNA genomic regions in the tumor and normal genomes of the three canine cancer cases.