Abstract
Pseudomonas aeruginosa, a Gram-negative bacterium, is a member of the ESKAPE pathogens and one of the leading causes of healthcare-associated infections worldwide. Aminoglycosides (AGs) are recognized for their efficacy against P. aeruginosa. The most common resistance mechanism against AGs is the acquisition of AG-modifying enzymes (AMEs) by the bacteria, including AG N-acetyltransferases (AACs), AG O-phosphotransferases (APHs), and AG O-nucleotidyltransferases (ANTs). In this study, we obtained 122 multidrug-resistant P. aeruginosa clinical isolates and evaluated the antibacterial effects of six AGs and two carbapenems alone against all clinical isolates, and in combination against eight selected strains. We further probed for four representatives of the most common AME genes [aac(6′)-Ib, aac(3)-IV, ant(2")-Ia, and aph(3′)-Ia] by polymerase chain reaction (PCR) and compared the AME patterns of these 122 clinical isolates to their antibiotic resistance profile. Among the diverse antibiotics resistance profile displayed by these clinical isolates, we found correlations between the resistance to various AGs as well as between the resistance to one AG and the resistance to carbapenems. PCR results revealed that the presence of aac(6′)-Ib renders these isolates more resistant to a variety of antibiotics. The correlation between resistance to various AGs and carbapenems partially reflects the complex resistance strategies adapted in these pathogens and encourages the development of strategic treatment for each P. aeruginosa infection by considering the genetic information of each isolated bacteria.
Keywords: : aminoglycoside-modifying enzymes (AMEs), drug combination, ESKAPE pathogens, resistance patterns
Introduction
Pseudomonas aeruginosa is a member of a group of Gram-negative (−) and Gram-positive (+) bacterial pathogens known as ESKAPE [where E stands for Enterococcus faecium (+), S for Staphylococcus aureas (+), K for Klebsiella pneumoniae (−), A for Acinetobacter baumannii (−), P for Pseudomonas aeruginosa (−), and E for Enterobacter species (−)] due to their ability to escape a broad array of antimicrobial agents.1 Its pathogenicity, transmission, and resistance have caused the currently available antibiotics to rapidly lose activity against these pathogens.2–7
As an opportunistic pathogen, P. aeruginosa is a leading cause of healthcare-associated infections and poses great threat in critically ill and immunocompromised patients.8,9 According to the Center for Diseases Control and Prevention, P. aeruginosa is estimated to cause 51,000 infections in United States every year among which 13% are caused by multidrug-resistant strains. It is the most common cause of nosocomial pneumonia (17%); third most common cause of urinary track infections (7%); fourth most common cause of surgical site infections (8%); as well as the fifth common isolate overall from all sites (9%).10 P. aeruginosa is also one of the greatest challenges faced by military healthcare professionals, since it is often isolated among antibiotic-resistant bacteria in such settings in patients with military injuries and wounds.11
Due to their excellent efficacy against Gram-negative bacteria, aminoglycosides (AGs) are widely used to treat infections caused by P. aeruginosa. In addition to amikacin (AMK), gentamicin (GEN), and tobramycin (TOB) that are currently approved in the United States for systemic administration, netilmicin (NET) and sisomicin (SIS) are widely employed in AG treatment regiments in European countries. Apramycin (APR), which is currently used in veterinary treatment, is another AG that has shown excellent antibacterial efficacy, limited toxicity, and was found to be less prone to the development of resistance over time.12–14 Nonetheless, resistance has quickly developed to these antimicrobial agents with a significant portion of clinical isolates identified now resistant to clinically used AG antibiotics.14–16
Resistance to AGs results from a variety of mechanisms including (1) changes in bacterial cell envelope composition, (2) modification on RNA target, (3) upregulation of efflux pumps, and (4) acquisition of AG-modifying enzymes (AMEs), with the last accounting for most of the resistance cases. AMEs comprise three different types of enzymes: AG N-acetyltransferases (AACs), AG O-nucleotidyltransferases (ANTs), and AG O-phosphotransferases (APHs). These enzymes transfer an acetyl, a nucleotidyl, or a phosphate group onto AGs, respectively. AACs, which target the 6′, 2′, and 3 amino moieties on AG molecules, are the most common AMEs in resistant organisms. A more recently discovered multiacetylating AAC, the enhanced intracellular survival (Eis) protein,17–22 could become highly problematic in the future with its ability to acetylate more than one site on AGs. Most APHs, on the other hand, attack the 3′ and 2′ positions. ANTs are the least common AMEs and attack the 4′ or the 2′ hydroxyl groups.23
Our laboratory, and many other research groups, is dedicated to developing AG antibiotics and understanding resistance mechanisms.24–39 Previously, researchers have observed discrepant results over whether AG and carbapenem combination therapies yield positive outcomes for the killing of P. aeruginosa.40–43
In an effort to understand in greater detail the underlying resistance mechanisms in P. aeruginosa, in this study, we obtained 122 multidrug-resistant P. aeruginosa clinical isolates from the University of Kentucky Hospital System and evaluated the in vitro activity of various clinically relevant AGs, including AMK, APR, GEN, NET, SIS, and TOB, and two carbapenem antibiotics, meropenem (MEM) and imipenem (IPM), against these clinical isolates. We evaluated the bactericidal effect of AG and carbapenem alone against all clinical isolates, and in combination against eight selected strains. We have also performed polymerase chain reaction (PCR) probing for four of the most common AME genes, aac(6′)-Ib, aac(3)-IV, ant(2")-Ia, and aph(3′)-Ia, and compared the AME patterns of these 122 clinical isolates to their antibiotic resistance profiles.
Materials and Methods
Materials and instrumentation
All P. aeruginosa strains involved in this study were obtained from the University of Kentucky Hospital System. All PCR reagents, including Phusion DNA polymerase, and ladders for DNA gel electrophoresis were purchased from New England BioLabs (NEB, Ipswich, MA). Primers used in PCR and Mueller Hinton broth were purchased from Sigma-Aldrich (Milwaukee, WI). InstaGene Matrix was manufactured by Bio-Rad® (Hercules, CA). All antibiotics used in this study, including AGs (AMK, APR, GEN, NET, SIS, and TOB) and carbapenems (MEM and IPM) were purchased from AK Scientific (Mountain View, CA). Data were analyzed and plotted in Sigma Plot 12.3 and Prism 7.
Susceptibility testing for AGs and carbapenems
The minimum inhibitory concentration (MIC) values of all AGs and MEM were determined against all 122 clinical isolates (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/mdr), and the MIC values of IPM were determined for a small portion of the clinical isolate library (Supplementary Table S2) comprising the strains used for MEM and IPM combination study with AGs.
Using the broth microdilution method according to guidelines from the Clinical and Laboratory Standards Institute (CLSI),44 a 100 μl aliquot of a bacterial culture in its exponential phase were incubated with 100 μl of the various antibiotics (final concentration range of 0.5–128 μg/ml) for 16 hours and then visually inspected for growth. The resistance breakpoint values for each of the various antibiotics against P. aeruginosa strains were as follows: AMK (S ≤ 16, I = 32, R ≥ 64), GEN (S ≤ 4, I = 8, R ≥ 16), NET (S ≤ 8, I = 16, R ≥ 32), TOB (S ≤ 4, I = 8, R ≥ 16), MEM (S ≤ 2, I = 4, R ≥ 8), and IPM (S ≤ 2, I = 4, R ≥ 8) according to the CLSI.45 The resistance cutoff values for SIS against P. aeruginosa have not been established in the United States and are considered to be S ≤ 4 and R > 4 in this study.46,47
Statistical analyses were performed by using Prism 7 and all values were transformed to log2 scale before analysis. Comparisons between groups of clinical isolates susceptible or resistant to a certain antibiotic were made by two-tailed Mann–Whitney test. Significance was defined as p ≤ 0.05. Significance levels were defined in further detail as 0.033, 0.002, <0.001, which were marked in figures by *, **, and ***, respectively. Please note that to use the Mann–Whitney test for continuous variable, we considered all MIC values that were >128 μg/ml to be 256 μg/ml for the sake of the test and all that were ≤0.25 μg/ml to be 0.25 μg/ml.
Screening for AME genes by PCR
The presence of aac(6′)-Ib, aac(3)-IV, ant(2″)-Ia, and aph(3′)-Ia genes was evaluated by PCR using specific primers, as previously described.48,49 A 40 μl aliquot of an overnight Mueller–Hinton liquid culture of each P. aeruginosa strain were centrifuged at 13,000 rpm for 1 minute to collect the bacteria, which were then mixed with 100 μl of InstaGene Matrix and heated up at 100°C for 30 minutes. The PCR program was also previously described.48,49 The expected sizes were 482, 230, 534, and 624 bp for aac(6′)-Ib, aac(3)-IV, ant(2″)-Ia, and aph(3′)-Ia, respectively (Supplementary Fig. S1 and Supplementary Table S3). Statistical analyses were performed by using GraphPad Instat and Prism 7 software as described in the section “Susceptibility testing for AGs and carbapenems”. Comparisons between two groups or three groups of clinical strains were made by two-tailed Mann–Whitney test or Kruskal–Wallis test.
Combination studies of AGs and carbapenem by checkerboard assays
To assess the interactions between AGs and carbapenem antibiotics, combination studies were performed using standard checkerboard assays with the concentration of AGs varied horizontally and that of MEM/IPM varied vertically. For each of AMK and TOB, we selected four distinct clinical isolates: one that was susceptible to both AG and MEM, one that was susceptible to AG and resistant to MEM, one that was resistant to AG and susceptible to MEM, and one that was resistant to both AG and MEM.
The concentrations of antibiotics for each strain varied as follows: #1 (AMK: 0.008–4 μg/ml, MEM: 0.03–2 μg/ml, IPM: 0.13–8 μg/ml), #26 (AMK: 4 μg/ml, MEM: 0.06–4 μg/ml, IPM: 0.13–8 μg/ml), #50 (AMK: 0.03–16 μg/ml, MEM: 2–128 μg/ml, IPM: 2–128 μg/ml), #28 (AMK: 0.13–64 μg/ml, MEM: 0.5–32 μg/ml, IPM: 1–64 μg/ml), #99 (TOB: 0.004–2 μg/ml, MEM: 0.06–4 μg/ml, IPM: 0.13–8 μg/ml), #116 (TOB: 0.06–32 μg/ml, MEM: 0.06–4 μg/ml), #33 (TOB: 0.004–2 μg/ml, MEM: 2–128 μg/ml, IPM: 1–64 μg/ml), and #15 (TOB: 0.25–128 μg/ml, MEM: 0.5–32 μg/ml, IPM: 1–64 μg/ml). The combination of TOB and IPM was not tested against strain #116 due to a higher than 128 μg/ml IPM MIC.
After 16 hours of incubation at 37°C, bacterial growth was inspected visually and fractional inhibitory concentration index (FICI) values were calculated by adding the fractions of MIC of each drug in combination over MIC value of that drug alone for each of the two drugs. Drug interaction is defined as synergistic if FICI ≤ 0.5, indifferent if 0.5 < FICI < 4, antagonistic if FICI > 4.50
Results and Discussion
Susceptibility testing of various antibiotics against 122 P. aeruginosa clinical isolates
We first determined the MIC values of various AGs (AMK, APR, NET, SIS, and TOB) and MEM against all 122 P. aeruginosa strains (Supplementary Table S1).45,46 The breakpoint concentrations for APR are yet to be determined. Therefore, for all analyses in this study, APR was not categorized based on susceptibility or resistance. As summarized in Table 1, 117 strains were susceptible to AMK and only 3 strains were resistant to AMK. The susceptibility of all strains showed a similar distribution to GEN, NET, SIS, and TOB: ∼55% of the strains susceptible and ∼40% resistant. Approximately 60% of these 122 strains were susceptible to MEM and <20% of them were resistant to MEM.
Table 1.
Susceptibility of 122 Pseudomonas aeruginosa Clinical Isolates to Various Antibiotics
| Susceptible (S) | Intermediate (I) | Resistant (R) | ||||
|---|---|---|---|---|---|---|
| Antibiotic | No. of strains | % | No. of strains | % | No. of strains | % |
| AMK | 117 | 95.9 | 2 | 1.0 | 3 | 2.5 |
| GEN | 68 | 55.8 | 5 | 4.1 | 49 | 40.2 |
| NET | 66 | 54.1 | 15 | 12.3 | 41 | 33.6 |
| SIS | 65 | 53.3 | — | — | 57 | 46.7 |
| TOB | 70 | 57.4 | 3 | 2.5 | 49 | 40.2 |
| MEM | 72 | 59.0 | 27 | 22.1 | 23 | 18.9 |
The resistance cutoff values (in μg/ml) for various antibiotics against P. aeruginosa are AMK (S ≤ 16, I = 32, R ≥ 64), GEN (S ≤ 4, I = 8, R ≥ 16), NET (S ≤ 8, I = 16, R ≥ 32), TOB (S ≤ 4, I = 8, R ≥ 16), and MEM (S ≤ 2, I = 4, R ≥ 8) according to the CLSI. The resistance cutoff values for SIS against P. aeruginosa are not established in the United States and are considered to be S ≤ 4 and R > 4 μg/ml in this study. APR is not included in this analysis due to lack of established resistance cutoff.
AMK, amikacin; APR, apramycin; CLSI, Clinical and Laboratory Standards Institute; GEN, gentamicin; MEM, meropenem; NET, netilmicin; SIS, sisomicin; TOB, tobramycin.
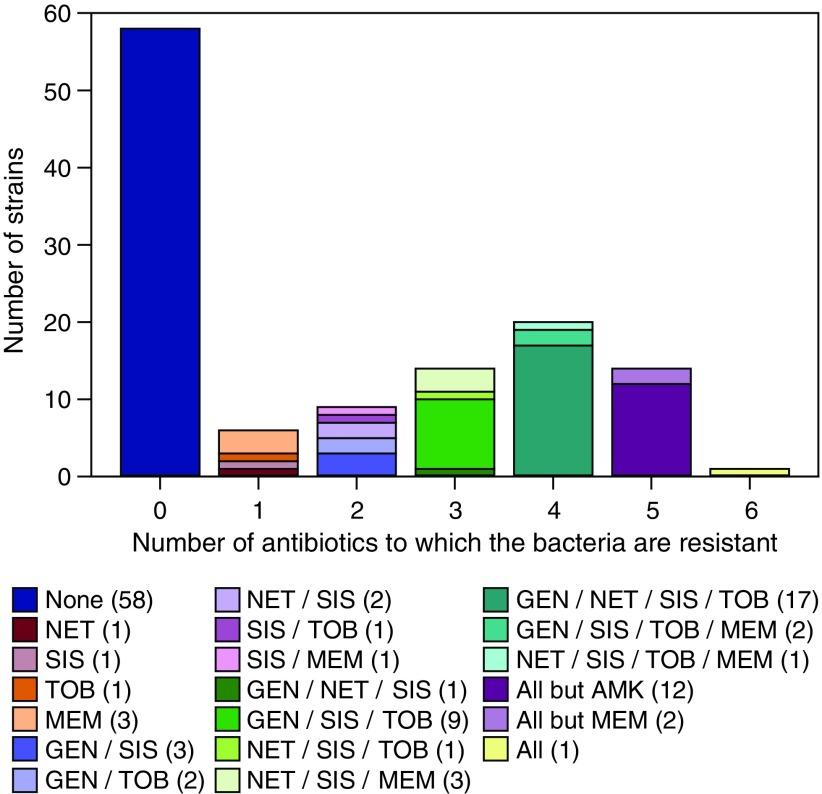
To further explore whether the resistance to these antibiotics occur mostly individually or in groups of antibiotics, we next categorized the P. aeruginosa strains studied based on the number of antibiotics that they were resistant to (Fig. 1). We found that over half of the strains (58 strains) were susceptible to all of the antibiotics tested in this study. We observed that simultaneous resistance to all six antibiotics is rare (one strain only). Moreover, we found 20 strains that concurrently resisted the action of four antibiotics.
FIG. 1.
Number of strains grouped by the number of antibiotics to which the Pseudomonas aeruginosa strains are resistant. The specific antibiotic to which each strain is resistant to are marked in the legend with the number of strains with each resistance profile in parentheses. The resistance cutoff values for each antibiotic are as follows: AMK (S ≤ 16, I = 32, R ≥ 64), GEN (S ≤ 4, I = 8, R ≥ 16), MEM (S ≤ 2, I = 4, R ≥ 8), NET (S ≤ 8, I = 16, R ≥ 32), and TOB (S ≤ 4, I = 8, R ≥ 16) according to the CLSI.45 The resistance cutoff values for SIS against P. aeruginosa are not established in the United States and are considered to be S ≤ 4 and R > 4 in this study based on Barry et al.46 AMK, amikacin; CLSI, Clinical and Laboratory Standards Institute; GEN, gentamicin; MEM, meropenem; NET, netilmicin; SIS, sisomicin; TOB, tobramycin. Color images available online at www.liebertpub.com/mdr
To establish the correlations between the susceptibility/resistance to one antibiotic to the resistance to another drug in these P. aeruginosa strains, we further categorized these clinical isolates based on their susceptibility/resistance to each individual antibiotic (AMK, GEN, NET, SIS, TOB, and MEM) and analyzed their resistance patterns to other antibiotics within this group (Table 2, Fig. 2, Supplementary Figs. S2 and S3).
Table 2.
Summary of the Median Minimum Inhibitory Concentration Values, Range of Minimum Inhibitory Concentration Values, and Rate of Resistance to Each Antibiotic Based on the Susceptibility to Amikacin, Gentamicin, Netilmicin, Sisomicin, Tobramycin, and Meropenem
| MIC (μg/ml) | MIC (μg/ml) | |||||
|---|---|---|---|---|---|---|
| Antibiotic | Median | Range | Resistance rate % | Median | Range | Resistance rate % |
| AMK susceptible (n = 117) | AMK resistant (n = 3) | |||||
| GEN | ≤2 | ≤2 to >8 | 38.5 | >8 | >8 | 100 |
| NET | 8 | ≤0.25 to >128 | 60.7 | >128 | >128 | 100 |
| SIS | 4 | ≤0.25 to >128 | 38.5 | >128 | >128 | 100 |
| TOB | 2 | ≤0.25 to >128 | 38.5 | >128 | ≥128 | 100 |
| MEM | 1 | ≤0.25 to >128 | 17.1 | 4 | 0.5 to >128 | 33 |
| GEN susceptible (n = 69) | GEN resistant (n = 49) | |||||
| AMK | 2 | 0.5 to 32 | 0 | 8 | 1 to 128 | 6.1 |
| NET | 4 | ≤0.25 to >128 | 10.1 | >128 | 2 to >128 | 67.4 |
| SIS | 2 | ≤0.25 to >128 | 14.5 | >128 | ≤0.25 to >128 | 95.9 |
| TOB | 0.5 | ≤0.25 to 64 | 6.3 | 64 | 2 to >128 | 91.8 |
| MEM | 0.5 | ≤0.25 to >128 | 11.6 | 4 | ≤0.25 to >128 | 30.6 |
| NET susceptible (n = 66) | NET resistant (n = 41) | |||||
| AMK | 2 | 0.5 to 8 | 0 | 8 | 1 to 128 | 7.3 |
| GEN | ≤2 | ≤2 to >8 | 12.1 | >8 | ≤2 to >8 | 80.5 |
| SIS | 2 | ≤0.25 to >128 | 12.1 | >128 | 1 to >128 | 97.6 |
| TOB | 0.75 | ≤0.25 to 128 | 10.6 | 64 | ≤0.25 to >128 | 12.2 |
| MEM | 0.5 | ≤0.25 to 64 | 6.1 | 4 | ≤0.25 to >128 | 41.5 |
| SIS susceptible (n = 65) | SIS resistant (n = 57) | |||||
| AMK | 2 | 0.5 to 16 | 0 | 8 | 1 to 128 | 5.3 |
| GEN | ≤2 | ≤2 to >8 | 3.1 | >8 | ≤2 to >8 | 82.5 |
| NET | 4 | ≤0.25 to >128 | 1.5 | >128 | 2 to >128 | 70.2 |
| TOB | 0.5 | ≤0.25 to >128 | 4.6 | 32 | ≤0.25 to >128 | 80.7 |
| MEM | 0.5 | ≤0.25 to 64 | 4.6 | 4 | ≤0.25 to >128 | 35.1 |
| TOB susceptible (n = 70) | TOB resistant (n = 49) | |||||
| AMK | 2 | 1 to 128 | 0 | 8 | 1 to 128 | 2.0 |
| GEN | ≤2 | ≤2 to >8 | 5.7 | >8 | ≤2 to >8 | 91.8 |
| NET | 4 | ≤0.25 to >128 | 7.1 | >128 | 2 to >128 | 57.6 |
| SIS | 2 | ≤0.25 to 128 | 11.4 | >128 | ≤0.25 to >128 | 93.9 |
| MEM | 0.5 | ≤0.25 to 64 | 5.7 | 4 | ≤0.25 to >128 | 32.7 |
| MEM susceptible (n = 73) | MEM resistant (n = 23) | |||||
| AMK | 4 | 0.5 to 128 | 1.4 | 8 | 0.5 to 64 | 4.4 |
| GEN | ≤2 | ≤2 to >8 | 20.6 | >8 | ≤2 to >8 | 65.2 |
| NET | 4 | ≤0.25 to >128 | 16.4 | 128 | ≤0.25 to >128 | 73.9 |
| SIS | 2 | ≤0.25 to >128 | 24.7 | >128 | ≤0.25 to >128 | 87.0 |
| TOB | 1 | ≤0.25 to >128 | 16.4 | 32 | ≤0.25 to >128 | 69.6 |
Intermediate (I) strains are not included in this analysis. Results of APR are not shown in this table due to lack of susceptibility cutoff standards.
MIC, minimum inhibitory concentration.
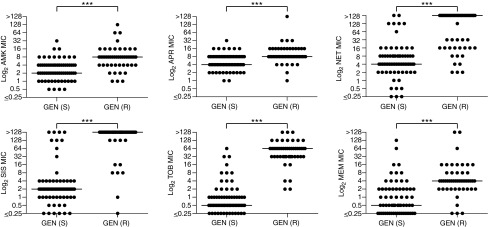
FIG. 2.
Distribution of MIC values of each antibiotic based on the susceptibility to GEN. The median MIC of each antibiotic is marked by a horizontal bar. Additional distribution plots of MIC values of each antibiotic based on the susceptibility to other antibiotics (NET and SIS, and TOB and MEM) are presented in Supplementary Figs. S2, S3. We did not plot the MIC values of each antibiotic based on the susceptibility to AMK as the vast majority of the 122 P. aeruginosa strains tested are susceptible to AMK and only 3 strains are resistant to this AG. Significance was defined as p value ≤0.05 (*** <0.001). AG, aminoglycoside; MIC, minimum inhibitory concentration.
First, we separated the 122 strains based on their susceptibility/resistance to a first antibiotic, then analyzed the median MIC values, range of MIC values, and the resistance rate for a second antibiotic within each group (Table 2). Then, we plotted the MIC values of the second antibiotic based on the susceptibility/resistance to the first antibiotic to see the distribution of the MIC values of all the clinical isolates. The data based on GEN susceptibility/resistance are presented in Fig. 2, whereas those based on the susceptibility/resistance to the rest of the antibiotics are displayed in Supplementary Figs. S2 and S3.
Taking GEN as an example, we found that within the GEN-susceptible group (n = 69 strains), the median MIC values for AMK, APR, NET, SIS, TOB, and MEM were 2, 4, 4, 2, 0.5, and 0.5 μg/ml, respectively, which were lower than those in the GEN-resistant group (n = 49 strains) where the median MIC values for the above antibiotics were 8, 8, >128, >128, 64, and 4 μg/ml, respectively (Table 2). We observed that the range of MIC values in GEN-susceptible and GEN-resistant categories did not differ much, potentially due to the large sample size in this study. However, the resistance rates to the other five antibiotics in the GEN-resistant groups were prominently higher than in the GEN-susceptible groups.
The differences in median MIC values and overall distributions of MIC values between the GEN-susceptible and GEN-resistant groups can be clearly observed in Fig. 2. For instance, the median MIC values for NET, SIS, TOB, and MEM were all significantly different (≥8-fold difference) between the GEN-susceptible (lower median MIC) and GEN-resistant (higher median MIC) groups. In contrast, the median AMK and APR MIC values between the GEN-susceptible and GEN-resistant groups only differed by fourfold and twofold, respectively.
Similarly, the overall distributions of MIC values (looking at individual dots on the plots) of NET, SIS, TOB, and MEM were significantly different between the GEN-susceptible and GEN-resistant groups, whereas these differences were less extensive for AMK and practically absent for APR.
It is important to note that the trends observed in resistance rates, median MIC values, and overall distributions of MIC values in the GEN-susceptible and GEN-resistant groups were also observed when the data were analyzed based on the susceptibility/resistance to all other antibiotics (Table 2 and Supplementary Figs. S2 and S3), with the exception of the resistance rates to TOB between the NET-susceptible (10.6%) and NET-resistant (12.2%) groups, which were not significantly different from each other (Table 2). Furthermore, in the case of AMK, we observed that the median MIC values and the resistance rates to all other antibiotics in the AMK-resistant group looked profoundly different from those in the AMK-susceptible group. However, the small sample size of the AMK-resistant group (three strains) would not necessarily allow us to generalize this observation to most P. aeruginosa strains.
Based on the results from Table 2, Fig. 2, Supplementary Figs. S2, and S3, we postulated that the correlations observed between the susceptibility/resistance to one AG with that of other AGs could be explained by different resistance mechanisms affecting multiple AGs, which was not surprising given that the presence of AMEs is the most common resistance mechanism against AG antibiotics. For instance, if the bacteria acquired an AME, the most common mechanism of resistance to AGs, then it would likely be resistant to any AGs that can be inactivated by this particular AME due to the structural similarities between the AG molecules.
Furthermore, given that all AGs tested in this study possess a 6′ amino group, except for APR, it is not hard to explain what we observed in Table 2, Fig. 1, Supplementary Figs. S2, and S3, where we found that the strains that are resistant to one AG also have higher resistance rates and overall higher MIC values to other AGs.
Moreover, the correlations established between the resistance to MEM and those to AGs, which are two distinct classes of antibiotics with discrete chemical structures, although not surprising, may suggest the presence of other resistance mechanisms that are less structurally specific, and therefore, work against different classes of antibiotics. Another possibility to explain this phenomenon is the resistance elements to various antibiotics that could have accumulated simultaneously in bacteria, that is, the resistance element to carbapenems could have accumulated inside these P. aeruginosa isolates at the same time as resistance elements to AG antibiotics develop.51 Furthermore, since both classes of antibiotics are often used in patients infected with P. aeruginosa, the resistance to both classes of antibiotics could potentially result from previous antibiotics exposure.52–54
Screening for AME genes by PCR
Having established that half of the P. aeruginosa clinical isolates studied displayed resistance to AGs, we further investigated whether the observed AG resistance could be explained by the presence of one or multiple AME resistance genes. Therefore, we performed PCR with lysed bacteria probing for four of the most common AME genes [aac(6′)-Ib, aac(3)-IV, ant(2")-Ia, and aph(3′)-Ia] as previously described.48 A representative PCR result for each gene is presented in Supplementary Fig. S1 and a summary of the AMEs present in all clinical isolates is presented in Supplementary Table S3.
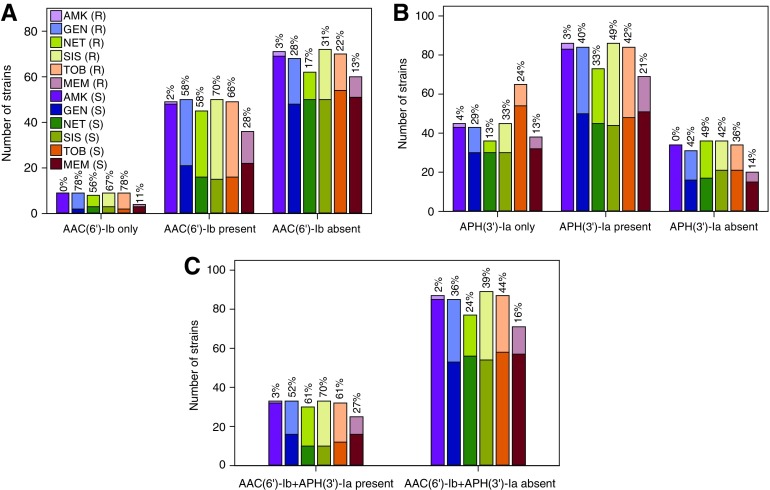
Overall, 50 (41.0%), 13 (10.7%), 7 (5.7%), and 86 (70.5%) of the 122 P. aeruginosa clinical isolates possessed aac(6′)-Ib, aac(3)-IV, ant(2")-Ia, and aph(3′)-Ia genes, respectively. These statistics agree with the common understanding of aac and aph being the most common AME genes and ant being the least common. Contrary to what was observed in a previous study that identified aac(6′)-Ib as the most common AME in K. pneumoniae strains,48 we found aph(3′)-Ia to be the most common AME gene harbored by the P. aeruginosa clinical isolates in our study. The 122 clinical isolates displayed 11 different AME patterns (Table 3). There were 17 strains (13.9%) that contained no AME genes. Fifty-eight strains (47.5%) contained one AME gene only, among which, 9 (7.4%), 4 (3.3%), and 45 (36.9%) strains contained aac(6′)-Ib only, aac(3)-IV only, and aph(3′)-Ia only, respectively.
Table 3.
The Median Minimum Inhibitory Concentration Values of Each Antibiotic Based on the Presence and Absence of Different Aminoglycoside-Modifying Enzyme Gene Patterns
| AMK | APR | GENa | NET | SIS | TOB | MEM | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median MIC (μg/ml) | Median MIC (μg/ml) | Median MIC (μg/ml) | Median MIC (μg/ml) | Median MIC (μg/ml) | Median MIC (μg/ml) | Median MIC (μg/ml) | |||||||||
| AME pattern | No. of strains | Patten present | Pattern absent | Patten present | Pattern absent | Patten present | Pattern absent | Patten present | Pattern absent | Patten present | Pattern absent | Patten present | Pattern absent | Patten present | Pattern absent |
| No AMEs | 17 | 2 | 4 | 4 | 8 | ≤2 | ≤2 | 4 | 8 | 2 | 4 | 1 | 4 | 0.5 | 2 |
| AAC(6′)-Ib only | 9 | 8 | 4 | 8 | 8 | >8 | ≤2 | >128 | 8 | >128 | 4 | 32 | 1 | 4 | 2 |
| AAC(3)-IV only | 4 | 3 | 4 | 8 | 8 | ≤2 | ≤2 | 4 | 8 | 2 | 4 | 1 | 2 | 0.5 | 2 |
| APH(3)-Ia only | 45 | 4 | 4 | 8 | 8 | ≤2 | 4 | 8 | 8 | 2 | 16 | 1 | 8 | 1 | 2 |
| AAC(6′)-Ib+AAC(3)-IV | 4 | 8 | 4 | 8 | 8 | >8 | ≤2 | >128 | 8 | >128 | 4 | 64 | 2 | 16 | 2 |
| AAC(6′)-Ib+APH(3′)-Ia | 33 | 4 | 4 | 8 | 8 | >8 | ≤2 | 128 | 8 | >128 | 2 | 32 | 1 | 2 | 1 |
| AAC(3)-IV+ANT(2″)-Ia | 1 | 1 | 4 | 4 | 8 | >8 | ≤2 | 2 | 8 | >128 | 4 | 64 | 2 | 4 | 2 |
| AAC(3)-IV+APH(3′)-Ia | 3 | 1 | 4 | 2 | 8 | ≤2 | ≤2 | ≤0.25 | 8 | ≤0.25 | 4 | 0.5 | 2 | 0.5 | 2 |
| ANT(2″)-Ia+APH(3′)-Ia | 2 | 2 | 4 | 8 | 8 | >8 | ≤2 | 18 | 8 | — | 4 | 96 | 2 | 6 | 2 |
| AAC(6′)-Ib+AAC(3)-IV+ANT(2″)-Ia | 1 | 4 | 4 | 8 | 8 | ≤2 | ≤2 | 8 | 8 | 1 | 4 | 1 | 2 | 2 | 2 |
| AAC(6′)-Ib+ANT(2″)-Ia+APH(3′)-Ia | 3 | 8 | 4 | 16 | 8 | >8 | ≤2 | 16 | 8 | 128 | 4 | 64 | 2 | 4 | 2 |
Most GEN MIC values were obtained from the University of Kentucky Hospital System, ranging from ≤2 to >8 μg/ml.
—, Indicates that it was impossible to determine the median MIC value as the values involved were a combination of ≤0.25 and >128 in μg/ml.
AAC, aminoglycoside N-acetyltransferase; AME, aminoglycoside-modifying enzyme; ANT, aminoglycoside O-nucleotidyltransferase; APH, aminoglycoside O-phosphotransferase.
Not surprisingly, zero strains contained ant(2")-Ia by itself. Furthermore, 43 (35.3%) strains contained 2 AME genes: 4 (3.3%) strains contained the combination of aac(6′)-Ib and aac(3)-IV, 33 (27.0%) strains contained aac(6′)-Ib and aph(3′)-Ia, 1 (0.8%) strain contained aac(3)-IV and ant(2")-Ia, 3 (2.5%) strains contained aac(3)-IV and aph(3′)-Ia, and 2 (1.6%) strains contained ant(2")-Ia and aph(3′)-Ia. There were also four (2.5%) strains that possessed three AME genes: either aac(6′)-Ib, aac(3)-IV, and ant(2")-Ia (1 (0.8%) strain; #31 in Supplementary Table S3 or aac(6′)-Ib, ant(2")-Ia, and aph(3′)-Ia (3 (2.5%) strains; #8, #105, and #115 in Supplementary Table S3. It is important to note that none of the clinical isolates studied contained all four AME genes probed for.
To determine whether the presence of the AME genes found in the clinical isolates correlated with the MIC values that were observed for the AGs and to see whether they had a potential influence on the MIC values of MEM, we categorized all P. aeruginosa strains based on their AME gene patterns and compared the median MIC values of each antibiotic between the pattern present and the pattern absent groups [e.g., for the group of strains that contained aph(3′)-Ia only, the pattern present group consisted of the 45 strains that possessed only aph(3′)-Ia, whereas the pattern absent group included the rest of the 77 P. aeruginosa strains that either did not contain this gene or contained other AME genes in addition to aph(3′)-Ia] (Table 3).
For the no AME group, the median TOB and MEM MIC values showed a mild difference (fourfold) between the pattern present and absent groups, not surprisingly with the pattern present (no AMEs) group showing lower MIC values. No significant difference (≥8-fold) was observed between the pattern present and absent groups for other antibiotics in this no AME group.
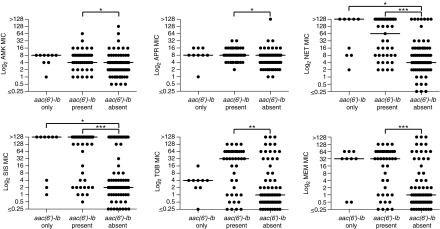
For the three different groups that possessed a single AME gene, the group that contained aac(6′)-Ib only showed significant differences (≥8-fold) in the median MIC values of GEN, NET, SIS, and TOB between the pattern present and absent groups, with the median MIC values of the pattern absent group for each of the four antibiotics lying in the susceptible range and the pattern present median MIC values falling in the resistant range for each antibiotics. The other single AME gene pattern groups did not display as significant a contrast with the exceptions of a mild difference (fourfold) in the median MEM MIC values between the aac(3)-IV only pattern present and absent groups, and eightfold differences in the median MIC values of SIS and TOB between the aph(3′)-Ia only pattern present and absent groups. Counterintuitively, the aph(3′)-Ia only pattern present group displayed lower MIC values than those in the pattern absent group for SIS and TOB. These interesting observations may suggest that AAC(6′)-Ib could be the most significant AME contributing to the resistance to AGs in this study.
Additionally, considering the subtle differences in the chemical structures of these AGs, AMK and APR are the only two AGs in this study that possess 3′ hydroxyl groups to be targeted by APH(3′)-Ia, yet, differences in the median MIC values in the aph(3′)-Ia only pattern present and absent groups were not observed for those AGs. However, for SIS and TOB, which contain hydrogen atoms at their 3′ position, and therefore, do not allow inactivation by the APH(3′)-Ia enzyme, the eightfold lower in the median MIC values observed between the aph(3′)-Ia only pattern present and absent groups potentially indicate that other resistance mechanisms are contributing to resistance to these AG antibiotics in the P. aeruginosa clinical isolates tested.
In the five different groups that possessed two AME genes, substantial differences could be observed between the pattern present and absent groups for GEN, NET, SIS, TOB, and MEM, with the exceptions of the median MEM MIC values between the pattern present and absent groups harboring aac(6′)-Ib and aph(3′)-Ia or aac(3)-IV and ant(2")-Ia, the median GEN MIC values for the groups harboring aac(3)-IV and aph(3′)-Ia, and the median NET and SIS MIC values for the groups harboring ant(2")-Ia and aph(3′)-Ia. As for the two groups that contained three AME genes, the only significant differences between the pattern present and absent groups were observed in median GEN, SIS, and TOB MIC values for the P. aeruginosa strains that contained aac(6′)-Ib, ant(2")-Ia, and aph(3′)-Ia.
Another interesting observation was that the one strain containing aac(6′)-Ib, aac(3)-IV, and ant(2")-Ia showed slightly lower MIC to SIS and TOB in the pattern present group when compared to the pattern absent group. However, the differences between the MIC values were not significant (≤4-fold difference) to experimentally equivalent (fourfold difference). Additionally, having only one strain in the pattern present group also made it hard to generalize this observation to all P. aeruginosa strains harboring this particular AME pattern.
In addition to the above observations, it is also important to note that none of these AME patterns correlated with significant differences in median AMK and APR MIC values between the pattern present and absent groups, which agrees with previous studies suggesting that AMK and APR are less vulnerable to AMEs and, therefore, less prone to developing resistance due to the acquisition of AMEs.48 These two AGs can, therefore, greatly help patients infected with resistance microorganisms. Furthermore, the much weaker correlation between MEM MIC values and AME gene patterns could easily be explained by the structural difference between AGs and carbapenems and the fact that AMEs do not chemically modify carbapenems.
Although these P. aeruginosa clinical isolates possessed diverse patterns of AME genes, we next decided to further analyze the clinical isolates harboring three of the most significant/common AME patterns: aac(6′)-Ib, aph(3′)-Ia, and aac(6′)-Ib+aph(3′)-Ia. We first separated all the clinical isolates based on the exclusive (gene only) and nonexclusive (gene present) presence and absence (gene absent) of the three gene patterns above and compared the MIC value distributions of each antibiotic (Figs. 3 and 4, and Supplementary Fig. S4). These findings are in agreement with the previous observations from Table 3 and with a previous report stating that aph(3′)-Ia in combination with another AME correlated with higher resistance rates against various antibiotics.48 It is also important to note that the presence of AME genes studied does not necessarily equate to the expression and function of the AME proteins. Therefore, the presence of certain AME genes, if the gene was not expressed, would not contribute to a resistant phenotype.
FIG. 3.
Distribution of MIC values of each antibiotic grouped by the presence or absence of the aac(6′)-Ib gene. The median MIC value in each group, aac(6′)-Ib only, aac(6′)-Ib present, and aac(6′)-Ib absent, is represented by a horizontal bar. Plots depicting the distribution of MIC values of each antibiotic group by the presence and absence of aph(3′)-Ia, and aac(6′)-Ib+aph(3′)-Ia genes are presented in Supplementary Fig. S4. Significance was defined as p-value ≤0.05 (*0.033, **0.002, *** <0.001).
FIG. 4.
Number of P. aeruginosa clinical isolates harboring various AME gene patterns. (A) aac(6′)-Ib, (B) aph(3′)-Ia, and (C) aac(6′)-Ib+aph(3′)-Ia. The percentage of P. aeruginosa strains, which are resistant to each antibiotic within each AME pattern group are marked at the top of each column. AME, aminoglycoside-modifying enzyme. Color images available online at www.liebertpub.com/mdr
Overall, the P. aeruginosa clinical isolates involved in this study showed great AME gene diversity and resistance profiles to various antibiotics. Recently, research groups have directed a lot of effort to the establishment of combination therapies and seen promising results in killing various ESKAPE pathogens.55–57 We selected four P. aeruginosa strains that were susceptible to AMK and MEM, susceptible to AMK and resistant to MEM, susceptible to MEM and resistant to AMK, and resistant to both AMK and MEM, and four that were susceptible to TOB and MEM, susceptible to TOB and resistant to MEM, susceptible to MEM and resistant to TOB, and resistant to both TOB and MEM, and performed combination studies to investigate potential synergistic effects between AGs (AMK and TOB) and carbapenems (MEM and IPM) (Note: MIC values for IPM alone are presented in Supplementary Table S2) via standard checkerboard assays.
Our results, however, suggested no synergistic effects between these two classes of antimicrobial agents with the FICI ranging from 0.625 to 2. This result added on to the existing dispute on whether AG and carbapenem combination therapy could be potentially helpful to patients with P. aeruginosa infections, and the answer is embedded in the intrinsic diversity of P. aeruginosa and the resistance mechanism(s) that it harbors.40–43
Conclusions
By evaluating the resistance profile of various AGs and carbapenems of 122 P. aeruginosa clinical isolates, we found that the correlation in the resistance between two AGs, and the correlation between the resistance to AGs and that to carbapenems suggested a variety of antibiotic resistance mechanisms in addition to the presence of AMEs. Specifically, we discovered that the presence of aac(6′)-Ib, whether alone or in the presence of other AMEs, led to higher rates of resistance to various antibiotics involved in this study, whereas the presence of aph(3′)-Ia did not, even though the latter was present at a higher frequency in these P. aeruginosa strains. However, these observations only revealed a small portion of the complex resistance mechanisms to various antibiotics. Thus, our study confirmed the unlikeliness of a single “standard treatment” for all P. aeruginosa infections. Instead, personalized therapies that take advantage of the genetic information of the isolated bacteria, such as resistance gene screening, should be utilized to aid in the selection of antimicrobial agents for determination of treatment regiment to account for the complex nature of P. aeruginosa.
Supplementary Material
Acknowledgments
This work was supported by NIH grant AI090048 (to S.G.-T.). S.Y.L.H. was in part supported by a University of Kentucky Presidential Fellowship. We thank Prof. David S. Burgess for sharing with us the clinical isolates that we used in this study.
Disclosure Statement
The authors declare no conflicts of interest.
References
- 1.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B., Scheld M., Spellberg B., and Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 2.Narten M., Rosin N., Schobert M., and Tielen P. 2012. Susceptibility of Pseudomonas aeruginosa urinary tract isolates and influence of urinary tract conditions on antibiotic tolerance. Curr. Microbiol. 64:7–16 [DOI] [PubMed] [Google Scholar]
- 3.Obritsch M.D., Fish D.N., MacLaren R., and Jung R. 2005. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy. 25:1353–1364 [DOI] [PubMed] [Google Scholar]
- 4.Pendleton J.N., Gorman S.P., and Gilmore B.F. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11:297–308 [DOI] [PubMed] [Google Scholar]
- 5.Pobiega M., Maciag J., Chmielarczyk A., Romaniszyn D., Pomorska-Wesolowska M., Ziolkowski G., Heczko P.B., Bulanda M., and Wojkowska-Mach J. 2015. Molecular characterization of carbapenem-resistant Pseudomonas aeruginosa strains isolated from patients with urinary tract infections in Southern Poland. Diagn. Microbiol. Infect. Dis. 83:295–297 [DOI] [PubMed] [Google Scholar]
- 6.Qiu X., Kulasekara B.R., and Lory S. 2009. Role of horizontal gene transfer in the evolution of Pseudomonas aeruginosa virulence. Genome Dyn. 6:126–139 [DOI] [PubMed] [Google Scholar]
- 7.Rice L.B. 2010. Progress and challenges in implementing the research on ESKAPE pathogens. Infect. Control Hosp. Epidemiol. 31 Suppl 1:S7–S10 [DOI] [PubMed] [Google Scholar]
- 8.Hirsch E.B., and Tam V.H. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 10:441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Nosocomial Infection Surveillance (NNIS) system report: data summary from January 1992 through June 2003. Available at www.cdc.gov/ncidod/dhqp/pdf (accessed July13, 2017). [DOI] [PubMed]
- 10.Fujitani S., Moffett K.S., and Yu V.L. 2017. Pseudomonas aeruginosa, antimicrobe: infectious disease & antimicrobial agents. Available at www.antimicrobe.org/new/b112.asp (accessed July13, 2017)
- 11.Calhoun J.H., Murray C.K., and Manring M.M. 2008. Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin. Orthop. Relat. Res. 466:1356–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fosso M.Y., Li Y., and Garneau-Tsodikova S. 2014. New trends in aminoglycosides use. MedChemComm 5:1075–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matt T., Ng C.L., Lang K., Sha S.H., Akbergenov R., Shcherbakov D., Meyer M., Duscha S., Xie J., Dubbaka S.R., Perez-Fernandez D., Vasella A., Ramakrishnan V., Schacht J., and Bottger E.C. 2012. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc. Natl. Acad. Sci. U. S. A. 109:10984–10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang A.D., Smith K.P., Eliopoulos G.M., Berg A.H., McCoy C., and Kirby J.E. 2017. In vitro apramycin activity against multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Diagn. Microbiol. Infect. Dis. 88:188–191 [DOI] [PubMed] [Google Scholar]
- 15.Karlowsky J.A., Hoban D.J., Hackel M.A., Lob S.H., and Sahm D.F. 2017. Antimicrobial susceptibility of Gram-negative ESKAPE pathogens isolated from hospitalized patients with intra-abdominal and urinary tract infections in Asia-Pacific countries: SMART 2013–2015. J. Med. Microbiol. 66:61–69 [DOI] [PubMed] [Google Scholar]
- 16.Biedenbach D.J., Giao P.T., Hung Van P., Su Minh Tuyet N., Thi Thanh Nga T., Phuong D.M., Vu Trung N., and Badal R.E. 2016. Antimicrobial-resistant Pseudomonas aeruginosa and Acinetobacter baumannii from patients with hospital-acquired or ventilator-associated pneumonia in Vietnam. Clin. Ther. 38:2098–2105 [DOI] [PubMed] [Google Scholar]
- 17.Chen W., Biswas T., Porter V.R., Tsodikov O.V., and Garneau-Tsodikova S. 2011. Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. Proc. Natl. Acad. Sci. U. S. A. 108:9804–9808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W., Green K.D., Tsodikov O.V., and Garneau-Tsodikova S. 2012. Aminoglycoside multiacetylating activity of the enhanced intracellular survival protein from Mycobacterium smegmatis and its inhibition. Biochemistry 51:4959–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green K.D., Chen W., and Garneau-Tsodikova S. 2012. Identification and characterization of inhibitors of the aminoglycoside resistance acetyltransferase Eis from Mycobacterium tuberculosis. ChemMedChem 7:73–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houghton J.L., Green K.D., Pricer R.E., Mayhoub A.S., and Garneau-Tsodikova S. 2013. Unexpected N-acetylation of capreomycin by mycobacterial Eis enzymes. J. Antimicrob. Chemother. 68:800–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houghton J.L., Biswas T., Chen W., Tsodikov O.V., and Garneau-Tsodikova S. 2013. Chemical and structural insights into the regioversatility of the aminoglycoside acetyltransferase Eis. ChemBioChem 14:2127–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsodikov O.V., Green K.D., and Garneau-Tsodikova S. 2014. A random sequential mechanism of aminoglycoside acetylation by Mycobacterium tuberculosis Eis protein. PLoS One 9:e92370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez M.S., and Tolmasky M.E. 2010. Aminoglycoside modifying enzymes. Drug Resist. Updat. 13:151–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houghton J.L., Green K.D., Chen W., and Garneau-Tsodikova S. 2010. The future of aminoglycosides: the end or renaissance? ChemBioChem 11:880–902 [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Green K.D., Johnson B.R., and Garneau-Tsodikova S. 2015. Inhibition of aminoglycoside acetyltransferase resistance enzymes by metal salts. Antimicrob. Agents Chemother. 59:4148–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaul P., Green K.D., Rutenberg R., Kramer M., Berkov-Zrihen Y., Breiner-Goldstein E., Garneau-Tsodikova S., and Fridman M. 2011. Assessment of 6′- and 6′′′-N-acylation of aminoglycosides as a strategy to overcome bacterial resistance. Org. Biomol. Chem. 9:4057–4063 [DOI] [PubMed] [Google Scholar]
- 27.Green K.D., Chen W., and Garneau-Tsodikova S. 2011. Effects of altering aminoglycoside structures on bacterial resistance enzyme activities. Antimicrob. Agents Chemother. 55:3207–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandrika Thamban N., Green K.D., Houghton J.L., and Garneau-Tsodikova S. 2015. Synthesis and biological activity of mono- and di-N-acylated aminoglycosides. ACS Med. Chem. Lett. 6:1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fong D.H., Xiong B., Hwang J., and Berghuis A.M. 2011. Crystal structures of two aminoglycoside kinases bound with a eukaryotic protein kinase inhibitor. PLoS One 6:e19589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez C., Arivett B.A., Actis L.A., and Tolmasky M.E. 2015. Inhibition of AAC(6′)-Ib-mediated resistance to amikacin in Acinetobacter baumannii by an antisense peptide-conjugated 2′,4′-bridged nucleic acid-NC-DNA hybrid oligomer. Antimicrob. Agents Chemother. 59:5798–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shakya T., Stogios P.J., Waglechner N., Evdokimova E., Ejim L., Blanchard J.E., McArthur A.G., Savchenko A., and Wright G.D. 2011. A small molecule discrimination map of the antibiotic resistance kinome. Chem. Biol. 18:1591–1601 [DOI] [PubMed] [Google Scholar]
- 32.Welch K.T., Virga K.G., Whittemore N.A., Ozen C., Wright E., Brown C.L., Lee R.E., and Serpersu E.H. 2005. Discovery of non-carbohydrate inhibitors of aminoglycoside-modifying enzymes. Bioorg. Med. Chem. 13:6252–6263 [DOI] [PubMed] [Google Scholar]
- 33.Zhang W., Chen Y., Liang Q., Li H., Jin H., Zhang L., Meng X., and Li Z. 2013. Design, synthesis, and antibacterial activities of conformationally constrained kanamycin A derivatives. J. Org. Chem. 78:400–409 [DOI] [PubMed] [Google Scholar]
- 34.Fair R.J., Hensler M.E., Thienphrapa W., Dam Q.N., Nizet V., and Tor Y. 2012. Selectively guanidinylated aminoglycosides as antibiotics. ChemMedChem 7:1237–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai A., Uemura S., Johansson M., Puglisi E.V., Marshall R.A., Aitken C.E., Korlach J., Ehrenberg M., and Puglisi J.D. 2013. The impact of aminoglycosides on the dynamics of translation elongation. Cell Rep. 3:497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llano-Sotelo B., Hickerson R.P., Lancaster L., Noller H.F., and Mankin A.S. 2009. Fluorescently labeled ribosomes as a tool for analyzing antibiotic binding. RNA 15:1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang C.W., Fosso M., Kawasaki Y., Shrestha S., Bensaci M.F., Wang J., Evans C.K., and Takemoto J.Y. 2010. Antibacterial to antifungal conversion of neamine aminoglycosides through alkyl modification. Strategy for reviving old drugs into agrofungicides. J. Antibiot. 63:667–672 [DOI] [PubMed] [Google Scholar]
- 38.Maianti J.P., Kanazawa H., Dozzo P., Matias R.D., Feeney L.A., Armstrong E.S., Hildebrandt D.J., Kane T.R., Gliedt M.J., Goldblum A.A., Linsell M.S., Aggen J.B., Kondo J., and Hanessian S. 2014. Toxicity modulation, resistance enzyme evasion, and A-site X-ray structure of broad-spectrum antibacterial neomycin analogs. ACS Chem. Biol. 9:2067–2073 [DOI] [PubMed] [Google Scholar]
- 39.Duscha S., Boukari H., Shcherbakov D., Salian S., Silva S., Kendall A., Kato T., Akbergenov R., Perez-Fernandez D., Bernet B., Vaddi S., Thommes P., Schacht J., Crich D., Vasella A., and Bottger E.C. 2014. Identification and evaluation of improved 4′-O-(alkyl) 4,5-disubstituted 2-deoxystreptamines as next-generation aminoglycoside antibiotics. mBio. 5:e01827-01814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura A., Hosoda M., Kato T., Yamada Y., Itoh M., Kanazawa K., and Nouda H. 2000. Combined effects of meropenem and aminoglycosides on Pseudomonas aeruginosa in vitro. J. Antimicrob. Chemother. 46:901–904 [DOI] [PubMed] [Google Scholar]
- 41.Ferrara A., Grassi G., Grassi F.A., Piccioni P.D., and Gialdroni Grassi G. 1989. Bactericidal activity of meropenem and interactions with other antibiotics. J. Antimicrob. Chemother. 24 Suppl A:239–250 [DOI] [PubMed] [Google Scholar]
- 42.Kropec A., Lemmen S., Wursthorn M., and Daschner F.D. 1994. Combination effect of meropenem with aminoglycosides and teicoplanin on Pseudomonas and enterococci. Infection 22:306–308 [DOI] [PubMed] [Google Scholar]
- 43.Oie S., Sawa A., Kamiya A., and Mizuno H. 1999. In-vitro effects of a combination of antipseudomonal antibiotics against multi-drug resistant Pseudomonas aeruginosa. J. Antimicrob. Chemother. 44:689–691 [DOI] [PubMed] [Google Scholar]
- 44.Clinical and Laboratory Standards Institute. 2012. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Approved Standard. 9th ed. CLSI document M07-A9; Wayne, PA [Google Scholar]
- 45.Clinical and Laboratory Standards Institute. 2014. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI document M100-S24; Wayne, PA [Google Scholar]
- 46.Barry A.L., Miller G.H., Hare R.S., Jones R.N., and Thornsberry C. 1984. Modification of interpretive breakpoints for netilmicin disk susceptibility tests with Pseudomonas aeruginosa. J. Clin. Microbiol. 19:311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barry A.L., Thornsberry C., Jones R.N., and Gerlach E.H. 1981. Gentamicin, tobramycin, and sisomicin disc susceptibility tests. Revised zone standards for interpretation. Am. J. Clin. Pathol. 75:524–531 [DOI] [PubMed] [Google Scholar]
- 48.Almaghrabi R., Clancy C.J., Doi Y., Hao B., Chen L., Shields R.K., Press E.G., Iovine N.M., Townsend B.M., Wagener M.M., Kreiswirth B., and Nguyen M.H. 2014. Carbapenem-resistant Klebsiella pneumoniae strains exhibit diversity in aminoglycoside-modifying enzymes, which exert differing effects on plazomicin and other agents. Antimicrob. Agents Chemother. 58:4443–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watkins D., Kumar S., Green K.D., Arya D.P., and Garneau-Tsodikova S. 2015. Influence of linker length and composition on enzymatic activity and ribosomal binding of neomycin dimers. Antimicrob. Agents Chemother. 59:3899–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meletiadis J., Mouton J.W., Meis J.F., and Verweij P.E. 2003. In vitro drug interaction modeling of combinations of azoles with terbinafine against clinical Scedosporium prolificans isolates. Antimicrob. Agents Chemother. 47:106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hawkey P.M., and Jones A.M. 2009. The changing epidemiology of resistance. J. Antimicrob. Chemother. 64 Suppl 1:i3-i10 [DOI] [PubMed] [Google Scholar]
- 52.Grall N., Lazarevic V., Gaia N., Couffignal C., Laouenan C., Ilic-Habensus E., Wieder I., Plesiat P., Angebault C., Bougnoux M.E., Armand-Lefevre L., Andremont A., Duval X., and Schrenzel J. 2017. Unexpected persistence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in the faecal microbiota of hospitalised patients treated with imipenem. Int. J. Antimicrob. Agents. 50:81–87 [DOI] [PubMed] [Google Scholar]
- 53.Hansen K.C.M.Schwensen S. A. F., Henriksen D.P., Justesen U.S., and Sydenham T.V. 2017. Antimicrobial resistance in the Bacteroides fragilis group in faecal samples from patients receiving broad-spectrum antibiotics. Anaerobe 47:79–85 [DOI] [PubMed] [Google Scholar]
- 54.Chuanchuen R., Beinlich K., Hoang T.T., Becher A., Karkhoff-Schweizer R.R., and Schweizer H.P. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 45:428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yadav R., Landersdorfer C.B., Nation R.L., Boyce J.D., and Bulitta J.B. 2015. Novel approach to optimize synergistic carbapenem-aminoglycoside combinations against carbapenem-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 59:2286–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clock S.A., Tabibi S., Alba L., Kubin C.J., Whittier S., and Saiman L. 2013. In vitro activity of doripenem alone and in multi-agent combinations against extensively drug-resistant Acinetobacter baumannii and Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 76:343–346 [DOI] [PubMed] [Google Scholar]
- 57.Principe L., Capone A., Mazzarelli A., D'Arezzo S., Bordi E., Di Caro A., and Petrosillo N. 2013. In vitro activity of doripenem in combination with various antimicrobials against multidrug-resistant Acinetobacter baumannii: possible options for the treatment of complicated infection. Microb. Drug Resist. 19:407–414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.